Abstract
KCNE4 is a membrane protein belonging to a family of single transmembrane domain proteins known to have dramatic effect on the gating of certain potassium channels. However, no functional role of KCNE4 has been suggested so far. In the present paper we demonstrate that KCNE4 is an inhibitory subunit to KCNQ1 channels. Co-expression of KCNQ1 and KCNE4 in Xenopus oocytes completely inhibited the KCNQ1 current. This was reproduced in mammalian CHO-K1 cells. Experiments with delayed expression of mRNA coding for KCNE4 in KCNQ1-expressing oocytes suggested that KCNE4 exerts its effect on KCNQ1 channels already expressed in the plasma membrane. This notion was supported by immunocytochemical studies and Western blotting, showing no significant difference in plasma membrane expression of KCNQ1 channels in the presence or absence of KCNE4. The impact of KCNE4 on KCNQ1 was specific since no effect of KCNE4 could be detected if co-expressed with KCNQ2-5 channels or hERG1 channels. RT-PCR studies revealed high KCNE4 expression in embryos and adult uterus, where significant expression of KCNQ1 channels has also been demonstrated.
The KCNE family is a group of small, non-conducting, single transmembrane domain proteins known to modulate certain voltage-gated potassium channels (Takumi et al. 1988; Abbott et al. 1999). Five different KCNE proteins have been described and functional roles for three of these (KCNE1-3) have been determined. The KCNE proteins function as accessory β-subunits. Until now, interactions with the following potassium channels have been described: KCNQ1 (modulated by KCNE1-3), KCNQ4 (modulated by KCNE3), Kv3.4 (modulated by KCNE3), Kv4.2 and Kv4.3 (modulated by KCNE2), HCN1 and HCN2 (modulated by KCNE2), and hERG (modulated by KCNE1-3). KCNE1 (minK) expressed together with KCNQ1 (KvLQT1) produces a current that resembles the slow delayed rectifier current (IKs) found in cardiac myocytes, KCNE2 (MiRP1) changes the gating properties of hERG, Kv4.2 and Kv4.3 as well as KCNQ1 channels, and enhances expression levels and activation kinetics for HCN1 and HCN2 channels. Finally, KCNE3 (MiRP2) co-expressed with KCNQ1 mediates a constitutively open potassium channel, and when KCNE3 is expressed together with Kv3.4 it resembles current recorded from native skeletal muscle (Barhanin et al. 1996; Sanguinetti et al. 1996; McDonald et al. 1997; Piccini et al. 1999; Schroeder et al. 2000; Tinel et al. 2000; Abbott et al. 1999, 2001; Yu et al. 2001; Zhang et al. 2001). With respect to the KCNE4 (MiRP3) subunit no function or physiological role has yet been suggested.
KCNQ1 is a six transmembrane domain voltage-gated K+ channel which plays an important role in controlling the potassium current in several tissues. KCNQ1 mRNA has been found in heart, kidney, liver, lungs, gastro-intestinal organs, thymus, exocrine pancreas, salivary glands, uterus and stria vascularis in the cochlea (Barhanin et al. 1996; Sanguinetti et al. 1996; Neyroud et al. 1997; Gould & Pfeifer, 1998; Jentsch, 2000). KCNQ1 expression induces a voltage-dependent, slowly activating and slowly deactivating outward potassium current.
When KCNQ1 is expressed together with KCNE1 the activation kinetics are considerably slowed and the voltage dependency shifts towards more positive potentials, giving a current resembling the slow delayed rectifier cardiac current IKs (Sanguinetti & Jurkiewicz, 1990; Barhanin et al. 1996; Sanguinetti et al. 1996). The functional interaction between KCNQ1 and KCNE1 subunits also clearly emerges from studying patients in whom mutations in either KCNQ1 or KCNE1 genes can lead to prolonged cardiac QT intervals (Romano Ward syndrome, autosomal dominant) or prolonged QT intervals combined with congenital deafness (Jervell and Lange Nielsen syndrome, autosomal recessive) (Wang et al. 1996; Neyroud et al. 1997; Splawski et al. 1997; Tyson et al. 1997; Duggal et al. 1998; Priori et al. 1999; Jentsch, 2000).
Recently, Tinel et al. (2000) demonstrated that co-expression of KCNQ1 with KCNE2 modifies the KCNQ1 characteristics, leading to currents with an apparently instantaneous activation, a rapid deactivation process, and a linear current-voltage relationship. KCNE2 is like KCNE1 found in the heart and a potential function of the KCNQ1 -KCNE2 complex may therefore be to modulate the resting potential. Co-expression of KCNQ1 and KCNE3 forms a constitutively open potassium channel at all membrane potentials and may be important for chloride secretion in the intestine where both subunits are expressed (Schroeder et al. 2000).
KCNE4 was first described by Abbott et al. (1999) but the function of this subunit remains unknown so far. Here we demonstrate that co-expression of KCNE4 and KCNQ1 in both Xenopus oocytes and CHO-K1 cells results in an almost total inhibition of the KCNQ1 current at all membrane potentials. Furthermore, we find that KCNE4 does not alter the currents generated by KCNQ2-5 or hERG1 channels. KCNE4 mRNA is found predominantly in embryo and adult uterus where KCNQ1 expression has also been demonstrated (Gould & Pfeifer, 1998).
METHODS
Molecular biology
To obtain robust expression in Xenopus laevis oocytes, cDNAs coding for KCNQ1-5, KCNE1 (kindly provided by S. Goldstein, Yale University School of Medicine, New Haven, CT, USA), mKCNE3 (accession no. W71921, NCBI), mKCNE4 (accession no. 391797, NCBI) and hERG1 (kindly provided by G. A. Robertson, University of Wisconsin-Madison Medical School, WI, USA) were subcloned into a dual function oocyte and mammalian expression vector named pXOOM, containing 5′ and 3′ untranslated regions for the β-globin gene together with a poly-A sequence (Krieg & Melton, 1984; Jespersen et al. 2002).
For expression in mammalian CHO cells, KCNQ1 (from pCl-KvLQT1, kindly provided by M. Lazdunski, Institute de Pharmacologie Moleculaire et Cellulaire, CNRS, Valbonne, France) and the internal ribosome entry site (IRES) from EMCV (from pIRES-puro, Clontech, Palo Alto, CA, USA) was PCR amplified, linked together by overlap extension and subcloned into the mammalian expression vector pNS2n, a custom-designed derivative of pcDNA3neo (Invitrogen, Carlsbad, CA, USA) expressing a neo-EGFP fusion gene, to give pIRES-KCNQ1. The nucleotide linker between IRES and KCNQ1 was designed to have an optimal sequence for initiation of translation (ACACGATAATACCATG) (Morgan et al. 1992). pKCNE4-KCNQ1 was constructed by inserting the KCNE4 gene upstream of the IRES element in pIRES-KCNQ1. Use of an IRES element for bicistronic expression ensures expression of both KCNE4 and KCNQ1 proteins. The integrity of all PCR-generated constructs was confirmed by sequencing.
RT-PCR
Tissues were isolated from 3-week-old female NMRI mice, which had been killed by cervical dislocation in accordance with Danish ethical legislation. Total RNA was extracted by the RNAqueous-Midi kit (Ambion, Austin, TX, USA) according to the manufacturer's instructions. To avoid DNA contamination, the obtained RNA was treated with DNaseI followed by phenol-chloroform extraction. cDNA synthesis was performed with M-MLV reverse transcriptase (Promega, Madison, WI, USA) and a dN6 random primer, according to the manufacturer's instructions. The used PCR primers are complementary to the 5′ and 3′-end of the KCNE4 coding sequence. Taq polymerase (Amersham Biosciences) was used in the PCR under the following conditions: 95 °C for 30 s, 65 °C for 15 s and 74 °C for 1 min, preceded by 2 min at 95 °C and followed by 5 min at 74 °C.
In vitro transcription and capping were performed using the mCAP mRNA capping kit (Stratagene, La Jolla, CA, USA). mRNA was phenol-chloroform extracted, ethanol precipitated and dissolved in TE buffer (10 mm Tris-HCl, 1mm EDTA, pH 7.5) to a concentration of ∼0.1 μg μl−l. The integrity of the transcripts was confirmed by agarose gel electrophoresis, and mRNA was stored at -80 °C until injection.
Transient expression in Xenopus oocytes and CHO-K1 cells
Xenopus laevis surgery and oocyte treatment were performed as previously described (Grunnet et al. 2001). Xenopus oocytes were collected under anaesthesia from frogs that were humanely killed after the final collection. These experiments were performed according to Danish ethical legislation. Oocytes were kept in Kulori medium (90 mm NaCl, 1 mm KCl, 1 mm MgCl2, 1 mm CaCl2, 5 mm Hepes, pH 7.4) for 24 h at 19 °C before injection of 50 nl mRNA (≈5 ng). For co-expression of K+ channels and KCNE subunits, mRNA was mixed in an approximate 1:1 molar ratio. All injections were performed using a Nanoject microinjector (Drummond Scientific Co., Broomall, PA, USA, USA). Oocytes were kept at 19 °C in Kulori medium for 2-7 days before measurements were performed. The Kulori medium was changed once a day.
CHO-K1 cells (American Type Culture Collection, Manassas, VA, USA) were grown in Dulbecco's modified Eagle's medium (DMEM; Life Technologies) supplemented with 10 % fetal calf serum (FCS; Life Technologies) and 40 mg l−1l-proline (Life Technologies) at 37 °C, in 5 % CO2. One day prior to transfection, 2 × 106 cells were plated in a cell culture T75 flask (Nunc). Cells were transfected with 2 μg of either pIRES-KCNQ1 or pKCNE4-KCNQ1, using Lipofectamine (Life Technologies) according to the manufacturer's instructions. Electrophysiological studies were performed 2 days post transfection.
Electrophysiology
Oocytes
Current through expressed KCNQ1 channels was recorded using a two-electrode voltage-clamp amplifier (Dagan CA-1B, Minneapolis, MN, USA). Electrodes were pulled from borosilicate glass capillaries on a horizontal patch electrode puller (DMZ universal puller, Zeitz instruments, Munich, Germany) and had tip resistances between 0.3 and 2.0 MΩ when filled with 1 m KCl. During the experiments oocytes were placed in a small chamber (volume, 200 μl) connected to a continuous flow system (flow, 6 ml min−1). KCNQ1 channels were activated by membrane depolarization and channel activity was measured in Kulori solution consisting of (mm): 90 NaCl, 1 KCl, 1 MgCl2, 1 CaCl2, 5 Hepes; pH was 7.4. All experiments were performed at room temperature. The condition of each single oocyte was controlled before measurements by recording membrane potentials. Only oocytes with membrane potentials below -40 mV were used for current recordings. An exception was oocytes co-expressing KCNQ1 and KCNE4 where a consistent change to slightly more depolarized membrane potentials was observed. For these experiments only oocytes with membrane potential below -20 mV were used for current recordings.
CHO-K1 cells
Experiments were performed in whole-cell patch-clamp configuration at room temperature with an EPC-9 amplifier (HEKA Elektronik, Lambrecht, Germany). Pipettes were pulled from thin-walled borosilicate glass (ModelOhm, Copenhagen, Denmark) and had a resistance between 1.5 and 2.5 MΩ. A custom-made perfusion chamber (volume, 15 μl) with a fixed AgCl-Ag pellet electrode was mounted on the stage of an inverted microscope.
Coverslips with transient transfected CHO cells were transferred to the perfusion chamber and superfused with a physiological solution consisting of (mm): 150 NaCl, 4 KCl, 2 CaCl2, 1 MgCl2 and 10 Hepes (pH 7.4 with NaOH).
Pipettes were filled with solutions consisting of (mm): 144 KCl, 1 EGTA and 10 Hepes (pH 7.2 with KOH). CaCl2 and MgCl2 were added in concentrations calculated (EqCal, BioSoft, Cambridge, UK) to give a free Mg2+ concentration of 1 mm and free Ca2+ concentrations of 100 nm.
No zero current or leak current subtraction was performed during the experiments. Cell capacitance and series resistance was updated before each pulse application. Series resistance values were between 2.5 and 10.0 MΩ and only experiments where the resistance remained constant during the experiments were analysed. Series resistance was compensated by 75 %. Current signals were low-pass filtered at 3 kHz and acquired using Pulse software (HEKA Elektronik).
Cell surface biotinylation
CHO-K1 cells
The plasma membrane was labelled using the membrane-impermeant biotinylation reagent, sulfo-NHS-SS-biotin (Pierce, Rockford, IL, USA). CHO-K1 cells on coverslips transfected with KCNQ1 or the biscistronic KCNE4-KCNQ1 were rinsed three times at 4 °C with a modified PBS medium called PBS2+ containing CaCl2 and MgCl2 (composition, mm: 136 NaCl, 2.5 KCl, 1.5 KH2PO4, 6.5 NaHPO4, 1 CaCl2, 1 MgCl2) and washed for 5 min. The buffer was removed and biotinylation was performed in 35 mm dishes by incubation in 1 mg ml−l sulfo-NHS-SS-biotin in PBS2+ medium for 2 × 30 min at 4 °C. The biotinylation was terminated by removal of the medium and by five washes (2 min each) in 2 mg ml−l bovine serum albumin (BSA) in PBS2+ medium (BSA/PBS2+ medium) at 4 °C. The cells were then processed for fluorescence microscopy.
Xenopus oocytes
Oocytes injected with KCNQ1 or KCNE4 and KCNQ1 mRNA were rinsed three times at 4 °C in Kulori solution, and washed for 5 min. The buffer was removed and biotinylation was performed in 35 mm dishes by incubation in 1.5 mg ml−l sulfo-NHS-SS-biotin (Pierce) in Kulori solution for 2 × 20 min at 4 °C. The biotinylation was terminated by a 20 min incubation in Kulori solution containing 100 mm glycine.
Preparation of membranes from Xenopus oocytes
For membrane preparation, batches of 50 oocytes injected with KCNQ1 or KCNQ1 and KCNE4 or non-injected oocytes were homogenized in 10 % sucrose dissolved in homogenization buffer (600 mm KCl, 5 mm Mops, 100 μM PMSF, 1 μM pepstatin A, 1 μM p-aminobenzamide, 1 μg ml−l aprotinine and 1 μg ml−l leupeptin pH 6.8) in a volume of 10 μl per oocyte with 10 strokes at 1000 r.p.m. in a glass/Teflon homogenizer (Braun-Melsungen) at 0 °C. The homogenate was placed on top of a step gradient consisting of 7 ml of 50 % sucrose and 3.5 ml of 20 % sucrose in homogenization buffer and centrifuged at 67 000 g for 30 min at 4 °C in a Beckman SW 40 rotor. The interface (between 20 % and 50 % sucrose) was collected and subjected to centrifugation at 84 000 g for 30 min at 4 °C in a Beckman Ti 70.1 rotor. The supernatant was discarded, and the pellet resuspended in 200 μl of medium containing 300 mm sucrose, 100 mm KCl, 5 mm Mops, pH 6.8 and stored at -80 °C until use.
Fluorescence and confocal analysis
CHO-K1 cells were fixed in 4 % (w/v) paraformaldehyde in phosphate-buffered saline (PBS; pH 7.4) for 15 min, washed in PBS and quenched for 20 min with 50 mm NH4Cl in PBS. After washing in PBS, cells were permeabilized with PBS containing 0.1 % (w/v) Triton X-100 and 0.12 % (w/v) fish skin gelatin for 8 min. To reduce non-specific labelling, cells were blocked with 0.25 % (w/v) gelatin in PBS for 30 min. The cells were then exposed to a primary antibody against KCNQ1 (Santa Cruz Biotechnology) for 1 h followed by an incubation with Alexa 488-streptavidin and Alexa 568-donkey anti-goat IgG (Molecular Probes, Leiden, The Netherlands) for 45 min. After washing, the coverslips were mounted in Prolong antifade (Molecular Probes, Leiden, The Netherlands).
The fluorescent-labelled samples were examined by sequential laser scanning confocal microscopy using a LEICA TCS SP2 equipped with argon and helium-neon lasers.
Streptavidin-coupled agarose adsorption
All steps were performed on ice unless otherwise indicated. Extracts of purified oocyte membranes were diluted in 0.5 vol of a 1:1 slurry of streptavidin-coupled agarose beads (Sigma) in binding buffer (1 % TX-100, 150 mm NaCl, 5 mm EDTA, 50 mm Tris-HCl, pH 7.5). Samples were incubated overnight at 4 °C with end-over-end rolling mixing. The beads were then pelleted by centrifugation for 2 min at 367 g. They were washed three times in binding buffer, twice in binding buffer containing 0.5 m NaCl and once in 50 mm Tris (pH 7.5). The pelleted beads were resuspended in 50 mm Tris (pH 7.5) and Laemmli sample buffer (0.0625 m Tris, 9.9 % glycerol, 2 % SDS, 5 % β-mercaptoethanol, 0.00125 % Bromophenol Blue) added. Biotinylated proteins were eluted by heating for 4 min on a heating block at 100 °C, and were immediately collected using a syringe. The eluates were analysed using SDS-PAGE followed by immunoblotting.
Immunoblotting
SDS-PAGE (8 % acrylamide) was performed using the Bio-Rad Laboratories minigel system (Hercules, CA, USA). Proteins were transferred onto a hybond-P PVDF transfer membrane (Amersham Biosciences, 0.45 μm) in 25 mm Tris base, 200 mm glycine, 20 % methanol using a mini trans-blot (Bio-Rad Laboratories). After transfer, the membrane was incubated for 1 h at room temperature in blocking buffer (PBS containing 5 % low-fat milk powder and 0.1 % Tween-20). The membrane was incubated overnight in blocking buffer containing an antibody directed against KCNQ1 (1/100 dilution, Santa Cruz Biotechnology, Santa Cruz, CA, USA). After washing, bound antibody was revealed with HRP-conjugated donkey anti-goat IgG antibody (1/10000, Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) in blocking buffer for 30 min, followed by visualization with the ECLPlus detection system (Amersham Biosciences) according to manufacturer's instructions. Immunoblots were exposed on hyperfilm ECL (Amersham Biosciences).
Analysis of data
Data analysis and drawings were performed using IGOR software (WaveMetrics, Lake Oswego, OR, USA) or GraphPad Prism (GraphPad Software, San Diego, CA, USA) software. All deviations of calculated mean values are given as s.e.m. values.
RESULTS
KCNE4 inhibits KCNQ1 current
To investigate a potential role for KCNE4 subunits we expressed either KCNQ1 channels alone or KCNQ1 channels together with KCNE4 subunits in oocytes from Xenopus laevis. As expected, KCNQ1-expressing oocytes showed a slowly activating voltage-dependent current, which was activated at potentials more positive than -50 mV (Fig. 1A). In contrast, the presence of KCNE4 resulted in a dramatic decrease in the measured KCNQ1 current (Fig. 1B). The corresponding averaged I-V curves for 55 and 37 oocytes are shown in Fig. 1C and D, respectively. For oocytes expressing only KCNQ1 channels, the average maximal current measured at +60 mV after 72 h was 3.40 ± 0.8 μA (n = 55). In KCNQ1-KCNE4-expressing oocytes maximum current recorded 72 h after injection was 0.23 ± 0.12 μA (n = 37). To determine if the observed reduction in KCNQ1 current in the presence of KCNE4 reflected a complete inhibition, step protocols were performed with KCNQ1-KCNE4-expressing oocytes in the presence or absence of 5 mm extracellular Ba2+, which inhibits KCNQ1 channels but has a lower affinity for endogenous oocyte currents. The current observed for KCNQ1 and KCNE4 in the presence of Ba2+ was 0.25 ± 0.09 μA (n = 7). This result is not different from that observed in the absence of Ba2+, indicating that KCNE4 completely inhibits the KCNQ1 current.
Figure 1. KCNE4 subunits inhibit KCNQ1 current.
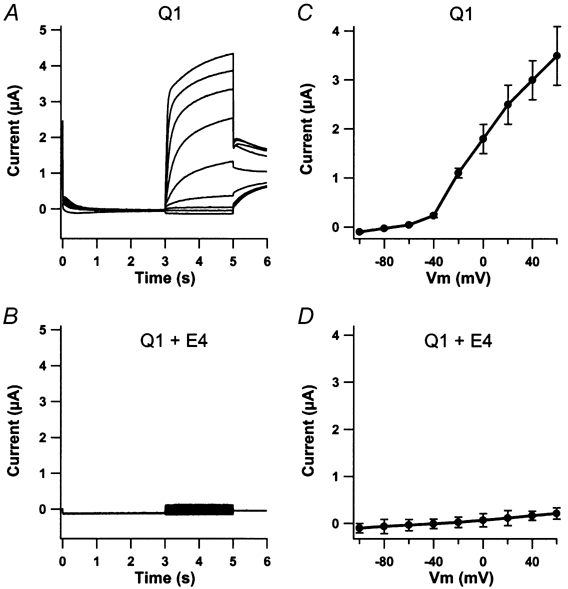
Oocytes were clamped at -80 mV for 3 s and current traces were elicited by 2 s voltage steps for potentials ranging from -100 to +60 mV in 20 mV increments. Tail currents were recorded at -30 mV. Maximal current levels were measured at the end of the 2 s voltage step. Oocytes expressing only KCNQ1 channels (A) responded to this voltage protocol with a slowly activating current having a maximum level at 3.40 ± 0.8 μA recorded at +60 mV (n = 55). In contrast, oocytes injected with both KCNQ1 and KCNE4 mRNA (B) showed only a very limited response to the applied voltage protocol with a maximum current level of 0.28 ± 0.12 μA recorded at +60 mV (n = 37). The corresponding averaged I-V curves revealed a voltage-gated current activated at approximately -50 mV for KCNQ1-expressing oocytes (C) and a linear I-V curve for KCNQ1 -KCNE4 expressing oocytes (D) without any obvious voltage dependency.
It has been demonstrated that KCNE subunits affect the sensitivity of KCNQ1 channels to Ca2+, pH and certain drugs (Unsold et al. 2000; Boucherot et al. 2001). To investigate if the presence of KCNE4 confers the requirement for a specific activator to the KCNQ1 channel, the nature of the KCNE4 inhibition was examined in a number of experiments in the presence or absence of various compounds with possible ion channel modulator effects. In summary, none of the following treatments reversed the KCNE4 inhibition of KCNQ1 current: changes in external pH (6.0 to 8.0, n = 3), increased intracellular Ca2+ (1 μM ionomycin in the extracellular solution, n = 5), increased intracellular cAMP (10 μM forskolin and 500 μM 3-isobutyl-1-methylxanthine (IBMX) in the extracellular solution, n = 11), simultaneously increased intracellular Ca2+ and cAMP (1 μM ionomycin, 10 μM forskolin and 500 μM IBMX in the extracellular solution, n = 5), decreased intracellular ATP (5 min incubation in 3 mm NaAzid, n = 3) or inhibition of cyclic nucleotide-gated kinases and protein kinase C (injection of H-7 (Sigma I 7016); final concentration 0.5 μM, n = 6).
KCNQ1 current is modified by delayed injection of KCNE subunits
To further characterize the inhibitory properties of KCNE4, we performed experiments with delayed injection of KCNE subunits (Fig. 2A). Oocytes were initially injected with KCNQ1 mRNA and specific current was detectable after approximately 36 h. The expression level was followed until an average maximum current level of approximately 3 μA was reached (day 3 after initial injection, recorded at +60 mV). Oocytes were then separated into five groups and four of these were injected with mRNA for KCNE1, KCNE3 or KCNE4, or H2O, and channel activity was measured every sixth hour after injection. As seen in Fig. 2A, oocytes injected with KCNQ1 alone continued to increase expression until a maximal current of approximately 8 μA was reached after 6 days. Delayed injection of mRNA for KCNE1 and KCNE3 modulated the KCNQ1 current. Both subunits augmented the maximum current significantly. For oocytes injected with KCNE1 mRNA this was to be expected since this subunit has been shown to promote membrane translocation of KCNQ1 channels (Romey et al. 1997). In contrast, oocytes re-injected with KCNE4 mRNA showed a drastic decrease in maximum current reaching total inhibition of the KCNQ1 current in approximately 18 h. Oocytes re-injected with water showed, as expected, no difference in KCNQ1 current compared with KCNQ1-expressing oocytes that were not re-injected (data not shown). The present results demonstrate that KCNE subunits can be delayed expressed and still exert their effect on KCNQ1 channels. For the KCNE4 subunit this could mean that the inhibitory nature is due to a fast turnover of KCNQ1 channels, an internalization of KCNQ1 channels already present in the plasma membrane, or an inhibition of KCNQ1 channels in the plasma membrane.
Figure 2. KCNQ1 current can be modified by delayed injection of KCNE subunits.
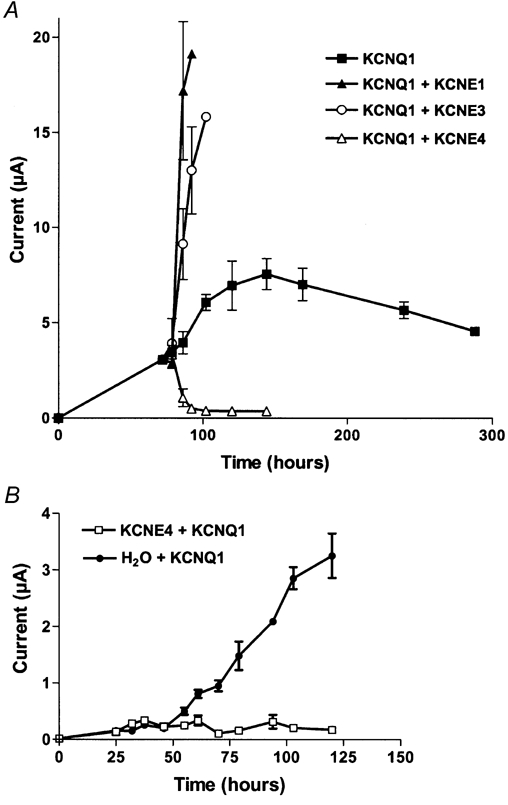
A, oocytes injected with KCNQ1 mRNA alone were incubated for 72 h to secure a stable current level at approximately 3 μA recorded at +60 mV (▪). KCNE1, 3 or 4 were subsequently injected and the current level at +60 mV was measured every 6 h. Both KCNE1 (▴)- and KCNE3 (○)-injected oocytes responded with a large augmentation of the maximal current. In contrast, in KCNE4-injected oocytes (▵) the KCNQ1 current decreased significantly. KCNQ1-expressing oocytes which were not further injected with any KCNE subunits continued to increase expression until a maximal current of approximately 8 μA was reached after 6 days. B, KCNE4-expressing oocytes were injected with KCNQ1 mRNA (□) or H2O (•) after 24 h delay, and KCNQ1 current was subsequently followed. No specific KCNQ1 current could be observed at any time in the presence of KCNE4 indicating that this subunit was not able to increase the turnover of KCNQ1 channels. For every point four individual oocytes were measured and the data averaged.
To determine if KCNE4 inhibits KCNQ1 currents by increasing the turnover rate of conducting KCNQ1 channels, oocytes already expressing KCNE4 were subsequently injected with KCNQ1 mRNA. This experiment is shown in Fig. 2B. Oocytes expressing KCNE4 were further injected 24 h later with KCNQ1 mRNA, and the current level observed at +60 mV was followed for the next 95 h. As can be seen in Fig. 2B, KCNQ1 current could not be observed at any time in the presence of KCNE4 subunits, while non-injected control oocytes subsequently injected with KCNQ1 showed specific current after approximately 48 h. This result speaks in favour of the idea that KCNE4 does not inhibit KCNQ1 channels by increasing the turnover rate of the channel. If this had been the case, it would have been expected that functional KCNQ1 channels were observed for a short period of time in the oocytes pre-injected with KCNE4 subunits. Since this was not observed, a more likely explanation for the observed inhibitory effect of KCNE4 is that KCNE4 subunits interact with the gating of KCNQ1 channels or mediate internalization of functional KCNQ1 channels.
Delayed injection of KCNE subunits not only changed the maximal current level of KCNQ1-expressing oocytes but also time constants for activation and the I-V curves (Fig. 3). As expected, the current in oocytes expressing KCNQ1- KCNE1 had slower activation and the current measured at the end of the 2 s pulses showed slightly outward rectification and a maximal current amplitude of 18.5 ± 1.8 μA, n = 4 (Fig. 3B and F). KCNQ1-KCNE3-expressing oocytes showed a linear I-V curve, a maximal current amplitude of 9.4 ± 1.3 μA, n = 4, and a reversal potential around -95 mV (Fig. 3C and G). KCNQ1-KCNE4-expressing oocytes showed a complete inhibition of the current (Fig. 3D and H, n = 4).
Figure 3. Corresponding current traces and I-V curves from the four groups of oocytes described in Fig. 2.
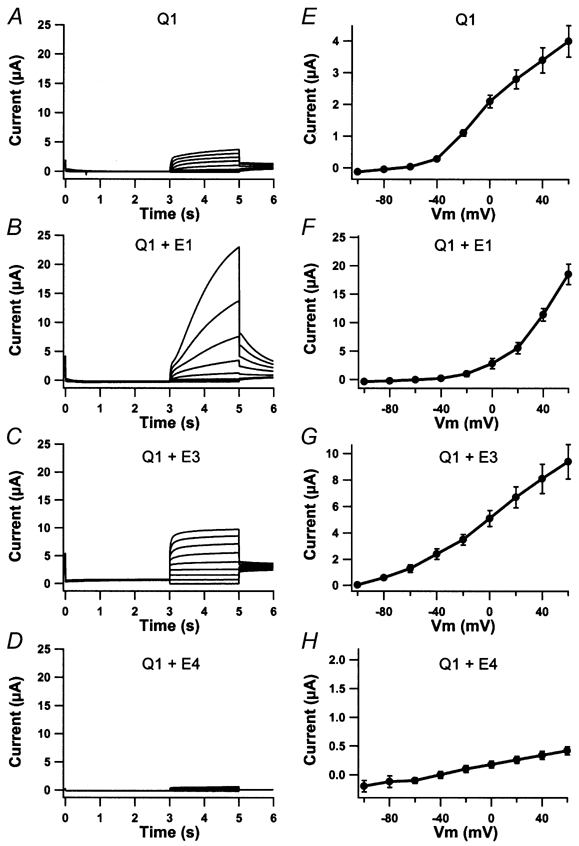
KCNQ1-expressing oocytes in the absence of KCNE injection (A) or after delayed injection with KCNE1 (B), KCNE3 (C) or KCNE4 (D) were activated by voltage steps at potentials ranging from -100 to +60 mV with 20 mV increments. Tail currents were recorded at -30 mV and oocytes were clamped at -80 mV for 3 s between each voltage ramp. The corresponding averaged I-V curves are shown in E-H. Recordings were performed 84 h after KCNQ1 injection and 12 h after KCNE injection.
Taken together, these observations suggest that it is a common mechanism for KCNE subunits to modify the gating of KCNQ1 channels already present in the membrane.
Since KCNE subunits all have a severe impact on KCNQ1 gating, we wished to investigate the effect of expressing more than one KCNE subunit together with KCNQ1. mRNAs coding for KCNE1, KCNE3 and KCNE4 were injected in different combinations in KCNQ1-expressing oocytes. Co-expression of KCNE3 and KCNE4 completely blocked the KCNQ1 activity, demonstrating that KCNE4 subunits are able to compete with the interactions between KCNQ1 and KCNE3 (n = 10). For KCNE1 and KCNE4 a more complex current response was observed: 2 of 11 oocytes responded to voltage activation with a current resembling a reduced KCNQ1-KCNE1 current (maximum current recorded at +60 mV was 5.2 and 3.4 μA, respectively). The rest of the oocytes responded with an inhibition of the KCNQ1 gating which was not different from that observed with KCNE4 in the absence of the KCNE1 subunit (maximum current amplitude recorded at +60 mV, 0.37 ± 0.20 μA).
KCNE4 interacts specifically with KCNQ1
Having established the inhibitory impact of KCNE4 subunits on KCNQ1 current, the specificity of this property was investigated. Oocytes were injected with either KCNQ1, 2, 3, 4 or 5, or hERG1 mRNA and 72 h after injection a stable current level was observed for all constructs (Fig. 4). The maximal current level recorded at +60 mV was: for KCNQ1 channels 3.9 ± 0.5 μA, for KCNQ2 channels 0.69 ± 0.17 μA, for KCNQ2+3 channels 13.6 ± 1.0 μA, for KCNQ4 channels 6.00 ± 2.73 μA and for KCNQ5 channels 10.76 ± 1.54 μA. Maximal current for hERG1 channels was recorded at 0 mV and was 2.55 ± 0.8 μA. KCNQ channels all responded to voltage ramps with slowly activating, non-inactivating currents and hERG1 channels with slowly activating current reaching maximum current amplitude at 0 mV. KCNE4 was subsequently injected into oocytes already expressing each of these channel types and a potential effect on the different KCNQ and hERG currents was recorded 24 h later. As illustrated in Fig. 4, KCNE4 injection only showed an effect on KCNQ1-expressing oocytes. No significant effect on either maximal current or activation/deactivation kinetics was observed for any of the other potassium channels tested. The maximal currents observed approximately 24 h after KCNE4 expression was: for KCNQ1 channels 0.39 ± 0.15 μA, for KCNQ2 channels 1.20 ± 0.50 μA, for KCNQ2+3 channels 12.10 ± 0.48 μA, for KCNQ4 channels 7.40 ± 0.90 μA and for KCNQ5 channels 10.11 ± 2.67 μA. Maximal current for hERG channels was recorded at 0 mV and was 2.24 ± 0.32 μA. These results indicate a specific KCNQ1-KCNE4 interaction. For all different constructs, n = 4.
Figure 4. KCNE4 interacts specifically with KCNQ1.
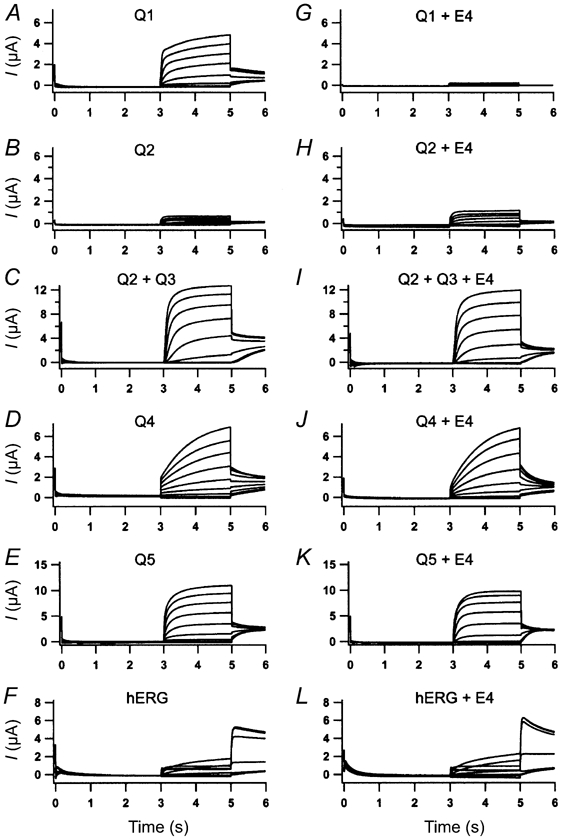
Oocytes injected with KCNQ1 (A), KCNQ2 (B), KCNQ2+3 (C), KCNQ4 (D), KCNQ5 (E) or hERG1 (F) mRNA were subject to voltage activation. From a holding potential of -80 mV that lasted for 3 s, current traces were elicited by 2 s voltage steps ranging from -100 to +60 mV in 20 mV increments. Tail current was recorded at -30 mV. All oocytes responded as expected and were subsequently injected with KCNE4 mRNA. After 24 h incubation, oocytes were again subjected to the above-mentioned voltage protocol. KCNQ1 -KCNE4-expressing oocytes (G) responded with a markedly reduced current while KCNQ2 -KCNE4 (H)-, KCNQ2+3 -KCNE4 (I)-, KCNQ4 -KCNE4 (J)-, KCNQ5 -KCNE4 (K)- and hERG1 -KCNE4 (L)-expressing oocytes showed no change in their response KCNE4. For all panels n = 4.
The inhibitory property of KCNE4 on KCNQ1 channels is similar in oocytes and mammalian CHO-K1 cells
To examine if the inhibitory effect of KCNE4 could be an artifact of the expression system, KCNQ1 or KCNQ1 + KCNE4 were expressed in mammalian CHO-K1 cells (Fig. 5). When activated by voltage ramps, cells transfected with KCNQ1 expressed a slowly activating, non-inactivating, voltage-dependent current (Fig. 5A). The kinetic of this current was not different from KCNQ1 channels expressed in oocytes. When CHO-K1 cells were transfected with a bicistronic construct containing both KCNQ1 and KCNE4, activation by voltage ramps revealed a current response, which was not different from non-transfected CHO-K1 cells (Fig. 5B). This complete inhibition of the KCNQ1 current, as a consequence of the presence of the KCNE4 subunit, is comparable to results obtained in the oocyte system. Corresponding I-V curves are shown in Fig. 5C and D. Together these results indicate that the inhibitory nature of KCNE4 is a general phenomenon rather than an expression system-related incident. The average maximal current density, recorded at +60 mV, for KCNQ1 channels expressed in CHO-K1 cells was 79.3 ± 27 pA pF−1 (n = 5). In KCNQ1-KCNE4-expressing CHO-K1 cells the average maximal current density recorded at +60 mV was 10.3 ± 4 pA pF−1 (n = 6).
Figure 5. KCNE4 inhibition of KCNQ1 current after expression in a mammalian expression system.
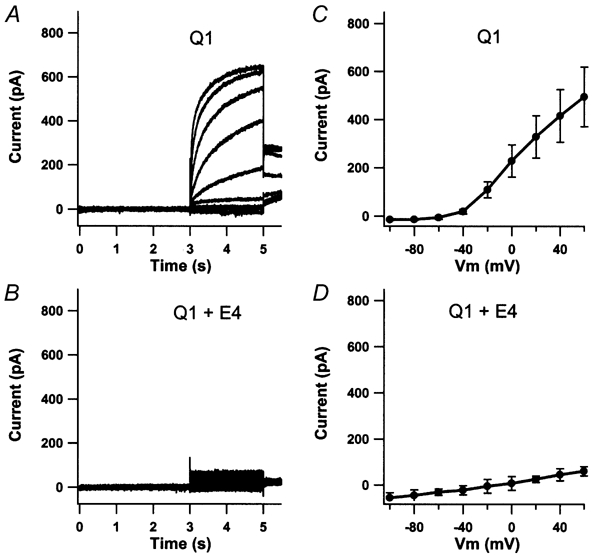
Traces represent whole-cell currents recorded during application of voltage ramps. CHO-K1 cells were clamped at -80 mV for 3 s and current traces elicited by 2 s voltage ramps for potentials from -100 mV to +60 mV in 20 mV increments. Tail currents were recorded at -30 mV. In CHO-K1 cells only expressing KCNQ1, this voltage protocol resulted in a slowly activating, non-inactivating voltage-dependent current (A). The average current amplitude recorded at +60 mV was 79.3 ± 27 pA pF−1 (n = 5). In contrast, co-expression of KCNQ1 and KCNE4 resulted in a dramatic reduction in the observed maximum current with a maximum average current amplitude of 10.3 ± 4 pA pF−1 (n = 6) (B). The responding I-V curves are shown in C and D, respectively.
Cellular distribution of KCNQ1 is unaltered in the presence of KCNE4 in CHO-K1 cells
Experiments with delayed injection of either KCNE4 in KCNQ1-expressing oocytes or KCNQ1 in KCNE4-expressing oocytes indicated that the inhibition due to the KCNE4 subunit is likely to take place in the plasma membrane even though it can still be speculated that KCNE4 works by an internalization of KCNQ1 channels. To obtain additional support for the idea that KCNE4 actually interacts with KCNQ1 in the plasma membrane, immunocytochemistry was performed on transfected CHO-K1 cells. These experiments were carried out by biotinylation of cells transfected either with a construct containing KCNQ1 alone or with a biscistronic construct containing both KCNE4 and KCNQ1. Subsequently, transfected cells were stained both with an antibody raised against KCNQ1 channels and with a fluorescence-labelled streptavidin analogue. KCNQ1 protein was detected both intracellularly and in the plasma membrane, independent of the presence of KCNE4 (Fig. 6). Since streptavidin interacts with the biotinylated plasma membrane this staining can be used as a membrane marker. When an image of the labelling was superimposed onto an image of the KCNQ1 staining, co-localization was detected both in the presence or absence of KCNE4. These results support the idea that the KCNE4 subunit does not change the cellular distribution of KCNQ1 channels and that KCNE4 actually interacts with KCNQ1 channels present in the plasma membrane.
Figure 6. Confocal fluorescence analysis of the subcellular localization of exogenous KCNQ1 in CHO-K1 cells.
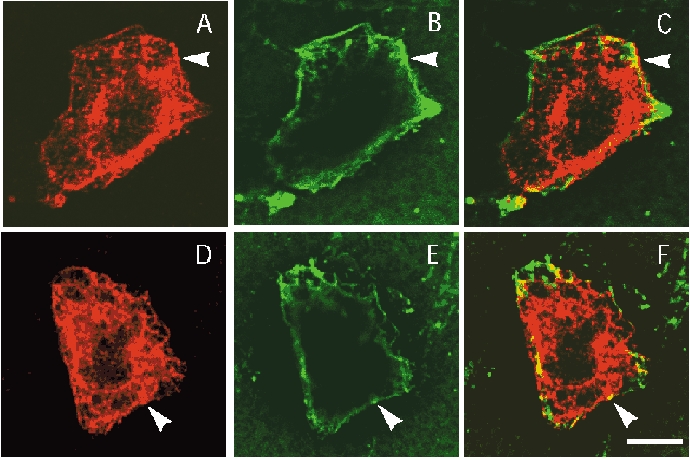
Cells were transfected with KCNQ1 (A-C) or KCNQ1-KCNE4 (D-F) and the cell surface was biotinylated. After biotinylation cells were fixed and permeabilized, then double-labelled with anti-KCNQ1 antibody (red) and Alexa 488-streptavidin (green). In both cases KCNQ1 (A and D) displayed membrane labelling (arrowheads) as demonstrated by the colocalization with Alexa 488-streptavidin staining (B and E). KCNQ1 labelling was also observed in intracellular structures probably due to the high level of expression. C shows images A and B overlaid and F shows D and E overlaid. Scale bar, 8 μm.
To obtain an estimate for the amount of KCNQ1 channels in the plasma membrane in the presence or absence of KCNE4 subunits, localisation experiments were also performed with oocytes expressing either KCNQ1 alone or KCNQ1 and KCNE4. Intact oocytes were biotinylated and their membranes subsequently purified. Biotinylated plasma membrane proteins were isolated, and KCNQ1 channels were visualized by Western blotting using a specific antibody. As seen in Fig. 7, specific staining was observed in membranes from oocytes expressing KCNQ1 channels alone or both KCNQ1 channels and KCNE4 subunits. Together these results demonstrate that the KCNE4 subunit does not modulate the membrane expression of KCNQ1 channels either in mammalian cells or in Xenopus oocytes.
Figure 7. KCNE4 can interact with KCNQ1 present in the plasma membrane.
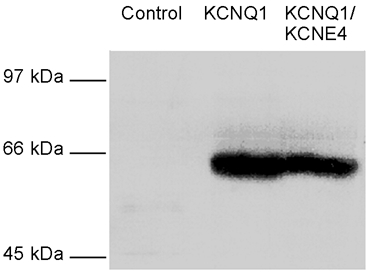
Oocytes were biotinylated with sulfo-NHS-SS-biotin and membranes subsequently purified by differential centrifugation. Biotinylated proteins were isolated on streptavidin-coupled agarose beads. Following SDS-PAGE and electroblotting, transferred biotinylated proteins were stained with the KCNQ1 antibody and revealed using ECL. KCNQ1 was expressed at comparable levels in the plasma membrane in the absence and presence of KCNE4.
Tissue distribution of KCNE4
To obtain information about the distribution of KCNE4 Northern blot analysis was performed on RNA isolated from different NMRI mouse tissues. Unfortunately, the expression levels in the chosen tissues were too low to be detected by Northern blotting so RT-PCR analyses were therefore performed (Fig. 8). RNA extraction was performed in three independent experiments and PCR amplification subsequently performed nine times, all providing consistent results. After 29 PCR cycles clear bands were recognized in uterus and 14-day-old embryo tissue, and faint bands were identified in several other tissues such as kidney, small intestine, lung and heart (Fig. 8A). If the PCR cycle number was increased to 35, clear bands could be detected in all tissues, but not with cDNA prepared from HEK 293 or CHO-K1 cells (Fig. 8B). To achieve equal amounts of cDNA for all PCR reactions, α-tubulin was included as an internal standard (Fig. 8C).
Figure 8. Tissue distribution of KCNE4.
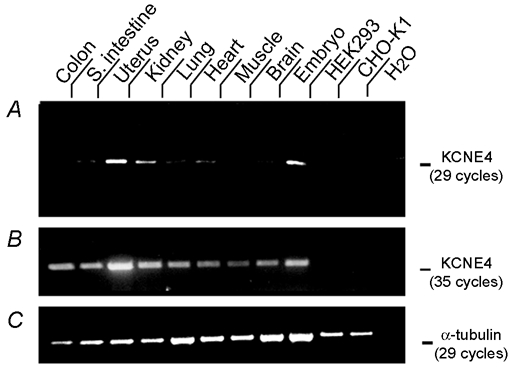
A, RT-PCR analyses of total RNA extracted from NMRI mouse tissue, 14-day-old embryo Swiss Webster mice (Ambion) and HEK 293 and CHO-K1 cells were performed using primers flanking the entire KCNE4 coding region, thereby giving rise to the 487 bp band on an ethidium bromide-stained agarose gel. When 29 PCR cycles were performed, intense labelling was observed in uterus and embryonic tissue. Increasing the number of PCR cycles to 35, gave specific bands for all tissue tested (B). To secure equal amounts of cDNA in each reaction α-tubulin was included as an internal standard. These results are shown in C.
DISCUSSION
KCNQ1 channels are found in a number of non-neuronal tissues as well as in the inner ear and serve functions as diverse as repolarization of heart myocytes and salt secretion in epithelial tissue. A number of recent papers have pointed out that subunits from the KCNE family (KCNE1-3) are necessary and sufficient to provide heterologously expressed KCNQ1 channels with characteristics resembling native cardiac and epithelial currents, demonstrating that KCNQ1 current is strictly modulated through a number of different interactions with KCNE subunits (Sanguinetti et al. 1996; Abbott et al. 1999; Schroeder et al. 2000). The present paper is, to our knowledge, the first report attempting to reveal the function of the KCNE4 subunit. A complete reduction in KCNQ1 current in the presence of the KCNE4 subunit was observed, and this seems to be a phenomenon independent of the applied expression system, since inhibition was observed in both Xenopus oocytes and CHO-K1 cells.
It is known that in addition to their effect on gating, KCNE1 and KCNE3 subunits also alter the sensitivity of KCNQ1 channels to Ca2+ and pH (Unsold et al. 2000; Boucherot et al. 2001). To examine if the efficient inhibition observed with KCNE4 could be abolished by recruitment of such modulators, different attempts to release the inhibition by manipulating the concentration of several intracellular molecules such as Ca2+, H+, cAMP, cGMP and ATP were attempted. Increasing and/or decreasing the concentration of any of these molecules had absolutely no effect on the inhibitory effect of co-expressed KCNE4 subunits on KCNQ1 channels.
The inhibitory property of KCNE4 on KCNQ1 current seems to be very efficient as well as specific since KCNQ2, 3, 4, 5 and hERG1 channels were unaffected by co-expression of KCNE4 when expressed in oocytes. In co-expression studies with KCNQ1 and delayed injection of equal amounts of KCNE3 and KCNE4, all oocytes tested developed current responses to voltage ramps similar to those seen in oocytes injected simultaneously with KCNQ1 and KCNE4. The more complex results obtained from delayed injections of equal amounts of KCNE1 and KCNE4 mRNA in KCNQ1-expressing oocytes are interesting. In these experiments it was possible to obtain both KCNQ1- KCNE1- and KCNQ1-KCNE4-like currents, suggesting that it could be possible that more than one KCNE subunit at a time can interact with the KCNQ1 channel complex.
When KCNE4 mRNA was injected into KCNQ1-expressing oocytes, a fast inhibitory effect (detectable after 12 h) on the KCNQ1 current was observed. A fast change of KCNQ1 gating was also seen after delayed expression of KCNE1 or KCNE3 subunits. This phenomenon indicates that KCNQ1 channels might constitute a baseline current that in certain tissues may be modulated by interaction with newly synthesized KCNE subunits. In the oocyte expression system this idea is supported by the relatively slow turnover of KCNQ1 channels, reaching a maximum after 6 days, and the very fast translation and translocation of KCNE subunits, being able to exert their effect on KCNQ1 channels within 12 h after mRNA injection.
The fact that KCNQ1 channels can be inhibited by delayed injection of KCNE4 subunits is in contrast to what has been reported for another potassium channel β-subunit, namely Kvβ4. This cytoplasmatic β-subunit has been demonstrated to specifically increase the surface expression of Kv2.2 channels without affecting any kinetic properties of these channels (Fink et al. 1996). However, the impact of Kvβ4 is significantly correlated to the timing of RNA injection. With simultaneous injection of Kv2.2 and Kvβ4 mRNA, surface expression of Kv2.2 is increased up to 6-fold. In contrast, delayed injection of Kvβ4 only increased Kv2.2 surface expression slightly, pointing to the fact that the two proteins have to assemble early in the secretory pathway (Fink et al. 1996). Shi et al. (1996) have confirmed this idea. By applying pulse-chase experiments in combination with immunoprecipitation studies it was shown that Kvβ4 interacts with the N-terminus of Kv2.2 while translation is still in progress (Shi et al. 1996). Since results in this paper demonstrated that delayed injection of KCNE4 mRNA is able to inhibit KCNQ1 channels, it may be suggested that this inhibitory property is not dependent on an early assembly in the secretory pathway but could take place in the plasma membrane.
Having demonstrated the time scale for the KCNE4- KCNQ1 interaction, further experiments were conducted to examine if this interaction takes place in the plasma membrane. Two different strategies were applied. In CHO-K1 cells, distribution of KCNQ1 channels in the presence or absence of KCNE4 protein was investigated. In addition, biotinylation of surface proteins was performed to obtain information about both intracellular distribution and membrane distribution of KCNQ1 channels. No obvious difference in cellular distribution was observed after introduction of KCNE4 in these cells, indicating that KCNE4 does not exert its inhibitory effect by a marked redistribution of KCNQ1 channels. To clarify if KCNE4 actually inhibits KCNQ1 channels located in the plasma membrane, without changing the number of channels in the membrane, experiments with biotinylated and purified plasma membranes from oocytes were performed. In this study we have demonstrated that the number of KCNQ1 channels in the plasma membrane is not significantly changed in the presence of KCNE4. Together these results suggest that KCNE4 inhibits KCNQ1 channels in the plasma membrane.
These observations, in combination with the specificity of KCNE4 and the time scale for the KCNE4-KCNQ1 interaction, are interesting since they point to a possible physiological role of the KCNE4 subunit. The highest level of KCNE4 expression was detected in uterus where a significant amount of KCNQ1 mRNA also has been detected (Gould & Pfeifer, 1998). This colocalization of KCNQ1 and KCNE4 in the same tissue could indicate a physiological role for the demonstrated interaction between the two proteins, for example under hormonal regulation of uterus function. This idea is tempting since hormones regulate the expression level of KCNE1, which is also present in the uterus (Felipe et al. 1994). The time scale for KCNE4 interaction with KCNQ1 is compatible with a hormonal regulation. KCNQ1 expression has also been demonstrated in many other tissues such as lung and intestine where the channel plays a role in secretion (Lock & Valverde, 2000; Mall et al. 2000; Demolombe et al. 2001; Kunzelmann et al. 2001). It could be speculated that KCNE4 is used as a regulatory subunit under physiological or pathophysiological conditions where secretion is reduced since low levels of KCNE4 RNA could be detected in these tissues by RT-PCR. However, there is at present no experimental data to support this idea.
KCNE subunits do not always affect KCNQ1 gating identically in oocyte expression systems and mammalian cells. An example is co-expression of KCNQ1 together with the subunit KCNE2 which reveals a linear I-V relationship when expressed in COS cells (Tinel et al. 2000), but not when expressed in oocytes (Abbott et al. 1999). KCNQ1 and KCNE4 were therefore co-expressed in mammalian CHO-K1 cells. As demonstrated, no difference in the ability of KCNE4 to inhibit the KCNQ1 channel could be found, indicating that the ability of KCNE4 to block KCNQ1 channels is a general phenomenon independent of the expression system.
In summary, the present paper demonstrates inhibitory properties of the KCNE4 subunit on KCNQ1 gating. The presented results indicate that this inhibition is very likely to take place in the plasma membrane. These findings further add to the complex regulation of KCNQ1 channels by interaction with different KCNE subunits, but the exact nature of the interaction between KCNQ1 channels and KCNE4 subunits is still an open question.
Acknowledgments
We thank Birthe Lynderup, Tove Soland and Inge Kjeldsen for competent technical assistance. This work was supported by the Danish Heart Association, John and Birthe Meyer Foundation, The Medical Research Council, The Novo Nordisk Foundation, The Velux Foundation and The Natural Science Research Council.
REFERENCES
- Abbott GW, Butler MH, Bendahhou S, Dalakas MC, Ptacek LJ, Goldstein SA. MiRP2 forms potassium channels in skeletal muscle with Kv3. 4 and is associated with periodic paralysis. Cell. 2001;104:217–231. doi: 10.1016/s0092-8674(01)00207-0. [DOI] [PubMed] [Google Scholar]
- Abbott GW, Sesti F, Splawski I, Buck ME, Lehmann MH, Timothy KW, Keating MT, Goldstein SA. MiRP1 forms IKr potassium channels with HERG and is associated with cardiac arrhythmia. Cell. 1999;97:175–187. doi: 10.1016/s0092-8674(00)80728-x. [DOI] [PubMed] [Google Scholar]
- Barhanin J, Lesage F, Guillemare E, Fink M, Lazdunski M, Romey G. KVLQT1 and lsK (minK) proteins associate to form the I(Ks) cardiac potassium current. Nature. 1996;384:78–80. doi: 10.1038/384078a0. [DOI] [PubMed] [Google Scholar]
- Boucherot A, Schreiber R, Kunzelmann K. Regulation and properties of KCNQ1 (KVLQT1) and impact of the cystic fibrosis transmembrane conductance regulator. Journal of Membrane Biology. 2001;182:39–47. doi: 10.1007/s00232-001-0030-4. [DOI] [PubMed] [Google Scholar]
- Demolombe S, Franco D, de Boer P, Kuperschmidt S, Roden D, Pereon Y, Jarry A, Moorman AF, Escande D. Differential expression of KvLQT1 and its regulator IsK in mouse epithelia. American Journal of Physiology – Cell Physiology. 2001;280:C359–372. doi: 10.1152/ajpcell.2001.280.2.C359. [DOI] [PubMed] [Google Scholar]
- Duggal P, Vesely MR, Wattanasirichaigoon D, Villafane J, Kaushik V, Beggs AH. Mutation of the gene for IsK associated with both Jervell and Lange-Nielsen and Romano-Ward forms of Long-QT syndrome. Circulation. 1998;97:142–146. doi: 10.1161/01.cir.97.2.142. [DOI] [PubMed] [Google Scholar]
- Felipe A, Knittle TJ, Doyle KL, Snyders DJ, Tamkun MM. Differential expression of Isk mRNAs in mouse tissue during development and pregnancy. American Journal of Physiology. 1994;267:C700–705. doi: 10.1152/ajpcell.1994.267.3.C700. [DOI] [PubMed] [Google Scholar]
- Fink M, Duprat F, Lesage F, Heurteaux C, Romey G, Barhanin J, Lazdunski M. A new K+ channel β subunit to specifically enhance Kv2. 2 (CDRK) expression. Journal of Biological Chemistry. 1996;271:26341–26348. doi: 10.1074/jbc.271.42.26341. [DOI] [PubMed] [Google Scholar]
- Gould TD, Pfeifer K. Imprinting of mouse Kvlqt1 is developmentally regulated. Human Molecular Genetics. 1998;7:483–487. doi: 10.1093/hmg/7.3.483. [DOI] [PubMed] [Google Scholar]
- Grunnet M, Jensen BS, Olesen SP, Klaerke DA. Apamin interacts with all subtypes of cloned small-conductance Ca2+-activated K+ channels. Pflügers Archiv. 2001;441:544–550. doi: 10.1007/s004240000447. [DOI] [PubMed] [Google Scholar]
- Jentsch TJ. Neuronal KCNQ potassium channels: physiology and role in disease. Nature Reviews Neuroscience. 2000;1:21–30. doi: 10.1038/35036198. [DOI] [PubMed] [Google Scholar]
- Jespersen T, Grunnet M, Angelo K, Klaerke DA, Olesen SP. Dual-function vector for protein expression in both mammalian cells and Xenopus laevis oocytes. Biotechniques. 2002 doi: 10.2144/02323st05. in the Press. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Research. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg PA, Melton DA. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Research. 1984;12:7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunzelmann K, Bleich M, Warth R, Levy-Holzman R, Garty H, Schreiber R. Expression and function of colonic epithelial KvLQT1 K+ channels. Clinical and Experimental Pharmacology and Physiology. 2001;28:79–83. doi: 10.1046/j.1440-1681.2001.03407.x. [DOI] [PubMed] [Google Scholar]
- Lock H, Valverde MA. Contribution of the IsK (MinK) potassium channel subunit to regulatory volume decrease in murine tracheal epithelial cells. Journal of Biological Chemistry. 2000;275:34849–34852. doi: 10.1074/jbc.C000633200. [DOI] [PubMed] [Google Scholar]
- Mall M, Wissner A, Schreiber R, Kuehr J, Seydewitz HH, Brandis M, Greger R, Kunzelmann K. Role of KVLQT1 in cyclic adenosine monophosphate-mediated Cl− secretion in human airway epithelia. American Journal of Respiratory Cell and Molecular Biology. 2000;23:283–289. doi: 10.1165/ajrcmb.23.3.4060. [DOI] [PubMed] [Google Scholar]
- McDonald TV, Yu Z, Ming Z, Palma E, Meyers MB, Wang KW, Goldstein SA, Fishman GI. A minK-HERG complex regulates the cardiac potassium current IKr. Nature. 1997;388:289–292. doi: 10.1038/40882. [DOI] [PubMed] [Google Scholar]
- Melman YF, Domenech A, de La LS, McDonald TV. Structural determinants of KvLQT1 control by the KCNE family of proteins. Journal of Biological Chemistry. 2001;276:6439–6444. doi: 10.1074/jbc.M010713200. [DOI] [PubMed] [Google Scholar]
- Morgan RA, Couture L, Elroy-Stein O, Ragheb J, Moss B, Anderson WF. Retroviral vectors containing putative internal ribosome entry sites: development of a polycistronic gene transfer system and applications to human gene therapy. Nucleic Acids Research. 1992;20:1293–1299. doi: 10.1093/nar/20.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyroud N, Tesson F, Denjoy I, Leibovici M, Donger C, Barhanin J, Faure S, Gary F, Coumel P, Petit C, Schwartz K, Guicheney P. A novel mutation in the potassium channel gene KVLQT1 causes the Jervell and Lange-Nielsen cardioauditory syndrome. Nature Genetics. 1997;15:186–189. doi: 10.1038/ng0297-186. [DOI] [PubMed] [Google Scholar]
- Piccini M, Vitelli F, Seri M, Galietta LJ, Moran O, Bulfone A, Banfi S, Pober B, Renieri A. KCNE1-like gene is deleted in AMME contiguous gene syndrome: identification and characterization of the human and mouse homologs. Genomics. 1999;60:251–257. doi: 10.1006/geno.1999.5904. [DOI] [PubMed] [Google Scholar]
- Priori SG, Barhanin J, Hauer RN, Haverkamp W, Jongsma HJ, Kleber AG, McKenna WJ, Roden DM, Rudy Y, Schwartz K, Schwartz PJ, Towbin JA, Wilde AM. Genetic and molecular basis of cardiac arrhythmias: impact on clinical management parts I and II. Circulation. 1999;99:518–528. doi: 10.1161/01.cir.99.4.518. [DOI] [PubMed] [Google Scholar]
- Romey G, Attali B, Chouabe C, Abitol I, Guillemare E, Barhanin J, Lazdunski M. Molecular mechanism and functional significance of the MinK control of the KvLQT1 channel activity. Journal of Biological Chemistry. 1997;272:16713–16716. doi: 10.1074/jbc.272.27.16713. [DOI] [PubMed] [Google Scholar]
- Sanguinetti MC, Curran ME, Zou A, Shen J, Spector PS, Atkinson DL, Keating MT. Coassembly of KVLQT1 and minK (IsK) proteins to form cardiac IKs potassium channel. Nature. 1996;384:80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- Sanguinetti MC, Jurkiewicz NK. Two components of cardiac delayed rectifier K+ current. Differential sensitivity to block by class III antiarrhythmic agents. Journal of General Physiology. 1990;96:195–215. doi: 10.1085/jgp.96.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder BC, Waldegger S, Fehr S, Bleich M, Warth R, Greger R, Jentsch TJ. A constitutively open potassium channel formed by KCNQ1 and KCNE3. Nature. 2000;403:196–199. doi: 10.1038/35003200. [DOI] [PubMed] [Google Scholar]
- Shi G, Nakahira K, Hammond S, Rhodes KJ, Schechter LE, Trimmer JS. β subunits promote K+ channels surface expression through effects early in biosynthesis. Neuron. 1996;16:843–852. doi: 10.1016/s0896-6273(00)80104-x. [DOI] [PubMed] [Google Scholar]
- Splawski I, Tristani-Firouzi M, Lehmann MH, Sanguinetti MC, Keating MT. Mutations in the hminK gene cause long QT syndrome and suppress IKs function. Nature Genetics. 1997;17:338–340. doi: 10.1038/ng1197-338. [DOI] [PubMed] [Google Scholar]
- Takumi T, Ohkubo H, Nakanishi S. Cloning of a membrane protein that induces a slow voltage-gated potassium current. Science. 1988;242:1042–1045. doi: 10.1126/science.3194754. [DOI] [PubMed] [Google Scholar]
- Tinel N, Diochot S, Borsotto M, Lazdunski M, Barhanin J. KCNE2 confers background current characteristics to the cardiac KCNQ1 potassium channel. EMBO Journal. 2000;19:6326–6330. doi: 10.1093/emboj/19.23.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson J, Tranebjaerg L, Bellman S, Wren C, Taylor JF, Bathen J, Aslaksen B, Sorland SJ, Lund O, Malcolm S, Pembrey M, Bhattacharya S, Bitner-Glindzicz M. IsK and KvLQT1: mutation in either of the two subunits of the slow component of the delayed rectifier potassium channel can cause Jervell and Lange-Nielsen syndrome. Human Molecular Genetics. 1997;6:2179–2185. doi: 10.1093/hmg/6.12.2179. [DOI] [PubMed] [Google Scholar]
- Unsold B, Kerst G, Brousos H, Hubner M, Schreiber R, Nitschke R, Greger R, Bleich M. KCNE1 reverses the response of the human K+ channel KCNQ1 to cytosolic pH changes and alters its pharmacology and sensitivity to temperature. Pflügers Archiv. 2000;441:368–378. doi: 10.1007/s004240000434. [DOI] [PubMed] [Google Scholar]
- Wang Q, Curran ME, Splawski I, Burn TC, Millholland JM, VanRaay TJ, Shen J, Timothy KW, Vincent GM, de Jager T, Schwartz PJ, Toubin JA, Moss AJ, Atkinson DL, Landes GM, Connors TD, Keating MT. Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nature Genetics. 1996;12:17–23. doi: 10.1038/ng0196-17. [DOI] [PubMed] [Google Scholar]
- Yu H, Wu J, Potapova I, Wymore RT, Holmes B, Zuckerman J, Pan Z, Wang H, Shi W, Robinson RB, El Maghrabi MR, Benjamin W, Dixon J, McKinnon D, Cohen IS, Wymore R. MinK-related peptide 1: A beta subunit for the HCN ion channel subunit family enhances expression and speeds activation. Circulation Research. 2001;88:E84–87. doi: 10.1161/hh1201.093511. [DOI] [PubMed] [Google Scholar]
- Zhang M, Jiang M, Tseng GN. minK-related peptide 1 associates with Kv4. 2 and modulates its gating function: potential role as beta subunit of cardiac transient outward channel? Circulation Research. 2001;88:1012–1019. doi: 10.1161/hh1001.090839. [DOI] [PubMed] [Google Scholar]


