Abstract
Heart development is a precisely coordinated process of cellular proliferation, migration, differentiation, and integrated morphogenetic interactions, and therefore it is highly susceptible to developmental anomalies such as the congenital heart disease (CHD). One of the major causes of CHD has been shown to be the mutations in key cardiac transcription factors, including nkx2.5. Here, we report the analysis of zebrafish mutant ftk that showed a progressive heart malformation in the later stages of heart morphogenesis. Our analyses revealed that the cardiac muscle maturation and heart morphogenesis in ftk mutants were impaired because of the disorganization of myofibrils. Notably, we found that the expression of nkx2.5 was down-regulated in the ftk heart despite the normal expression of gata4 and tbx5, suggesting a common mechanism for the occurrence of ftk phenotype and CHD. We identified ftk to be a loss-of-function mutation in a connexin gene, cx36.7/early cardiac connexin (ecx), expressed during early heart development. We further showed by a rescue experiment that Nkx2.5 is the downstream mediator of Ecx-mediated signaling. From these results, we propose that the cardiac connexin Ecx and its downstream signaling are crucial for establishing nkx2.5 expression, which in turn promotes unidirectional, parallel alignment of myofibrils and the subsequent proper heart morphogenesis.
Keywords: cardiomyocyte, congenital heart disease, trabeculation, futka, positional cloning
Normal development and function of the heart are indispensable for the survival of vertebrates. Cardiogenesis involves a precisely coordinated process of cellular proliferation, migration, and differentiation, and integrated morphogenetic interactions. Because of the complexity of these embryonic processes, heart development is highly susceptible to developmental anomalies collectively known as congenital heart diseases (CHD). The syndrome threatens the lives of as many as 1% of newborns (1, 2). Studies have uncovered that several transcription factors, including TBX5, NKX2.5, GATA4, and SALL4, cooperate to direct the cardiac cell lineage commitment and/or morphogenesis through the regulation of proteins characteristic of cardiomyocytes and that mutations in these transcription factors are responsible for many cases of CHD (3–7). Recent data have also suggested the significant roles of these transcription factors for maintenance of the functional heart in postnatal life as well (8–12). In the past decade, zebrafish (Danio rerio) has emerged as a valuable model system to study many developmental processes, including heart development, and a number of important insights have been gained from analyses of zebrafish mutants (13–17). In addition to the early determination and differentiation of heart primordium, zebrafish mutations are also expected to contribute to the clarification of a molecular mechanism that governs myofibrillogenesis and morphogenesis of the heart. The unidirectional alignment of myofibrils is essential for proper functioning of striated muscle such as skeletal and heart muscles (18). Despite a well characterized morphology, the molecular mechanisms of myofibril formation are poorly understood (19).
Here, we report the analysis of futka (ftk), a newly identified zebrafish mutant that displays abnormalities in myofibrillogenesis and morphogenesis of the heart. We show that the characteristics of ftk phenotypes, including progressive abnormalities in cardiac muscle maturation and heart morphogenesis, are in common with some types of CHD. In particular, the expression of key cardiac transcription factor, nkx2.5, which is one of the responsible genes in CHD, is severely down-regulated in the ftk heart that exhibits anisotropy of myofibrils. We identify that the ftk mutation is a loss-of-function allele of the connexin gene cx36.7/early cardiac connexin (ecx), expressed during early heart development. Furthermore, we show that the forced expression of nkx2.5 can rescue all ftk phenotypes, thus demonstrating that Nkx2.5 is the downstream mediator of Ecx function required for proper heart morphogenesis and maturation. Our analysis of ftk mutant reveals that Ecx is a unique regulator for establishing nkx2.5 expression at an early stage of heart development, which is essential for heart morphogenesis, including ordered alignment of myofibrils in cardiomyocytes and trabeculation of heart muscles.
Results and Discussion
Progressive Heart Phenotype in ftk Mutant.
A number of zebrafish heart mutations have been generated and analyzed so far (13–17, 20, 21); however, only a relatively small number of mutants have shown specific phenotypes in the later processes of heart formation, such as cardiomyocyte maturation, trabeculation of the cardiac chamber walls, and coordination of heartbeats, which are processes that occur after the initial formation of basic body plan of the heart. In our small-scale haploid mutant screening, we identified the zebrafish mutant ftk, which had no recognizable abnormality during early development and morphogenesis but showed progressive development of abnormalities in heart morphogenesis and function (Fig. 1). Early development of ftk embryos seemed to be normal until ≈24 hours postfertilization (hpf), i.e., around the time when the heart has completed tube formation and starts beating. The ftk phenotype was first recognized as a growing, but subtle, pericardiac edema at ≈28 hpf, which became clearer by 36 hpf [see supporting information (SI) Fig. 6]. After 48 hpf, ftk mutants developed severe heart phenotypes such as dilated heart chambers (Fig. 1 B and D). In addition to their abnormal heart morphology, blood flow was abnormal such that it often flowed back to adverse direction. Moreover, heartbeats were also irregular and slower in ftk mutants; the number of heartbeats was less than half of wild-type (Table 1 and SI Movies 1 and 2). Despite these heart anomalies, the ftk mutant could survive until ≈7 dpf.
Fig. 1.
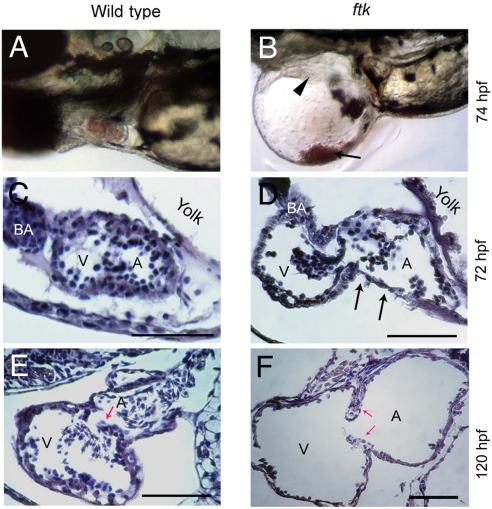
Progressive heart malformation in zebrafish ftk mutant. (A and B) Gross heart morphology of ftk mutant. Wild-type (A) and ftk mutant embryos (B) at 74 hpf. Arrowhead indicates heart perforations in the ftk mutant; arrow points to blood cells accumulated in the pericardiac cavity. (C–F) Sagittal sections of the heart. At 72 hpf (C and D), a dilated atrium (A) and ventricle (V) are evident in the ftk mutant. The cardiac muscles appear to be thinner (arrows in D), and the dilation of heart chambers is under way. At 120 hpf (E and F), the heart chamber dilation becomes more severe in ftk mutants with thinner ventricular and atrial walls. Red arrows indicate the valve leaflets. (Scale bars: 150 μm.)
Table 1.
Abnormal heartbeat in ftk mutant
| Larvae | n | 48 hpf |
72 hpf |
||
|---|---|---|---|---|---|
| Atrium | Ventricle | Atrium | Ventricle | ||
| Wild-type | 15 | 114 ± 3 | 144 ± 3 | 206 ± 4 | 206 ± 5 |
| ftk* | 12 | 108 ± 5 | 59 ± 5 | 80 ± 6 | 62 ± 4 |
| ftk morphant | 12 | 114 ± 5 | 54 ± 5 | 83 ± 5 | 57 ± 5 |
Quantitative analysis of heartbeat (per minute) at 48 hpf and 72 hpf at 28°C is shown. Measurement was performed by using the recorded movies.
*Compared with wild-type larvae, ftk mutants show unsynchronized contractions of the atrium and ventricle in addition to the slower heart beating.
In ftk mutants, circulating blood cells often leaked into the pericardiac cavity (Fig. 1B), resulting in a lack of circulating blood cells in the mutants. By close examination, the blood leakage was caused by perforations in the heart wall (Fig. 1B), suggesting that the mutant hearts were fragile and were not resistant to the blood pressure. These defects could account for the pericardiac edema phenotype in ftk mutants and progressive increase of its severity.
To evaluate further the defects in the mutant hearts, we examined histological sections. It has been reported that the trabeculation in ventricle occurs between 72 and 120 hpf of zebrafish heart (22). Consistent with this report, the ventricle wall was found gradually thickened from 72 to 120 hpf for the wild-type larvae (Fig. 1 C and E). In contrast, the thickness of cardiac chamber wall in ftk mutants was unsynchronized and partly thinner (Fig. 1D). At 120 hpf, the mutant heart walls became thinner and severely dilated, and proper heart chamber morphogenesis was not accomplished (Fig. 1 E and F). Thus, the process of cardiac muscle maturation and heart morphogenesis appeared to be impaired in ftk mutants.
The backflow of bloodstream raised the possibility that the development of valve leaflets could be deformed in ftk mutants, which might have affected the blood flow and in turn resulted in the abnormal heart morphogenesis. However, we did not observe any significant defects in the valve leaflets of atrioventricular valves and bulbus arteriosus in larvae at 5 days postfertilization (dpf) (Fig. 1F and data not shown), in which the leaflets are clearly recognized in the atrioventricular region around this time (Fig. 1E) (17). This finding suggests that the dilated abnormal chamber morphology may be the cause of blood backflow. Taken together, from gross morphological inspection, the initial basic design of the heart such as the chamber specification and valve formation appeared to be normal in ftk mutants, but the mutant heart chambers were thinner and dilated, lacking the trabeculation of heart muscles. Thus, the ftk mutation appeared to affect primarily the maturation of cardiac muscles and subsequent proper heart morphogenesis.
Abnormal Myofibril Organization in ftk Cardiac Muscles.
To characterize the abnormalities in the ftk cardiomyocytes further, we examined the subcellular ultrastructure of cardiomyocytes. It had been reported that the ordered pattern of myofibers becomes evident in wild-type zebrafish cardiomyocytes at ≈48 hpf (23); therefore, we performed the transmission electron microscopy (TEM) analysis at ≈58 hpf. Normally, mature cardiomyocytes at this stage of development contain highly developed mitochondria and a well organized thick assembly of myofibrils (Fig. 2A). When sections were made along the longitudinal axis of heart, we observed that most of the myofibrils in a single cell were organized in the same orientation in this stage of wild-type larvae, and the myofibers in adjacent cardiac muscles were also in the same orientation (Fig. 2 A and B). However, in ftk mutants, we found that the myofibril organization was irregular such that the myofibril bundles were oriented randomly (Fig. 2 C and D; n = 10). In sections along the longitudinal axis of heart, many myofibril bundles, each of which seemed to be smaller than those of wild-type, run perpendicular to the plane of section (asterisks in Fig. 2D) in addition to many fiber bundles running in the plane of section (double-headed arrows in Fig. 2D).
Fig. 2.
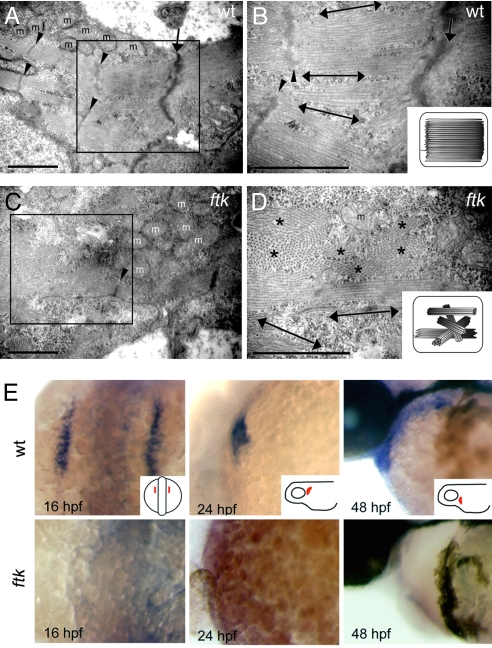
ftk mutation primarily affected the myofibril organization and nkx2.5 expression. (A–D) Myofibril organization examined by TEM analysis. In wild-type embryos at 58 hpf (A and B), myofibrils assemble as thick parallel bundles with orientation toward the longitudinal direction of the cardiomyocyte (double-headed arrows). In contrast, myofibrils in the ftk heart (C and D) are oriented in irregular directions. Double-headed arrows and asterisks indicate the myofibril bundles sectioned parallel and perpendicular to the long axis of the cells, respectively. Arrowheads, Z-lines; arrows, cell–cell contact sites; m, mitochondria. (Scale bars: 500 nm.) (B and D Insets) Schematic drawings of myofibril organization. (E) Whole-mount in situ hybridization analysis of nkx2.5 expression. In wild-type zebrafish embryos (Upper) The expression is detected in the developing heart; however, it is not detectable in ftk mutants (Lower). Mutants at early stages were genotyped by RFLP analysis. (Insets) Orientations of embryos.
Aside from the disorganized myofibril orientation, other subcellular structures characteristic of the cardiac muscles seemed to be normal. Intriguingly, in contrast to the disorganized heart myofibrils, the myofibrils themselves in the mutant skeletal muscles were assembled normally (SI Fig. 7), indicating that ftk mutation affected only myofibrils in the cardiac muscles. Overall, it is reasonable to assume that the myofibril organization may affect the heartbeat, cardiac muscle trabeculation, and heart morphogenesis; thus, our observations suggested that the ftk phenotypes were derived from a primary defect in myofibril organization.
Down-Regulation of Nkx2.5 Transcription in ftk Mutants.
We further investigated the ftk phenotype at the molecular level by looking at the expression of genes for cardiac structural proteins such as Cmlc2 and Vmhc, signaling molecules such as Bmp2b, Bmp4, and Notch1b, and transcription factors such as MyoD, Gata2, Gata4, Gata5, Tbx5, Mef2c, NFATc1, and Nkx2.5. Surprisingly, only nkx2.5 expression was severely down-regulated in ftk mutants from the early stage of heart development (Fig. 2E), whereas the expression of other cardiac genes was not affected (data not shown).
A number of studies have established that Nkx2.5 is a key transcription factor that regulates the differentiation, maturation, and maintenance of cardiomyocytes throughout life (24) and that the gene is mutated in a class of human CHD (4–7, 12, 25). Although the nkx2.5 gene knockout mice showed a severer phenotype than ftk in terms of cardiomyocyte commitment (26, 27), the overall phenotypes of ftk, including the loss of Nkx2.5, were reminiscent of those of nkx2.5 knockout mice. To confirm that ftk phenotype is caused by the loss of nkx2.5 expression, we examined the phenotype produced by the knockdown of nkx2.5 (SI Fig. 8A and SI Table 2). The cardiac phenotypes, including the severe pericardiac edema, irregular heartbeat, reverse blood flow (SI Movie 3), and disorganized myofibril orientation (SI Fig. 8B; n = 8), were similar to those in ftk mutants, demonstrating that the entire cardiac phenotype of ftk can be explained by loss of nkx2.5 expression. Taken together, our observations suggest a link between phenotypes of the ftk mutant and CHD; and hence we considered that the zebrafish ftk mutant could serve as a useful model for studying the mechanism of CHD and heart morphogenesis.
The ftk Locus Encodes a Unique Cardiac Connexin Gene.
In exploring the molecular basis of the ftk mutant, we decided to determine the ftk gene by the positional cloning approach. By using single-strand conformation polymorphism analysis, we mapped the ftk locus to a region on zebrafish linkage group (LG) 7 (Fig. 3A) that houses six putative ORFs. The region neither contained nkx2.5 gene, which is on LG 14, nor any similar nkx family genes. We amplified cDNAs of these ORFs from ftk mutants and wild-type embryos and determined their sequences. By sequencing analysis, we found that an adenine (A) to thymine (T) missense mutation, which results in a substitution of an aspartic acid (D) residue with a valine (V) at position 12 in the N-terminal domain, occurred in the connexin gene (Figs. 3B and 4 A and B). We designated this connexin as Cx36.7 according to its calculated molecular weight. To date, the connexin gene family comprises 20 members in humans, 19 in mice, and 37 in zebrafish (28). Judging from the phylogeny, the zebrafish Cx36.7 appeared to be the close homolog of human CX31.9 or CX40.1 (SI Fig. 9A), and it is more likely to be the ortholog of CX31.9 from the synteny conservation between corresponding genomic regions of human and zebrafish chromosomes (SI Fig. 9B). Although it has been shown that the CX31.9 gene is expressed in adult human heart (29), the expression during heart development has not been demonstrated.
Fig. 3.
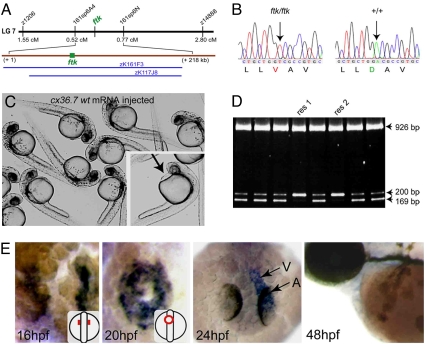
Molecular characterization of ftk gene. (A) Chromosome map of the ftk locus on LG 7 between markers z1206 and z14888. An enlarged map (brown line) and overlapping BAC clones (blue lines) are shown below. (B) Missense mutation in the zebrafish cx36.7 gene of the ftk mutant. (C and D) Rescue of the ftk phenotype by cx36.7 mRNA injection. Embryos obtained from crosses of ftk heterozygotes and injected with cx36.7 mRNA (30 pg) did not show the ftk phenotype at 36 hpf (C). (Inset) Sibling uninjected ftk mutant at 28 hpf, which displays the growing pericardiac edema (arrow). Genotyping of mRNA-injected embryos by RFLP analysis confirmed that two of eight embryos were genotypically ftk mutants (res1, res2) (D). (E) Whole-mount in situ hybridization analysis of cx36.7 expression during heart development. (Insets) Orientation of embryos.
Fig. 4.
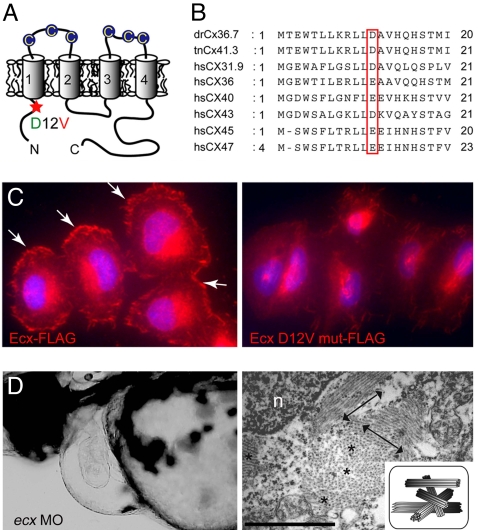
ftk is a loss-of-function mutation of ecx/cx36.7. (A) Putative topology of Ecx protein. Extracellular cysteine residues are indicated by circles. The D12V mutation located in the N-terminal intracellular region of Ecx is indicated with a red asterisk. (B) Alignment of the N-terminal domain of zebrafish Ecx with Tetraodon Cx41.3 and six human connexins. (C) Subcellular localization of FLAG-tagged Ecx proteins in HeLa cells. Wild-type Ecx is localized at the cell surface (Left, arrows), whereas the mutant protein is not recruited to the membrane surface (Right). Nuclei were stained with Hoechst 33342 (blue). (D) Production of ftk phenotype with the ecx MO. (Left) Severe pericardiac edema and dilated heart chambers are observed in larvae injected with ecx MO. Lateral view at 56 hpf is shown. (Right) Disorganized myofibrils as seen in ftk mutants were also caused by the ecx MO. Double-headed arrows and asterisks indicate the myofibril bundles sectioned parallel and perpendicular to the long axis of the cells, respectively. (Scale bar: 500 nm.) n, nucleus. (Inset) Schematic of myofibril organization.
To confirm that cx36.7 and ftk were the same gene, we tested whether or not the cx36.7 expression could rescue the ftk phenotypes. When the mRNA encoding the wild-type Cx36.7 was injected into ftk embryos, the ftk phenotype, including the heart dilation and severe pericardiac edema, was rescued at doses >30 pg of mRNA without affecting other parts of embryos (Fig. 3 C and D and SI Table 3), thus establishing that cx36.7 is the responsible gene for the ftk mutant.
To verify the direct role of ftk in the developing heart further, we determined the expression of cx36.7 during stages between 30% epiboly to 5 dpf. The expression was detectable in the ventral half of the marginal zone in 50% epiboly stage embryo (SI Fig. 10) and confined to the region of cardiac precursors in the following stages. By 16 hpf, the expression was localized in the myocardial precursor cells on both sides of the midline (Fig. 3E) and distributed to the whole heart in the later stages. Such expression of cx36.7 is similar to that of nkx2.5 expression (30); however, the expression started to decline by 36 hpf and was barely detectable at 48 hpf. Thus, the zebrafish Cx36.7 appeared to function in the developing heart, with a peak of expression corresponding to the time when the ftk phenotypes arise. We also observed that cx36.7 gene was normally expressed in ftk mutants. For its unique expression in developing heart, we called this connexin as “early cardiac connexin” (Ecx) and use this name in the following sections.
The ftk Mutation Abolishes the Connexin Function.
Connexins oligomerize to form gap junction, a cell surface channel involved in communication between neighboring cells and/or between the extracellular–intracellular milieus (31); however, they are atypical membrane proteins lacking an apparent signal peptide, and their translocation mechanism to plasma membrane is poorly understood. When the amino acid sequences of Ecx and several connexins were compared, the amino acid residue at position 12, corresponding to the mutated residue in the ftk mutant, was conserved or substituted with another acidic residue, glutamic acid (Fig. 4 A and B). It has been shown that this residue is crucial for the connexin function (32); therefore, it is reasonable to postulate that the D12V mutation may be a loss-of-function mutation of Ecx. To confirm this hypothesis, we introduced the C-terminally FLAG-tagged Ecx proteins in HeLa cells. Remarkably, the mutant Ecx did not reach the cell surface, in contrast to the dense cell surface distribution of the wild-type Ecx protein (Fig. 4C), supporting our notion that the mutant ecx could be a loss-of-function allele caused by the inability of protein trafficking to the cell surface.
To confirm further that ftk was a loss-of-function mutation of ecx, we knocked down the ecx gene expression by injecting an antisense morpholinooligonucleotide (MO) for this gene into wild-type embryos. The resultant morphant displayed ftk-like phenotypes, such as the heart chamber dilation and irregular and slower heartbeat, which were indistinguishable from those of ftk (97%; n = 720; Fig. 4D, Table 1, and SI Movie 4). These data strongly supported that the ftk mutation was a loss-of-function phenotype. Importantly, we also confirmed by the TEM analysis that the ecx-morphants had disorganized myofibril bundles as in the ftk mutant (Fig. 4D, Right; n = 10) and that nkx2.5 expression in the morphant heart was down-regulated as in ftk mutants (SI Fig. 11). Taken together, these facts indicated that Ecx function was required for the formation of ordered and parallel myofibrils in cardiomyocytes.
Connexin Function Is Directly Required for Heart Morphogenesis Through the Regulation of Nkx2.5 Expression.
Connexins have a major function in cell–cell coupling of cardiac muscles, which raises the possibility that loss-of-Ecx function could affect primarily the cardiac conduction system and lead to the phenotypes in heartbeat, myofibrils, and heart morphogenesis. To check whether ftk mutation affected primarily the conduction system, we visualized the calcium wave in the heart by using the calcium sensitive fluorescent dye. In ftk mutants, the Ca2+ wave that represents the electrical conduction between the cardiac muscles normally and smoothly sweeps across the heart from the atrium terminal to the ventricle terminal despite the uncoordinated and irregular contraction of chambers of ftk mutant (SI Movies 5 and 6). This result suggested that the irregular heartbeat in ftk mutants is not caused by the impaired cellular coupling between cardiomyocytes but rather by the inability of cardiomyocytes themselves to respond properly to the electrical conduction.
Thus, our results suggested that connexin function in cell–cell coupling may not be responsible for the ftk phenotypes but rather that other connexin functions, such as hemichannel activity or nonchannel functions relating to interactions with cytoplasmic proteins, are required for proper myofibril organization and heart morphogenesis. Indeed, in accordance with the above data, we also found through a dye uptake assay that Ecx could function as a hemichannel, but the mutant Ecx did not (SI Fig. 12). As we have suggested, there are significant similarities between the ftk mutant and CHD in their anomalies in heart morphogenesis and loss of Nkx2.5 function. Additionally, previous studies in mice have shown that the null mutant mice of nkx2.5 exhibited an array of heart phenotypes such as impaired chamber morphogenesis and defective cardiomyocyte trabeculation (26, 27). These studies raise the possibility that Nkx2.5 is a downstream mediator of Ecx-derived signaling during heart morphogenesis.
To test further the possibility that ftk phenotypes are caused solely by the loss of Nkx2.5, we examined the ability of Nkx2.5 to compensate the loss of Ecx. We coinjected nkx2.5 mRNA along with 2 ng of ecx MO, which was sufficient to cause the ftk phenotype in nearly all embryos (SI Table 4), and assayed the ftk phenotypes. Remarkably, as small as 10 pg of nkx2.5 mRNA was enough to prevent all ftk phenotypes, including the heart morphogenesis (Fig. 5A and SI Table 4), myofibril organization (Fig. 5B; n = 8), and slower heartbeat (SI Movies 7 and 8). Similarly, nkx2.5 mRNA injection could rescue ftk mutants obtained by crosses of ftk heterozygotes (SI Table 4). More intriguingly, nkx2.5 mRNA injection rescued a significant percentage (≈50%) of ftk mutants to viability up to 21 dpf (SI Table 5), suggesting that the early nkx2.5 expression is sufficient for proper heart formation. Possibly, signaling mechanisms other than that of Ecx may maintain nkx2.5 expression at later stages. From these data, we concluded that Nkx2.5 was the downstream mediator of Ecx-derived signaling and responsible for all ftk phenotypes that were seen during heart morphogenesis.
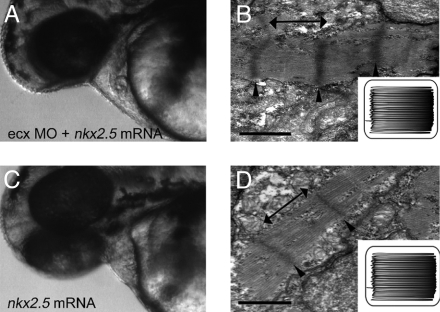
Fig. 5.
Rescue of ftk phenotype by nkx2.5 expression. (A–D) Embryos injected with nkx2.5 mRNA plus ecx MO (A and B) or nkx2.5 mRNA alone (C and D). Embryos were examined for their external phenotypes at 48 hpf (A and C; lateral views are shown in the lower right) and myofibril organization at 56 hpf by TEM analysis (B and D). The ftk phenotypes in the heart caused by ecx MO were apparently attenuated by coinjection with nkx2.5 mRNA. Coinjection of nkx2.5 mRNA also rescued the ftk-like abnormality in myofibril organization. Arrowheads, Z-lines; double-headed arrows, myofibril bundles. (Insets, B and D) Schematic of myofibril organization.
Concluding Remarks.
Our work revealed a role of connexin in cardiomyocyte maturation through the establishment of nkx2.5 expression. Without the Ecx function and the Ecx-mediated signal, the consequent down-regulation of Nkx2.5 leads to the abnormalities of myofibril organization, cardiomyocyte maturation, and heart morphogenesis. However, in contrast to sustained expression of nkx2.5 in the heart, ecx is only expressed before 48 hpf (Fig. 3E), hence some other mechanism may be required for the sustained expression of nkx2.5. Studies have suggested that Bmp2 and Bmp4 (33, 34), Fgf (35), and phosphatidylinositol 3-kinase signaling (36) may also be the upstream regulators of nkx2.5 transcription. In particular, Bmp signaling may induce cardiac nkx2.5 expression through the activation of the composite enhancer by Smads in collaboration with Gata4 (37). These factors and/or signals may have a role for the later regulation of nkx2.5 transcription in combination with Ecx-derived signal.
In addition, an epigenetic regulation for sustained nkx2.5 transcription is also suggested (38). In mice deficient for Polycomb group gene Rae 28, Nkx2.5 expression was initiated normally but not sufficiently sustainable. Furthermore, a possible mechanism for the maintenance of nkx2.5 may be that other cardiac connexins expressed in the subsequent stages may take over the role of Ecx in later stages and maintain the nkx2.5 expression (39, 40). Intriguingly, connexins such as cx40 and cx43 have been suggested to be the downstream target of Nkx2.5 (27, 41). A feedback mechanism between connexins and Nkx2.5 might exist for the sustained nkx2.5 expression. Such mechanisms regulating the nkx2.5 transcription may be included in the machinery that controls and maintains the functional heart.
An important question remained is what signaling molecule(s), regulated by Ecx, modulate(s) early transcription of nkx2.5. Because Ecx can act as a functional hemichannel and gap junction (data not shown), we suspect that Ecx may mediate extracellular and/or intercellular signaling through molecules such as Ca2+ or inositol 1,4,5-trisphosphate (42, 43) to activate the nkx2.5 transcription. Further comprehensive studies will be needed to elucidate the mechanism for early induction and establishment of nkx2.5 expression in the heart. The zebrafish ftk mutant may serve as a useful model for understanding the signaling mechanisms underlying proper morphogenesis and maintenance of a functional heart, the dysfunction of which leads to CHD.
Materials and Methods
Zebrafish Strains.
The wild-type strain TL was used for mutagenesis with N-ethyl-N-nitrosourea. Tü and WIK strains were used for genetic mapping. The ftk mutant, jk65, was identified by a small-scale haploid mutant screen performed at the University of Tokyo (A.K., unpublished data). From the mutant morphology, which is similar to blown pufferfish, we termed the ftk mutant as futka, a local name of pufferfish in the Ganges basin from which the zebrafish originated.
Positional Cloning.
Positional cloning was performed by a standard method by scoring 1,500 mutant embryos. Briefly, we mapped the ftk mutation to the vicinity of z8252 on LG 7. We further performed fine mapping and identified two closely linked markers, z1206 and z14462. By chromosomal walking with the bacterial artificial chromosome (BAC) library, we identified zK161F3 as a clone that covered the entire ftk locus. The whole genomic sequence of zK161F3 was obtained from the assembled zebrafish genome sequence. The putative candidate cDNAs encoding MARCH III, gap junction connexin, SH3 containing the Grb2 SH3 domain, ferritin, serum amyloid A, and 60S acidic ribosomal protein P2 were amplified by RT-PCR to identify the mutation. The sequence and mutation were verified in genomic sequences.
Transfection.
Constructs with 3×FLAG tags at the C terminus were made by subcloning the wild-type and mutant ecx into pcDNA3.1 (Invitrogen). The constructs were used to transfect HeLa cells, and the cells were fixed and subjected to immunological staining with anti-FLAG antibody (Sigma). Nuclei were stained with Hoechst 33342.
Electron Microscopy.
Embryos were fixed at ≈58 hpf with 2.5% glutaraldehyde and 4% paraformaldehyde in PBS (pH 7.0). After fixation was carried out in 0.5% OsO4 and 0.8% K3Fe(CN)6, the specimens were processed by a standard procedure for TEM. Ultrathin sections (80 nm) were stained with uranyl acetate and lead citrate and analyzed with an electron microscope.
In Situ Hybridization.
Whole-mount in situ hybridization was carried out by using a standard protocol (44). Respective plasmids for probes were made by the PCR based on the reported sequences in the GenBank and cloned into pBluescript.
Knockdown.
Antisense ecx MO (5′-GGAGAGCGAAGGTGCCATCACTGCT-3′) and nkx2.5 MO (5′-CATTTGGCTAGAGAACATTGCCATG-3′) (Gene Tools) targeting the ATG initiation sites of respective transcripts were used. The MOs were dissolved in 1× Danieau buffer, and aliquots of 2 ng (ecx MO) or the concentrations indicated in SI Table 2 (nkx2.5 MO) were injected into embryos at the one- to two-cell stage.
mRNA Injections.
mRNAs were synthesized in vitro by using an mMESSAGE mMACHINE kit (Ambion) and injected into one- to two-cell stage embryos. Some embryos were genotyped by Hin1I restriction fragment-length polymorphism (RFLP) analysis.
Dye-Uptake Assay.
The dye-uptake assay for Ecx-expressing cultured cells was performed according to the procedure as described in ref. 45 with some minor modification. Stocks of fluorescent dyes were prepared in PBS (pH 7.4) at 2 mg/ml for Lucifer yellow CH lithium salt (557 molecular weight; Invitrogen) and 25 mg/ml for the dextran–rhodamine (10,000 molecular weight, Invitrogen), which was used for labeling the dead cells. The HeLa cells (≈105) were transfected with the construct encoding Ecx (either wild-type or mutant). Transfection efficiency was evaluated by cotransfection with constructs encoding EGFP and was found to exhibit an expression efficiency of ≈80%. At 48 h after transfection, transfected cells were rinsed twice with PBS on ice and mechanically stimulated with a continuous drip of 800 μl of dye released from a height of ≈4.5 cm above the culture dish. The stimulation was repeated three times, and the dye-covered cells were incubated on ice for 5 min. Cultures were washed 10 times with ice-cold PBS and examined under a fluorescent microscope. Phase-contrast and fluorescent images were collected around the drip target site. Four independent transfections were assayed for dye uptake.
Supplementary Material
Acknowledgments.
We thank Akira Kato, Yoko Yamamoto, Silvia Penuela, and Qing Shao for technical assistance and Setsuko Sato for secretarial assistance. This work was supported by grants-in-aid for scientific research from the Ministry of Education, Culture, Sport, Science, and Technology of Japan (MEXT) and by funding from the 21st Century COE Program of MEXT.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB274833).
This article contains supporting information online at www.pnas.org/cgi/content/full/0708451105/DC1.
References
- 1.D'Alton ME, DeCherney AH. Prenatal diagnosis. N Engl J Med. 1993;328:114–120. doi: 10.1056/NEJM199301143280208. [DOI] [PubMed] [Google Scholar]
- 2.Ransom J, Srivastava D. The genetics of cardiac birth defects. Semin Cell Dev Biol. 2007;18:132–139. doi: 10.1016/j.semcdb.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Akazawa H, Komuro I. Roles of cardiac transcription factors in cardiac hypertrophy. Circ Res. 2003;92:1079–1088. doi: 10.1161/01.RES.0000072977.86706.23. [DOI] [PubMed] [Google Scholar]
- 4.Hatcher CJ, Diman NY, McDermott DA, Basson CT. Transcription factor cascades in congenital heart malformation. Trends Mol Med. 2003;9:512–515. doi: 10.1016/j.molmed.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Akazawa H, Komuro I. Cardiac transcription factor Csx/Nkx2-5: Its role in cardiac development and diseases. Pharmacol Ther. 2005;107:252–268. doi: 10.1016/j.pharmthera.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Clark KL, Yutzey KE, Benson DW. Transcription factors and congenital heart defects. Annu Rev Physiol. 2006;68:97–121. doi: 10.1146/annurev.physiol.68.040104.113828. [DOI] [PubMed] [Google Scholar]
- 7.Oka T, Xu J, Molkentin JD. Reemployment of developmental transcription factors in adult heart disease. Semin Cell Dev Biol. 2007;18:117–131. doi: 10.1016/j.semcdb.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson JT, Rackley MS, O'Brien TX. Up-regulation of the cardiac homeobox gene Nkx2-5 (CSX) in feline right ventricular pressure overload. Am J Physiol. 1998;274:H1569–H1573. doi: 10.1152/ajpheart.1998.274.5.H1569. [DOI] [PubMed] [Google Scholar]
- 9.Saadane N, Alpert L, Chalifour LE. Expression of immediate early genes, GATA-4, and Nkx-2.5, in adrenergic-induced cardiac hypertrophy and during regression in adult mice. Br J Pharmacol. 1999;127:1165–1176. doi: 10.1038/sj.bjp.0702676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toko H, et al. Csx/Nkx2-5 is required for homeostasis and survival of cardiac myocytes in the adult heart. J Biol Chem. 2002;277:24735–24743. doi: 10.1074/jbc.M107669200. [DOI] [PubMed] [Google Scholar]
- 11.Jay PY, et al. Nkx2-5 mutation causes anatomic hypoplasia of the cardiac conduction system. J Clin Invest. 2004;113:1130–1137. doi: 10.1172/JCI19846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pashmforoush M, et al. Nkx2-5 pathways and congenital heart disease: Loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell. 2004;117:373–386. doi: 10.1016/s0092-8674(04)00405-2. [DOI] [PubMed] [Google Scholar]
- 13.Stainier DY, et al. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development. 1996;123:285–292. doi: 10.1242/dev.123.1.285. [DOI] [PubMed] [Google Scholar]
- 14.Stainier DY. Zebrafish genetics and vertebrate heart formation. Nat Rev Genet. 2001;2:39–48. doi: 10.1038/35047564. [DOI] [PubMed] [Google Scholar]
- 15.Glickman NS, Yelon D. Cardiac development in zebrafish: Coordination of form and function. Semin Cell Dev Biol. 2002;13:507–513. doi: 10.1016/s1084952102001040. [DOI] [PubMed] [Google Scholar]
- 16.Auman HJ, Yelon D. Vertebrate organogenesis: Getting the heart into shape. Curr Biol. 2004;14:R152–R153. [PubMed] [Google Scholar]
- 17.Beis D, et al. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 2005;132:4193–4204. doi: 10.1242/dev.01970. [DOI] [PubMed] [Google Scholar]
- 18.Sanger JW, et al. Myofibrillogenesis in skeletal muscle cells. Clin Orthop Relat Res. 2002:S153–S162. doi: 10.1097/00003086-200210001-00018. [DOI] [PubMed] [Google Scholar]
- 19.Sanger JW, et al. How to build a myofibril. J Muscle Res Cell Motil. 2005;26:343–354. doi: 10.1007/s10974-005-9016-7. [DOI] [PubMed] [Google Scholar]
- 20.Trinh LA, Yelon D, Stainier DY. Hand2 regulates epithelial formation during myocardial diferentiation. Curr Biol. 2005;15:441–446. doi: 10.1016/j.cub.2004.12.083. [DOI] [PubMed] [Google Scholar]
- 21.D'Amico L, Scott IC, Jungblut B, Stainier DY. A mutation in zebrafish hmgcr1b reveals a role for isoprenoids in vertebrate heart-tube formation. Curr Biol. 2007;17:252–259. doi: 10.1016/j.cub.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 22.Hu N, Sedmera D, Yost HJ, Clark EB. Structure and function of the developing zebrafish heart. Anat Rec. 2000;260:148–157. doi: 10.1002/1097-0185(20001001)260:2<148::AID-AR50>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 23.Xu X, et al. Cardiomyopathy in zebrafish due to mutation in an alternatively spliced exon of titin. Nat Genet. 2002;30:205–209. doi: 10.1038/ng816. [DOI] [PubMed] [Google Scholar]
- 24.Kasahara H, et al. Cardiac and extracardiac expression of Csx/Nkx2.5 homeodomain protein. Circ Res. 1998;82:936–946. doi: 10.1161/01.res.82.9.936. [DOI] [PubMed] [Google Scholar]
- 25.McElhinney DB, et al. NKX2.5 mutations in patients with congenital heart disease. J Am Coll Cardiol. 2003;42:1650–1655. doi: 10.1016/j.jacc.2003.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Lyons I, et al. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeobox gene Nkx2-5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka M, et al. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- 28.Eastman SD, et al. Phylogenetic analysis of three complete gap junction gene families reveals lineage-specific duplications and highly supported gene classes. Genomics. 2006;87:265–274. doi: 10.1016/j.ygeno.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 29.Nielsen PA, et al. Molecular cloning, functional expression, and tissue distribution of a novel human gap junction-forming protein, connexin-31.9: Interaction with zona occludens protein-1. J Biol Chem. 2002;277:38272–38283. doi: 10.1074/jbc.M205348200. [DOI] [PubMed] [Google Scholar]
- 30.Chen JN, Fishman MC. Zebrafish tinman homolog demarcates the heart field and initiates myocardial differentiation. Development. 1996;122:3809–3816. doi: 10.1242/dev.122.12.3809. [DOI] [PubMed] [Google Scholar]
- 31.van Veen TA, van Rijen HV, Jongsma HJ. Physiology of cardiovascular gap junctions. Adv Cardiol. 2006;42:18–40. doi: 10.1159/000092560. [DOI] [PubMed] [Google Scholar]
- 32.Lagree V, et al. Specific amino acid residues in the N terminus and TM3 implicated in channel function and oligomerization compatibility of connexin43. J Cell Sci. 2003;116:3189–3201. doi: 10.1242/jcs.00604. [DOI] [PubMed] [Google Scholar]
- 33.Schultheiss TM, Burch JB, Lassar AB. A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes Dev. 1997;11:451–462. doi: 10.1101/gad.11.4.451. [DOI] [PubMed] [Google Scholar]
- 34.Monzen K, et al. Bone morphogenetic proteins induce cardiomyocyte differentiation through the mitogen-activated protein kinase kinase kinase TAK1 and cardiac transcription factors Csx/Nkx-2.5 and GATA-4. Mol Cell Biol. 1999;19:7096–7105. doi: 10.1128/mcb.19.10.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alsan BH, Schultheiss TM. Regulation of avian cardiogenesis by Fgf8 signaling. Development. 2002;129:1935–1943. doi: 10.1242/dev.129.8.1935. [DOI] [PubMed] [Google Scholar]
- 36.Naito AT, et al. Early stage-specific inhibitions of cardiomyocyte differentiation and expression of Csx/Nkx-2.5 and GATA-4 by phosphatidylinositol 3-kinase inhibitor LY294002. Exp Cell Res. 2003;291:56–69. doi: 10.1016/s0014-4827(03)00378-1. [DOI] [PubMed] [Google Scholar]
- 37.Brown CO, III, et al. The cardiac determination factor, Nkx2-5, is activated by mutual cofactors GATA-4 and Smad1/4 via a novel upstream enhancer. J Biol Chem. 2004;279:10659–10669. doi: 10.1074/jbc.M301648200. [DOI] [PubMed] [Google Scholar]
- 38.Shirai M, et al. The Polycomb-group gene Rae28 sustains Nkx2.5/Csx expression and is essential for cardiac morphogenesis. J Clin Invest. 2002;110:177–184. doi: 10.1172/JCI14839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon AM, McWhorter AR. Vascular abnormalities in mice lacking the endothelial gap junction proteins connexin37 and connexin40. Dev Biol. 2002;251:206–220. doi: 10.1006/dbio.2002.0826. [DOI] [PubMed] [Google Scholar]
- 40.Simon AM, et al. Heart and head defects in mice lacking pairs of connexins. Dev Biol. 2004;265:369–383. doi: 10.1016/j.ydbio.2003.09.036. [DOI] [PubMed] [Google Scholar]
- 41.Kasahara H, et al. Nkx2.5 homeoprotein regulates expression of gap junction protein connexin 43 and sarcomere organization in postnatal cardiomyocytes. J Mol Cell Cardiol. 2003;35:243–256. doi: 10.1016/s0022-2828(03)00002-6. [DOI] [PubMed] [Google Scholar]
- 42.Bennett MV, Contreras JE, Bukauskas FF, Saez JC. New roles for astrocytes: Gap junction hemichannels have something to communicate. Trends Neurosci. 2003;26:610–617. doi: 10.1016/j.tins.2003.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goodenough DA, Paul DL. Beyond the gap: Functions of unpaired connexon channels. Nat Rev Mol Cell Biol. 2003;4:285–294. doi: 10.1038/nrm1072. [DOI] [PubMed] [Google Scholar]
- 44.Thisse CTB. High resolution whole-mount in situ hybridization. Zebrafish Sci Monitor. 1998;5:8–9. [Google Scholar]
- 45.Penuela S, et al. Pannexin 1 and pannexin 3 are glycoproteins that exhibit many distinct characteristics from the connexin family of gap junction proteins. J Cell Sci. 2007;120:3772–3783. doi: 10.1242/jcs.009514. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.