Abstract
Transcription of metabolic genes is transiently induced during recovery from exercise in skeletal muscle of humans. To determine whether pre-exercise muscle glycogen content influences the magnitude and/or duration of this adaptive response, six male subjects performed one-legged cycling exercise to lower muscle glycogen content in one leg and then, the following day, completed 2.5 h low intensity two-legged cycling exercise. Nuclei and mRNA were isolated from biopsies obtained from the vastus lateralis muscle of the control and reduced glycogen (pre-exercise glycogen = 609 ± 47 and 337 ± 33 mmol kg−1 dry weight, respectively) legs before and after 0, 2 and 5 h of recovery. Exercise induced a significant (P < 0.05) increase (2- to 3-fold) in transcription of the pyruvate dehydrogenase kinase 4 (PDK4) and uncoupling protein 3 (UCP3) genes in the reduced glycogen leg only. Although PDK4, lipoprotein lipase (LPL) and hexokinase II (HKII) mRNA were elevated in the reduced glycogen leg before exercise, no consistent difference was found between the two legs in response to exercise. In a second study, six subjects completed two trials (separated by 2 weeks) consisting of 3 h of two-legged knee extensor exercise with either control (398 ± 52 mmol kg−1 dry weight) or low (240 ± 38 mmol kg−1 dry weight) pre-exercise muscle glycogen. Exercise induced a significantly greater increase in PDK4 transcription in the low glycogen (> 6-fold) than in the control (< 3-fold) trial. Induction of PDK4 and UCP3 mRNA in response to exercise was also signficantly higher in the low glycogen (11.4- and 3.5-fold, respectively) than in the control (5.0- and 1.7-fold, respectively) trial. These data indicate that low muscle glycogen content enhances the transcriptional activation of some metabolic genes in response to exercise, raising the possibility that signalling mechanisms sensitive to glycogen content and/or FFA availability may be linked to the transcriptional control of exercise-responsive genes.
In human skeletal muscle, endurance exercise activates the transcription of several genes that encode for various regulatory and metabolic proteins (Pilegaard et al. 2000). For many genes, transcriptional activation occurs primarily during the initial few hours of recovery, returning to basal levels within ≈24 h after exercise. These recent data (Pilegaard et al. 2000), as well as other previous findings (Neufer & Dohm, 1993; O'Doherty et al. 1994, 1996; Seip et al. 1997; Neufer et al. 1998; Tsuboyama-Kasaoka et al. 1998; Kraniou et al. 2000), have led to the suggestion that the metabolic adaptations in skeletal muscle associated with exercise training may be generated by the cumulative effects of transient increases in transcription during recovery from each exercise bout (Williams & Neufer, 1996; Pilegaard et al. 2000). However, the cellular and molecular mechanisms underlying the acute transcriptional regulation of gene expression during recovery from exercise remain undefined.
Recovery from exercise is characterized by a number of changes in skeletal muscle metabolism that persist for several hours after exercise. Both glucose uptake and glycogen synthase (GS) activity are increased following exercise, presumably as a direct consequence of decreased muscle glycogen content (Wojtaszewski et al. 2000; Nielsen et al. 2001). The sensitivity of muscle to insulin is also increased after exercise (Ivy & Holloszy, 1981; Garetto et al. 1984; Richter et al. 1984), providing a second mechanism for enhancing glucose uptake and GS activity. In fact, both non-insulin- and insulin-mediated increases in glucose uptake and GS activity are inversely related to muscle glycogen concentration immediately after exercise (Wojtaszewski et al. 2000, 2001), are highest in the first 2-3 h of recovery, and reverse progressively as glycogen content is restored over the course of 18-24 h (Young et al. 1983; Garetto et al. 1984; Cartee et al. 1989). Taken together these findings provide a clear indication of the high metabolic priority given to glycogen resynthesis during recovery from exercise (Richter et al. 2001). The similar time courses over which the restoration of glycogen and the transcriptional activation of exercise-responsive genes occur raise the intriguing possibility that both processes may be linked by common signalling and/or regulatory mechanisms.
The purpose of the present study was to determine the potential influence of pre-exercise muscle glycogen content on the exercise-induced transcriptional regulation of metabolic genes in humans. Two studies were conducted. In Study A, muscle glycogen content was reduced in one leg on the day before the experiment by a combination of one-legged cycling exercise and carbohydrate restriction. On the day of the experiment, two-legged cycling exercise was performed comparing responses in the reduced glycogen and control legs. In Study B, two trials were conducted (separated by 2 weeks), each consisting of two-legged cycling exercise the day before the experiment to lower muscle glycogen stores followed by either a carbohydrate-rich (control trial) or carbohydrate-restricted (low glycogen trial) diet. On the day of the experiment for both trials, identical two-legged dynamic knee extensor exercise was performed, comparing responses in the control and low glycogen trials. In both studies, the effect of varied pre-exercise muscle glycogen content on the molecular responses to exercise was determined by measuring the transcriptional activity and mRNA content of selected metabolic genes.
METHODS
Subjects
Twelve healthy physically active male volunteers ranging in age from 22 to 33 years, with an average height of 183 cm (range 167-193 cm) and a mean weight of 79 kg (range 70-93 kg) participated in the study. The subjects gave their informed consent after having been informed of the experimental procedures and any risk and discomfort associated with the experiment. The study was approved by the Copenhagen and Frederiksberg Ethics Committee, Denmark and the Human Investigations Committee, Yale University, USA, and conformed to the Declaration of Helsinki.
Experimental design
Study A
Six subjects participated in this study. Maximal oxygen uptake (VO2max,two legs) was determined by an incremental cycle ergometer test 1-2 weeks before the day of the experiment and found to average 4.6 l min−1 (range 4.0-5.5 l min−1). The VO2max,one leg was calculated as 74 % of the VO2max,two legs (Pernow & Saltin, 1971). Late in the afternoon of the day before the experiment, the subjects completed ≈90 min of one-legged cycling exercise to lower muscle glycogen stores of that leg using the following protocol: 20 min continuous one-legged cycling at ≈75 % of VO2max,one leg followed by intermittent (90 s cycling-90 s rest) one-legged cycling beginning at an intensity corresponding to 90 % of VO2max,one leg and then progressively decreasing the load (to 85 %, 80 %, 75 %, 70 %, 65 %, 60 % and 55 % VO2max,one leg) until the intensity could not be sustained for 90 s. Finally, an all-out one-legged cycling bout was performed at 85 % VO2max,one leg. In order to further lower glycogen stores in the liver, the subjects then completed 30 min of two-arm cycling exercise. After the exercise protocol, carbohydrate intake was restricted overnight.
In the 2 days preceding the experiment, subjects were provided with dietary instructions such that a minimum of 500 g of carbohydrates per day was consumed to ensure adequate muscle glycogen storage. After the glycogen depletion exercise, a high fat evening meal was provided to the subjects which contained 4.3 MJ and less than 22 g of carbohydrate. On the morning of the day of the experiment, a high fat breakfast meal (2.7 MJ and less than 22 g of carbohydrate) was provided 1 h prior to initiation of the experimental protocol. The subjects then completed 2.5 h of two-legged cycling at ≈45 % VO2max. Muscle biopsies were obtained from the middle portion of the vastus lateralis muscle of both the reduced glycogen and the control glycogen leg before exercise (pre-exercise), immediately after exercise (time 0) and after 2 h and 5 h of recovery, using the percutaneous needle biopsy technique with suction (Bergstrom, 1962). After the exercise bout, subjects were provided with high carbohydrate food containing a total of 6.7 MJ and 309 g of carbohydrate (3.9 ± 0.3 g carbohydrate kg−1 body weight) over the 5 h recovery period as meals immediately after exercise and after 1 h and 3 h of recovery. No other food or beverage (other than water) was allowed.
Study B
The subjects (n = 6) completed, in a randomized order and separated by at least 2 weeks, two trials of two-legged knee extensor exercise on a modified Krogh cycle ergometer (Andersen et al. 1985). The trials were identical in design except that muscle glycogen content was either normal (control trial) or lowered (low glycogen trial) prior to exercise. One week before the first trial, a two-legged knee extensor exercise test was performed to determine the maximal work load. Resistance was increased every 2 min until a kicking frequency of 60 extensions min−1 could no longer be maintained. The highest work load that the subjects could maintain for 2 min was set as the maximum work load.
On the afternoon before each trial, the subjects completed 60 min of cycling exercise at a work load equivalent to ≈70 % of their maximal work load. To further lower liver glycogen stores, the subjects completed an additional 60 min of two-arm cycling exercise. After this glycogen depletion exercise protocol, subjects consumed either a low carbohydrate (low glycogen trial) or a high carbohydrate (control trial) meal in isocaloric amounts. The subjects reported to the laboratory the following day after an overnight fast and a muscle sample (pre-exercise) was obtained from the middle portion of the vastus lateralis muscle. A femoral arterial catheter was inserted under local anaesthesia (lidocaine, 20 mg ml−1). The subjects then performed two-legged knee extensor exercise for 3 h at ≈60 % of their maximum 2 min work load. Additional muscle biopsies were taken after 1.5 and 3 h of exercise and at 2 h of recovery. Blood samples were drawn at rest before exercise, after 0.5, 1, 1.5, 2, and 3 h of exercise as well as after 1 and 2 h of recovery. Whole body oxygen uptake (VO2) and carbon dioxide (VCO2) production were determined by a standard open circuit technique (Oxygen Analyzer S-3A/I, Ametek, USA; LB 2, Beckham, USA; VRDC/HC 1, Parvo Medics, USA) at rest before exercise, after 0.5 h, 1 h, 1.5 h, 2 h and 3 h of exercise and after 1 and 2 h of recovery. The respiratory exchange ratio (RER) was calculated as VCO2/ VO2.
Blood analyses
Plasma was immedialtely separated from blood cells by centrifugation, and stored at −80 °C until analysed. Plasma NEFA concentration was determined using a Wako NEFA-c kit (Wako Chemical, Neuss, Germany), plasma glucose was analysed with a Glucose HK 125 kit (ABX Diagnostics, Montpellier, France) and plasma glycerol was determined enzymatically according to the method of Wieland (1974). All three were measured on an automated analyser (Cobas Fara, Roche, Switzerland).
Muscle biopsies
Biopsy samples were immediately placed on an ice-cold glass plate, cleaned of connective tissue and blood, weighed, and separated for isolation of nuclei (120-130 mg) or frozen in liquid nitrogen (20-40 mg) for mRNA and glycogen determination.
Isolation of nuclei
Nuclei were isolated from muscle biopsies as previously described (Pilegaard et al. 2000). In brief, fresh muscle tissue was placed in 35 ml of ice-cold buffer A (15 mm Hepes, pH 7.5, 60 mm KCl, 3 mg ml−1 bovine serum albumin (BSA), 300 mm sucrose, 5 mm each of EDTA and EGTA, 1 mm dithiothreitol (DTT), 0.5 mm spermidine, 0.15 mm spermine, 2 μg ml−1 leupeptin and 1 mm phenylmethylsulfonyl floride (PMSF)), thoroughly minced, rotated for 5 min at 4 °C, and gently homogenized for 20 s (setting 15, Kinematica Polytron PT2100, Kinematica AG, Switzerland). Samples were allowed to settle on ice for 5 min and then centrifuged at 700 g for 10 min at 4 °C. The crude nuclear pellets were gently resuspended in 10 ml of buffer B (15 mm Hepes, 60 mm KCl, 3 mg ml−1 BSA, 300 mm sucrose, 0.1 mm each of EDTA and EGTA, 0.5 % Triton X-100, 1 mm DTT, 0.5 mm spermidine, 0.15 mm spermine, 2 μg ml−1 each of leupeptin and aprotinin) and filtered through pre-wetted cheesecloth. Nuclei were repelleted (700 g, 10 min, 4 °C), gently resuspended in 10 ml of buffer C (15 mm Hepes, 60 mm KCl, 5 mm magnesium acetate, 3 mg ml−1 BSA, 300 mm sucrose, 0.1 mm each of EDTA and EGTA, 1 mm DTT, 0.5 mm spermidine, 0.15 mm spermine, 2 μg ml−1 each of leupeptin and aprotinin) and repelleted (700 g, 10 min, 4 °C). Final nuclear pellets were resuspended in 200 μl of storage buffer (75 mm Hepes, 60 mm KCl, 15 mm NaCl, 5 mm magnesium acetate, 0.1 mm each of EDTA and EGTA, 40 % glycerol, 1 mm DTT, 0.5 mm spermidine, 0.15 mm spermine, 2 μg ml−1 each of leupeptin and aprotinin), quick-frozen in liquid nitrogen, and stored at −80 °C.
Nuclear run-on
Relative transcription of genes was determined by a RT-PCR-based nuclear run-on technique as previously described (Hildebrandt & Neufer, 2000; Pilegaard et al. 2000). Briefly, incomplete transcripts were allowed to proceed to completion in the presence of the non-radioactive nucleotides CTP, GTP, UTP and ATP. After lysing the nuclei, nuclear proteins and genomic DNA were enzymatically digested (proteinase K and DNase I) and nascent RNA transcripts isolated by extraction (TriZol, Gibco-BRL, Rockville, MD, USA) and precipitation. Nascent RNA transcripts were then subjected to a second DNase digestion, precipitated, thoroughly rinsed, and resuspended overnight (4 °C) in 22 μl DEPC-treated water containing 10 mm Tris (pH 8.0) and 0.1 mm EDTA.
Isolation of genomic DNA
Genomic DNA (gDNA) was isolated from 20 μl of each preparation of nuclei as previously described (Hildebrandt & Neufer, 2000; Pilegaard et al. 2000). Final gDNA pellets were resuspended overnight (4 °C) in 50 μl nuclease-free water containing 10 mm Tris (pH 8.0) and 0.1 mm EDTA. The relative gDNA content of the nuclei samples was determined by PCR amplification of the β-actin gene (described below under Reverse transcription and PCR), and used to adjust the volumes of the nascent RNA transcript (from nuclear run-on) samples for differences in nuclei content prior to the nuclear run-on reaction.
RNA isolation
Total RNA was isolated from ≈20-25 mg of tissue by a modified guanidinium thiocyanate (GT)-phenol-chloroform extraction method adapted from Chomczynski & Sacchi (1987) and as described previously (Pilegaard et al. 2000). In order to facilitate localization of the RNA pellet, 100 μg of yeast tRNA was added to the aqueous phase during the procedure. RNA was resuspended overnight (4 °C) in 2 μl mg−1 original tissue weight in DEPC-treated water containing 0.1 mm EDTA.
Reverse transcription and PCR
Reverse transcription (RT) of both nascent RNA from the nuclear run-on reactions and total RNA samples was performed using the Superscript II RNase H− system (Gibco-BRL) as previously described (Hildebrandt & Neufer, 2000; Pilegaard et al. 2000). RT products of nascent nuclear run-on RNA were diluted with nuclease-free water based on the relative gDNA content of each nuclei preparation (see above) with an average volume set to 75 μl. A further dilution was then made based on PCR amplification of β-actin. For RT products of total RNA (RT-RNA), samples were first diluted to a total of 110 μl, and relative differences in total RNA yield among samples were determined by PCR amplification of β-actin mRNA in each RT-RNA sample. The final volumes were then adjusted based on the relative β-actin mRNA content with the average total volume set to 220 μl.
Transcription and mRNA content of a given gene (and β-actin content in the gDNA samples) were determined by PCR (in duplicate) using 2.5 μl of diluted RT products (i.e. from the run-on RNA, total RNA) or gDNA samples in a total volume of 25 μl of reaction mixture containing 10 mm Tris (pH 9.0), 50 mm KCl, 0.1 % Triton X-100, 1.5-2.5 mm MgCl2, 0.3 mm each of PCR-grade dATP, dCTP, dGTP and dTTP, 0.25 μg each of forward and reverse primers, and 0.75 units of recombinant Taq DNA polymerase (Gibco-BRL). PCR was performed in a PTC dual block DNA engine (MJ Research Inc., Watertown, MA, USA) using the general cycle profile: 94 °C for 2 min, [94 °C for 30 s, annealing temperature for 50 s, 72 °C for 50 s] × 10 + [94 °C for 30 s, annealing temperature for 50 s, 72 °C for 50 s + 20 s extension/cycle] × remaining number of cycles. PCR primer pairs (Table 1) were constructed from human specific sequence data (Entrez-NIH) using DNA analysis computer software (Lazergene, DNASTAR, Inc., Madison, WI, USA). Prior testing of annealing temperature, MgCl2 concentration and number of PCR cycles was performed to establish optimal conditions within the linear range for PCR amplification. PCR products were separated by gel (2.5 % agarose) electrophoresis, stained with ethidium bromide and visualized by UV exposure using a CCD integrating camera (Gel Doc, Bio-Rad, Hercules, CA, USA), and quantified under non-saturated conditions using analysis software (Molecular Analyst, Bio-Rad). For each subject, samples from the low glycogen and control conditions (all time points) were run simultaneously to allow for relative comparisons.
Table 1.
Primers and reaction conditions used for PCR
| Gene | Forward primer | Reverse primer | AT(°C) | [MgCl2](mM) | Size(bp) |
|---|---|---|---|---|---|
| UCP3 | 5′AGAACCATCGCCAGGGAGGAAGGA3′ | 5′CACCGGGGAGGCCACCACTGT3′ | 52 | 2.0 | 216 |
| PDK4 | 5′CGGCTTGCCAATTTCTCGTCTGTA3′ | 5′TGATCCCGTAAAGTGTCCTGAGTG3′ | 53 | 2.0 | 272 |
| HKII | 5′TCAACCCCGGCAAGCAGAGG3′ | 5′CCGCCGGGCCACCACAGT3′ | 58 | 1.5 | 287 |
| LPL | 5′GTCCGCGGGCTACACCAAACT3′ | 5′ACTCGGGGCTTCTGCATACTCAAA3′ | 57 | 1.5 | 220 |
| BA | 5′CCCAAGGCCAACCGCGAGAAGAT3′ | 5′GTCCCGGCCAGCCAGGTCCAG3′ | 61 | 1.5 | 219 |
AT, annealing temperature; UCP3, uncoupling protein 3; PDK4, pyruvate dehydrogenase kinase 4; HKII, hexokinase II; LPL, lipoprotein lipase; BA, β-actin.
Glycogen determination
Frozen muscle samples (10-20 mg) were freeze-dried, dissected free of connective tissue, weighed and hydrolysed in 1 m HCl. Glycogen concentrations were determined by a standard enzymatic technique with fluorometric detection (Passonneau & Lauderdale, 1974).
Statistics
Transcriptional activity and mRNA contents were normalized to β-actin transcription and mRNA levels respectively, and samples were expressed relative to the corresponding resting (pre) control sample, which was set to 1.0. Values are means ± s.e.m. Two-way ANOVA for repeated measures was used to evaluate the main effects of time and muscle glycogen content for each gene. To determine the specific effect of time (exercise and recovery) within each leg (Study A) or trial (Study B), one-way ANOVA for repeated measures was performed. Student-Newman-Keuls post hoc tests were used to locate specific differences between treatments (glycogen content) and across time. Differences were considered significant at P < 0.05.
RESULTS
Study A
Glycogen
At rest before exercise, muscle glycogen content averaged 609 ± 47 mmol kg−1 dry weight in the control leg and 337 ± 33 mmol kg−1 dry weight in the reduced glycogen leg (i.e. 55 % of control) (P < 0.05, Table 2). During exercise the glycogen content of the control leg decreased (P < 0.05) by 31 % (187 mmol kg−1 dry weight), whereas only 9 % of the glycogen (32 mmol kg−1 dry weight) was used in the reduced glycogen leg. Nevertheless, glycogen content remained significantly lower in the reduced glycogen leg immediately after exercise and through 5 h of recovery (Table 2).
Table 2.
Muscle glycogen (mmol kg−1 dry weight)
| Time points | Control | Low glycogen |
|---|---|---|
| Study A | ||
| Pre-exercise | 609 ± 47 | 337 ± 33† |
| 0 h recovery | 423 ± 43* | 306 ± 36† |
| 5 h recovery | 521 ± 55 | 368 ± 26† |
| Study B | ||
| Pre-exercise | 398 ± 52 | 240 ± 38† |
| 1.5 h exercise | 272 ± 44* | 168 ± 41† |
| 3 h exercise | 153 ± 50* | 101 ± 43 |
| 2 h recovery | 234 ± 41* | 215 ± 49 |
Values are means ± s.e.m.
Significantly (P < 0.05) different from pre-exercise.
Significantly (P < 0.05) different from control.
Transcription and mRNA
Before exercise, transcription of the PDK4, UCP3, HKII and LPL genes was similar in the control and reduced glycogen legs (Fig. 1). In response to exercise, transcription of UCP3 increased 2- to 3-fold in both the control and reduced glycogen legs, although only increases in the reduced glycogen leg reached statistical significance (P < 0.05). There was also a trend for an increase in transcription of PDK4 (≈4-fold, P = 0.057) and HKII (≈3.5-fold, P = 0.062) in the reduced glycogen leg only. LPL transcription did not increase in response to exercise in either leg. Transcription of all genes returned to near pre-exercise levels within 2-5 h after exercise.
Figure 1. Study A: effect of reduced pre-exercise muscle glycogen on transcription of metabolic genes in reponse to 2.5 h of two-legged cycling exercise.
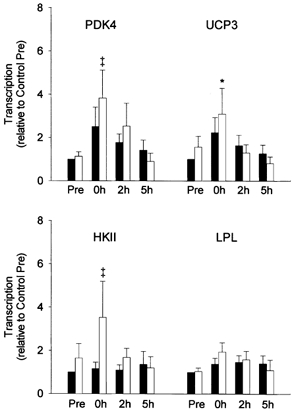
Summary data (mean ± s.e.m.) from the control (▪) and reduced glycogen (□) legs showing fold change in transcription of the pyruvate dehydrogenase kinase 4 (PDK4), uncoupling protein 3 (UCP3), hexokinase II (HKII) and lipoprotein lipase (LPL) genes (relative to β-actin) in response to 2.5 h of two-legged cycling exercise. Muscle glycogen content was reduced in one leg by one-legged cycling exercise and a low carbohydrate diet overnight. Nuclei were isolated from muscle of both the control and reduced glycogen legs before exercise (Pre), immediately after exercise (0h), and after 2 h and 5 h of recovery. Significant (P < 0.05) main effects for time were found for PDK4 and UCP3. * Significantly (P < 0.05) different from Pre in the same leg. ‡ Indicates a P value of 0.057 for PDK4 and 0.062 for HKII relative to Pre in the same leg.
Prior to exercise, PDK4, HKII and LPL mRNA levels were significantly elevated in the reduced glycogen compared with the control leg (Fig. 2), possibly reflecting residual effects from the glycogen depletion protocol of the previous day. In the reduced glycogen leg, exercise did not induce any significant further increase in PDK4, HKII or LPL mRNA. However, in the control leg, exercise elicited 2- to 7-fold (P < 0.05) increases in PDK4, HKII and LPL mRNA to levels similar to those observed in the reduced glycogen leg. In fact, only HKII mRNA remained signifcantly higher during recovery (0 and 5 h recovery time points, Fig. 2) in the reduced glycogen vs.the control leg. Exercise did not induce a detectable change in UCP3 mRNA in either leg.
Figure 2. Study A: effect of reduced pre-exercise muscle glycogen on mRNA content of metabolic genes in reponse to 2.5 h of two-legged cycling exercise.
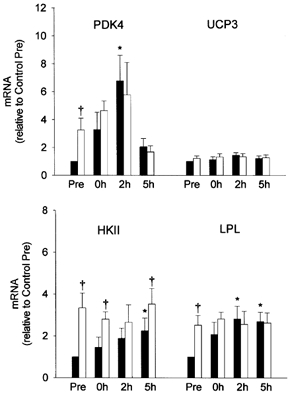
Summary data (mean ± s.e.m.) from the control (▪) and reduced glycogen (□) legs showing fold change in mRNA content of the pyruvate dehydrogenase kinase 4 (PDK4), uncoupling protein 3 (UCP3), hexokinase II (HKII) and lipoprotein lipase (LPL) genes (relative to β-actin) in response to 2.5 h of two-legged cycling exercise. Muscle glycogen content was reduced in one leg by one-legged cycling exercise and a low carbohydrate diet overnight. Total RNA was isolated from muscle of both the control and reduced glycogen legs before exercise (Pre), immediately after exercise (0h), and after 2 and 5 h of recovery. Significant (P < 0.05) main effects for time and glycogen content were found for PDK4 and HKII, respectively. * Significantly (P < 0.05) different from Pre in the same leg. † Significantly (P < 0.05) different from the corresponding time point in control leg.
Study B
Glycogen
Subjects in this study completed two separate trials, one in which muscle glycogen levels were normal (398 ± 52 mmol kg−1 dry weight; control trial) and one in which muscle glycogen levels were lowered to 60 % of control (240 ± 38 mmol kg−1 dry weight; low glycogen trial). During the 3 h of exercise, glycogen utilization was 245 mmol kg−1 dry weight in the control trial and 139 mmol kg−1 dry weight in the low glycogen trial, a relative decrease of ≈60 % in both trials. Glycogen levels were not different between trials after 3 h of exercise or after 2 h of recovery (Table 2).
Respiratory exchange ratio (RER) and plasma glucose, FFA and glycerol concentrations
The differences in muscle glycogen content between the two trials resulted in significant differences in plasma substrate concentrations (Table 3). Before exercise, plasma glucose, FFA and glycerol were not different between trials, although glucose concentration tended to be lower (P = 0.051) in the low glycogen trial. Glucose concentration was signficantly lower in the low glycogen trial at all time points during and after exercise, reaching a low of 3.14 ± 0.10 mmol l−1 in the low glycogen trial and 4.33 ± 0.38 mmol l−1 in the control trial after 3 h of exercise. Exercise also induced significantly greater increases in plasma FFA and glycerol in the low glycogen trial than in the control trial. Accordingly, RER tended to be lower throughout exercise and recovery in the low glycogen trial, although differences did not reach statistical significance.
Table 3.
Arterial plasma glucose, free fatty acids (FFA) and glycerol concentrations (mM) and respiratory exchange ratio (RER) before, during and after exercise
| Glucose | FFA | Glycerol | RER | |||||
|---|---|---|---|---|---|---|---|---|
| Time | CON | LG | CON | LG | CON | LG | CON | LG |
| Pre | 5.08 ± 0.09 | 4.68 ± 0.12† | 0.43 ± 0.07 | 0.64 ± 0.05 | 0.04 ± 0.01 | 0.05 ± 0.01 | 0.82 ± 0.01 | 0.73 ± 0.04 |
| 0.5 h ex | 4.87 ± 0.12 | 4.34 ± 0.15* | 0.60 ± 0.11 | 1.27 ± 0.08* | 0.11 ± 0.02 | 0.22 ± 0.02* | 0.89 ± 0.02 | 0.82 ± 0.06 |
| 1 h ex | 4.64 ± 0.11 | 3.95 ± 0.17* | 0.71 ± 0.12 | 1.61 ± 0.18* | 0.12 ± 0.01 | 0.28 ± 0.02* | 0.88 ± 0.02 | 0.79 ± 0.04 |
| 1.5 h ex | 4.67 ± 0.15 | 3.58 ± 0.21* | 1.11 ± 0.13 | 2.02 ± 0.20* | 0.18 ± 0.02 | 0.35 ± 0.02* | 0.87 ± 0.02 | 0.79 ± 0.06 |
| 2 h ex | 4.65 ± 0.26 | 3.32 ± 0.27* | 1.30 ± 0.13 | 2.34 ± 0.22* | 0.22 ± 0.02 | 0.40 ± 0.02* | 0.88 ± 0.01 | 0.78 ± 0.07 |
| 3 h ex | 4.33 ± 0.38 | 3.14 ± 0.10* | 1.93 ± 0.23 | 2.62 ± 0.29* | 0.32 ± 0.03 | 0.48 ± 0.04* | 0.83 ± 0.02 | 0.77 ± 0.09 |
| 1 h rec | 4.33 ± 0.21 | 3.73 ± 0.17* | 1.24 ± 0.22 | 1.80 ± 0.20* | 0.08 ± 0.01 | 0.11 ± 0.02 | 0.73 ± 0.02 | 0.70 ± 0.03 |
| 2 h rec | 4.56 ± 0.15 | 3.95 ± 0.19* | 1.52 ± 0.25 | 1.64 ± 0.23 | 0.10 ± 0.01 | 0.11 ± 0.01 | 0.79 ± 0.03 | 0.73 ± 0.01 |
Values are means ±s.e.m. Blood was drawn before exercise (Pre), after 0.5 h (0.5 h ex), 1 h (1 h ex), 1.5 h (1.5 h ex), 2 h (2 h ex) and 3 h (3 h ex) of two-legged kicking exercise at 60 % of the resistance which could be sustained for 2 min and 1 h (1 h rec) and 2 h (2 h rec) after the end of exercise. For plasma data n = 6 except at 3 h of exercise where n = 4 and for RER n = 3–6.
Significantly different from control, P < 0.05.
P = 0.051.
Transcription and mRNA
Transcription data for the PDK4 gene was obtained from four subjects and is summarized in Fig. 3. In the control trial, exercise elicited a progressive increase in PDK4 transcription peaking 2 h after exercise (3.5-fold, P < 0.05). In the low glycogen trial, PDK4 transcription was slightly elevated prior to exercise and increased (P < 0.05) further during exercise to a level more than 6 times higher than the control pre-exercise level. At both the 3 h exercise and 2 h recovery time points, transcription of the PDK4 gene was significantly higher in the low glycogen than in the control trial. Transcription was not determined for the UCP3, HKII or LPL genes.
Figure 3. Study B: effect of reduced pre-exercise muscle glycogen on transcription of PDK4 in reponse to 3 h of two-legged dynamic knee-extensor exercise.
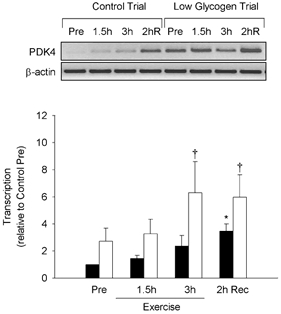
Top, negative images of PCR products (ethidium bromide-stained) generated from RT-PCR-based nuclear run-on analysis of pyruvate dehydrogenase kinase 4 (PDK4) and β-actin genes in response to 3 h of two-legged dynamic knee-extensor exercise. Pre-exercise muscle glycogen content was manipulated by glycogen depletion exercise the previous day (both trials) followed by either a normal (control trial) or low (low glycogen trial) carbohydrate diet overnight. Nuclei were isolated from muscle samples obtained before exercise (Pre), after 1.5 and 3 h of exercise, and after 2 h of recovery. Bottom, summary data (means ± s.e.m.) from the control (▪) and low glycogen (□) trials showing fold change in transcription of the PDK4 gene (relative to β-actin). Significant (P < 0.05) main effects for glycogen (control vs. low) and time (exercise and recovery) were found. * Significantly (P < 0.05) different from Pre within the same trial. † Significantly (P < 0.05) different from the control trial at the same time point.
Glycogen depletion affected the mRNA content of several genes prior to exercise; PDK4, UCP3 and HKII mRNA levels were all significantly elevated (1.5- to 7.6-fold) before exercise in the low glycogen compared with the control trial (Fig. 4 and Fig. 5). In general, this difference between trials persisted during exercise. After 3 h of exercise, PDK4 and UCP3 mRNA were increased 11.4- and 3.5-fold, respectively, compared with the control pre-exercise level (P < 0.05) in the low glycogen trial, but only 5.0- and 1.7-fold, respectively, in the control trial (P > 0.05). During the 2 h recovery period that followed, PDK4 mRNA remained significantly elevated in the low glycogen trial (≈11-fold) but increased sharply in the control trial (≈12-fold over pre-exercise level, P < 0.05), eliminating the difference in PDK4 mRNA between trials. UCP3 mRNA increased by ≈55 % in both trials during the 2 h recovery period and thus remained higher (P < 0.05) in the low glycogen trial. LPL and HKII mRNA increased 2.0- to 2.3-fold in both trials (P < 0.05) with no differences between trials.
Figure 4. Study B: effect of reduced pre-exercise muscle glycogen on mRNA content of metabolic genes in reponse to 3 h of two-legged dynamic knee-extensor exercise.
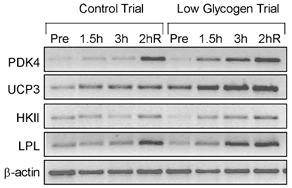
Negative images of PCR products (ethidium bromide-stained) generated from RT-PCR-based mRNA analysis from a single subject for the pyruvate dehydrogenase kinase 4 (PDK4), uncoupling protein 3 (UCP3), hexokinase II (HKII), lipoprotein lipase (LPL) and β-actin genes in response to 3 h of two-legged knee-extensor exercise. Pre-exercise muscle glycogen content was manipulated by glycogen depletion exercise the previous day (both trials) followed by either a normal (control trial) or low (low glycogen trial) carbohydrate diet overnight. Total RNA was isolated from muscle biopsies before exercise (Pre), after 1.5 h and 3 h of exercise, and after 2 h of recovery (2hR).
Figure 5. Study B: effect of reduced pre-exercise muscle glycogen on mRNA content of metabolic genes in reponse to 3 h of two-legged dynamic knee-extensor exercise.
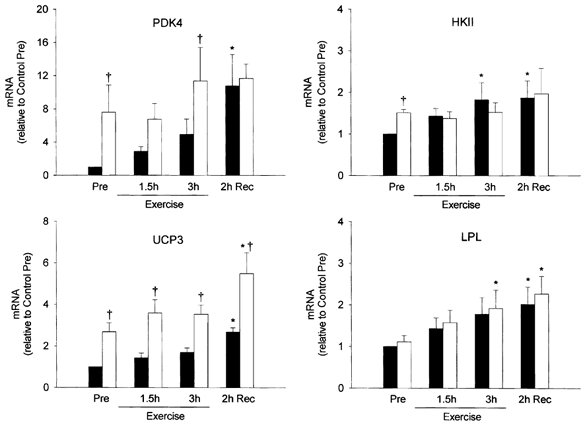
Summary data (means ± s.e.m.) from the control (▪) and low glycogen (□) trials showing fold change in mRNA content of the pyruvate dehydrogenase kinase 4 (PDK4), uncoupling protein 3 (UCP3), hexokinase II (HKII) and lipoprotein lipase (LPL) genes (relative to β-actin) in response to 3 h of two-legged knee-extensor exercise. Pre-exercise muscle glycogen content was manipulated by glycogen depletion exercise the previous day (both trials) followed by either a normal (control trial) or low (low glycogen trial) carbohydrate diet overnight. Total RNA was isolated from muscle biopsies obtained before exercise (Pre), after 1.5 and 3 h of exercise, and after 2 h of recovery. Significant (P < 0.05) main effects of glycogen (control vs. low) were found for PDK4 and UCP3, and of time (exercise and recovery) for PDK4, UCP3 and LPL. * Significantly (P < 0.05) different from Pre in the same trial. † Significantly (P < 0.05) different from the corresponding time point in the control trial.
DISCUSSION
Low intensity (1.5-4 h) exercise activates the transcription of several metabolic genes in skeletal muscle of humans (Pilegaard et al. 2000). Although transcription of some genes is elevated immediately after exercise, the activation of most genes studied thus far occurs primarily during recovery from exercise, peaking within 1-4 h and returning to pre-exercise levels within 24 h after exercise. The timing and transient nature of this response raises the possibility that the regulation of transcription may be linked to other aspects of recovery from exercise. In rats for example, non-insulin-mediated glucose transport and glycogen synthase activity (Ivy & Holloszy, 1981; Garetto et al. 1984; Richter et al. 1984), as well as the metabolic sensitivity to insulin (Mikines et al. 1988; Perseghin et al. 1996), remain elevated in muscle for several hours after exercise, probably reflecting the high priority given to glycogen resynthesis during recovery from exercise (Richter et al. 2001). In both humans and rats, this enhanced muscle insulin sensitivity is highest in the first 2 h after exercise, then progressively reverses as glycogen content is restored (Ivy & Holloszy, 1981; Garetto et al. 1984; Richter et al. 1984; Mikines et al. 1988; Perseghin et al. 1996). However, when muscle glycogen resynthesis is limited by restricting carbohydrate intake, muscle insulin sensitivity remains elevated for at least 48 h (Young et al. 1983; Garetto et al. 1984; Cartee et al. 1989). Although factors associated with elevated FFA levels and/or elevated counterregulatory hormones may play a role triggering the heightened sensitivity to insulin after exercise, recent evidence indicates that this response is a direct consequence of low muscle glycogen content (Nielsen et al. 2001). The major finding of the present study is that the transcriptional activation of PDK4 in response to exercise is enhanced in skeletal muscle when glycogen content is low. Exercise-induced increases in PDK4 and UCP3 mRNA were also enhanced under conditions of low muscle glycogen content. Collectively, these observations suggest that mechanisms involved in substrate utilization during exercise and/or the restoration of muscle glycogen after exercise may be linked to the transcriptional activation of selected metabolic genes in response to exercise.
The glycogen depletion protocols employed in Study A and Study B produced markedly different muscle glycogen concentrations prior to the exercise trials. In Study A, subjects were given specific dietary instructions for the two days preceding the experiment to insure adequate carbohydrate intake (500 g day−1) and muscle glycogen storage. Although the one-legged glycogen depletion protocol carried out on the day before the experiment was effective in generating a difference between the two legs in pre-exercise glycogen concentration (45 % difference, Table 2), absolute glycogen content was relatively high (609 mmol kg−1 dry weight) in the control leg and only slightly lower than normal in the reduced glycogen leg (337 mmol kg−1 dry weight). Exercise (2.5 h of two-legged cycling) increased transcription of the PDK4 and UCP3 genes in both legs (≈2- to 4-fold); however, only responses in the reduced glycogen leg were statistically significant. In Study B, subjects were instructed to consume their normal diet in the days preceding each trial with no specific instruction given concerning carbohydrate intake. Identical exercise-glycogen depletion protocols were followed on the day preceding both the control and low glycogen trials, varying only carbohydrate intake overnight. Using this protocol, pre-exercise muscle glycogen concentration averaged 398 mmol kg−1 dry weight in the control trial and 240 mmol kg−1 dry weight in the low glycogen trial, a difference of 42 % (nearly identical to Study A). The lower absolute glycogen concentration elicited a 2-fold greater increase in PDK4 transcription in response to exercise in the low glycogen trial (> 6-fold) than in the control trial (3-fold). The effect of low pre-exercise glycogen on transcription was confirmed at the mRNA level as PDK4 and UCP3 mRNA were significantly elevated in the low glycogen relative to the control trial. In addition, from this same study (Study B), we have recently reported that low glycogen content also dramatically enhances the exercise-induced transcriptional activation of the IL-6 gene in muscle (Keller et al. 2001). The results from the present study, together with our previous findings (Keller et al. 2001), provide evidence that the transcriptional activation of selected genes in skeletal muscle of humans is increased in response to exercise when pre-exercise muscle glycogen concentration is low.
The results from the present study, along with the well-established inverse relationship between glycogen concentration and insulin action (Richter et al. 2001), suggest that either the absolute level of glycogen or the amount of glycogen used during exercise may be a critical factor sensed and responded to within muscle. In the present study, glycogen utilization during exercise was highest in the two control conditions (control leg in Study A and control trial in Study B), clearly indicating that glycogen breakdown per se was not the key factor influencing the regulation of transcription. In Study B, muscle glycogen concentration decreased to similar levels by the end of exercise in the two trials (≈100-150 mmol kg−1 dry weight; Table 1), whereas PDK4 transcription, as well as PDK4 and UCP3 mRNA, was significantly elevated in the low glycogen trial compared with the control trial. The similar absolute muscle glycogen concentration in the two trials at a time when gene expression was clearly different between trials may argue against a potential link between glycogen content and the regulation of transcription. However, it is important to note that glycogen content was significantly lower in the low glycogen trial throughout the entire exercise bout and that this coincided with increased levels of both PDK4 and UCP3 mRNA. These findings therefore raise the possibility that the influence of glycogen content on the regulation of metabolic gene expression in skeletal muscle may be a gradual response to glycogen utilization, possibly intensifying as the absolute level of glycogen declines during exercise. In fact, consistent with our previous findings (Pilegaard et al. 2000), the induction of PDK4 and UCP3 in the control trial occurred primarily during the recovery period after exercise. Whether this pattern of gene expression is indicative of a regulatory mechanism sensitive to a critically low or ‘threshold’ level of glycogen remains to be determined.
Glycogen metabolism is largely controlled by the actions of the glycogen synthase and glycogen phosphorylase enzymes, both of which are tightly regulated by allosteric effectors and covalent modification via phosphorylation (Roach, 1990, 1991). Dephosphorylation of regulatory sites within the glycogen synthase enzyme, along with glycogen phosphorylase and phosphorylase kinase, serves to coordinate the stimulation of glycogen synthesis. In skeletal muscle, this process is catalysed by PP1G, a glycogen-bound form of protein phosphatase-1 (PP1) that consists of a catalytic subunit, PP1c, complexed with one of three glycogen-targeting subunits, PTG, R6, or RGL (also known as GM) (Stralfors et al. 1985; Armstrong et al. 1997; Printen et al. 1997). Interestingly, a number of glycogen regulatory enzymes, including PP1c and glycogen synthase kinase 3 (GSK3), bind not only with targeting proteins specific for glycogen (Stralfors et al. 1985; Tang et al. 1991; Doherty et al. 1995, 1996; Armstrong et al. 1997; Printen et al. 1997), but also the sarcoplasmic reticulum, myofibrils and the nucleus (Hubbard & Cohen, 1993; Allen et al. 1998), indicating that the intracellular distribution of regulatory enzymes such as PP1c and GSK3, via interaction with specific non-catalytic targeting proteins, plays an important role in determining substrate specificity and enzymatic activity (Hubbard & Cohen, 1993; Ragolia & Begum, 1998). It is therefore possible that, as glycogen content declines during prolonged exercise, enzymes typically bound to the glycogen scaffold (e.g. PP1c, GSK3) may be released and become free to associate with different targeting proteins that direct enzyme activity to other parts of the cell. Likewise, during recovery from exercise, interaction of glycogen-regulating enzymes with glycogen would be expected to increase as glycogen content rises, thereby progressively limiting targeted enzyme activity within other parts of the cell. Whether such a mechanism plays a role in coordinating glycogen resynthesis with the transient regulation of transcription during recovery from exercise warrants further research.
Another possibility is that the observed effects of low muscle glycogen content on gene expression may be related to signalling events triggered by elevated FFA levels and/or utilization. In accordance with this idea, it is interesting to note that, in Study A, a single trial was conducted in which both the control and reduced glycogen legs were exposed to the same FFA concentration, whereas in Study B, two separate trials were performed with the low glycogen trial characterized by significantly higher plasma FFA and glycerol concentrations compared with the control trial. Thus, the more marked induction of PDK4 and UCP3 in the low glycogen trial of Study B compared with the low glycogen leg in Study A may have been due at least in part to differences in FFA concentration between the control and low glycogen trials in Study B. It is well established that FFAs, via their interaction with the peroxisome proliferator activator receptor (PPAR) family of transcription factors, contribute to the regulation of lipid metabolism genes (Latruffe & Vamecq, 1997). For example, fasting elicits marked increases in the expression of both PDK4 and UCP3 in skeletal muscle of rats (Samec et al. 1998; Wu et al. 1999; Hildebrandt & Neufer, 2000; Sugden et al. 2000), due in part to FFA-mediated activation of PPARα (Wu et al. 1999). Interestingly, however, administration of the PPARα agonist, WY-14 643, to fed animals evokes only a partial increase in muscle PDK4 expression (Sugden et al. 2001). Moreover, blocking the increase in serum FFA during starvation by injection of nicotinic acid fails to prevent the fasting-induced increase in UCP3 mRNA in gastrocnemius and tibialis anterior muscle of rats (Samec et al. 1998). Taken together, these findings imply that additional factors unrelated to FFA signalling are also involved in the regulation of PDK4 and UCP3 gene expression in skeletal muscle. The extent to which FFA signalling may contribute specifically to the regulation of transcription in response to exercise has not been determined.
In summary, the results of the present study provide evidence that low muscle glycogen content enhances the transcriptional activation and/or mRNA content of the PDK4 and UCP3 genes in response to exercise in humans. These data raise the possibility that the transient transcriptional activation of selected metabolic genes in response to exercise may be coordinately linked to signalling mechanisms sensitive to glycogen content and/or FFA availability.
Acknowledgments
The authors wish to thank the subjects who participated in this study for their extraordinary effort. The technical assistance of Audrey Hildebrandt, Carsten Bo Nielsen and Hans Søndergaard is gratefully acknowledged. This study was supported by grants from the Danish National Research Foundation (504-14), the National Institute of Arthritis and Musculoskeletal and Skin Diseases (AR-45372), Bethesda, MD, USA, and Team Denmark and The Ministry of Culture Committee on Sports Research.
REFERENCES
- Allen PB, Kwon YG, Nairn AC, Greengard P. Isolation and characterization of PNUTS, a putative protein phosphatase 1 nuclear targeting subunit. Journal of Biological Chemistry. 1998;273:4089–4095. doi: 10.1074/jbc.273.7.4089. [DOI] [PubMed] [Google Scholar]
- Andersen P, Adams RP, Sjogaard G, Thorboe A, Saltin B. Dynamic knee extension as model for study of isolated exercising muscle in humans. Journal of Applied Physiology. 1985;59:1647–1653. doi: 10.1152/jappl.1985.59.5.1647. [DOI] [PubMed] [Google Scholar]
- Armstrong CG, Browne GJ, Cohen P, Cohen PT. PPP1R6, a novel member of the family of glycogen-targetting subunits of protein phosphatase 1. FEBS Letters. 1997;418:210–214. doi: 10.1016/s0014-5793(97)01385-9. [DOI] [PubMed] [Google Scholar]
- Bergstrom J. Muscle electrolytes in man. Scandinavian Journal of Clinical Laboratory Investigation. 1962;68:1–110. [Google Scholar]
- Cartee GD, Young DA, Sleeper MD, Zierath J, Wallberg-Henriksson H, Holloszy JO. Prolonged increase in insulin-stimulated glucose transport in muscle after exercise. American Journal of Physiology. 1989;256:E494–499. doi: 10.1152/ajpendo.1989.256.4.E494. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Analytical Biochemistry. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Doherty MJ, Moorhead G, Morrice N, Cohen P, Cohen PT. Amino acid sequence and expression of the hepatic glycogen-binding (GL)-subunit of protein phosphatase-1. FEBS Letters. 1995;375:294–298. doi: 10.1016/0014-5793(95)01184-g. [DOI] [PubMed] [Google Scholar]
- Doherty MJ, Young PR, Cohen PT. Amino acid sequence of a novel protein phosphatase 1 binding protein (R5). which is related to the liver- and muscle-specific glycogen binding subunits of protein phosphatase 1. FEBS Letters. 1996;399:339–343. doi: 10.1016/s0014-5793(96)01357-9. [DOI] [PubMed] [Google Scholar]
- Garetto LP, Richter EA, Goodman MN, Ruderman NB. Enhanced muscle glucose metabolism after exercise in the rat: the two phases. American Journal of Physiology. 1984;246:E471–475. doi: 10.1152/ajpendo.1984.246.6.E471. [DOI] [PubMed] [Google Scholar]
- Hildebrandt AL, Neufer PD. Exercise attenuates the fasting-induced transcriptional activation of metabolic genes in skeletal muscle. American Journal of Physiology - Endocrinology and Metabolism. 2000;278:E1078–1086. doi: 10.1152/ajpendo.2000.278.6.E1078. [DOI] [PubMed] [Google Scholar]
- Hubbard MJ, Cohen P. On target with a new mechanism for the regulation of protein phosphorylation. Trends in Biochemical Sciences. 1993;18:172–177. doi: 10.1016/0968-0004(93)90109-z. [DOI] [PubMed] [Google Scholar]
- Ivy JL, Holloszy JO. Persistent increase in glucose uptake by rat skeletal muscle following exercise. American Journal of Physiology. 1981;241:C200–203. doi: 10.1152/ajpcell.1981.241.5.C200. [DOI] [PubMed] [Google Scholar]
- Keller C, Steensberg A, Pilegaard H, Osada T, Saltin B, Pedersen BK, Neufer PD. Transcriptional activation of the IL-6 gene in human contracting skeletal muscle: influence of muscle glycogen content. FASEB Journal. 2001;15:2748–2750. doi: 10.1096/fj.01-0507fje. [DOI] [PubMed] [Google Scholar]
- Kraniou Y, Cameron-Smith D, Misso M, Collier G, Hargreaves M. Effects of exercise on GLUT-4 and glycogenin gene expression in human skeletal muscle. Journal of Applied Physiology. 2000;88:794–796. doi: 10.1152/jappl.2000.88.2.794. [DOI] [PubMed] [Google Scholar]
- Latruffe N, Vamecq J. Peroxisome proliferators and peroxisome proliferator activated receptors (PPARs) as regulators of lipid metabolism. Biochimie. 1997;79:81–94. doi: 10.1016/s0300-9084(97)81496-4. [DOI] [PubMed] [Google Scholar]
- Mikines KJ, Sonne B, Farrell PA, Tronier B, Galbo H. Effect of physical exercise on sensitivity and responsiveness to insulin in humans. American Journal of Physiology. 1988;254:E248–259. doi: 10.1152/ajpendo.1988.254.3.E248. [DOI] [PubMed] [Google Scholar]
- Neufer PD, Dohm GL. Exercise induces a transient increase in transcription of the GLUT-4 gene in skeletal muscle. American Journal of Physiology. 1993;265:C1597–1603. doi: 10.1152/ajpcell.1993.265.6.C1597. [DOI] [PubMed] [Google Scholar]
- Neufer PD, Ordway GA, Williams RS. Transient regulation of c-fos, alpha B-crystallin, and hsp70 in muscle during recovery from contractile activity. American Journal of Physiology. 1998;274:C341–346. doi: 10.1152/ajpcell.1998.274.2.C341. [DOI] [PubMed] [Google Scholar]
- Nielsen JN, Derave W, Kristiansen S, Ralston E, Ploug T, Richter EA. Glycogen synthase localization and activity in rat skeletal muscle is strongly dependent on glycogen content. Journal of Physiology. 2001;531:757–769. doi: 10.1111/j.1469-7793.2001.0757h.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Doherty RM, Bracy DP, Granner DK, Wasserman DH. Transcription of the rat skeletal muscle hexokinase II gene is increased by acute exercise. Journal of Applied Physiology. 1996;81:789–793. doi: 10.1152/jappl.1996.81.2.789. [DOI] [PubMed] [Google Scholar]
- O'Doherty RM, Bracy DP, Osawa H, Wasserman DH, Granner DK. Rat skeletal muscle hexokinase II mRNA and activity are increased by a single bout of acute exercise. American Journal of Physiology. 1994;266:E171–178. doi: 10.1152/ajpendo.1994.266.2.E171. [DOI] [PubMed] [Google Scholar]
- Passonneau JV, Lauderdale VR. A comparison of three methods of glycogen measurement in tissues. Analytical Biochemistry. 1974;60:405–412. doi: 10.1016/0003-2697(74)90248-6. [DOI] [PubMed] [Google Scholar]
- Pernow B, Saltin B. Availability of substrates and capacity for prolonged heavy exercise in man. Journal of Applied Physiology. 1971;31:416–422. doi: 10.1152/jappl.1971.31.3.416. [DOI] [PubMed] [Google Scholar]
- Perseghin G, Price TB, Petersen KF, Roden M, Cline GW, Gerow K, Rothman DL, Shulman GI. Increased glucose transport-phosphorylation and muscle glycogen synthesis after exercise training in insulin-resistant subjects. New England Journal of Medicine. 1996;335:1357–1362. doi: 10.1056/NEJM199610313351804. [DOI] [PubMed] [Google Scholar]
- Pilegaard H, Ordway GA, Saltin B, Neufer PD. Transcriptional regulation of gene expression in human skeletal muscle during recovery from exercise. American Journal of Physiology - Endocrinology and Metabolism. 2000;279:E806–814. doi: 10.1152/ajpendo.2000.279.4.E806. [DOI] [PubMed] [Google Scholar]
- Printen JA, Brady MJ, Saltiel AR. PTG, a protein phosphatase 1-binding protein with a role in glycogen metabolism. Science. 1997;275:1475–1478. doi: 10.1126/science.275.5305.1475. [DOI] [PubMed] [Google Scholar]
- Ragolia L, Begum N. Protein phosphatase-1 and insulin action. Molecular and Cellular Biochemistry. 1998;182:49–58. [PubMed] [Google Scholar]
- Richter EA, Derave W, Wojtaszewski JF. Glucose, exercise and insulin: emerging concepts. Journal of Physiology. 2001;535:313–322. doi: 10.1111/j.1469-7793.2001.t01-2-00313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter EA, Garetto LP, Goodman MN, Ruderman NB. Enhanced muscle glucose metabolism after exercise: modulation by local factors. American Journal of Physiology. 1984;246:E476–482. doi: 10.1152/ajpendo.1984.246.6.E476. [DOI] [PubMed] [Google Scholar]
- Roach PJ. Control of glycogen synthase by hierarchal protein phosphorylation. FASEB Journal. 1990;4:2961–2968. [PubMed] [Google Scholar]
- Roach PJ. Multisite and hierarchal protein phosphorylation. Journal of Biological Chemistry. 1991;266:14139–14142. [PubMed] [Google Scholar]
- Samec S, Seydoux J, Dulloo AG. Interorgan signaling between adipose tissue metabolism and skeletal muscle uncoupling protein homologs: is there a role for circulating free fatty acids? Diabetes. 1998;47:1693–1698. doi: 10.2337/diabetes.47.11.1693. [DOI] [PubMed] [Google Scholar]
- Seip RL, Mair K, Cole TG, Semenkovich CF. Induction of human skeletal muscle lipoprotein lipase gene expression by short-term exercise is transient. American Journal of Physiology. 1997;272:E255–261. doi: 10.1152/ajpendo.1997.272.2.E255. [DOI] [PubMed] [Google Scholar]
- Stralfors P, Hiraga A, Cohen P. The protein phosphatases involved in cellular regulation. Purification and characterisation of the glycogen-bound form of protein phosphatase-1 from rabbit skeletal muscle. European Journal of Biochemistry. 1985;149:295–303. doi: 10.1111/j.1432-1033.1985.tb08926.x. [DOI] [PubMed] [Google Scholar]
- Sugden MC, Bulmer K, Holness MJ. Fuel-sensing mechanisms integrating lipid and carbohydrate utilization. Biochemical Society Transactions. 2001;29:272–278. doi: 10.1042/0300-5127:0290272. [DOI] [PubMed] [Google Scholar]
- Sugden MC, Kraus A, Harris RA, Holness MJ. Fibre-type specific modification of the activity and regulation of skeletal muscle pyruvate dehydrogenase kinase (PDK) by prolonged starvation and refeeding is associated with targeted regulation of PDK isoenzyme 4 expression. Biochemical Journal. 2000;346:651–657. [PMC free article] [PubMed] [Google Scholar]
- Tang PM, Bondor JA, Swiderek KM, Depaoli-Roach AA. Molecular cloning and expression of the regulatory (RG1) subunit of the glycogen-associated protein phosphatase. Journal of Biological Chemistry. 1991;266:15782–15789. [PubMed] [Google Scholar]
- Tsuboyama-Kasaoka N, Tsunoda N, Maruyama K, Takahashi M, Kim H, Ikemoto S, Ezaki O. Up-regulation of uncoupling protein 3 (UCP3). mRNA by exercise training and down-regulation of UCP3 by denervation in skeletal muscles. Biochemical and Biophysical Research Communications. 1998;247:498–503. doi: 10.1006/bbrc.1998.8818. [DOI] [PubMed] [Google Scholar]
- Wieland O, editor. Glycerol Assay. New York: Academic Press; 1974. [Google Scholar]
- Williams RS, Neufer PD. Regulation of gene expression in skeletal muscle by contractile activity. In: Rowell LB, Shepherd JT, editors. Handbook of Physiology, section 12, Exercise: Regulation and Integration of Multiple Systems. Bethesda, MD, USA: American Physiological Society; 1996. pp. 1124–1150. [Google Scholar]
- Wojtaszewski JF, Hansen BF, Gade, Kiens B, Markuns JF, Goodyear LJ, Richter EA. Insulin signaling and insulin sensitivity after exercise in human skeletal muscle. Diabetes. 2000;49:325–331. doi: 10.2337/diabetes.49.3.325. [DOI] [PubMed] [Google Scholar]
- Wojtazsewski JF, Nielsen P, Kiens B, Richter EA. Regulation of glycogen synthase kinase-3 in human skeletal muscle: effects of food intake and bicycle exercise. Diabetes. 2001;50:265–269. doi: 10.2337/diabetes.50.2.265. [DOI] [PubMed] [Google Scholar]
- Wu P, Inskeep K, Bowker-Kinley MM, Popov KM, Harris RA. Mechanism responsible for inactivation of skeletal muscle pyruvate dehydrogenase complex in starvation and diabetes. Diabetes. 1999;48:1593–1599. doi: 10.2337/diabetes.48.8.1593. [DOI] [PubMed] [Google Scholar]
- Young JC, Garthwaite SM, Bryan JE, Cartier LJ, Holloszy JO. Carbohydrate feeding speeds reversal of enhanced glucose uptake in muscle after exercise. American Journal of Physiology. 1983;245:R684–688. doi: 10.1152/ajpregu.1983.245.5.R684. [DOI] [PubMed] [Google Scholar]


