Abstract
Kv channel interacting proteins (KChIPs) are Ca2+-binding proteins with four EF-hands. KChIPs modulate Kv4 channel gating by slowing inactivation kinetics and accelerating recovery kinetics. Thus, KChIPs are believed to be important regulators of Kv4 channels underlying transient outward K+ currents in many excitable cell types. We have cloned a structurally minimal KChIP2 isoform (KChIP2d) from ferret heart. KChIP2d corresponds to the final 70 C-terminal amino acids of other KChIPs and has only one EF-hand. We demonstrate that KChIP2d is a functional KChIP that both accelerates recovery and slows inactivation kinetics of Kv4.3, indicating that the minimal C-terminus can maintain KChIP regulatory properties. We utilize KChIP2d to further demonstrate that: (i) the EF-hand modulates effects on Kv4.3 inactivation but not recovery; (ii) Ca2+-dependent effects on Kv4.3 inactivation are mediated through a mechanism reflected in the slow time constant of inactivation; and (iii) a short stretch of amino acids exclusive of the EF-hand partially mediates Ca2+-independent effects on recovery. Our results demonstrate that distinct regions of a KChIP molecule are involved in modulating inactivation and recovery. The potential ability of KChIP EF-hands to sense intracellular Ca2+ levels and transduce these changes to alterations in Kv4 channel inactivation kinetics may serve as a mechanism allowing intracellular Ca2+ transients to modulate repolarization. KChIP2d is a valuable tool for elucidating structural domains of KChIPs involved in Kv4 channel regulation.
Compelling evidence from transgenic mice, immunoprecipitation data, gene expression analyses and functional heterologous expression studies supports the role of Kv channel interacting proteins (KChIPs) as modulators of Kv4 channels underlying the cardiac transient outward current and neuronal A-type current (An et al. 2000; Rosati et al. 2001; Kuo et al. 2001; Bähring et al. 2001; Guo et al. 2002a; Patel et al. 2002). KChIPs are calcium-binding proteins with four EF-hands that slow inactivation kinetics and accelerate recovery kinetics of Kv4 channels. Although it is presumed that KChIP EF-hands play an important role, little is understood of molecular mechanisms by which KChIPs modulate Kv4 channel gating.
Unlike Shaker potassium channels, Kv4 channels appear not to inactivate by well defined N- and C-type mechanisms (Jerng & Covarrubias, 1997; Jerng et al. 1999). Neither deleting regions of the N- and C-termini nor mutating regions within the pore removes Kv4 channel inactivation. Similarly, while it is known that KChIPs bind to the Kv4 channel N-terminus (An et al. 2000; Bähring et al. 2001), it is unclear how such an interaction produces modulatory effects. The domains of both KChIPs and Kv4 channels involved in these processes are also unknown.
In this study we explore the KChIP interactions modulating Kv4.3 inactivation and recovery kinetics through a novel, structurally minimal KChIP2 isoform, KChIP2d, cloned from ferret heart. KChIP2d corresponds to only the C-terminal 70 amino acids of other KChIP2 isoforms and has only one EF-hand, yet maintains functional effects. KChIP2d is thus an ideal tool for determining critical domains of the KChIP molecule involved in mediating function. We utilize KChIP2d to differentiate Ca2+-dependent and Ca2+-independent functional effects on Kv4.3 gating. In addition, we demonstrate that different regions of the KChIP2d molecule modulate effects on inactivation and recovery kinetics. These results not only elucidate potential mechanisms by which KChIPs modulate Kv4.3 channel function, but also provide insight into the physiological consequences of Ca2+-dependent regulation of Kv4.3.
Methods
Animal protocols
Ferret and Xenopus laevis protocols were conducted under NIH approved guidelines of the Institutional Animal Care and Use Committee, University at Buffalo, SUNY. Eight- to twelve-week-old male ferrets were anaesthetized intraperitoneally (sodium pentobarbital, 35 mg kg−1) and hearts were removed (Brahmajothi et al. 1999). Xenopus oocytes were collected under anaesthetic (0.75 g l−1 3-aminobenzoic acid ethyl ester) from frogs that were humanely killed after the final collection.
Cloning KChIP2d
KChIP2d was cloned from ferret heart using RT-PCR with the primers 5′-CGGCTCTGCGCTCACCTACTG-3′ in the 5′ untranslated region and 5′-CTAGATGACATTGTCAAAGAGCTGC-3′ at the 3′ end of the coding sequence. The multiple PCR products were ligated into pBSII-KS+ (Stratagene, La Jolla, CA, USA) and sequenced. This strategy identified previously reported KChIP clones (Patel et al. 2002) as well as KChIP2d (GenBank AF538875).
RNase protection assays
The KChIP2d RNase protection assay probe representing the first 280 base pairs of KChIP2d was constructed by truncating KChIP2d at the Nde I restriction enzyme site and subcloning into pBSII-KS+. The cyclophilin control probe was prepared as previously described (Patel et al. 2002). Linearized plasmids were transcribed in the presence of α-[32P]rCTP to generate high specific activity probes (MaxiScript Kit, Ambion Inc., Austin, TX, USA). Ferret heart RNA was isolated using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA). RNase protection assay hybridizations and digestions were performed with the RPA III Kit (Ambion).
Site-directed mutagenesis
KChIP2d-mutEF and KChIP2d-Frq constructs were prepared using PCR mutagenesis techniques utilizing primers incorporating the desired nucleotide mutations. A single mutagenic primer, 5′-AGATGGCCAGGAACAAGGATGCCGTGGT-3′, was used for KChIP2d-mutEF and overlapping mutagenic primers, 5′-TGATGGGCAATTATGTCGAACTTGCAGAAGAAGAGGAGGCCCCAA-3′ and 5′-GCCTCCTCTTCTTCTGCAAGTTCGACATAATTGCCCATCATGTCA-3′ were used for KChIP2d-Frq. Incorporation of mutations was verified by DNA sequencing.
In vitro transcribed cRNA
Ferret Kv4.3 (Patel et al. 2002), KChIP2d and derivative constructs were subcloned into the pGEM-HE5 oocyte expression vector. Plasmids were linearized and transcribed with the T7 mMESSAGE mMACHINE Kit (Ambion). RNA quality was checked by glyoxal-agarose gel electrophoresis and concentration was determined by spectrophotometry.
Electrophysiology
Xenopus oocytes, prepared as previously described (Comer et al. 1994), were injected with Kv4.3 cRNA or a 1:1 molar ratio of Kv4.3 and KChIP cRNA. Two-microelectrode voltage-clamp recordings (Gene Clamp 500B, Axon Instruments, Union City, CA, USA) were performed 3–7 days after cRNA injection. Currents were filtered at 1 kHz and digitized at 5 kHz (Digidata 1320A, Axon Instruments). Microelectrodes were filled with 3 m KCl and 10 mm Hepes (pH 7.40) with resistances of 0.8-3.0 MΩ. Recordings (22 ± 2 °C) were conducted in ND 96 solution (mm: 96 NaCl, 2 KCl, 1 MgSO4, 1.8 CaCl2, 5 Hepes, pH 7.40) or in low-chloride ND 96 solution (same composition as above with 96 mm sodium aspartate in place of NaCl) to reduce endogenous chloride currents expressed in certain batches of oocytes.
BAPTA/AM experiments
Three to five days after cRNA injection, following control recordings in ND 96 solution with 0.1 % DMSO, oocytes were superperfused with ND 96 solution plus 0.1 % DMSO and 100 μm BAPTA/AM (Calbiochem, La Jolla, CA, USA) for 10–30 min and a second set of recordings was performed.
Analysis
Data from a minimum of two independently isolated batches of oocytes were acquired and analysed using pCLAMP 8.0 (Axon Instruments) and Microcal Origin (Microcal Software, Inc., Northampton, MA, USA). Determination of statistical significance was conducted using t test analysis, paired and independent as appropriate. Statistical significance was taken at P < 0.01.
Results
Cloning and cardiac expression of KChIP2d
Ferret KChIP2d represents a novel KChIP2 isoform. The cDNA sequence begins in the 5′ untranslated region, omits the sequence region of the start codon utilized by other KChIP2 isoforms, and rejoins in the 3′ region of the coding sequence (Fig. 1A). The sequence deletion results in a truncated protein representing only the C-terminal 70 amino acids common to other KChIP2 isoforms. KChIP2d thus contains only one EF-hand compared to the four present in other KChIPs (Fig. 1B). KChIP2d is thus an ideal candidate for KChIP structure-function analysis.
Figure 1. KChIP2d sequence and cardiac expression.
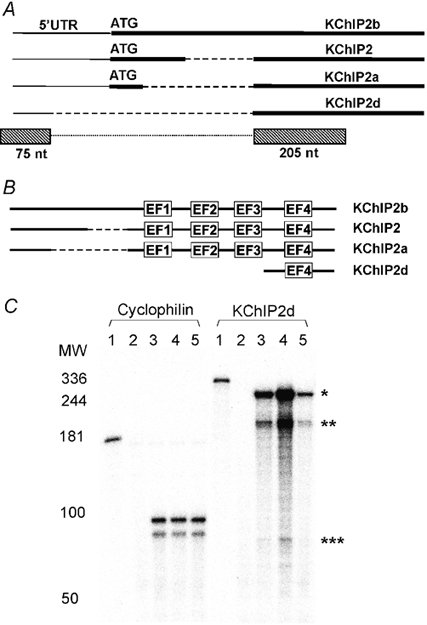
A, schematic diagram (not to scale) of KChIP nucleotide sequences. Thick lines represent coding sequence, thin lines 5′ untranslated region and dashed lines omitted regions. ATG start codons are marked. KChIP2d RNase protection assay probes represented by boxes below the schematic diagram. The dotted line connecting the boxes indicates that sequence regions in the boxes are part of the same probe without a gap in between. B, schematic diagram (not to scale) of KChIP amino acid sequences. Dashed lines represent omitted regions. EF-hands are indicated. C, RNase protection assay hybridizations of cyclophilin and KChIP2d probes (full length probes, lane 1) to yeast RNA (lane 2), ferret heart right ventricular RNA (lane 3), left ventricular epicardial RNA (lane 4) and left ventricular endocardial RNA (lane 5). Five micrograms of ferret heart RNA were hybridized to the cyclophilin control probe and 10 μg to the KChIP2d probe. Double bands in cyclophilin control hybridizations are accounted for by mismatches in the sequence of the probe (Patel et al. 2002). KChIP2d probe protected fragments: KChIP2d (*280 nt); KChIP2+2a+2b (** 205 nt and *** 75 nt). The gradient of KChIP2d expression across the ventricles parallels the distribution of the cardiac transient outward current (Näbauer et al. 1996; Brahmajothi et al. 1999).
To verify expression of KChIP2d in ferret heart, we performed RNase protection assays utilizing an antisense probe designed to the first 280 nucleotides of KChIP2d. This probe would hybridize to one fragment of 280 nucleotides for KChIP2d and two fragments of 205 and 75 nucleotides for other KChIP2 isoforms. Hybridization of the probe to RNA isolated from ferret heart revealed all three bands (Fig. 1C). The band representing KChIP2d was most intense, suggesting a high level of KChIP2d expression.
Ca2+ modulation of KChIP2d function
The most distinct functional effects of KChIPs on Kv4 channels include slowing inactivation kinetics and accelerating recovery kinetics. We therefore determined whether KChIP2d maintained these functions by coexpressing Kv4.3 and KChIP2d in Xenopus oocytes. Surprisingly, similar to other KChIPs, KChIP2d both slowed inactivation and accelerated recovery of Kv4.3 (Fig. 2A).
Figure 2. Ca2+ regulation of KChIP2d.
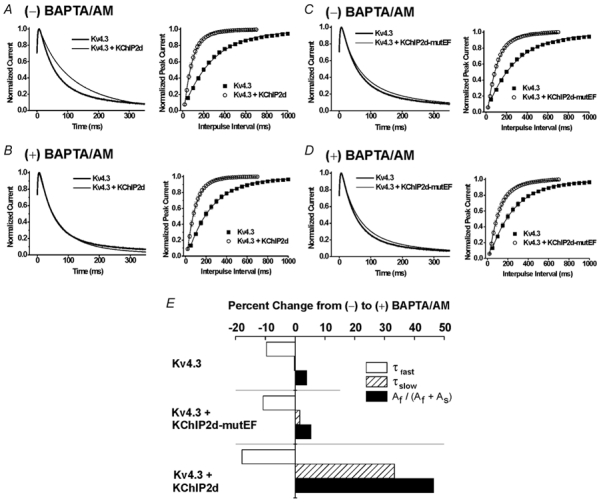
(-) BAPTA/AM indicates results obtained under initial control conditions; (+) BAPTA/AM indicates results recorded in the presence of 100 μm BAPTA/AM. Left panels: representative current recordings of Kv4.3 expressed alone (thick lines) or coexpressed with either KChIP2d or KChIP2d-mutEF (thin lines). Currents were recorded (2000 ms pulses to +50 mV from a holding potential (Vh) of −100 mV, normalized to peak and final steady-state values for comparative purposes) from the same oocytes before and after BAPTA/AM application. First 350 ms illustrated. Right panels: mean recovery kinetics (± s.e.m.; error bars not apparent when smaller than symbol) measured using a double-pulse P1-P2 protocol (P1 and P2 were 1000 ms pulses to +50 mV separated by variable interpulse intervals at Vh = −100 mV). A and B, Kv4.3 alone and Kv4.3+KChIP2d under control conditions (A) and after BAPTA/AM application (B). C and D, Kv4.3 alone and Kv4.3+KChIP2d-mutEF under control conditions (C) and after BAPTA/AM application (D). E, percentage change in time constants of inactivation from (-) to (+) BAPTA/AM. Kv4.3 expressed alone and expressed with either KChIP2d or KChIP2d-mutEF. Bars illustrate mean percentage change of τfast, τslow, and fractional contribution of the fast component of inactivation (Af/(Af + As)). Percentage changes calculated from mean values summarized in Table 1.
Inactivation kinetics of Kv4.3 and Kv4.3 plus KChIP2d during 2 s pulses to +50 mV were well described as the sum of two exponential terms. In the presence of KChIP2d, the fast time constant (τfast) displayed a modest increase while the slow time constant (τslow) decreased two-fold compared to Kv4.3 expressed alone (Table 1). This resulted in an enhanced fractional contribution of the slow component. In summary, the data demonstrate that KChIP2d has two effects on Kv4.3 inactivation: (i) a change in the inherent gating kinetics of the two components of inactivation (slowing of τfast, acceleration of τslow); and (ii) a shift towards the slower component of inactivation (decreased Af/(Af + As), where Af and As are the initial amplitudes of the fast and slow components of inactivation, respectively).
Table 1.
Effects of 100 μm BAPTA/AM on Kv4.3 and Kv4.3 expressed with KChlp2d, KChlp2d-mutEF or KChlp2d-Frq
| (−)BAPTA/AM | (+)BAPTA/AM | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| τf,inact(ms) | τs,inact(ms) | Af/(Af + As) | τrec(ms) | τf,inact(ms) | τs,inact(ms) | Af/(Af + As) | τrec(ms) | n | |
| Kv4.3 | 54.4±0.9 | 425±17.6 | 0.822±0.004 | 293.9±18.4 | 49.1±1.2 | 424.1±12.1 | 0.854±0.006* | 249.6±19.0 | 5 |
| Kv4.3 + KChlp2d | 73.2±4.8 | 206.7±21.7 | 0.540±0.034 | 84.4±8.1 | 60.1±2.1 | 275.5±30.5* | 0.791±0.017 | 85.9±7.6 | 7 |
| Kv4.3 + KChlp2d-mutEF | 67.4±5.1 | 439.0±35.4 | 0.780±0.008 | 103.4±8.1 | 60.1±4.2* | 446.0±32.2 | 0.821±0.008* | 106.8±9.9 | 4 |
| Kv4.3 + KChlp2d-Frq | 54.2±3.1 | 390.3±21.8 | 0.810±0.009 | 194.9±16.2 | — | — | — | — | 12 |
Data are mean ± s.e.m. from the indicated number of oocytes. Inactivation kinetics τf,inact and τs,inact measured at +50mV. Recovery τrec measure at Vh = −100 mV. Asterisks indicate mean values that are significantly different (P < 0.01) between (−) and (+) BAPTA/AM.
A prominent structural feature of KChIP2d is the single EF-hand. To determine if Ca2+ binding to this EF-hand mediated effects on inactivation and recovery kinetics, we examined the effects of decreasing intracellular Ca2+ on these kinetic properties. When oocytes were exposed to BAPTA/AM, inactivation in the presence of KChIP2d resembled that of Kv4.3 alone. However, KChIP2d maintained regulatory effects on Kv4.3 recovery (Fig. 2B). We verified that these effects were a result of Ca2+ binding to the EF-hand by mutating the conserved aspartic acid and glycine at positions 1 and 6 of the EF-hand (Linse & Forsén, 1995) to alanines (KChIP2d-mutEF). As expected, KChIP2d-mutEF had only small effects on Kv4.3 inactivation but retained regulatory effects on recovery (Fig. 2C). In addition, application of BAPTA/AM had minimal effects on inactivation or recovery of Kv4.3 plus KChIP2d-mutEF (Fig. 2D). Thus, KChIP2d-mutEF did not retain residual Ca2+-mediated effects on Kv4.3 gating properties.
To further evaluate Ca2+-dependent KChI2d effects, we analysed the two time constants of inactivation, τfast and τslow, and their fractional contributions in the presence and absence of BAPTA/AM (Fig. 2E, Table 1). In the presence of BAPTA/AM, τfast decreased for Kv4.3 alone as well as for Kv4.3 plus KChIP2d or KChIP2d-mutEF. Because decreases in τfast were seen under all expression conditions, changes in τfast could not be attributed to specific changes mediated by Ca2+ binding to the EF-hand of KChIP2d. In contrast, changes produced by BAPTA/AM in both τslow and the fractional contribution of the two inactivation components (Af/(Af + As)) were most prominent in the presence of KChIP2d but were minimal for Kv4.3 expressed alone or with KChIP2d-mutEF. Specifically, in the presence of KChIP2d, BAPTA/AM increased both τslow and the fractional contribution of the fast component of inactivation. The KChIP2d-specific changes in τslow and fractional contribution in the presence of BAPTA/AM suggested that the KChIP2d EF-hand mediated these changes.
Alternate sites of KChIP2d mediate effects on recovery kinetics
Decreasing intracellular Ca2+ levels or mutating the KChIP2d EF-hand did not affect the ability of KChIP2d to accelerate Kv4.3 recovery kinetics. This suggested that a part of the KChIP2d molecule independent of the EF-hand might mediate the effects on recovery.
KChIPs belong to the neuronal calcium sensor subfamily of Ca2+-binding proteins (Nef, 1996; An et al. 2000). Another member of this family, human frequenin, binds to Kv4 channels and slows inactivation in a Ca2+-dependent manner (Nakamura et al. 2001). However, in contrast to KChIPs, frequenin either does not accelerate recovery or has minimal effects (Nakamura et al. 2001; Guo et al. 2002b). We compared the protein sequences of KChIPs with frequenin to look for regions that might mediate effects on recovery. We identified a stretch of eight amino acids that were highly conserved for all KChIPs but differed for frequenin (Fig. 3A). We mutated six of these amino acids in KChIP2d to the corresponding amino acids of frequenin (KChIP2d-Frq), hypothesizing that this might result in a complete or partial loss of regulatory effects of KChIP2d on recovery. Coexpression of Kv4.3 and KChIP2d-Frq did indeed result in Kv4.3 recovery kinetics that were ≈2.3-fold slower than those seen when KChIP2d and Kv4.3 were coexpressed (Fig. 3B, Table 1; τrec values significantly different, P < 0.01).
Figure 3. Sequence comparison of KChIPs and human frequenin for construction of the KChIP2d-Frq mutation.
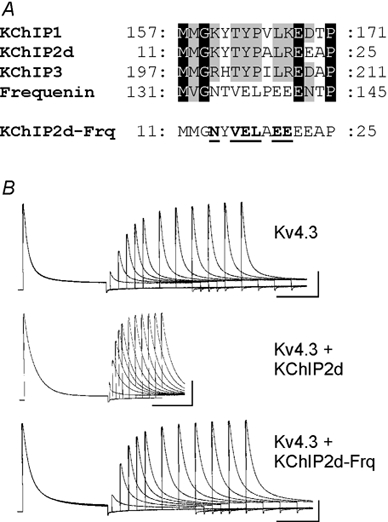
A, sequence alignment of KChIP1 (GenBank AAF33682), KChIP2d, KChIP3 (AAF33684) and frequenin (AF186409) representing the region mutated in KChIP2d-Frq. Black shading, amino acids identical in all sequences. Grey shading, amino acids conserved in at least three of the sequences. Bottom sequence illustrates the KChIP2d-Frq mutant. Mutated amino acids are bold and underlined. B, representative recovery waveforms (Vh = −100 mV; same protocol as in Fig. 2) for Kv4.3, Kv4.3 + KChIP2d, and Kv4.3 + KChIP2d-Frq. Calibration bars: 1 μA, 500 ms.
Interestingly, KChIP2d-Frq also abolished regulatory effects on both τfast and τslow of inactivation. We cannot rule out the possibility that this mutation may be altering binding of the KChIP2d-Frq molecule to Kv4.3. Nonetheless, our data support continued interaction since recovery of KChIP2d-Frq, although significantly slower than KChIP2d, is still significantly faster (P < 0.01) than Kv4.3 alone.
Discussion
KChIP2d, the smallest identified KChIP isoform, maintains regulatory effects on Kv4.3 channels similar to those produced by larger KChIP family members. A minimal region of the common C-terminus, as exemplified by KChIP2d, can therefore mediate regulation of Kv4.3 channel function. Our analysis reveals that the single EF-hand of KChIP2d produces Ca2+-dependent slowing of Kv4.3 inactivation. However, acceleration of Kv4.3 recovery is not dependent upon the EF-hand, but involves other domains of the KChIP2d molecule. Therefore, distinct regions of a KChIP molecule mediate effects on specific Kv4.3 gating properties.
Proposed KChIP model: extrapolation from the crystal structure of human frequenin
KChIPs and human frequenin belong to the same family of calcium-binding proteins (Guo et al. 2002b) and share 59 % amino acid sequence similarity. However, while frequenin slows inactivation of Kv4.3 channels, it has minimal effects on recovery (Nakamura et al. 2001; Guo et al. 2002b). We utilized these functional differences between frequenin and KChIPs in the construction of the KChIP2d-Frq mutant. Because the KChIP crystal structure is unknown, we utilized the crystal structure of human frequenin in its Ca2+-bound form as a model for interpretation of our results (Bourne et al. 2001). Three characteristics of frequenin that may be critical for comparison with KChIP structure-function are (i) a hydrophobic crevice, (ii) four EF-hands aligned on the face opposite the hydrophobic crevice, and (iii) a prominent loop protruding from the periphery of the molecule.
The hydrophobic crevice may exist in KChIPs and represent the site involved in binding to the Kv4 N-terminus (Bourne et al. 2001). This would allow Ca2+ binding to the EF-hand on the opposite face of the KChIP molecule to produce localized conformational changes without altering binding to the Kv4 N-terminus. Such EF-hand-mediated conformational changes may involve exposing hydrophobic and hydrophilic surfaces that interact with cytoplasmic domains of Kv4.3 involved in the slow component of inactivation (Ikura, 1996; Lewit-Bentley & Réty, 2000).
Since the EF-hand is not responsible for modulation of Kv4.3 recovery kinetics, another region of the KChIP2 molecule must be involved. Comparison to frequenin suggests that amino acids altered in the KChIP2d-Frq construct would correspond to a prominent loop protruding from the periphery of the molecule. The cytoplasmic accessibility of this loop makes it a credible candidate for KChIP interactions with a regulatory domain of Kv4.3. The substantially reduced acceleration of recovery produced by KChIP2d-Frq is consistent with this hypothesis. While verification of this model awaits determination of KChIP crystal structure and detailed site-directed mutagenesis analyses, our results are consistent with its predictions.
KChIP2d modulation of Kv4.3 inactivation
KChIP2d specifically promotes a more prominent slow component of Kv4.3 inactivation which is dependent upon Ca2+ binding to the EF-hand. The selective effects of intracellular Ca2+ on τslow and minimal effects on τfast suggest that the two components of inactivation are mediated by distinct molecular mechanisms. However, mutagenesis studies on Kv4 channels have not identified specific domains regulating these two components of inactivation, although evidence indicates that concerted actions of the cytoplasmic N- and C-termini and regions near the internal mouth of the pore are involved (Jerng & Covarrubias, 1997; Jerng et al. 1999).
KChIPs may serve as useful probes for elucidating inactivation mechanisms displayed by Kv4 family members. While all Kv4 channels inactivate as multi-exponential processes, fractional contributions of different components vary considerably amongst family members. Kv4.1 channel inactivation occurs predominantly through the slow component while Kv4.3 channels favour the fast component (Beck et al. 2002). Interestingly, a mutation near the inner mouth of the Kv4.1 pore dramatically slows inactivation (Jerng et al. 1999), while a similar mutation in Kv4.3 results in only modest effects (Wang et al. 2002). These observations suggest that this mutant targets the slow component of inactivation that is more prominent in Kv4.1 than Kv4.3. Of significance is the observation that KChIP and Kv4.3 pore mutant coexpression results in a dramatic slowing of inactivation similar to the effects seen with the Kv4.1 mutant alone (Wang et al. 2002). This is probably due to the KChIP-mediated shift towards the slow component of inactivation. The ability of KChIPs to produce shifts in fractional contributions of components of inactivation provides a powerful means for further evaluating specific Kv4 channel mutations designed to determine molecular mechanisms underlying these processes.
Physiological implications of Ca2+-dependent modulation of KChIP function
Kv4 channels underlie the transient outward current (Ito) recorded in cardiac ventricular myocytes (Nerbonne, 2002). Interesting physiological implications arise from KChIP2d's selective modulation of Kv4.3 inactivation through a Ca2+-dependent mechanism and attenuation of such effects in Xenopus oocytes under basal conditions by BAPTA/AM. In particular, recordings of Ito in cardiac myocytes are conducted in the presence of millimolar levels of EGTA or BAPTA in the pipette and blockers of the L-type Ca2+ current (Näbauer et al. 1996; Brahmajothi et al. 1999; Xu et al. 1999). These manoeuvres significantly attenuate the intracellular Ca2+ transient that occurs during the action potential. A common observation is that inactivation kinetics of Ito are faster than those of Kv4.3 channels expressed with KChIPs (Patel et al. 2002; Deschênes et al. 2002). Our results demonstrating that decreased intracellular Ca2+ levels accelerate inactivation in the presence of KChIPs provide a plausible explanation for this apparent discrepancy: the low [Ca2+]i conditions for myocyte Ito recordings may mimic the effects of BAPTA/AM on KChIPs in oocytes. Thus, Ito inactivation kinetics under normal physiological conditions, where Ca2+ levels increase during the action potential, may be significantly slower than those predicted by conventional patch clamp measurements. Ito may therefore play a much more prominent role in repolarization than presently believed.
The potential ability of KChIP EF-hands to sense intracellular Ca2+ levels and transduce these changes into alterations in Kv4 channel inactivation kinetics may serve as a negative feedback mechanism. This feedback mechanism would be similar to that provided by calmodulin in mediating Ca2+-dependent inactivation of the L-type calcium current (Peterson et al. 1999; Pitt et al. 2001). KChIPs may therefore play dual roles in regulating Kv4 currents in many excitable cell types: (i) Ca2+-independent modulation of recovery; and (ii) Ca2+-dependent modulation of inactivation - a dynamic process dependent upon the magnitude and kinetics of intracellular Ca2+ transients and relative Ca2+ affinities of KChIPs involved.
Acknowledgments
We would like to thank Rajarshi Parai for assistance. This work was supported by NIH grants to H.C.S. (HL52874, HL19216) and D.L.C. (HL58913) and an American Heart Association Established Investigator Award to D.L.C.
References
- An WF, Bowlby MR, Betty M, Cao J, Ling HP, Mendoza G, Hinson JW, Mattsson KI, Strassle BW, Trimmer JS, Rhodes KJ. Modulation of A-type potassium channels by a family of calcium sensors. Nature. 2000;403:553–556. doi: 10.1038/35000592. [DOI] [PubMed] [Google Scholar]
- BÄhring R, Dannenberg J, Peters HC, Leicher T, Pongs O, Isbrandt D. Conserved Kv4 N-terminal domain critical for effects of Kv channel-interacting protein 2. 2 on channel expression and gating. Journal of Biological Chemistry. 2001;276:23888–23894. doi: 10.1074/jbc.M101320200. [DOI] [PubMed] [Google Scholar]
- Beck EJ, Bowlby M, An WF, Rhodes KJ, Covarrubias M. Remodelling inactivation gating of Kv4 channels by KChIP1, a small-molecular-weight calcium-binding protein. Journal of Physiology. 2002;538:691–706. doi: 10.1113/jphysiol.2001.013127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne Y, Dannenberg J, Pollmann V, Marchot P, Pongs O. Immunocytochemical localization and crystal structure of human frequenin (neuronal calcium sensor 1) Journal of Biological Chemistry. 2001;276:11949–11955. doi: 10.1074/jbc.M009373200. [DOI] [PubMed] [Google Scholar]
- Brahmajothi MV, Campbell DL, Rasmusson RL, Morales MJ, Trimmer JS, Nerbonne JM, Strauss HC. Distinct transient outward potassium current (Ito) phenotypes and distribution of fast-inactivating potassium channel alpha subunits in ferret left ventricular myocytes. Journal of General Physiology. 1999;113:581–600. doi: 10.1085/jgp.113.4.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comer MB, Campbell DL, Rasmusson RL, Lamson DR, Morales MJ, Zhang Y, Strauss HC. Cloning and characterization of an Ito-like potassium channel from ferret ventricle. American Journal of Physiology. 1994;267:H1383–1395. doi: 10.1152/ajpheart.1994.267.4.H1383. [DOI] [PubMed] [Google Scholar]
- DeschÊnes I, DiSilvestre D, Juang GJ, Wu RC, An WF, Tomaselli GF. Regulation of Kv4. 3 current by KChIP2 splice variants: a component of native cardiac Ito. Circulation. 2002;106:423–429. doi: 10.1161/01.cir.0000025417.65658.b6. [DOI] [PubMed] [Google Scholar]
- Guo W, Li H, Aimond F, Johns DC, Rhodes KJ, Trimmer JS, Nerbonne JM. Role of heteromultimers in the generation of myocardial transient outward K+ currents. Circulation Research. 2002a;90:586–593. doi: 10.1161/01.res.0000012664.05949.e0. [DOI] [PubMed] [Google Scholar]
- Guo W, Malin SA, Johns DC, Jeromin A, Nerbonne JM. Modulation of Kv4-encoded K+ currents in the mammalian myocardium by neuronal calcium sensor-1. Journal of Biological Chemistry. 2002b;277:26436–26443. doi: 10.1074/jbc.M201431200. [DOI] [PubMed] [Google Scholar]
- Ikura M. Calcium binding and conformational response in EF-hand proteins. Trends in Biochemical Sciences. 1996;21:14–17. [PubMed] [Google Scholar]
- Jerng HH, Covarrubias M. K+ channel inactivation mediated by the concerted action of the cytoplasmic N- and C-terminal domains. Biophysical Journal. 1997;72:163–174. doi: 10.1016/S0006-3495(97)78655-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerng HH, Shahidullah M, Covarrubias M. Inactivation gating of Kv4 potassium channels: molecular interactions involving the inner vestibule of the pore. Journal of General Physiology. 1999;113:641–660. doi: 10.1085/jgp.113.5.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo HC, Cheng CF, Clark RB, Lin JJ, Lin JL, Hoshijima M, NguyÊÑ-TrÂn VT, Gu Y, Ikeda Y, Chu PH, Ross J, Giles WR, Chien KR. A defect in the Kv channel-interacting protein 2 (KChIP2). gene leads to a complete loss of Ito and confers susceptibility to ventricular tachycardia. Cell. 2001;107:801–813. doi: 10.1016/s0092-8674(01)00588-8. [DOI] [PubMed] [Google Scholar]
- Lewit-Bentley A, RÉty S. EF-hand calcium-binding proteins. Current Opinions in Structural Biology. 2000;10:637–643. doi: 10.1016/s0959-440x(00)00142-1. [DOI] [PubMed] [Google Scholar]
- Linse S, ForsÉn S. Determinants that govern high-affinity calcium binding. In: Means AR, editor. Calcium Regulation of Cellular Function. New York: Raven Press, Ltd; 1995. pp. 89–151. [DOI] [PubMed] [Google Scholar]
- NÄbauer M, Beuckelmann DJ, Überfuhr P, Steinbeck G. Regional differences in current density and rate-dependent properties of the transient outward current in subepicardial and subendocardial myocytes of human left ventricle. Circulation. 1996;93 doi: 10.1161/01.cir.93.1.168. [DOI] [PubMed] [Google Scholar]
- Nakamura TY, Pountney DJ, Ozaita A, Nandi S, Ueda S, Rudy B, Coetzee WA. A role for frequenin, a Ca2+-binding protein, as a regulator of Kv4 K+-currents. Proceedings of the National Academy of Sciences of the USA. 2001;98:12808–12813. 168–177. doi: 10.1073/pnas.221168498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nef P. Neuron specific calcium sensors (the NCS subfamily) In: Celio MR, Pauls T, Schwaller B, editors. Guidebook to the Calcium-Binding Proteins. Oxford: Oxford University Press; 1996. pp. 94–98. [Google Scholar]
- Nerbonne JM. Molecular analysis of voltage-gated K+ channel diversity and functioning in the mammalian heart. In: Page E, Fozzard HA, Solaro RJ, editors. Handbook of Physiology, section 2, The Cardiovascular System, The Heart. Vol. 1. New York: Oxford University Press; 2002. p. 594. [Google Scholar]
- Patel SP, Campbell DL, Morales MJ, Strauss HC. Heterogeneous expression of KChIP2 isoforms in the ferret heart. Journal of Physiology. 2002;539:649–656. doi: 10.1113/jphysiol.2001.015156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson BZ, Demaria CD, Adelman JP, Yue DT. Calmodulin is the Ca2+ sensor for Ca2+-dependent inactivation of L-type calcium channels. Neuron. 1999;22:549–558. doi: 10.1016/s0896-6273(00)80709-6. [DOI] [PubMed] [Google Scholar]
- Pitt GS, Zuhlke RD, Hudmon A, Schulman H, Reuter H, Tsien RW. Molecular basis of calmodulin tethering and Ca2+-dependent inactivation of L-type Ca2+ channels. Journal of Biological Chemistry. 2001;276:30794–30802. doi: 10.1074/jbc.M104959200. [DOI] [PubMed] [Google Scholar]
- Rosati B, Pan Z, Lypen S, Wang HS, Cohen I, Dixon JE, McKinnon D. Regulation of KChIP2 potassium channel β subunit gene expression underlies the gradient of transient outward current in canine and human ventricle. Journal of Physiology. 2001;533:119–125. doi: 10.1111/j.1469-7793.2001.0119b.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Patel SP, Qu Y, Hua P, Strauss HC, Morales MJ. Kinetic properties of Kv4. 3 and their modulation by KChIP2b. Biochemical and Biophysical Research Communications. 2002;295:223–229. doi: 10.1016/s0006-291x(02)00658-7. [DOI] [PubMed] [Google Scholar]
- Xu H, Guo W, Nerbonne JM. Four kinetically distinct depolarization-activated K+ currents in adult mouse ventricular myocytes. Journal of General Physiology. 1999;113:661–678. doi: 10.1085/jgp.113.5.661. [DOI] [PMC free article] [PubMed] [Google Scholar]


