Abstract
To further understand molecular mechanisms underlying skeletal muscle hypertrophy, expression profiles of translationally and transcriptionally regulated genes were characterized following an acute bout of maximally activated eccentric contractions. Experiments demonstrated that translational mechanisms contribute to acute gene expression changes following high resistance contractions with two candidate mRNAs, basic fibroblast growth factor (bFGF) and elongation factor-1 alpha (EF1α), targeted to the heavier polysomal fractions after a bout of contractions. Gene profiling was performed using Affymetrix Rat U34A GeneChips with either total RNA or polysomal RNA at one and six hours following contractions. There were 18 genes that changed expression at one hour and 70 genes that were different (60 genes increased:10 genes decreased)at six hours after contractions. The model from this profiling suggests that following high resistance contractions skeletal muscle shares a common growth profile with proliferating cells exposed to serum. This cluster of genes can be classified as ‘growth’ genes and is commonly associated with progression of the cell cycle. However, a unique aspect was that there was induction of a cluster of tumour suppressor or antigrowth genes. We propose that this cluster of ‘antigrowth’ genes is induced by the stress of contractile activity and may act to maintain skeletal muscle in the differentiated state. From the profiling results, further experiments determined that p53 levels increased in skeletal muscle at 6 h following contractions. This novel finding of p53 induction following exercise also demonstrates the power of expression profiling for identification of novel pathways involved in the response to muscle contraction.
It has been well established that high resistance exercise leads to an adaptive response in mammalian skeletal muscle which involves increases in myofibre size (hypertrophy) without increases in the number of fibres (hyperplasia; Booth & Thomason, 1991). However, there is still very little understood about the molecular mechanisms that mediate this adaptation. This is due, in part, to the fact that each bout of muscle contractions results in a complex series of both intra- and intercellular changes within skeletal muscle. Major challenges in understanding the molecular mechanisms underlying exercise-induced muscle adaptations are the complexity of the stimulus coupled with the diversity of phenotypic adaptations seen in skeletal muscle. Expression profiling is an emerging type of highly parallel ‘hypothesis-generating’ research, where expression changes across the entire genome can be assayed then fitted with existing knowledge of physiological pathways. Expression profiling can serve to dramatically expand existing models that have been developed using candidate gene and/or protein studies and can also point to novel physiological processes not previously identified.
Recently, studies of skeletal muscle hypertrophy have begun to identify specific signalling pathways and factors involved in the response to growth-inducing exercise (Baar & Esser, 1999; Dunn et al. 1999; Bodine et al. 2001). In particular, the prolonged activation of p70S6k is associated with exercise-induced muscle hypertrophy. One of the downstream functions of p70s6k has been shown to be the regulation of translation of a subset of mRNAs. These mRNAs contain a 5′ polypyrimidine tract (referred to as 5′ TOP) and they are found in mRNAs encoding translation factors (e.g. eEF1α) and ribosomal proteins (e.g. S6) (Meyuhas & Hornstein, 2000). Thus, when p70S6k is activated, 5′ TOP mRNAs are more efficiently translated, resulting in increased protein production without changes in steady state mRNA levels.
There has been increased interest in the contribution of post-transcriptional and/or translational regulation of gene expression in recent years. Post-transcriptional mechanisms have been shown to be important during development, in response to extracellular stimuli (e.g. insulin) and following viral infections. Most recently, it has been shown that approximately 20 % of the genes regulated following T-cell activation are changed due to translational control (Mathews et al. 2000; Pradet-Balade, 2001). Thus the overall aim of this project was to identify transcriptionally and translationally regulated genes in skeletal muscle following high resistance exercise using oligonucleotide gene array technology. The results from these series of experiments were used to test the following hypotheses: (1) Selective translation of subsets of mRNAs will contribute significantly to changes in gene expression in skeletal muscle following growth-inducing contractions. (2) High resistance contractions will lead to expression of a cluster of genes that are common to other cell types exposed to a growth stimulus. (3) Information from the expression profiling results will lead to the identification of novel pathway(s) involved in the response of skeletal muscle to contractile activity.
Methods
All procedures were approved by the Animal Care Committee of the University of Illinois at Chicago and were in accordance with the Guidelines for Care and Use of Laboratory Animals. Female Wistar rats (Charles River Laboratories, Wilmington, MA, USA) were maintained on a constant 12:12 h light-dark cycle. Upon arrival animals were allowed to acclimatize for six days before any intervention took place. Animals were age- (6-7 weeks) and weight- (208 ± 16 g) matched in all experiments. Food and water were available ad libitum. The surgical and electrical stimulation interventions as well as the tissue collections were performed under anaesthesia (sodium pentobarbital, 50 mg kg−1i.p.) and following tissue collection animals were killed by an intracardiac injection of saturated KCl. After electrode implantation, animals were housed individually and were allowed to recover for five days prior to the experimental exercise bout.
High resistance exercise protocol
The model of resistance exercise used was chosen based on its efficacy in inducing skeletal muscle hypertrophy (Baar & Esser, 1999). Multistrand electrodes (Medwire, Inc., Mount Vernon, NY, USA) were implanted on both sides of the right sciatic nerve above the anatomical branching point. Tetanic contractions were delivered using a Grass S5 Stimulator (Grass Instruments, Quincy, MA, USA) at a frequency of 100 Hz, 6–12 V, 1 ms duration, 9 ms delay for 10 sets of six repetitions, each repetition lasting 3 s. A 10 s recovery was given between repetitions and 1 min was allowed between sets, with the stimulation protocol lasting a total time of 20 min. At 1 and 6 h following the acute bout of contractions, experimental and contralateral control muscles were rapidly removed, weighed, frozen in liquid nitrogen and stored at −80 °C until processed.
Isolation of polysomal RNA and Northern blots
Polysome profiles were generated as described previously (Baar & Esser, 1999). Homogenates from extensor digitorum longus (EDL) and soleus muscles were loaded on a 0.5-1.2 m linear sucrose gradient. The gradients were spun for 120 min at 250 000 g (40 000 r.p.m.) and 2 °C. Following ultracentrifugation, the sample was pumped through a Spectra/Chrom flow-through UV monitor (A254) and fractionated into six equal volumes using a FRAC-100 fraction collector (Amersham Biosciences, Piscataway, NJ, USA). The fractions were incubated with 100 μg ml−1 proteinase K in 100 mm Tris-HCl (pH 7.4), 50 mm EDTA and 5 % SDS for 30 min at 37 °C with gentle agitation. RNA was isolated from the fractions by standard procedures using phenol/ chloroform treatment followed by ethanol precipitation. The RNA was resuspended in water for Northern analyses.
Standard Northern blotting procedures were performed. RNA was run on a 1 % formaldehyde-agarose gel, transferred to nitrocellulose using the Genie electrophoretic transfer system (Idea Scientific, Minneapolis, MN, USA) and crosslinked using a Stratalinker 1800 (Stratagene, La Jolla, CA, USA). Membranes were then incubated overnight in 4 × standard sodium phosphate EDTA buffer (SSPE; 3 m NaCl, 0.2 m sodium phosphate and 0.02 m EDTA), 5 × Denhardt's solution, 1 % SDS and 0.5 mg ml−1 salmon sperm DNA. DNA probes were synthesized using the StripEZ labelling kit (Ambion, Inc., Austin, TX, USA) and the blots hybridized overnight at 65 °C. Following washing, the blots were exposed to film and analysed using autoradiography followed by densitometric scans (AlphaImager 2200; Alpha Innotech Corp., CA, USA). The cDNA probes used were: 1.5 kb of the α-skeletal actin gene (Stratagene, La Jolla, CA, USA), 1 kb of the basic fibroblast growth factor (generously provided by Dr P. Wagner, University of San Diego, CA, USA) and a 680 bp region of the EF1α cDNA generated using RT-PCR with the primers 5′ GCA TCA CCA TCG ACA TCT CC 3′ and 5′ TGC ATT TCC ACA GAC TTC ACC 3′. The specificity of the EF1α probe was tested using mRNA from soleus, EDL, heart and liver (data not shown).
RNA isolation for expression profiling
Each RNA sample for the expression profiling experiments required pooling of muscle tissues from two animals because of the limited RNA yield from polysomes. For this study, two exercised tibialis anterior (TA) muscles were pooled and the unexercised TA muscles from the same two animals were pooled to generate the control/unexercised sample. The two muscles were powdered together under liquid N2. For the isolation of total RNA, 100–150 mg of the powdered muscle was used and total RNA extracted using the TRIZOL reagent (Invitrogen Corp., CA, USA). The remaining powdered muscle (≈0.7 g) was homogenized in three volumes of polysome buffer (25 mm Tris-HCl (pH 8.0), 250 mm KCl, 30 mm MgCl2, 1 mm DTT, with 1 mg ml−1 cycloheximide and 4 U ml−1 RNAsin) following the same protocol described earlier (Baar & Esser, 1999) and the total polysomal fraction was collected. This fraction was incubated with 100 μg ml−1 proteinase K in 100 mm Tris-HCl (pH 7.4), 50 mm EDTA and 5 % SDS for 30 min at 37 °C with gentle agitation. RNA was isolated from the polysome fraction by standard procedures using the TRIZOL reagent.
Expression profiling
Expression profiling was done with Affymetrix Rat Genome U34A GeneChips (48 chips for this study), containing ≈7000 full-length genes and ≈1000 expressed sequence tags (ESTs). Polysomal RNA and total RNA from unexercised TA muscles (control) and exercised contralateral muscles at 1 h (n = 2 samples from muscles from four rats, two muscles per sample) and 6 h (n = 3 samples from six rats) after an acute bout of exercise were generated for this analysis. Biotinylated cRNAs prepared from polysomal and total RNAs were hybridized to U34A GeneChips in duplicate. Procedures of cRNA preparation and microarray processing were performed as previously described (Chen et al. 2000).
Data analysis
Absolute analysis of Affymetrix image data was done using Affymetrix Microarray Suite 4.0 as previously described (Lockhart et al. 1996). Briefly, each gene was queried with 16 ‘perfect match’ 25 bp oligonucleotides (PM) and paired ‘mismatched’ oligonucleotides (MM) designed with a single mismatch in the centre position. Comparison of the hybridization signal from the PM and mm probes allows for a specificity measure of signal intensity and elimination of most non-specific cross-hybridization. Values of intensity differences as well as ratios of each probe pair are used for determination of whether a gene is called ‘present’ or ‘absent’.
The differentially expressed gene lists for each time point were generated using GeneSpring software (Silicon Genetics, CA, USA). In GeneSpring, those genes showing at least four Affymetrix ‘present’ calls were selected prior to further data processing. In other words, only genes that were detected as ‘present’ on at least four Affymetrix microarrays out of a total of 48 microarrays would be included for statistical analysis. This step increases the stringency and quality control of the analysis by taking advantage of the redundancy and bioinformatics inherent to the Affymetrix GeneChips and associated software to ascertain a ‘confidence level’ with regard to a gene's expression above background levels. Welch's t test was performed to calculate the probabilities of significant gene expression changes. Since the probe sets were tested multiple times, we used a highly stringent P value cut-off (P < 0.001) to reduce the number of false positives to less than 1 in 1000.
The criteria for categorizing differentially expressed genes into ‘translationally’, ‘transcriptionally’ and ‘transcriptionally and translationally’ regulated categories are the following: Translationally regulated genes are those in which the P value is less than 0.001 in the polysomal pools but larger than 0.05 in the total RNA pools. Transcriptionally regulated genes are those in which the P value is less than 0.001 in the total RNA pools but larger than 0.05 in the polysomal RNA pools. Transcriptionally and translationally regulated genes are those in which the P value is less than 0.001 in one of the two pools and less than 0.05 in the other.
Immunofluorescent staining
Immunofluorescent staining was performed on both control and exercised muscles following high resistance contractions, as described previously (Chen et al. 2000). Serial 4 μm-thick frozen muscle sections were cut with an IEC Minotome cryostat (Global Medical Instrumentation, Inc., MN, USA), mounted to Superfrost Plus Slides (Fisher Scientific, PA, USA) and fixed in cold anhydrous acetone. Sections were then blocked for 30 min in 1 × PBS containing 10 % horse serum and incubated with anti-c-fos primary antibody (1:10 dilution; Oncogene Research Products, MA, USA) for 3 h at room temperature. Washes were done with 1 × PBS containing 10 % horse serum and sections then incubated with secondary antibody for 1 h. Cy3-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch Laboratories, PA, USA) was diluted 1:500.
Western blot for p53
Skeletal muscle nuclear extracts were obtained as previously described (Blough et al. 1999). Protein concentrations were determined using the DC protein assay (Bio-Rad, Hercules, CA, USA). Fifty micrograms of the nuclear extract were separated by SDS-PAGE on a 12 % gel. After electrophoresis, the proteins were transferred to a nitrocellulose membrane (Bio-Rad). The membrane was blocked for 2 h at 4 °C with 5 % powdered milk in TBST (Tris-buffered saline + 0.1 % Tween). The membrane was then incubated overnight at 4 °C with anti-p53 primary antibody at 1:1000 (Cell Signaling Technology, Inc., Beverly, MA, USA). The membrane was then washed and incubated for 45 min at room temperature with peroxidase-conjugated horse anti-rabbit IgG secondary antibody at 1:5000 (Vector Laboratories, Burlingame, CA, USA). Antibody binding was detected using an enhanced chemiluminescence detection kit (Amersham Biociences). The membrane was then visualized by Ponceau S stain to confirm equal sample loading.
Results
Translation regulation of mRNA subsets following high resistance contractions
We have previously shown that p70S6k phosphorylation levels are elevated at 6 h following an acute bout of high resistance contractions (Baar & Esser, 1999). Since activation of the p70S6k pathway has been shown to modulate translational regulation of subsets of mRNAs in other cell types, we evaluated the distribution of selected mRNAs in the polysome pool in skeletal muscle following contractile activity. Extracts from muscles were separated by sucrose gradient ultracentrifugation and then fractionated into six samples. The six fractions isolated corresponded to: (1) untranslated mRNA and tRNAs; (2) preinitiation mRNA; (3) monosomal mRNA; (4) light polysomal mRNA; (5) middle polysomal mRNA; and (6) heavy polysomal mRNA. We used Northern blots to determine the distribution of two candidate mRNAs that are regulated at the level of translation initiation, bFGF and elongation factor 1 alpha (EF1α) and one mRNA that is not translationally regulated, α-actin (Jefferies & Thomas, 1994; Smith et al. 1999). To quantitatively assess specific mRNA distribution across the fractions we set the strongest band for each sample equal to 1 unit and normalized the signal in the other fractions of the same sample relative to this strongest signal. This analysis will not permit precise quantitative comparison of mRNA levels between the different Northern blots but it does provide a measure of the relative distribution of each mRNA across the different fractions within each sample.
As can be seen in Fig. 1A and B, the distributions of both EF1α and bFGF mRNAs changed at 6 h following one bout of contractions. For EF1α, very little change was seen in mRNA quantity in the untranslated and preinitiation pools (fractions 1 and 2). Levels of EF1α mRNA decreased in the monosome pool (fraction 3) following exercise and this was associated with a very large increase in the number of ribosomes loaded on each EF1α mRNA (see change in fraction 6). In contrast to the pattern for EF1α, the pattern of bFGF mRNA distribution in the control muscle was primarily associated with the monosome fraction, with very little detected in the untranslated, preinitiation and middle to heavy polysome pools. However, at 6 h following high resistance exercise there was a significant shift of bFGF mRNA from the monosome to the heavier polysomal pools, which reflected a greater number of ribosomes per mRNA. As a control for mRNA specificity, we determined the polysomal distribution of α-skeletal actin mRNA, which, based on 5′ untranslated region (UTR) sequence/structure, would not be differentially regulated in the polysome pool. These results are shown in Fig. 1C, with the distribution of actin mRNA not changing in polysomal profiles with exercise. It is important to note that we also determined the distribution of EF1α and bFGF mRNA in the polysome fractions from control and exercised soleus muscles at 6 h. Consistent with the observation that p70S6k does not change in soleus muscles following contractions (Baar & Esser, 1999), there was no change in mRNA distribution with exercise (data not shown).
Figure 1. mRNA distribution in polysomes following an acute bout of high resistance contractions.
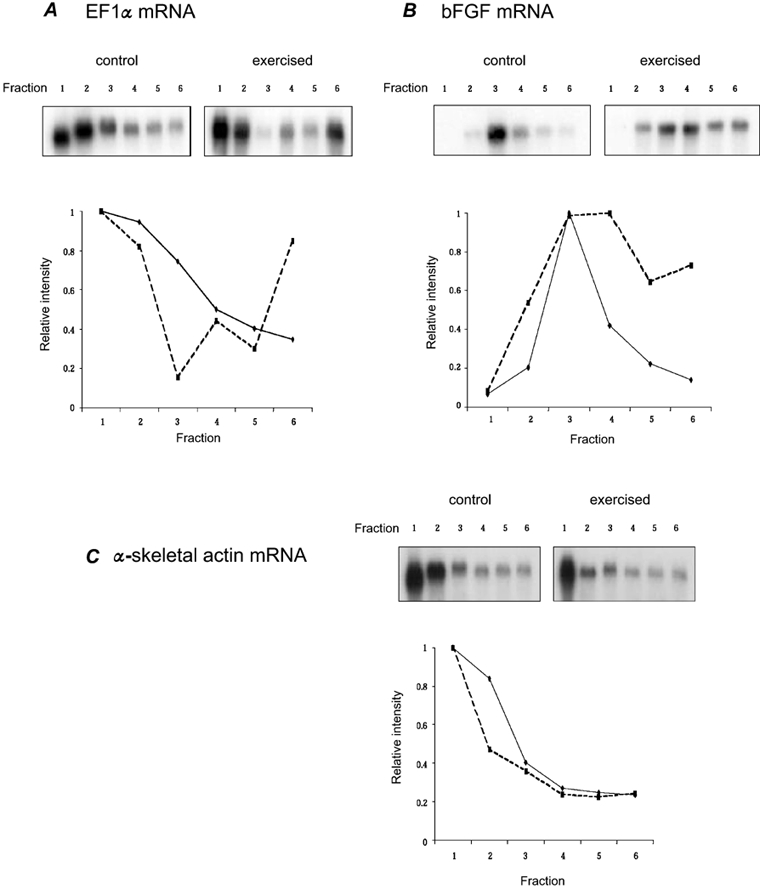
Each fraction represents the untranslated RNAs (fractions 1-3) and the heavier, translated mRNAs (fractions 4-6). Representative Northern blots are provided in the top panels with control muscle on the left and exercised muscle on the right. Results from the Northern blots were quantified using densitometry and the relative proportion of mRNA in each fraction has been quantified and presented graphically (lower panel). A, distribution of EF1α (elongation factor 1 α); B, distribution of basic fibroblast growth factor (bFGF); C, distribution of α-skeletal actin mRNA. For each graph, continuous line represents control and dashed line represents exercised.
Expression profiling of rat muscle following exercise
The highly redundant Affymetrix microarrays (Rat Genome U34A array) were used for the gene profiling in this study. By evaluating changes in total and polysomal RNA expression we identified genes that are regulated at the translational and/or transcriptional level following an acute bout of growth-inducing exercise. Approximately 7000 known full-length genes and 1000 ESTs were analysed, using 32 oligonucleotide probes (16 probe pairs) for each gene. Duplicate chips were tested for each RNA sample. Gene expression changes at 1 h and 6 h post-exercise that were more than two-fold and statistically significant (P < 0.001) are shown in Tables 1 and 2. The images in Fig. 2 provide examples of genes that were either translationally regulated or both transcriptionally and translationally regulated. The complete results of the profiling are available on our web site (http://microarray.cnmcresearch.org/resources.htm) under ‘Muscle’ and ‘Rat’. We found approximately 57 % (4995 probe sets) of all genes tested to be present in at least four chips.
Table 1.
Expression of transcriptionally and translationally regulated genes 1 h after acute muscle contractions
| P value | Average fold change | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1h | 6h | 1h | 6h | ||||||
| Probe accession number | Polysome | Total | Polysome | Total | Polysome | Total | Polysome | Total | Gene description |
| Translational regulation | |||||||||
| Rc_AA800551_at | 4.0×10−4 | — | 2.5×10−5 | 1.9×10−2 | 4.3 | — | 12.7 | 4.3 | AA800551 EST |
| Rc_AA892578_at | 9.1×10−4 | — | 7.3×10−3 | — | 4.2 | — | 2.0 | — | AA892578 EST |
| rc_AI639444_at | 3.9×10−4 | — | 4.4×10−6 | 1.0×10−3 | 3.0 | — | 2.2 | 3.0 | Sarcosin |
| rc_AI179610_at | 2.8×10−4 | — | 1.3×10−3 | 1.1×10−2 | 2.8 | — | 3.0 | 2.8 | Heme oxygenase 1 |
| rc_AI639317_at | 3.5×10−4 | — | — | 4.3×10−2 | −3.7 | — | — | −3.7 | AI639317 cDNA |
| Transcriptional regulation | |||||||||
| X86003_at | — | 1.2×10−4 | 7.4×10−4 | 5.0×10−1 | — | 9.3 | 2.5 | 3.4 | Neuron-derived orphan receptor-2 |
| U78102_at | — | 3.9×10−3 | 4.4×10−4 | 4.7×10−3 | — | 7.8 | 3.5 | 3.9 | Egr2 (krox20) |
| rc_AI169756_s_at | — | 3.3×10−4 | 2.6×10−4 | 6.5×10−2 | — | 2.6 | 3.1 | 1.5 | Gene 33 |
| AF030091UTR#1_g_at | — | 7.9×10−4 | 2.3×10−2 | 1.9×10−2 | — | 2.4 | 1.3 | 1.9 | Activity and neurotransmitter-induced early gene 6 (ania-6) |
| Transcriptional and translational regulation | |||||||||
| rc_AA819776_f_at | 6.8×10−4 | 4.2×10−2 | 7.1×10−4 | 9.4×10−3 | 8.4 | 1.8 | 10.3 | 8.4 | Similar to mouse HSP 86 |
| rc_AA944397_at | 7.3×10−4 | 1.9×10−2 | 6.8×10−3 | 3.1×10−2 | 5.2 | 2.6 | 10.5 | 5.2 | HSP86 |
| M24067_at | 6.1×10−3 | 2.2×10−4 | 8.0×10−3 | 1.8×10−2 | 2.7 | 4.0 | 2.8 | 2.7 | Plasminogen activator inhibitor-1 (PAI-1) |
| rc_AA891041_at | 1.7×10−2 | 1.8×10−4 | 1.4×10−2 | 3.8×10−2 | 15.6 | 26.9 | 19.0 | 15.6 | pjunB gene |
| X54686eds_at | 1.3×10−2 | 2.9×10−4 | — | 1.2×10−2 | 10.5 | 16.4 | — | 10.5 | pjunB |
| AF020618_g_at | 2.3×10−2 | 4.6×10−4 | 2.8×10−4 | 2.3×10−2 | 4.4 | 4.7 | 3.8 | 4.4 | Progression elevated gene 3 |
| X17053mRNA_s_at | 4.6×10−2 | 4.2×10−4 | 3.5×10−2 | 4.3×10−2 | 8.3 | 2.5 | 11.8 | 8.3 | Immediate-early serum-responsive JE gene |
Table 2.
Expression of transcriptionally and translationally regulated genes 6 h after acute muscle contractions
| P value | Average fold change | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1h | 6h | 1h | 6h | ||||||
| Probe set accession number | Polysome | Total | Polysome | Total | Polysome | Total | Polysome | Total | Gene description |
| Translational regulation | |||||||||
| X71898_at | — | — | 5.7×10−6 | — | — | — | 14.6 | — | uPAR-1 |
| U02553cds_s_at | — | 2.2×10−2 | 9.6×10−4 | — | — | 10.8 | 8.4 | — | MAP kinase phosphatase |
| rc_AI176710_at | — | 1.4×10−2 | 2.5×10−8 | — | — | 4.2 | 7.2 | — | NOR-1 |
| rc_AA859648_at | — | — | 7.6×10−5 | — | — | — | 6.8 | — | AA859648 similar to mouse heat shock protein 40 |
| M19651_at | — | — | 5.8×10−4 | — | — | — | 6.7 | — | Fra-1 |
| M61875_at | — | — | 8.1×10−4 | — | — | — | 5.8 | — | CD44 |
| L32591mRNA_g_at | — | — | 2.6×10−5 | — | — | — | 5.4 | — | GADD45 |
| L32591mRNA_at | — | — | 2.5×10−4 | — | — | — | 5.4 | — | GADD45 |
| rc_AI014163_at | — | 4.0×10−2 | 1.8×10−3 | — | — | 1.9 | 4.4 | — | PC4 |
| rc_AI169756_s_at | — | 3.3×10−4 | 2.6×10−4 | — | — | 2.6 | 3.1 | — | Gene 33 |
| AJ006295_at | — | — | 7.2×10−8 | — | — | — | 2.9 | — | AF-9 |
| rc_AA997968_at | — | — | 6.6×10−4 | — | — | — | 2.8 | — | RNB6 |
| S56464mRNA_at | — | 2.6×10−2 | 6.1×10−4 | — | — | 1.8 | 2.6 | — | Hexokinase II |
| D26393exin_s_at | — | — | 1.5×10−4 | — | — | — | 2.5 | — | Hexokinase II |
| rc_AA799396_g_at | — | — | 2.7×10−4 | — | — | — | 2.5 | — | AA799396 EST |
| X86003_at | — | 1.2×10−4 | 7.4×10−4 | — | — | 9.3 | 2.5 | — | Neuron-derived orphan receptor-2(NOR-2) |
| D86711_at | — | — | 2.5×10−4 | — | — | — | 2.3 | — | D86711 c DNA |
| U75899Mrna_at | — | — | 2.9×10−4 | — | — | — | 2.2 | — | HSPB2 |
| D90404_g_at | — | — | 9.0×10−4 | — | — | — | −2.6 | — | Cathepsin C |
| J04035_at | — | — | 5.0×10−4 | — | — | — | −2.2 | — | Tropoelastin |
| rc_AA85955_at | — | — | 9.1×10−4 | — | — | — | −2.2 | — | Eukaryotic elongation factor 2 kinase (Eef2k) |
| D86039_g_at | — | 2.6×10−2 | 3.5×10−4 | — | — | −2.2 | −2.1 | — | ATP-sensitive inwardly rectifying K+ channel, BIR(Kir6.2) |
| Transcriptional and translational regulation | |||||||||
| X06769cds_at | — | 4.5×10−3 | 6.5×10−4 | 3.7×10−2 | — | 43.6 | 33.9 | 29.7 | c-fos |
| X06769cds_g_at | — | 4.3×10−3 | 1.6×10−4 | 1.3×10−2 | — | 13.5 | 9.3 | 10.0 | c-fos |
| X03347cds_g_at | — | 6.7×10−3 | 3.3×10−4 | 5.9×10−2 | — | 12.3 | 6.9 | 10.2 | FBR-murine osteosarcoma provirus |
| Z75029_s_at | 3.3×10−3 | 2.1×10−2 | 3.7×10−5 | 2.0×10−2 | 8.8 | 9.8 | 26.4 | 8.8 | HSP70 |
| L12025_at | — | — | 1.4×10−4 | 5.0×10−4 | — | — | 23.8 | 10.9 | Tumour-associated glycoprotein E4 (Tage4) |
| U50736_s_at | — | — | 1.8×10−4 | 6.8×10−4 | — | — | 19.8 | 10.5 | CARP |
| M63282_at | — | 1.9×10−3 | 9.1×10−5 | 1.8×10−3 | — | 9.4 | 16.5 | 7.7 | Activating transcription factor 3(Atf3) |
| rc_AA800551_at | 4.0×10−4 | — | 2.5×10−5 | 1.9×10−2 | 4.3 | — | 12.7 | 4.3 | AA800551 similar to mouse DNAJ-like 2 |
| M60921_at | — | 7.2×10−3 | 1.1×10−4 | 1.5×10−2 | — | 11.3 | 10.8 | 5.9 | PC3 |
| rc_AA944156_s_at | — | 5.4×10−3 | 4.1×10−5 | 9.0×10−3 | — | 6.0 | 5.3 | 5.4 | PC3 |
| M60921_g_at | — | 5.7×10−3 | 9.5×10−6 | 2.7×10−2 | — | 8.8 | 5.3 | 3.2 | PC3 |
| rc_AA819776_f_at | 6.8×10−4 | 4.2×10−2 | 7.1×10−4 | 9.4×10−3 | 8.4 | 1.8 | 10.3 | 8.4 | Similar to mouse Hsp86-1 |
| L35271_at | — | — | 1.3×10−4 | 7.4×10−3 | — | — | 10.2 | 2.0 | AML1 |
| L05489_at | — | 1.6×10−2 | 5.5×10−4 | 3.4×10−2 | — | 6.7 | 9.4 | 7.0 | Heparin-binding EGF-like growth factor |
| U75397UTR#1_s_at | — | 2.0×10−3 | 8.8×10−4 | 4.9×10−3 | — | 29.1 | 8.8 | 21.7 | Egr1 |
| AF023087_s_at | — | 2.7×10−3 | 6.3×10−4 | 1.9×10−3 | — | 15.7 | 6.8 | 10.9 | Egr1 |
| rc_AA893280_at | — | — | 5.0×10−4 | 1.7×10−3 | — | — | 8.4 | 2.3 | AA893280 EST |
| rc_AI176546_at | 1.6×10−3 | 1.4×10−2 | 1.9×10−6 | 8.3×10−3 | 3.9 | 2.3 | 6.6 | 3.9 | HSP86 |
| rc_AA894318_at | — | — | 1.1×10−4 | 4.4×10−2 | — | — | 6.5 | 12.3 | AA894318 EST |
| X92.069_at | — | — | 4.9×10−5 | 5.5×10−3 | — | — | 6.4 | 3.1 | P2X5 |
| rc_AI043631_s_at | — | — | 2.5×10−4 | 1.5×10−2 | — | — | 5.6 | 2.9 | Antizyme inhibitor |
| D89983_at | — | 3.2×10−2 | 1.7×10−7 | 2.3×10−2 | — | 1.5 | 4.0 | −1.1 | Antizyme inhibitor |
| U53922_at | 4.3×10−3 | — | 2.0×10−6 | 1.6×10−2 | 2.6 | — | 5.1 | 2.6 | Dnaj-like protein (RDJI) |
| rc_AA799773_at | — | 2.4×10−3 | 8.6×10−5 | 3.9×10−4 | — | 2.3 | 5.1 | 2.4 | AA799773 EST |
| rc_AA799396_at | — | — | 2.5×10−5 | 2.5×10−3 | — | — | 5.0 | 5.4 | AA799396 EST |
| rc_AA859827_at | — | — | 6.2×10−4 | 1.1×10−3 | — | — | 5.0 | 8.9 | AA859827 similar to mouse uridine-cytidine kinase 2 |
| rc_AA799330_at | — | — | 2.3×10−9 | 1.9×10−2 | — | — | 4.9 | 1.9 | AA799330 EST similar to mouse Pelota |
| X81193_at | — | — | 1.2×10−4 | 4.9×10−3 | — | — | 4.9 | 4.3 | Muslce LIM |
| rc_AA799773_g_at | — | 4.8×10−3 | 2.4×10−4 | 3.9×10−2 | — | 2.0 | 4.6 | 2.5 | AA799773 EST |
| rc_AA892333_at | — | — | 6.5×10−4 | 1.4×10−2 | — | — | 4.4 | 4.6 | Alpha-tubulin |
| rc_AI070295_at | — | — | 9.5×10−4 | 5.6×10−3 | — | — | 4.2 | 1.5 | AI070295 similar to Mouse Gadd45 |
| rc_AI070295_g_at | — | — | 1.7×10−4 | 1.7×10−1 | — | — | 4.4 | 1.7 | AI070295 similar to Mouse Gadd45a |
| XI7053cds_s_at | 9.9×10−2 | 1.8×10−2 | 1.7×10−2 | 8.1×10−5 | 2.2 | 1.6 | 3.9 | 2.2 | Immediate-early serum responsive JE. |
| AF020618_g_at | — | — | 2.3×10−2 | 4.6×10−4 | 4.4 | 4.7 | 3.8 | 4.4 | Progression elevated gene 3 |
| U78102_at | — | 3.9×10−5 | 4.4×10−4 | 4.7×10−3 | — | 7.8 | 3.5 | 3.8 | Egr2 (krox20) |
| U03416_at | — | — | 2.3×10−6 | 9.1×10−3 | — | — | 3.5 | 4.9 | Neuronal olfactomedin-related ER localized protein (D2Sutle) |
| AF036548_g_at | — | — | 7.0 ×10−5 | 8.5×10−3 | — | — | 3.1 | 2.6 | RGC-32 |
| AF036548_at | — | — | 2.0×10−4 | 3.4×10−3 | — | — | 10.0 | 2.8 | RGC-32 |
| U17837cds_at | — | — | 1.2×10−4 | 2.1×10−3 | — | — | 2.9 | 3.2 | Zinc finger protein RIZ |
| Y00396mRNA_g_at | — | 4.4 × 4 10−2 | 5.3 × 10−4 | 2.8 × 10−2 | — | 1.7 | 2.8 | 1.6 | c-myc |
| AJ006295_g_at | 2.2×10−2 | — | 1.7×10−7 | 6.×10−3 | 2.2 | — | 2.7 | 2.2 | AF-9 |
| rc_AA892635_at | — | — | 1.9×10−4 | 3.8×10−3 | — | — | 2.7 | 1.4 | ras-like protein (Tc10) |
| M64733mRNA_s_at | 9.9×10−2 | — | 2.5×10−4 | 3.4×10−3 | 1.8 | — | 2.5 | 1.8 | TRPM-2 |
| ABoo3726_at | — | — | 1.9×10−6 | 4.0×10−3 | — | — | 2.3 | 3.1 | Vesl |
| M84176_at | 3.2×10−2 | — | 1.4×10−4 | 1.7×10−2 | 1.6 | — | 2.3 | 1.6 | MyoD |
| rc_AI639058_s_at | 5.0×10−2 | — | 4.5×10−4 | 3.1×10−2 | 2.8 | — | 2.3 | 2.8 | AI639058 cDNA |
| X05566_i_at | — | — | 7.7×10−5 | 1.1×10−2 | — | — | 2.3 | 2.0 | Myosin regulatory light chain (RLC) |
| rc_AI639444_g_at | 8.2×10−3 | — | 1.9×10−6 | 1.0×10−1 | 3.0 | — | 2.5 | 3.0 | Sarcosin |
| rc_AI639444_at | 3.9×10−4 | — | 4.4×10−6 | 1.0×10−3 | 3.0 | — | 2.2 | 3.0 | Sarcosin |
| rc_AA799545_at | — | — | 1.1×10−5 | 4.8×10−3 | — | — | 2.2 | 1.8 | AA799545 similar to mouse chaperonin subunit 3, gamma |
| rc_AA9455867_at | — | 7.6×10−3 | 3.2×10−5 | 2.2×10−2 | — | 4.0 | 2.2 | 1.7 | c-jun |
| rc_AA891107_at | — | — | 6.4×10−4 | 1.1×10−2 | — | — | 2.2 | 2.7 | Diphosphoinositol polyphosphate phosphohydolase type II (Nudt4) |
| U95001UTR#1_s_at | — | — | 2.9×10−4 | 1.8×10−3 | — | — | 2.1 | 1.7 | Developmentally-regulated cardiac factor (DRCF-5) |
| rc_AI011498_at | — | 9.7×10−4 | 2.0×10−2 | — | — | −7.7 | −2.4 | BAF60b | |
| M26715_at | — | — | 1.5×10−5 | 4.5×10−3 | — | — | −3.8 | −1.2 | Phosphodiesterase 4A (Pde4a) |
| rc_AA859631_at | 4.7×10−2 | — | 5.2×10−4 | 5.8×10−3 | −1.6 | — | −3.3 | −1.6 | AA859631 ESt |
| rc_AI639108_at | — | — | 1.3×10−4 | 2.7×10−2 | — | — | −2.4 | −2.3 | AI639108 ESt |
| rc_AI639507_at | 3.1×10−4 | 6.3×10−3 | 5.6×10−4 | 4.0×4 10−2 | −1.6 | −2.0 | −2.4 | −1.6 | AI639507 cDNa |
| U69272_at | — | — | 1.9×10−4 | 2.3×10−3 | — | — | −2.1 | −2.0 | Interleukin-15 |
Figure 2. Examples of hybridization patterns for specific genes regulated in response to acute muscle contractions.
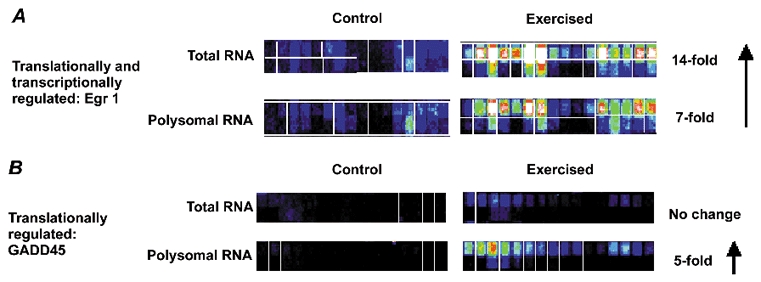
Each gene shown was queried with 16 ‘perfect match’ 25 bp oligonucleotides (top rows), with paired ‘mismatched’ oligonucleotides designed with a single mismatch in the centre (lower rows). Signal intensities are represented by different colour codes: (from high to low) white, red, yellow, green, blue and black A, hybridization of the Egr1 mRNA shows very low level in unexercised muscle. After a bout of exercise, Egr1 shows strong induction of expression, in both total RNA pools (transcriptional regulation) and polysomal pools (translational regulation). Hybridization only to the perfect match probes is seen, with relatively consistent hybridization signals to all 16 perfect match oligonucleotides. B, GADD45 shows strong translational induction after exercise. The gene does not show significant changes in the total RNA pools.
Comparison of RNA profiles from exercised and non-exercised muscle at 1 h after exercise identified 17 probe sets representing 15 known genes and ESTs that changed. (Table 1). Of the 15 genes that changed, 14 showed increases while one unknown cDNA clone showed a decrease in expression. As presented in Table 1, two known genes and three unknown ESTs were differentially expressed in the polysomal RNA pool. Four genes were changed in the total RNA pool and five genes were increased in both polysomal and total RNA pools. Sarcosin and haem oxygenase 1 (HO-1) were the known genes that changed levels only in the polysomal RNA pool indicating that those gene products are induced through translational mechanisms following exercise. Sarcosin is selectively expressed in human sarcomeric muscle and has been found up-regulated in human hypertrophic cardiomyopathy but its function is, as yet, not known (Taylor et al. 1998; Lim et al. 2001). HO-1, also translationally regulated, is responsible for the physiological breakdown of haem into equimolar amounts of biliverdin, carbon monoxide and iron. Increases in HO-1 mRNA levels have also been detected in skeletal muscle after more endurance type exercise (Essig et al. 1997; Pilegaard et al. 2000).
Several of the gene expression changes detected at 1 h were also seen at 6 h however the changes were not statistically significant (e.g. HO-1, ania-6, PAI-1, pJunB). This is due primarily to greater variability in expression levels at 6 h. It was interesting to note that three of the four genes that were changed only in the total RNA pool at 1 h (neuron-derived orphan receptor-2 (NOR-2), Egr2 (krox20) and gene 33) were detected in the polysomal pool at 6 h. This is consistent with a delay between transcript accumulation and ribosomal association.
As presented in Table 2, we identified 81 probe sets representing 70 known genes and ESTs that changed 6 h after exercise. Among them, 10 ESTs did not share high nucleotide sequence homology to any known gene by basic local alignment search tool (BLAST) analysis. Of the 70 genes that changed, 60 showed increases while 10 of the genes showed a decrease in expression. As presented in Table 2, about 65 % of the genes that changed following contractions were detected in both the total RNA and polysomal mRNA pools. This suggests that, in general, expression of these genes in skeletal muscle following an acute bout of contractions is probably regulated by changes in transcription and/or mRNA stability and these transcripts are readily mobilized to the polysomal fraction. There were, however, 25 genes that changed levels only in the polysomal RNA pool, indicating that those gene products are regulated through translational mechanisms following exercise. These genes included several involved in cell growth and/or differentiation (uPAR-1, MAP kinase phosphatase, Fra-1, PC4, neuron-derived orphan receptor-2 (NOR-2) and HSPB2) and anti-proliferation (GADD45, gene 33 and eukaryotic elongation factor 2 kinase (eEF2k)). It should be noted that we did not detect either bFGF or EF1α as translationally regulated genes, although the earlier experiments had shown that these two genes did show preferential distribution to heavier polysomal fractions by Northern blot analysis (Fig. 1, Table 1). For the microarray experiments we collected the total polysomal fraction and did not differentiate between light and heavy fractions because of the quantity of RNA required. Thus for translational regulation, the microarray experiments would only detect those mRNAs that were significantly re-distributed from the non-translated pools to the translated pools. Those mRNAs, like EF1α and bFGF, that demonstrate changes from the light polysomal fraction to the heavy fraction will not be detected. In addition, examination of the expression profiles for EF1α and bFGF showed that these two genes were judged ‘absent’ on the GeneChips in both exercised and control muscle; i.e. the hybridization pattern was not sufficiently above background to accurately measure RNA levels, making it more difficult to determine significant changes in expression.
When considering the genes that were changed at both transcriptional and translational levels, relatively few muscle-specific genes were detected (myoD, myosin light chain and sarcosin). Most of the genes that were up-regulated are associated with cell proliferation, including c-fos, c-jun, tumour-associated glycoprotein E4 (Tage4), activating transcription factor 3 (Atf3), heparin-binding EGF-like growth factor, Egr1, Egr2, antizyme inhibitor, RGC-32 and c-myc. All these growth-related genes were up-regulated as shown in Table 2. In addition, another up-regulated gene, AA799330 EST, is highly homologous to the Drosophila Pelota gene, which is implicated in mediating required translational regulation for proper cell cycle progression (Davis & Engebrecht, 1998). In contrast, five growth inhibition genes were also up-regulated (GADD45, TRPM-2, CARP, PC3 and RIZ) at 6 h after exercise. Three of these genes, GADD45, TRPM-2 and PC3, are commonly up-regulated at times when p53 is active and all five of these genes are believed to be important regulators of G1-S phase progression of the cell cycle (Carrier et al. 1994; Srivastava et al. 1998; Guardavaccaro et al. 2000).
The expression of five stress response genes was increased at either 1 h or 6 h post-exercise. Three of the five, HSP70, HSP 86 and HO-1, have previously been shown to be up-regulated after exercise (Locke & Noble, 1995; Essig et al. 1997). The other known stress response gene, RDj1, is a DnaJ-like protein, so is implicated in the hsc70-based chaperone system (Terada & Mori, 2000); this is the first report of its increase with contractile activity in skeletal muscle.
Localization of c-fos following exercise
One of the differentially expressed gene products in exercised and control muscle, c-fos, was localized using immunohistochemistry (Fig. 3A and B). Cryosections of the TA muscle following contractions were stained with antibodies to c-fos along with a nuclear marker (Hoechst 358; Fig. 3). A subset of myonuclei in exercised muscle, many of which are within the plasma membrane, were seen to stain strongly with c-fos (Fig. 3A). In control muscle, no nuclei were seen to stain positive for c-fos (Fig. 3B). The number of myonuclei immunostaining with c-fos following high resistance contractions varied considerably from region to region and muscle to muscle. The variability in c-fos localization within exercised muscle (both within and between myofibres) has been reported before and is an intriguing finding (Puntschart et al. 1998). This suggests that there may be differential loading across the myofibres resulting in a complex microenvironment following contractile activity.
Figure 3. Immunological confirmation of c-fos gene expression changes.
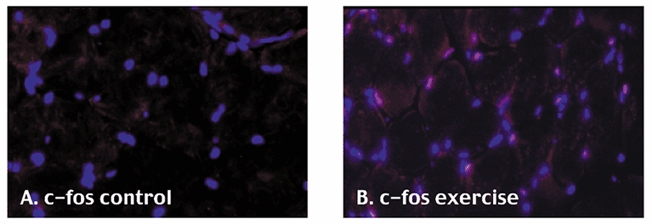
Cold acetone-fixed cryosections of control and exercised muscle are shown, immunostained with c-fos (Cy3; red signal) and Hoechst 358 (nuclear marker, blue signal).
Changes in p53 protein levels following contractions
The up-regulation of three growth-inhibitory genes associated with p53 in skeletal muscle at 6 h post-exercise was intriguing. From these observations we determined the levels of p53 protein in skeletal muscle following contractile activity. As can be seen in Fig. 4 there was a 49 ± 9 % increase in p53 levels in the exercised muscles (n = 3) compared to their contralateral controls (n = 3) (P < 0.05). Consistent with the change at 6 h we also detected a 57 ± 11 % increase in p53 levels in the nuclear extracts from exercise vs. control muscles at 1 h (data not shown).
Figure 4. Bar graph of means ± standard errors from densitometric scans of p53 Western blots.
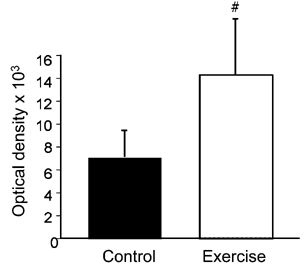
# statistically significant at P < 0.05: one-tailed t test.
Discussion
Skeletal muscle phenotype has long been known to be sensitive to changes in contractile activity patterns. From expression profiling experiments on both total RNA and translated polysomal RNA following one bout of high resistance contractions, we present for discussion four points that we found particularly reflective of the exercise-responsive gene expression clusters. The first point is the significant contribution of translational regulation to gene expression after contractions. Secondly, we have identified a cluster of genes, both up- and down-regulated, that contribute to net protein accumulation. Thirdly, the response of skeletal muscle to a growth stimulus shares common gene expression patterns with non-muscle cellular growth. Finally, the response to eccentric contractile activity includes the upregulation of genes likely to be involved in regeneration and repair. To try to summarize all the genes that change expression following contractile activity in this study we provide in Fig. 5 a potential cascade of transcriptionally and translationally regulated genes following a bout of eccentric contractile activity.
Figure 5. Proposed biochemical cascades in skeletal muscle tissue following a bout of high resistance exercise.
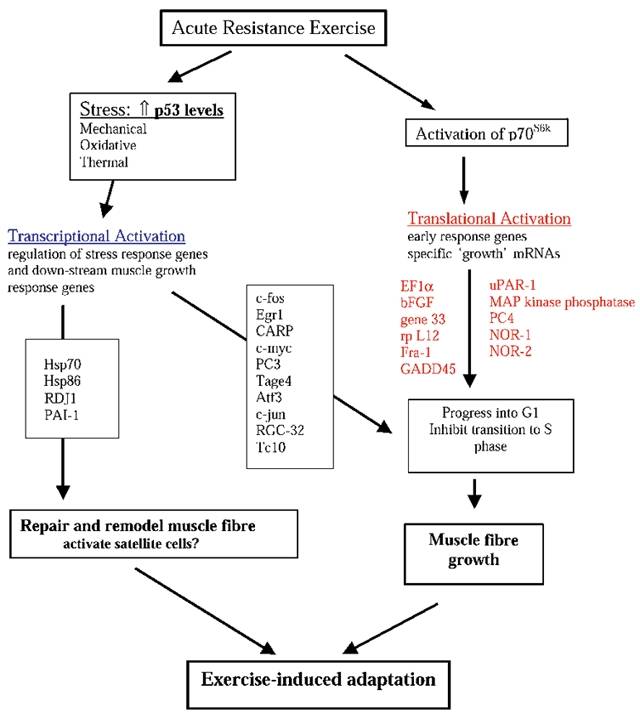
Previous work studying intracellular signalling pathways following one acute bout of high resistance contractions demonstrated that the p70S6k phosphorylation was significantly up-regulated after contractions and this elevation was maintained for up to 36 h after exercise (Baar & Esser, 1999). The critical role that this pathway plays in mediating skeletal muscle growth has more recently been supported by the work of Bodine et al. (2001). One aspect of gene expression that is linked to activation of the p70S6kpathway is the selective translation of a subset of mRNAs that contain terminal oligopyrimadine structures in their 5′UTRs (Meyuhas & Hornstein, 2000). Consistent with the increased phosphorylation of p70S6kin muscles at 6 h, approximately 25 genes were changed at the level of translation at 6 h post-contractions. This accounts for 30 % of the gene expression changes detected at this time point. These findings highlight the significant contribution that translational mechanisms make to the gene expression changes seen following muscle contractile activity. The role of translational regulation in skeletal muscle adaptation is not surprising as there is growing evidence that translational regulation is important for fundamental aspects of cell cycle progression and in response to cellular stresses (Pyronnet & Sonenberg, 2001).
One well established characteristic of the response of skeletal muscle to resistance exercise is the robust increase in protein synthesis for several hours after exercise. This has been documented in all models of resistance exercise across species ranging from mouse and chicken to human (Goldberg, 1968; Laurent et al. 1978; Smith et al. 1983; MacDougall et al. 1995). Consistent with the known changes in protein synthesis, the results of this study identified a significant cluster of gene expression changes that are consistent with net protein accumulation with increases in protein synthesis and decreases in protein degradation.
Three genes that showed increased expression following contractile activity are elongation factor 1α (EF1α), ribosomal protein L12 and c-myc. EF1α is a polypeptide elongation factor responsible for bringing the tRNAs to the ribosome. Recent studies have linked loss of EF1α protein with significant loss of skeletal muscle mass in the wasted mouse (Khalyfa et al. 2001). Ribosomal protein L12 is a component of the large 60S ribosomal subunit. The increase in ribosomal L12 mRNA is found in the polysomal pool at 6 h (it is not listed on Table 2 because the increase was less then 2-fold but the change was statistically significant: see web site listings). Recent studies have indicated that ribosomal protein L12 uses a distinct nuclear import pathway that may contribute to a mechanism for regulating ribosome synthesis and/or maturation (Plafker & Macara, 2002). Increases in both those proteins would result in increased capacity of the protein synthetic machinery in the cell. The increase in c-myc will also act to enhance protein synthesis, as recent studies have identified that c-myc acts to induce transcription of many of the ribosomal protein mRNAs (Kim et al. 2000; Neiman et al. 2001). Thus increases in expression of these three genes will synergistically act to increase many components of the ribosomal complex leading to increased synthetic capacity. There were also two genes that showed decreased expression in the polysome fraction following exercise. These are elongation factor 2 kinase and cathepsin C. Elongation factor 2 kinase phosphorylates and inactivates eukaryotic elongation factor 2, inhibiting protein synthesis (Wang et al. 2001). In this study, levels of eEF2k decreased in the polysomal pool following exercise, which would limit an inhibitor of protein synthesis. Another gene that was downregulated in the polysomal pool at 6 h was cathepsin C. Cathepsin C is a lysosomal protease and is associated with protein degradation. The changes determined for this group of five genes will clearly lead to a more anabolic state in the skeletal muscle following contractile activity.
The nuclei in mature mammalian skeletal muscle are post-mitotic, which results in muscle growth occurring by increasing fibre size and not increasing fibre or cell number. Thus, one of the goals of this study was to evaluate the gene expression changes determined during muscular hypertrophy and to compare them to known profiles from studies of cellular growth and/or proliferation. Two expression-profiling studies of the growth response of cultured fibroblasts after serum stimulation have recently been published (Iyer et al. 1999; Zong et al. 1999). It was surprising to find that several of the genes up-regulated in muscle following exercise were genes associated with cell cycle progression and proliferative growth. For example, Zong et al. (1999) analysed polysomal mRNAs following serum stimulation of fibroblasts and some of the genes increased in the polysomes from both studies included c-fos, Egr1, bFGF and EF1α (Iyer et al. 1999; Zong et al. 1999). This is consistent with the well-established role for selective translation of growth mRNAs in the regulation of cell cycle control (Pyronnet & Sonenberg, 2001). Iyer et al. (1999) studied gene expression profiles following serum stimulation of fibroblasts, focusing on transcriptional regulation (total RNA). The shared genes that were transcriptionally up-regulated in our in vivo muscle exercise study included c-myc, c-fos, Egr1 and antizyme inhibitor. Thus, it appears that many of the genes up-regulated following a resistance exercise stimulus can be defined as a general ‘growth response cluster’. This cluster is characterized by mRNAs that are induced after a growth stimulus, would probably be shared by many cell types and will be commonly associated with progression through the cell cycle.
At this time it is unclear which cell type(s) are contributing to the expression of the cell cycle progression genes. One possibility is that the satellite cells, which are myogenic cells lying outside the muscle membrane, are the cells expressing the majority of the cell cycle genes as they are induced to proliferate following the contractile activity. These cells are known to be activated following eccentric and concentric contractions and are implicated in skeletal muscle growth during development and exercise (Schultz, 1989; Allen & Rankin, 1990; Putman et al. 2000; Hawke & Garry, 2001). While this is a possibility, it should be noted that the satellite cells contribute approximately 1.8 % of the total nuclei within the TA muscle of rats in contrast to ≈43 % of the nuclei from the muscle fibres (Schmalbruch & Hellhammer, 1977; Hawke & Garry, 2001). In addition, not all satellite cells are activated following eccentric contractions. Therefore, if the cell cycle gene expression changes in this study come from the subset of satellite cells that are activated following contraction then those cells must be expressing those genes very highly to be detectable in a homogenate from whole TA muscle. Based on this argument, we suggest that the gene expression changes detected in this study at 1 and 6 h after contractions are probably reflecting changes in gene expression in the muscle fibre. Consistent with this argument, it is important to note that the increased c-fos expression in this study was detected in the nuclei within the muscle fibres and was not detected in satellite cells or other non-muscle cells.
In contrast, the ‘growth’ profile following high resistance contractions included a cluster of genes that can be categorized as antigrowth and/or tumour suppressor genes. This is very intriguing because muscle fibres are terminally differentiated multinucleated cells and myonuclei are mitotically inactive. Studies in vitro have demonstrated that serum stimulation will result in a G0 to G1 transition but these cells seem to be blocked prior to entering the S phase (Tiainen et al. 1996). Thus, we suggest that the expression of these antigrowth genes in skeletal muscle may act to counter the proliferative function of genes described above to maintain an inhibition of potential G1 to S phase transition.
The best-characterized antigrowth genes identified in this study include PC3 and GADD45 (Bradbury et al. 1991; Guardavaccaro et al. 2000; Sheikh et al. 2000). PC3 is a cell cycle inhibitor that, when overexpressed, has been shown to result in G1 arrest (Guardavaccaro et al. 2000). This gene is transcriptionally induced during terminal differentiation of pheochromocytoma (PC12) cells in response to nerve growth factor (NGF). We found 5- to10-fold increase in PC3 mRNA in polysomes (using two distinct probe sets), as well as 3- to 6-fold increase in total RNA. This large increase, especially in the translated polysomal pool, is consistent with a role for this protein in preventing the progression of myonuclei through the cell cycle. GADD45 (growth arrest DNA damage) is a well-characterized protein involved in cell cycle arrest at G1/S and/or G2/M through direct inhibition of cdc2a kinase (Agarwal et al. 1995; Jin et al. 2000; Taylor & Stark, 2001). We detected a 5-fold increase in GADD45 mRNA in the polysome fraction (using two probe sets). An additional gene that can be included in this ‘antigrowth’ category with PC3 and GADD45 is CARP, cardiac adriamycin responsive protein/cardiac ankyrin repeat protein (Jeyaseelan et al. 1997; Zou et al. 1997). Induction of CARP has been shown to be an early marker of cardiac and skeletal muscle hypertrophy (Aihara et al. 2000; Johnatty et al. 2000; Carson et al. 2002). Recent studies have shown that overexpression of CARP acts to decrease proliferation in smooth muscle vascular cells, suggesting it may function to maintain the differentiated state in heart and skeletal muscle during periods of growth (Kanai et al. 2001).
It was interesting to note that two antigrowth genes that were induced, PC3 and GADD45, are known targets for the stress responsive tumour suppressor gene, p53 (Carrier et al. 1994; Zhan et al. 1998; Guardavaccaro et al. 2000). Based on this observation we evaluated p53 protein levels in nuclear extracts from exercised and control muscles and found they were elevated by about 50 % at 1 h and 6 h. There are very few studies of p53 in adult skeletal muscle with the only induction seen in rat muscle following 18 days of spaceflight (Ohnishi et al. 2000). These findings are exciting because they illustrate the ability to use expression profiling to identify a novel, previously undefined pathway in skeletal muscle following contractile activity. We hypothesize that activation of p53 in skeletal muscle after resistance exercise is potentially an important mechanism by which the post-mitotic muscle cells maintain and/or repair any DNA damage that can occur in response to the expected oxidative, thermal and mechanical stresses associated with exercise. Based on this hypothesis we also suggest that the activation of p53 and induction of subsequent antigrowth genes following contractile activity would not be specific to high resistance contractions but would also be elevated following lower resistance endurance-type contractions.
One last exercise-related cluster we identified is likely to be associated with the fact that the muscles in this study underwent 60 eccentric or lengthening contractions and this type of contraction is well known to be associated with muscle damage. Cell damage typically induces thrombotic cascades, but such events could exacerbate muscle damage after acute exercise. We found two important proteins involved in thrombosis regulated in a complex manner in exercised muscle: plasminogen activator inhibitor (PAI-1) and urokinase plasminogen activator receptor (uPAR-1). We found the PAI-1 gene to be induced 2- to 4-fold in exercised muscle at 1 h post-contractions. PAI-1 is a serine protease inhibitor (SERPIN)-type inhibitor of tissue plasminogen activator (TPA) and activates thrombotic cascades through inhibiting the dissolution of small, localized clots in tissues (fibrinolysis) (Andreasen et al. 2000). We hypothesize that PAI-1 gene induction is part of a general cell damage cascade. In contrast to PAI-1, uPAR-1 showed a 15-fold induction in polysomal fractions but no significant increase in total RNA fractions at 6 h, suggesting that uPAR-1 mRNA is quickly targeted for translation after exercise. uPAR-1 is increasingly recognized to play a critical role in both fibrinolysis (anti-thrombosis) and in muscle regeneration (Fibbi et al. 2001; Lluis et al. 2001). Recent studies of murine knockouts for uPA have shown a lack of muscle regeneration and accumulation of fibrin in the degenerating muscle (Lluis et al. 2001). Additional studies have demonstrated production of uPA and uPAR-1 in muscle satellite cells, where the complex appears critical for migration of these cells (Fibbi et al. 2001). We hypothesize that uPAR-1 and its ligand uPA play a critical role in the response of muscle to acute exercise, both as an anti-thrombotic complex and as an extracellular proteolytic cascade critical for muscle regeneration and/or repair. Consistent with this key role, we hypothesize that uPAR-1 mRNA is kept pre-synthesized in the muscle, then immediately sent to the translational machinery after exercise.
In summary, we report expression profiling of an in vivo contraction-induced growth model in skeletal muscle. Results from this analysis suggest that translational regulation is an important mechanism of cellular adaptation to a complex physiological stimulus. In addition, the results suggest that skeletal muscle growth shares a common gene expression profile with other models of cellular growth. The growth cluster identified is characterized by the expression of genes that are commonly associated with progression of the cell cycle. However, it was interesting to detect an induction of a group of genes that are associated with tumour suppression or antigrowth response. Many of these antigrowth genes are likely to be induced by the rapid increase in p53 protein levels detected early after contraction and are probably not specific to high resistance contractions. There was also induction of a cluster of genes that are associated with repair and regeneration, which is an important process for skeletal muscle in response to eccentric contractions. We suggest that this balanced expression of these previously characterized growth and antigrowth genes with the repair and regeneration cluster results in the increase in cell size without the increases in cell number characteristic of normal skeletal muscle growth.
Acknowledgments
The authors would like to thank Drs Tim Koh and Shann Kim and the rest of the Esser lab for valuable suggestions on the manuscript. Supported by grants from the National Institutes of Health (NS29525 to E.P.H. and AR45617 to K.A.E.). Y.-W. Chen was supported by a DMDRC Post-doctoral Fellowship from Stichting Porticus. M. Fedele was supported by NIH training grant HHS HL07692-12.
References
- Agarwal ML, Agarwal A, Taylor WR, Stark GR. p53 controls both the G2/M and the G1 cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proceedings of the National Academy of Sciences of the USA. 1995;92:8493–8497. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aihara Y, Kurabayashi M, Saito Y, Ohyama Y, Tanaka T, Takeda S, Tomaru K, Sekiguchi K, Arai M, Nakamura T, Nagai R. Cardiac ankyrin repeat protein is a novel marker of cardiac hypertrophy: role of M-CAT element within the promoter. Hypertension. 2000;36:48–53. doi: 10.1161/01.hyp.36.1.48. [DOI] [PubMed] [Google Scholar]
- Allen RE, Rankin LL. Regulation of satellite cells during skeletal muscle growth and development. Proceedings of the Society for Experimental Biology and Medicine. 1990;194:81–86. doi: 10.3181/00379727-194-43060. [DOI] [PubMed] [Google Scholar]
- Andreasen PA, Egelund R, Petersen HH. The plasminogen activation system in tumor growth, invasion, and metastasis. Cellular and Molecular Life Sciences. 2000;57:25–40. doi: 10.1007/s000180050497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baar K, Esser K. Phosphorylation of p70(S6k) correlates with increased skeletal muscle mass following resistance exercise. American Journal of Physiology. 1999;276:C120–127. doi: 10.1152/ajpcell.1999.276.1.C120. [DOI] [PubMed] [Google Scholar]
- Blough E, Dineen B, Esser K. Extraction of nuclear proteins from striated muscle tissue. Biotechniques. 1999;26:202–204. doi: 10.2144/99262bm05. [DOI] [PubMed] [Google Scholar]
- Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yaucopoulos GD. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nature Cell Biology. 2001;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- Booth FW, Thomason DB. Molecular and cellular adaptation of muscle in response to exercise: perspectives of various models. Physiological Reviews. 1991;71:541–585. doi: 10.1152/physrev.1991.71.2.541. [DOI] [PubMed] [Google Scholar]
- Bradbury A, Possenti R, Shooter EM, Tirone F. Molecular cloning of PC3, a putatively secreted protein whose mRNA is induced by nerve growth factor and depolarization. Proceedings of the National Academy of Sciences of the USA. 1991;88:3353–3357. doi: 10.1073/pnas.88.8.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrier F, Smith ML, Bae I, Kilpatrick KE, Lansing TJ, Chen CY, Engelstein M, Friend SH, Henner WD, Gilmer TM, et al. Characterization of human Gadd45, a p53-regulated protein. Journal of Biological Chemistry. 1994;269:32672–32677. [PubMed] [Google Scholar]
- Carson JA, Nettleton D, Reecy JM. Differential gene expression in the rat soleus muscle during early work overload-induced hypertrophy. FASEB Journal. 2002;16:207–219. doi: 10.1096/fj.01-0544fje. [DOI] [PubMed] [Google Scholar]
- Chen YW, Zhao P, Borup R, Hoffman EP. Expression profiling in the muscular dystrophies: identification of novel aspects of molecular pathophysiology. Journal of Cell Biology. 2000;151:1321–1336. doi: 10.1083/jcb.151.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L, Engebrecht J. Yeast dom34 mutants are defective in multiple developmental pathways and exhibit decreased levels of polyribosomes. Genetics. 1998;149:45–56. doi: 10.1093/genetics/149.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn SE, Burns J, Michel R. Calcineurin is required for skeletal muscle hypertrophy. Journal of Biological Chemistry. 1999;274:21908–21912. doi: 10.1074/jbc.274.31.21908. [DOI] [PubMed] [Google Scholar]
- Essig DA, Borger DR, Jackson DA. Induction of heme oxygenase-1 (HSP32) mRNA in skeletal muscle following contractions. American Journal of Physiology. 1997;272:C59–67. doi: 10.1152/ajpcell.1997.272.1.C59. [DOI] [PubMed] [Google Scholar]
- Fibbi G, Barletta E, Dini G, Del Rosso A, Pucci M, Cerletti M, Del Rosso M. Cell invasion is affected by differential expression of the urokinase plasminogen activator/urokinase plasminogen activator receptor system in muscle satellite cells from normal and dystrophic patients. Laboratory Investigation. 2001;81:27–39. doi: 10.1038/labinvest.3780209. [DOI] [PubMed] [Google Scholar]
- Goldberg AL. Protein synthesis during work-induced growth of skeletal muscle. Journal of Cell Biology. 1968;36:653–658. doi: 10.1083/jcb.36.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardavaccaro D, Corrente G, Covone F, Micheli L, D'Agnano I, Starace G, Caruso M, Tirone F. Arrest of G(1)-S progression by the p53-inducible gene PC3 is Rb dependent and relies on the inhibition of cyclin D1 transcription. Molecular and Cellular Biology. 2000;20:1797–1815. doi: 10.1128/mcb.20.5.1797-1815.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawke TJ, Garry DJ. Myogenic satellite cells: physiology to molecular biology. Journal of Applied Physiology. 2001;91:534–551. doi: 10.1152/jappl.2001.91.2.534. [DOI] [PubMed] [Google Scholar]
- Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JC, Trent JM, Staudt LM, Hudson J, Jr, Boguski MS, Lashkari D, Shalon D, Botstein D, Brown PO. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- Jefferies HB, Thomas G. Elongation factor-1 alpha mRNA is selectively translated following mitogenic stimulation. Journal of Biological Chemistry. 1994;269:4367–4372. [PubMed] [Google Scholar]
- Jeyaseelan R, Poizat C, Baker RK, Abdishoo S, Isterabadi LB, Lyons GE, Kedes L. A novel cardiac-restricted target for doxorubicin. CARP, a nuclear modulator of gene expression in cardiac progenitor cells and cardiomyocytes. Journal of Biological Chemistry. 1997;272:22800–22808. doi: 10.1074/jbc.272.36.22800. [DOI] [PubMed] [Google Scholar]
- Jin S, Antinore MJ, Lung FD, Dong X, Zhao H, Fan F, Colchagie AB, Blanck P, Roller PP, Fornace AJ, Jr, Zhan Q. The GADD45 inhibition of Cdc2 kinase correlates with GADD45-mediated growth suppression. Journal of Biological Chemistry. 2000;275:16602–16608. doi: 10.1074/jbc.M000284200. [DOI] [PubMed] [Google Scholar]
- Johnatty SE, Dyck JR, Michael LH, Olson EN, Abdellatif M. Identification of genes regulated during mechanical load-induced cardiac hypertrophy. Journal of Molecular and Cellular Cardiology. 2000;32:805–815. doi: 10.1006/jmcc.2000.1122. [DOI] [PubMed] [Google Scholar]
- Kanai H, Tanaka T, Aihara Y, Takeda S, Kawabata M, Miyazono K, Nagai R, Kurabayashi M. Transforming growth factor-beta/Smads signaling induces transcription of the cell type-restricted ankyrin repeat protein CARP gene through CAGA motif in vascular smooth muscle cells. Circulation Research (Online) 2001;88:30–36. doi: 10.1161/01.res.88.1.30. [DOI] [PubMed] [Google Scholar]
- Khalyfa A, Bourbeau D, Chen E, Petroulakis E, Pan J, Xu S, Wang E. Characterization of elongation factor-1A (eEF1A-1) and eEF1A-2/S1 protein expression in normal and wasted mice. Journal of Biological Chemistry. 2001;276:22915–22922. doi: 10.1074/jbc.M101011200. [DOI] [PubMed] [Google Scholar]
- Kim S, Li Q, Dang CV, Lee LA. Induction of ribosomal genes and hepatocyte hypertrophy by adenovirus-mediated expression of c-Myc in vivo. Proceedings of the National Academy of Sciences of the USA. 2000;97:11198–11202. doi: 10.1073/pnas.200372597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent GJ, Sparrow MP, Millward DJ. Turnover of muscle protein in the fowl. Changes in rates of protein synthesis and breakdown during hypertrophy of the anterior and posterior latissimus dorsi muscles. Biochemical Journal. 1978;176:407–417. doi: 10.1042/bj1760407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DS, Roberts R, Marian A. Expression profiling of cardiac genes in human hypertrophic cardiomyopathy: insight into the pathogenesis of phenotypes. Journal of the American College of Cardiology. 2001;38:1175–1180. doi: 10.1016/s0735-1097(01)01509-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lluis F, Roma J, Suelves M, Parra M, Aniorte G, Gallardo E, Illa I, Rodriguez L, Hughes SM, Carmeliet P, Roig M, Munoz-Canoves P. Urokinase-dependent plasminogen activation is required for efficient skeletal muscle regeneration in vivo. Blood. 2001;97:1703–1711. doi: 10.1182/blood.v97.6.1703. [DOI] [PubMed] [Google Scholar]
- Locke M, Noble EG. Stress proteins: the exercise response. Canadian Journal of Applied Physiology. 1995;20:155–167. doi: 10.1139/h95-011. [DOI] [PubMed] [Google Scholar]
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, Brown EL. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nature Biotechnology. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- MacDougall JD, Gibala MJ, Tarnopolsky MA, MacDonald JR, Interisano SA, Yarasheski KE. The time course for elevated muscle protein synthesis following heavy resistance exercise. Canadian Journal of Applied Physiology. 1995;20:480–486. doi: 10.1139/h95-038. [DOI] [PubMed] [Google Scholar]
- Mathews M, Sonenberg N, Hershey J. Origins and Principles of Translational Control. In: Sonenberg N, Hershey J, Mathews M, editors. Translational Control of Gene Expression. New York: Cold Spring Harbor Press; 2000. pp. 1–32. [Google Scholar]
- Meyuhas O, Hornstein E. Translational control of TOP mRNAs. In: Sonenberg N, Hershey J, Mathews M, editors. Translational Control of Gene Expression. New York: Cold Spring Harbor Press; 2000. pp. 671–693. [Google Scholar]
- Neiman PE, Ruddell A, Jasoni C, Loring G, Thomas SJ, Brandvold KA, Lee R, Burnside J, Delrow J. Analysis of gene expression during myc oncogene-induced lymphomagenesis in the bursa of Fabricius. Proceedings of the National Academy of Sciences of the USA. 2001;98:6378–6383. doi: 10.1073/pnas.111144898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Wang X, Fukuda S, Takahashi A, Ohnishi K, S N. Accumulation of tumor suppressor p53 in rat muscle after a space flight. Advances in Space Research. 2000;25:2119–2122. doi: 10.1016/s0273-1177(99)01063-7. [DOI] [PubMed] [Google Scholar]
- Pilegaard H, Ordway GA, Saltin B, Neufer PD. Transcriptional regulation of gene expression in human skeletal muscle during recovery from exercise. American Journal of Physiology - Endocrinology and Metabolism. 2000;279:E806–814. doi: 10.1152/ajpendo.2000.279.4.E806. [DOI] [PubMed] [Google Scholar]
- Plafker SM, Macara IG. Ribosomal protein L12 uses a distinct nuclear import pathway mediated by importin 11. Molecular and Cellular Biology. 2002;22:1266–1275. doi: 10.1128/MCB.22.4.1266-1275.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradet-Balade B. Translation control: bridging the gap between genomics and proteomics. Trends in Biochemical Sciences. 2001;26:225–229. doi: 10.1016/s0968-0004(00)01776-x. [DOI] [PubMed] [Google Scholar]
- Puntschart A, Wey E, Jostarndt K, Vogt M, Wittwer M, Widmer HR, Hoppeler H, Billeter R. Expression of fos and jun genes in human skeletal muscle after exercise. American Journal of Physiology. 1998;274:C129–137. doi: 10.1152/ajpcell.1998.274.1.C129. [DOI] [PubMed] [Google Scholar]
- Putman CT, Dusterhoft S, Pette D. Satellite cell proliferation in low frequency-stimulated fast muscle of hypothyroid rat. American Journal of Physiology - Cell Physiology. 2000;279:C682–690. doi: 10.1152/ajpcell.2000.279.3.C682. [DOI] [PubMed] [Google Scholar]
- Pyronnet S, Sonenberg N. Cell-cycle-dependent translational control. Current Opinion in Genetics and Development. 2001;11:13–18. doi: 10.1016/s0959-437x(00)00150-7. [DOI] [PubMed] [Google Scholar]
- Schmalbruch H, Hellhammer U. The number of nuclei in adult rat muscles with special reference to satellite cells. Anatomical Record. 1977;189:169–175. doi: 10.1002/ar.1091890204. [DOI] [PubMed] [Google Scholar]
- Schultz E. Satellite cell behavior during skeletal muscle growth and regeneration. Medicine and Science in Sports and Exercise. 1989;21:S181–186. [PubMed] [Google Scholar]
- Sheikh MS, Hollander MC, Fornance AJ., Jr Role of Gadd45 in apoptosis. Biochemical Pharmacology. 2000;59:43–45. doi: 10.1016/s0006-2952(99)00291-9. [DOI] [PubMed] [Google Scholar]
- Smith CW, Klaasmeyer JG, Woods TL, Jones SJ. Effects of IGF-I, IGF-II, bFGF and PDGF on the initiation of mRNA translation in C2C12 myoblasts and differentiating myoblasts. Tissue and Cell. 1999;31:403–412. doi: 10.1054/tice.1999.0033. [DOI] [PubMed] [Google Scholar]
- Smith RH, Palmer RM, P J Reed PJ. Protein synthesis in isolated rabbit forelimb muscles. The possible role of metabolites of arachidonic acid in the response to intermittent stretching. Biochemical Journal. 1983;214:153–161. doi: 10.1042/bj2140153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava P, Russo J, Mgbonyebi O, Russo I. Growth inhibition and activation of apoptotic gene expression by human chorionic gonadotropin in human breast epithelial cells. Anticancer Research. 1998;18:4003–4010. [PubMed] [Google Scholar]
- Taylor AK, Obholz K, Linden G, Sadiev S, Klaus S, Carlson KO. DNA sequence and muscle-specific expression of human sarcosin transcripts. Molecular and Cellular Biochemistry. 1998;183:105–112. doi: 10.1023/a:1006824331819. [DOI] [PubMed] [Google Scholar]
- Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene. 2001;20:1803–1815. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- Terada K, Mori M. Human DnaJ homologs dj2 and dj3, and bag-1 are positive cochaperones of hsc70. Journal of Biological Chemistry. 2000;275:24728–24734. doi: 10.1074/jbc.M002021200. [DOI] [PubMed] [Google Scholar]
- Tiainen M, Pajalunga D, Ferrantelli F, Soddu S, Salvatori G, Sacchi A, Crescenzi M. Terminally differentiated skeletal myotubes are not confined to G0 but can enter G1 upon growth factor stimulation. Cell Growth and Differentiation. 1996;7:1039–1050. [PubMed] [Google Scholar]
- Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO Journal. 2001;20:4370–4379. doi: 10.1093/emboj/20.16.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Q, Chen IT, Antinore MJ, Fornace AJ., Jr Tumor suppressor p53 can participate in transcriptional induction of the GADD45 promoter in the absence of direct DNA binding. (erratum appears in Molecular and Cellular Biology 18, 5620) Molecular and Cellular Biology. 1998;18:2768–2778. doi: 10.1128/mcb.18.5.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Q, Schummer M, Hood L, Morris DR. Messenger RNA translation state: the second dimension of high-throughput expression screening. Proceedings of the National Academy of Sciences of the USA. 1999;96:10632–10636. doi: 10.1073/pnas.96.19.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Evans S, Chen J, Kuo H, Harvey R, Chien K. CARP, a cardiac ankyrin repeat protein, is downstream in the Nkx2-5 homeobox gene pathway. Development. 1997;124:793–804. doi: 10.1242/dev.124.4.793. [DOI] [PubMed] [Google Scholar]


