Abstract
Thioredoxins (Trxs) are ubiquitous small proteins with a redox-active disulfide bridge. In their reduced form, they constitute very efficient protein disulfide oxidoreductases. In chloroplasts, two types of Trxs (f and m) coexist and play central roles in the regulation of the Calvin cycle and other processes. Here, we identified a class of Trx targets in the inner plastid envelope membrane of chloroplasts that share a CxxC motif ≈73 aa from their carboxyl-terminal end. Members of this group belong to a superfamily of Rieske iron–sulfur proteins involved in protein translocation and chlorophyll metabolism. These proteins include the protein translocon protein TIC55, the precursor NADPH:protochlorophyllide oxidoreductase translocon protein PTC52, which operates as protochlorophyllide a-oxygenase, and the lethal leaf spot protein LLS1, which is identical with pheophorbide a oxygenase. The role of these proteins in dark/light regulation and oxidative control by the Trx system is discussed.
Keywords: protein translocation, tetrapyrrole biosynthesis, Tigrina d12, FLU mutant, photooxidative stress
Thioredoxins (Trxs) are small multifunctional redox-active proteins that are widely, if not universally, distributed among living organisms (1–3). In chloroplasts and other plastid types, two main types of Trxs (f and m) operate in disulfide/dithiol interchange and thereby act as powerful regulators of enzyme activity. Trxs catalyze two successive one-electron transfer reactions. Themselves, they are reduced via ferredoxin with electrons provided by photosynthetic electron transport. Trx targets involve enzymes of the Calvin cycle and oxidative pentose phosphate cycle that often contain redox-sensitive C(x)nC pairs (n = 2–8, or longer spacing between the two Cys residues; see ref. 1 for review). The mechanism requires the formation of a transient heterodisulfide bond between the reduced Trx and the oxidized enzyme before complete reduction of the target disulfide (4–7). Affinity purification procedures take advantage of the fact that mutation in one of the redox-active cysteines in the Trx active site (the one buried in the molecule) stabilizes the normally transient heterodisulfide, thereby covalently linking the target protein to Trx via a bond that can be cleaved by DTT (e.g., refs. 4, 6, 8, and 9).
Using this approach, we identified three inner plastid envelope membrane proteins from barley chloroplasts as novel Trx targets: the protein translocon protein TIC55 (10), the precursor NADPH:protochlorophyllide (Pchlide) oxidoreductase A (pPORA) translocon protein PTC52 (11), and pheophorbide a oxygenase [PAO; originally named lethal leaf spot protein (LLS1) (12, 13)]. All three proteins contain redox-active CxxC motifs and interacted with Trxs f and m. By contrast, the related Rieske nonheme oxygenase family member chlorophyllide a-oxygenase (CAO) (14), which lacks the CxxC motiv (15), was found not to be a Trx target. Together, our findings reveal a role of Trxs in dark/light regulation of protein translocation and chlorophyll (Chl) metabolism and provoke a scenario of deterrence to pigment-sensitized oxidative stress in chloroplasts.
Results
Affinity Purification of Plastid Envelope Proteins Interacting with Trx f and Trx m.
Chloroplast Trxs contain two neighboring Cys residues in a conserved motif, Trp-Cys-Gly-Pro-Cys (in some cases Trp-Cys-Pro-Pro-Cys), that is referred to as the Trx signature (summarized in refs. 1–3). We began by designing mutant Trx f and Trx m molecules in which the buried Cys was exchanged by a Ser or Ala residue. These were bound on Sepharose 4B and tested for their capability to bind known Trx target proteins present in stromal extracts of barley chloroplasts. Fig. 1A (lane 1) shows proteins that could be eluted with DTT. Protein mass spectrometry verified the presence of sedoheptulose-1,7-bisphosphate bisphosphatase (75 kDa), phosphoribulokinase (80 kDa), and NADP-dependent malate dehydrogenase (85 kDa), which are known to be Trx targets in spinach chloroplasts (1, 3).
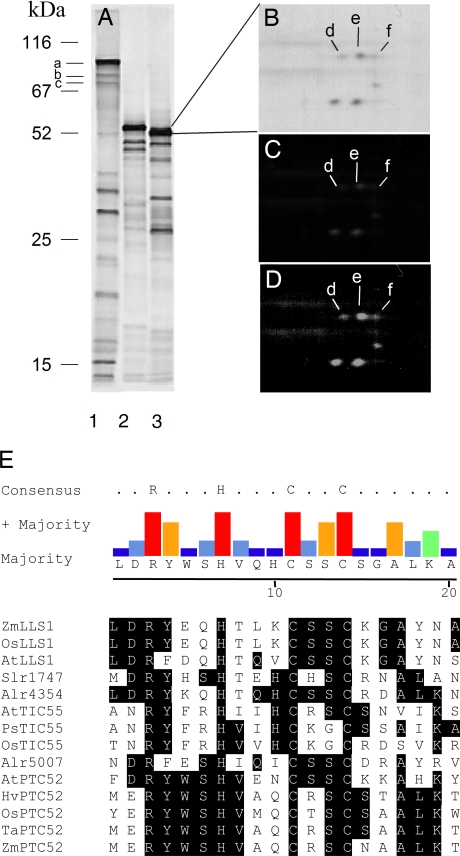
Fig. 1.
Identification of outer and inner plastid envelope membrane proteins interacting with Trx. (A) Identification of Trx targets in stromal extracts (lane 1) and solubilized outer (lane 2) and inner (lane 3) plastid envelope membranes of barley chloroplasts. The bands marked a, b, and c represent NADP-dependent malate dehydrogenase (85 kDa), phosphoribulokinase (80 kDa), and sedoheptulose-1,7-bisphosphate bisphosphatase (75 kDa), respectively. (B) Re-electrophoresis of the inner plastid envelope membrane proteins of ≈52 kDa obtained as in A. Protein spots were subjected to mass spectrometry and identified by sequence comparisons to be caused by TIC55 (d), PTC52 (e), and PAO (f). (C) Monobromobimane labeling of TIC55 (d), PTC52 (e), and PAO (f) before reduction with bacterial Trx. (D) Monobromobimane labeling of TIC55 (d), PTC52 (e), and PAO (f) after reduction with bacterial Trx. (E) Presence of a conserved CxxC motif in TIC55, PTC52, and PAO/LLS1 and related proteins from Hordeum vulgare (H.v.), Zea mays (Z.m.), Oryza sativa (O.s.), Triticum aestivum (T.a.), Arabidopsis thaliana (A.t.), and cyanobacteria (ORFs slr1747, alr4354, alr5007). Biocomputational methods and sequence data sources can be found in SI Text.
In a next step, outer and inner plastid envelope membranes were isolated from barley chloroplasts and solubilized in buffer containing 3% SDS. Each protein extract in turn was passed over the Trx affinity column, and protein was bound to Trx with eluted with DTT. The outer plastid envelope fraction gave rise to three main and a few minor bands (Fig. 1A, lane 2). Mass spectrometry identified the 55-kDa band as a protein related to Erv1 and Erv2, enzymes implicated in disulfide bridge formation in mitochondria and bacteria (16–19). The role of this protein needs to be established. For the inner plastid envelope protein fraction, five major bands were obtained, of which the 52-kDa band was by far more abundant than the other bands (Fig. 1A, lane 3). Several additional minor bands appeared that were not characterized further. Upon 2D separation of proteins in the 50- to 52-kDa range, six spots of different isoelectric points and molecular masses were found (Fig. 1B). Microsequence analysis identified three of the spots to be caused by TIC55, PTC52, and PAO (Fig. 1B, spots d-f). To confirm their role as potential Trx targets, the eluates were reduced by Trx, and the change in redox state was measured by fluorescence coupled with gel electrophoresis. Free SH groups were first blocked with N-ethylmaleimide, then the eluates were treated with the NADP/Trx system to reduce disulfide bonds and newly formed SH groups labeled with monobromobinane. Using nonequilibrium pH gradient electrophoresis and SDS/PAGE (20), we found that all three proteins (TIC55, PTC52, and PAO) showed, relative to the untreated control, an increase in fluorescence and thus qualified as Trx targets (Fig. 1 D versus C). Aside from the presence of conserved Rieske and 2Fe-2S clusters, TIC55, PTC52, and PAO share CxxC motifs at ≈73 aa from their COOH-terminal ends (Fig. 1E).
Effect of Trx on PTC52 and PAO Activity.
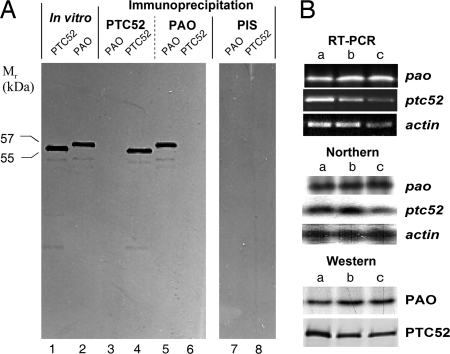
Given that no full-length cDNAs were available for PTC52 and PAO of barley, we used corresponding EST clones for barley PTC52 and a partial cDNA encoding lls1 of Zea mays for hybrid-selected translation and subsequent activity measurements. Fig. 2A shows that PTC52 and PAO could be detected in the mRNA of light-grown barley plants. The size of the detected precursors was slightly different and presumably reflected differences in the length of their transit peptides. Immunoprecipitations using monospecific (PTC52) (11) or monoclonal (PAO) (15) antibodies confirmed the identity of the different bands. Northern blotting revealed differences in the abundance of ptc52 and pao transcripts in dark-grown, light-exposed, and light-grown plants. Whereas ptc52 mRNA was most abundant in dark-grown seedlings, the pao mRNA was present at similar levels in all samples (Fig. 2B).
Fig. 2.
Expression of PTC52 and PAO. (A) Identification of PTC52 and PAO among the messenger products of 5-day-old dark-grown barley plants that had been exposed to light for 24 h, by hybrid-selected translation in the presence of [35S]methionine and immunoprecipitation using corresponding antisera and preimmune sera (PIS). (B) Expression of PTC52 and PAO in 5-day-old etiolated barley plants (lane a), 5-day-old dark-grown plants that had been exposed to light for 24 h (lane b), and 5-day-old light-grown plants (lane c), as revealed by RT- PCR, Northern hybridization, and Western blotting. As control, expression of actin was assayed.
PTC52 is normally active in the pPORA translocon where it operates as Pchlide a oxygenase (11). In contrast to this anabolic role of PTC52 in the C5 pathway, PAO acts catabolically in Chl breakdown (13). The reaction catalyzed by PAO is coupled to that of red Chl catabolite (RCC) reductase, leading to the loss of pigment color during leaf senescence. In vitro, the intermediate of the coupled reaction, RCC, does not accumulate to substantial amounts such that pFCC1 is the only detectable product (13, 21).
To test whether the activity of PTC52 and PAO may be sensitive to regulation by Trx, activity measurements were performed with established procedures (13, 14, 22). PTC52 and PAO of barley produced by hybrid-selected translation were incubated with Pchlide a and pheophorbide a. As controls, in vitro-translated and purified PTC52-(His)6 and PAO-(His)6 of Arabidopsis thaliana were used. After incubation, the assays were subjected to a step of gel filtration to remove unbound pigments. Protein–pigment complexes eluted with the flow-through were supplemented with O2, Fd, and a Fd-reducing system comprising glucose-6-phosphate, NADPH, glucose-6-phosphate dehydrogenase, Fd:NADPH oxidoreductase, and wheat germ extract. In case of PAO, in vitro-expressed RCC reductase was added. Fig. 3A shows representative HPLC tracings of pigments formed in the presence of PTC52 at time 0 (solid line) and after 15 min (dashed line). Pchlide a has a retention time of 15 min at the C18 RP material, whereas Pchlide b elutes at 12.5 min. Because the absorption coefficients of Pchlide a and Pchlide b are 5-fold different at 455 nm, the measured decrease in Pchlide a levels after incubation correlated with an apparent large, 5-fold increase in the amount of Pchlide b (Fig. 3A). Pchlide a to Pchlide b conversion required O2 and a Fd-reducing system. In the absence of these factors, no Pchlide b was produced (data not shown). When wheat germ lysate was omitted, low PTC52 activity was measurable, suggesting the presence of a factor that promoted the folding and assembly of the Rieske and 2Fe-2S clusters. In the absence of PTC52 protein, no Pchlide b was formed (data not shown).
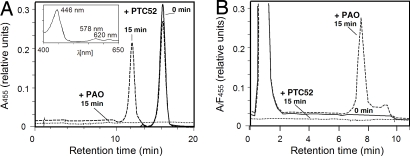
Fig. 3.
In vitro activity of PTC52 and PAO. (A) PTC52 was expressed as described in Fig. 2A, and activity measurements were performed with Pchlide a, O2, and an energy-regenerating system (see Materials and Methods). Pigments formed at time point zero (solid line) and after 15 min (dashed line) were extracted with acetone and subjected to RP-HPLC. Pigment detection was made by absorbance measurements at 455 nm, and the identity of the different peaks was confirmed by absorption spectrometry (Inset) and mass spectrometry (data not shown). (Inset) Shown is the spectrum of the peak that eluted at 12.5 min, which identified this compound as Pchlide b. As a control, in vitro-expressed PAO was used that failed to bind Pchlide a (stippled line). (B) The same as A, but showing activity measurements for PAO (F455) in the presence of pheophorbide a, O2, and the same energy-regenerating system as described before, at time point zero (solid line) and after 15 min (dashed line). Controls with PTC52 showed a lack of pheophorbide a binding (A455) and thus proved the specificity of the enzyme assay (stippled line).
No binding of Pchlide a was detectable with the PAO protein, and consequently no Pchlide b was produed (Fig. 3A, stippled line). Conversely, no binding of pheophorbide a was achieved for PTC52 under conditions that led to the production of pFCC-1 (Fig. 3B). pFCC-1 formation by PAO required RCCR, Fd, a Fd-reducing system, and the substrates pheophorbide a and O2.
We next tested the effect of stromal Trx f and Trx m on PTC52 and PAO activity. Either Trx was expressed in Escherichia coli, purified, and added to the in vitro-expressed proteins. Then the activity measurements were repeated as before. In a parallel sample, the effect of DTT was studied. Table 1 shows that addition of Trx m or addition of DTT led to 12- and 15-fold increases in PTC52 and PAO activity, respectively. These data suggested that PTC52 and PAO may undergo reversible oxidation/reduction reactions involving redox-active protein SH groups that in turn affected their activities, the reduced form being more active than the oxidized form.
Table 1.
The effects of Trx m and DTT on PTC52 and PAO activities
| Protein | Enzyme activity, nkat ·mg−1 protein |
|||
|---|---|---|---|---|
| −Trx m | +Trx m | −DTT | +DTT | |
| PTC52 | 0.68 | 8.20 | 0.70 | 8.40 |
| PAO | 1.04 | 15.60 | 1.08 | 16.20 |
PTC52 and PAO were tested for their Pchlide a-oxygenase and pheophorbide a-oxygenase activity, respectively, as described in Fig. 3. In two parallel sets of samples, the effects of isolated Trx m and DTT were examined.
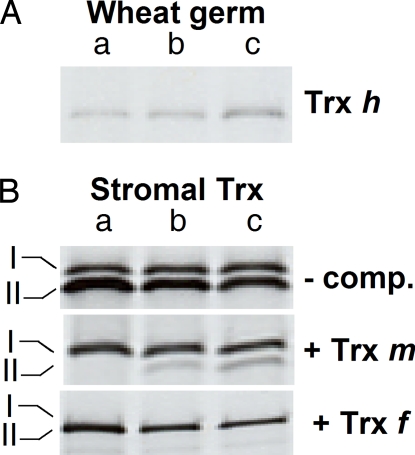
TIC55, PTC52, and PAO are localized in the inner envelope membrane of chloroplasts (10, 11, 23, 24), and all contain the CxxC motif that could have interacted with Trx. To confirm this hypothesis, hexa-histidine-tagged versions of Arabidopsis TIC55, PTC52, and PAO were produced containing an Ala residue in place of the first or second Cys residue in the CxxC motif. When in vitro-translated and Ni-NTA-purified proteins were used as bait, small amounts of a low molecular mass, ≈12-kDa wheat germ protein were recovered for all three proteins (Fig. 4A). DTT addition eluted this band. Protein mass spectrometry and Western blotting identified this band to be caused by wheat germ Trx h (data not shown). Upon testing stromal extract of barley chloroplast for binding, two protein bands were found for TIC55, PTC52, and PAO representing Trx f (Fig. 4B, top band, band I) and Trx m (Fig. 4B, bottom band, band II). Consistent with this view, band II was sensitive to excess peptide mimicking the presence of the redox-active sites of Trx m (Fig. 4B, +Trx m), whereas band I was blocked by competition by Trx f-specific peptide probe (Fig. 4B, +Trx f). These results showed that TIC55, PTC52, and PAO had no preference with regard to either Trx m or Trx f in the in vitro assay. Similar conclusions have been drawn from previous studies (8).
Fig. 4.
Verification of TIC55, PTC52, and PAO as Trx targets. TIC55- (lanes a), PTC52- (lanes b), and PAO-(His)6 (lanes c) mutant proteins containing engineered CxxC motifs with Cys-Ala exchanges replacing the first Cys residue were produced by coupled in vitro transcription/translation in wheat germ lysates, purified and used for Trx affinity chromatography. (A) Western blot of Trx-related proteins bound to immobilized TIC55-, PTC52-, and PAO-(His)6 in wheat germ extracts. (B) Western blot of Trx-related proteins bound to immobilized TIC55-, PTC52-, and PAO-(His)6 in stromal extract of barley chloroplasts. After treatment with DTT and extensive washing of the same affinity matrices used in A, stromal extract of isolated barley chloroplasts was passed over the columns. Binding reactions were carried out either in the absence of competitor (−comp.) or in the presence of competitors mimicking the thiol-reactive sites of Trx m and Trx f.
Role of Trxs in the Regulation of Protein Import.
It has been proposed that Trx could play a role in oxidative regulation (3). Taking into account the findings reported thus far, we assumed that Trx could regulate the activity of TIC55, PTC52, and PAO in response to oxidative stress. A way to test this hypothesis was provided by previous work on mutants that are de-regulated in porphyrin biosynthesis such as the fluorescent (FLU) mutant of Arabidopsis (25, 26) and its ortholog in barley, tigrina d12 (27), and other species. FLU and tigrina d12 seedlings overproduce free, nonprotein-bound protochlorophyllide (Pchlide) in darkness. Upon illumination, Pchlide operates as photosensitizer and gives rise to singlet oxygen (28). In seedlings kept under continuous white light, Pchlide is constantly converted to chlorophyllide by the NADPH:Pchlide oxidoreductase such that no singlet oxygen is produced. Mature plants cultivated under alternative dark/light cycles or subjected to nonpermissive light-to-dark-to-light shifts suffer from the same, pigment-sensitized photooxidative response as etiolated plants after illumination (25), allowing oxidative regulation of TIC55 and PTC52 activity to be tested by in planta and in vitro experiments. As a measure for this regulation, protein import was explored given that TIC55, PTC52, and the related Rieske protein family member CAO (and perhaps also PAO) have established roles in protein translocation across the inner plastid envelope membranes (10, 11, 24, 29).
Tigrina d12 seeds of barley were germinated in continuous white light for 5 days. Chloroplasts were isolated from primary leaves of such plants and tested for their in vitro-import capability for the small subunit precursor of ribulose-1,5-bisphosphate carboxylase/oxygenase (pSSU; a TIC55 import substrate; refs. 10 and 24), pPORA (a PTC52 import substrate; ref. 11), and the precursor to the major light-harvesting Chl a/b-binding protein of photosystem II (PSII) (pLHCII, a CAO import substrate; ref. 29). As controls, the precursors to the photosynthetic ferredoxin (pFdI) and nonphotosynthetic ferredoxin (pFdIII) were used. As shown previously, pFdIII is mis-sorted to the intermembrane space of chloroplasts in white light but it is imported into the stroma and processed in darkness (30). In contrast, pFdI is faithfully taken up under either condition (30). To see whether light-dark regulation and/or oxidative control would happen to the TIC55-, PTC52-, and CAO-containing import machineries, one pot of the originally light-grown tigrina d12 seedlings was transferred to darkness for 14 h, whereas another pot was shifted to a nonpermissive 10 h dark/4 h light shift.
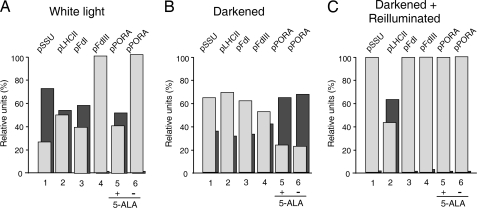
Fig. 5A summarizes the results of three independent import experiments. They showed that pSSU import, as measured as the amount of mature protein inside thermolysin-treated, repurified, intact chloroplasts, was higher for light-grown than for darkened plants. In plants subjected to the nonpermissive dark-to-light shift, no import was detectable. Similar results were obtained for pFdI, the photosynthetic ferredoxin. Consistent with previous reports, its nonphotosynthetic counterpart was undetectable as processed enzyme in import assays containing chloroplasts from light-grown plants but accumulated in a thermolysin-resistant, processed, mature form after import into chloroplasts from darkened plants (Fig. 5 and data not shown). In the case of pPORA, higher import rates were measured for chloroplasts isolated from darkened as compared with light-grown plants. Chloroplasts from light-grown plants imported the precursor only in the presence of the Pchlide precursor 5-aminolevulinic acid, a result that is consistent with our previous findings using in vitro-translated precursor (31) but is at variance with the results of Kim and Apel (32) using in planta assays. This issue needs to be resolved in subsequent work. With chloroplasts from dark-shifted plants, the endogenously produced Pchlide substituted for the exogenous 5-ALA-derived Pchlide. Upon the nonpermissive dark-to-light shift, no import of pPORA occurred. For pLHCII, the CAO import substrate, a different result was obtained. Whereas chloroplasts from light-grown plants imported more pLHCII than chloroplasts from darkened plants, no deleterious effect on import was apparent after the nonpermissive dark-to-light shift (Fig. 5).
Fig. 5.
In vitro protein import to monitor dark/light and oxidative regulation of TIC55, PTC52, and CAO activity. (A) 35S-labeled precursors were synthesized by coupled in vitro transcription/translation of cDNA clones and imported into chloroplasts of 5-day-old, light-grown tigrina d12 mutant plants of barley in a standard import reaction containing 2.5 mM Mg-ATP. (B and C) In two additional sets of incubation, chloroplasts were used from plants that had been shifted to a 14-h dark period before plastid isolation (B) or to a nonpermissive 10-h dark to 4-h light shift (C). To study substrate-dependent import of pPORA, aliquots of the plastid suspension were pretreated with 5-aminolevulinic acid (+ 5-ALA) or phosphate buffer (−5-ALA) and Mg-ATP, giving rise to intraplastidic Pchlide synthesis. Parallel assays were conducted that were treated with or without thermolysin after import. Levels of precursor and mature proteins were determined by SDS/PAGE and autoradiography. Percentages refer to the sum of precursor (light-gray columns) and mature proteins (dark-gray columns) in the assays.
Discussion
In the present study, a family of inner plastid envelope Trx targets was discovered. This family comprises three proteins sharing conserved Rieske and mononuclear iron binding domains and CxxC motifs (TIC55, PTC52, and PAO). Similar Rieske and mononuclear iron binding motifs are present in choline monooxygenase CMO and CAO, but the latter lack the redox-active CxxC motif present in TIC55, PTC52, and PAO and were not purified by the established affinity purification procedure.
At least two of the inner plastid envelope Trx targets, , namely TIC55 and PTC52, have roles in protein import. TIC55 is part of the general protein import machinery that is responsible for the transport of pSSU and perhaps other photosynthetic precursors into the chloroplast (10, 24). PTC52 establishes a distinctive translocon. This translocon is most abundantly expressed in etiolated plants and is involved in the Pchlide-dependent import of pPORA (11). Despite a light-induced decline in the amount of OEP16 (33) and PTC52 (this study) the PTC complex remains active in chloroplasts. The observed dark/light regulation could ensure that maximum amounts of pPORA are imported via the PTC complex at the end of the night where Pchlide reaccumulates (34). The expression of TIC55 may be constitutive, but no data have been published regarding the role of TIC55 at different stages of development to our knowledge. Both TIC55 and PTC52 were sensitive to oxidative control and were inactivated in reilluminated tigrina d12 plants, presumably by a porphyrin (Pchlide)-sensitized mechanism. The same holds true for PAO, the third identified Trx target, which is senescence-induced with low basal protein and mRNA levels in presenescent plants (13, 35, 36). The observed large ≈60-fold increase in PAO activity, which is at variance with the only ≈6-fold raise in pao mRNA level in senescent plants (13) suggests a posttranscriptional mode of control that could involve redox control of enzyme activity. It is tempting to hypothesize that PAO, in addition to its role in Chl breakdown, may be involved in the regulated import of senescence-induced gene products into the chloroplast, although hard biochemical evidence for the existence of a PAO-containing import apparatus is still lacking.
CAO, which lacks the CxxC motif and was not a Trx target, establishes a third TIC subcomplex (29). This complex was involved in the regulated import of the Chl b-containing light-harvesting proteins LHCII and CP29 (29). The in vitro import experiments shown here revealed that pLHCII import was subject to dark/light regulation but did not respond to oxidative control elicited by free, photoexcited Pchlide molecules accumulating in tigrina d12 plants. This result may be explained by the lack of the CxxC motif that could be oxidized by singlet oxygen. The rate of LHCII import may be controlled in a different way. Yamasato et al. (37) identified a pigment sensor domain in the intermembrane part of CAO that could operate as relay in providing Chlide b to incoming pLHCII and pCP29 apoproteins. Only when Chlide b is produced would pLHCII and pCP29 import occur.
The Chl b-containing light-harvesting proteins have important functions not only in light trapping and energy transduction but also in photoprotection (38). It is attractive to hypothesize that oxidative stress may signal a requirement for more LHC proteins and, therefore, no block but even an elevation of pLHCII and pCP29 import could be observed in reilluminated tigrina d12 plants. Once imported and processed, the mature LHCII normally associates with PSII but, during state transition, its location changes toward PSI (39). There is some evidence that the relocation of LHCII during state transition may be regulated by a PSI thiol-based mechanism that could involve Trx, in addition to the one depending on plastoquinone and protein phosphorylation (40). It is attractive to hypothesize that perhaps a need to tie Trx action to state transition may have led to a loss of Trx responsiveness of the CAO-containing TIC import machinery.
Conclusions
The results of the present study strongly suggest two major mechanisms of Trx-dependent control of protein import. The first mechanism is dark/light regulation that could help adjust the amounts of freshly imported proteins to the photosynthetic needs. Examples for this type of control include pSSU and pFdI that were imported into chloroplasts with higher rates in the light than in the dark. By contrast, pPORA was less efficiently imported into chloroplasts from light-grown versus darkened plants. This result may be explained by the fact that pPORA-bound Pchlide b serves as light trap (41) and could impede normal light harvesting during photosynthesis. For pFdIII, a different type of regulation was observed. pFdIII was mis-sorted to the inter envelope membrane space of chloroplasts of light-grown plants but was faithfully imported into the stroma and processed in chloroplasts of darkened plants. Although much less is known about the potential physiological background of this control, Hirohashi et al. (30) proposed that the limitation of pFdIII import may be a means to prevent potential interferences with normal electron transport involving FdI.
The second type of control in which Trx may be involved is oxidative control. Chloroplasts are a source of reactive oxygen species that very easily and irreversibly inactivate proteins and enzyme activities by modifying the reduction state of disulfide bridges, either established intramolecularly or intermolecularly. Especially the risk of singlet oxygen formation by virtue of photoreactive tetrapyrroles, such as Chl, Pchlide and pheophorbide a, that accumulate at different stages of plant development, may require such mechanism. In fact, plastids need to adapt their metabolism to different requirements at the beginning of greening, in light-adapted plants, and during leaf senescence. According to recent work (42), even amyloplasts, the yellow, starch-accumulating type of plastids, contain a full set of Trxs and Trx:ferredoxin reductases, highlighting the ubiquitous role of the Trx system for plastid metabolism and plant development.
Materials and Methods
Identification of Proteins Interacting with Trx f and Trx m.
Chloroplasts were isolated from 5-day-old light-grown barley (Hordeum vulgare L. Carina) and fractionated into stroma and membranes (11). The membranes were further separated into outer and inner plastid envelope membranes (11), and protein of each of these fractions was solubilized with 3% SDS. The resulting protein extracts in turn were subjected to Trx m affinity column by using wild-type Trx f and Trx m and their mutant f C49S and m C40A derivatives immobilized on Sepharose 4B (Amersham Pharmacia Biosciences) (8). Bound outer and inner envelope membrane proteins were subsequently eluted with 10 mM DTT, dialyzed, concentrated, and subjected to SDS/PAGE. The amount of protein collected on addition of DTT to the mutated Trx f and Trx m columns was similar (≈120–150 μg, depending in the actual experiment). Similar binding and elution steps were performed with activated thiol Sepharose columns that, however, yielded an extremely low amount of protein (≈5–10 μg).
Peptides mimicking the thiol-active sites of Trx f and Trx m (in bold) used for competitive binding assays were as follows: Trx f, NH2-26WPIVKAAGDKPVVLDMFTQWCGPCKAMAPKYE57-COOH and Trx m, NH2-18KEFVLESEVPVMVDFWAPWCGPCKLIAPVIDE49-COOH.
Confirmation of Trx Targets.
After elution from the Trx affinity columns with DTT, excess DTT was removed by extensive dialysis against 50 mM Tris·HCl, pH 7.5. Subsequently, the samples were treated with 1 mM N-ethylmaleimide to block free SH groups. Excess N-ethylmaleimide was removed by addition of 1 mM 2-mercaptopethanol. Half of the final samples was reduced with bacterial (N-ethylmaleimide) or stromal (barley) Trx, whereas the remaining halves were incubated with water. The reduced and the control samples in turn were treated with monobromobimane to label newly formed SH groups (43, 44).
2D Gel Separation.
Isoelectric focusing and SDS/PAGE were performed according to Scharf and Nover (45) using a pH range from 3 to 10 and 10–20% polyacrylamide gradients containing SDS. To separate inner envelope proteins, nonequilibrium pH gradient electrophoresis was used (45). Proteins were stained with Coomassie brilliant blue R-250 or subjected to Western blotting.
Protein Sequencing.
Individual protein spots were excised and in-gel digested with trypsin, and the resulting peptide mixtures were subjected to protein mass spectrometry. The obtained partial amino acid sequences were aligned with other proteins using the SEQUEST algorithm and the nonredundant protein database from the National Center for Biotechnology Information (8) and other standard computer programs [ref. 15; see supporting information (SI) Text].
Activity Measurements of PTC52 and PAO.
Activity measurements were carried out according to Oster et al. (14) and Pruzinska et al. (13), respectively. Final 50 μl-assays contained the following supplements: 2 mM Pchlide a or pheophorbide a, 10 μg of Fd (Sigma) and a Fd-reducing system [2 mM glucose-6-phosphate; 1 mM NADPH; 50 milliunits of glucose-6-phosphate dehydrogenase; 10 milliunits of Fd-NADPH-oxidoreductase (Sigma)]. Assays for testing PAO activity likewise contained RCC reductase, allowing conversion of pheophorbide a to be measured by the appearance of pFCC-1 (13). As a source of RCC reductase, the Arabidopsis enzyme was used that had been expressed from a respective cDNA in E. coli as described (13). Separation and identification of pigments by RP-HPLC has been described (13, 22). The production of pFCC-1 from pheophorbide a was linear for >1 h of incubation at 22°C and followed Michaelis-Menten kinetics with an apparent Km value of 6 μM, consistent with previous results (13).
Protein Import.
Import assays were carried out by using established procedures and 35S-labeled, wheat germ-translated precursors (11). Plastids were isolated from tigrina d12 plants that had been grown in continuous white light for 5 days or subjected to prior 14 h dark or 10-h dark/4-h white light shifts. Protein detection and quantification was made by SDS/PAGE and autoradiography or with a PhosphorImager.
Miscellaneous.
RNA preparation, Northern hybridization, and hybrid-selected translation using filter-bound pao- and ptc52-specific DNA probes were carried out as described (46).
Supplementary Material
Acknowledgments.
This is scientific paper no. 0201-08 from the College of Agricultural, Human, and Natural Resource Sciences of Washington State University. We thank D. J. Schnell (University of Massachusetts, Amherst), F. Kessler (Université de Neuchâtel, Neuchâtel, Switzerland), and J. Soll (University of Munich, Munich, Germany) for gifts of cDNA clones and antisera and B. Buchanan (University of California, Berkeley) for critical reading of the manuscript, support, and advice with the experiments. This study was supported by a Chaire d'Excellence program of the French Ministry of Research (C.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800378105/DC1.
References
- 1.Schürmann P, Jacquot J-P. Plant thioredoxin systems revisited. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:371–400. doi: 10.1146/annurev.arplant.51.1.371. [DOI] [PubMed] [Google Scholar]
- 2.Buchanan BB, Schürmann P, Wolosiuk RA, Jacquot J-P. The thioredoxin/ferredoxin system: From discovery to molecular structures and beyond. Photosynth Res. 2002;73:215–222. doi: 10.1023/A:1020407432008. [DOI] [PubMed] [Google Scholar]
- 3.Buchanan BB, Balmer Y. Redox regulation: A broadening horizon. Annu Rev Plant Biol. 2005;56:187–220. doi: 10.1146/annurev.arplant.56.032604.144246. [DOI] [PubMed] [Google Scholar]
- 4.Brandes HK, Larimer FW, Hartman FC. The molecular pathway for the regulation of phosphoribulokinase by thioredoxin f. J Biol Chem. 1996;271:3333–3335. doi: 10.1074/jbc.271.7.3333. [DOI] [PubMed] [Google Scholar]
- 5.Verdoucq L, Vignols F, Jacquot J-P, Chartier Y, Meyer Y. In vivo characterization of a thioredoxin h target protein defines a new peroxiredoxin family. J Biol Chem. 1999;274:19714–19722. doi: 10.1074/jbc.274.28.19714. [DOI] [PubMed] [Google Scholar]
- 6.Balmer Y, Schürmann P. Heterodimer formation between thioredoxin f and fructode-1,6-bisphosphatase from spinach chloroplasts. FEBS Lett. 2001;492:58–61. doi: 10.1016/s0014-5793(01)02229-3. [DOI] [PubMed] [Google Scholar]
- 7.Goyer A, et al. The internal Cys207 of Sorghum leaf NADP-malate dehydrogenase can form mixed disulfides with thioredoxin. FEBS Lett. 2002;444:165–169. doi: 10.1016/s0014-5793(99)00051-4. [DOI] [PubMed] [Google Scholar]
- 8.Balmer Y, et al. Proteomics gives insights into the regulatory function of chloroplast thioredoxins. Proc Natl Acad Sci USA. 2003;100:370–375. doi: 10.1073/pnas.232703799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hisabori T, et al. Thioredoxin affinity chromatography: A useful method for further understanding the thioredoxin network. J Exp Bot. 2003;56:1463–1468. doi: 10.1093/jxb/eri170. [DOI] [PubMed] [Google Scholar]
- 10.Caliebe A, et al. The chloroplastic protein import machinery contains a Rieske-type iron-sulfur cluster and mononuclear iron-binding protein. EMBO J. 1997;16:7342–7350. doi: 10.1093/emboj/16.24.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reinbothe S, Quigley F, Gray J, Schemenewitz A, Reinbothe C. Identification of plastid envelope proteins required for import of protochlorophyllide oxidoreductase A into the chloroplast of barley. Proc Natl Acad Sci USA. 2004;101:2197–2202. doi: 10.1073/pnas.0307284101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray J, Close PS, Briggs SP, Johal GS. A novel suppressor of cell death in plants encoded by the LLs1 gene of maize. Cell. 1997;89:25–31. doi: 10.1016/s0092-8674(00)80179-8. [DOI] [PubMed] [Google Scholar]
- 13.Pruzinska A, Tanner G, Anders I, Boca M, Hörtensteiner S. Chlorophyll breakdown: Pheophorbide a oxygenase is a Rieske-type iron–sulfur protein, encoded by the accelerated cell death 1 gene. Proc Natl Acad Sci USA. 2003;100:15259–15264. doi: 10.1073/pnas.2036571100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oster U, Tanaka R, Tanaka A, Rüdiger W. Cloning and functional expression of the gene encoding the key enzyme for chlorophyll b biosynthesis (CAO) from Arabidopsis thaliana. Plant J. 2000;21:305–310. doi: 10.1046/j.1365-313x.2000.00672.x. [DOI] [PubMed] [Google Scholar]
- 15.Gray J, et al. A small family of LLS1-related nonheme oxygenases in plants with an origin amongst oxygenic photosynthesizers. Plant Mol Biol. 2004;54:39–54. doi: 10.1023/B:PLAN.0000028766.61559.4c. [DOI] [PubMed] [Google Scholar]
- 16.Lee J, Hofhaus G, Lisowsky T. Erv1p from Saccharomyces cerevisiae is a FAD-linked sulfhydryl oxidase. FEBS Lett. 2000;477:62–66. doi: 10.1016/s0014-5793(00)01767-1. [DOI] [PubMed] [Google Scholar]
- 17.Gerber J, Muhlkenhoff U, Hofhaus G, Lill R, Lisowsky T. Yeast ERV2p is the first microsomal FAD-linked sulhydryl oxidase of the Erv1p/Alrp protein family. J Biol Chem. 2001;276:23486–23491. doi: 10.1074/jbc.M100134200. [DOI] [PubMed] [Google Scholar]
- 18.Mesecke N, et al. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121:1059–1069. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 19.Tokatlidis K. A disulfide relay system in mitochondria. Cell. 2005;121:965–967. doi: 10.1016/j.cell.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 20.Yano H, Wong JH, Lee YM, Cho M-J, Buchanan BB. A strategy for the identification of proteins targeted by thioredoxin. Proc Natl Acad Sci USA. 2001;98:4794–4799. doi: 10.1073/pnas.071041998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodoni S, et al. Chlorophyll breakdown in senescent chloroplasts: Cleavage of pheophorbide a in two enzymic steps. Plant Physiol. 1997;115:669–676. doi: 10.1104/pp.115.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reinbothe S, Pollmann S, Reinbothe C. In situ conversion of protochlorophyllide b to protochlorophyllide a in barley. Evidence for a novel role of 7-formyl reductase in the prolamellar body of etioplasts. J Biol Chem. 2003;278:800–806. doi: 10.1074/jbc.M209737200. [DOI] [PubMed] [Google Scholar]
- 23.Matile P, Schellenberg M. The cleavage of phaeophorbide a is located in the envelope of barley gerontoplasts. Plant Physiol Biochem. 1994;34:55–59. [Google Scholar]
- 24.Küchler M, Decker S, Hörmann F, Soll J, Heins L. Protein import into chloroplasts involves redox-regulated proteins. EMBO J. 2003;21:6136–6145. doi: 10.1093/emboj/cdf621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meskauskiene R, et al. FLU: A negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2001;98:12826–12831. doi: 10.1073/pnas.221252798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meskauskiene R, Apel K. TIGRINA d12, required for regulating the biosynthesis of tetrapyrroles in barley, is an ortholog of the FLU gene of Arabidopsis thaliana. FEBS Lett. 2002;553:119–124. doi: 10.1016/s0014-5793(03)00983-9. [DOI] [PubMed] [Google Scholar]
- 27.von Wettstein D, Kahn A, Nielsen OF, Gough S. Genetic regulation of chlorophyll synthesis analyzed with mutants in barley. Science. 1974;184:800–802. doi: 10.1126/science.184.4138.800. [DOI] [PubMed] [Google Scholar]
- 28.op den Camp R, et al. Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell. 2004;15:2320–2332. doi: 10.1105/tpc.014662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reinbothe C, et al. A role for chlorophyllide a-oxygenase in the regulated import and stabilization of light-harvesting chlorophyll a/b proteins in chloroplasts. Proc Natl Acad Sci USA. 2006;103:4777–4782. doi: 10.1073/pnas.0511066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirohashi T, Hase T, Nakai M. Maize nonphotosynthetic ferredoxin precursor is missorted to the intermembrane space of chloroplasts in the presence of light. Plant Physiol. 2001;125:2154–2163. doi: 10.1104/pp.125.4.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reinbothe S, Mache R, Reinbothe C. A second, substrate-dependent site of protein import into chloroplasts. Proc Natl Acad Sci USA. 2000;97:9795–9800. doi: 10.1073/pnas.160242597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim C, Apel K. Substrate-dependent and organ-specific chloroplast protein import in planta. Plant Cell. 2004;16:88–98. doi: 10.1105/tpc.015008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reinbothe S, Quigley F, Springer A, Schemenewitz A, Reinbothe C. The outer plastid envelope protein Oep16: Role as precursor translocase in import of protochlorophyllide oxidoreductase A. Proc Natl Acad Sci USA. 2004;101:2203–2208. doi: 10.1073/pnas.0301962101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Griffiths WT, Kay SA, Oliver RP. The presence of photoregulation of protochlorophyllide reductase in green tissues. Plant Mol Biol. 1985;4:13–22. doi: 10.1007/BF02498711. [DOI] [PubMed] [Google Scholar]
- 35.Ginsburg S, Schellenberg M, Matile P. Cleavage of chlorophyll-porphyrin (requirement for reduced ferredoxin and oxygen) Plant Physiol. 1994;105:545–554. doi: 10.1104/pp.105.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hörtensteiner S, Vicentini F, Matile P. Chlorophyll breakdown in senescent cotyledons of rape, Brassica napus L.: Enzymatic cleavage of phaeophorbide a in vitro. New Phytol. 1995;129:237–246. doi: 10.1111/j.1469-8137.1995.tb04293.x. [DOI] [PubMed] [Google Scholar]
- 37.Yamasato A, Nagata N, Tanaka R, Tanaka A. The N-terminal domain of chlorophyllide a oxygenase confers protein instability in response to chlorophyll b accumulation in Arabidopsis. Plant Cell. 2005;17:1585–1597. doi: 10.1105/tpc.105.031518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kühlbrandt W, Wang DN, Fujiyoshi Y. Atomic model of plant light-harvesting complex by electron crystallography. Nature. 1994;367:614–621. doi: 10.1038/367614a0. [DOI] [PubMed] [Google Scholar]
- 39.Allen JF. Protein phosphorylation in regulation of photosynthesis. Biochim Biophys Acta. 1992;1098:275–335. doi: 10.1016/s0005-2728(09)91014-3. [DOI] [PubMed] [Google Scholar]
- 40.Aro EM, Ohad I. Redox regulation of thylakoid protein phosphorylation. Antioxid Redox Signal. 2003;5:55–67. doi: 10.1089/152308603321223540. [DOI] [PubMed] [Google Scholar]
- 41.Reinbothe C, Lebedev N, Reinbothe S. A protochlorophyllide light-harvesting complex involved in de-etiolation of higher plants. Nature. 1999;397:80–84. [Google Scholar]
- 42.Balmer Y, et al. A complete ferredoxin/thioredoxin system regulates fundamental processes in amyloplasts. Proc Natl Acad Sci USA. 2006;103:298–2993. doi: 10.1073/pnas.0511040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Besse I, Wong JH, Kobrehel K, Buchanan BB. Thiocalsin: A thioredoxin-linked, substrate-specific protease dependent on calcium. Proc Natl Acad Sci USA. 1996;93:3169–3175. doi: 10.1073/pnas.93.8.3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balmer Y, et al. Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc Natl Acad Sci USA. 2004;101:2642–2647. doi: 10.1073/pnas.0308583101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scharf K-D, Nover L. Heat shock-induced alterations of ribosomal protein phosphorylation in plant cell cultures. Cell. 1982;30:427–437. doi: 10.1016/0092-8674(82)90240-9. [DOI] [PubMed] [Google Scholar]
- 46.Reinbothe C, Ortel B, Parthier B, Reinbothe S. Cytosolic and plastid forms of 5-enolpyruvylshikimate-3-phosphate synthase in Euglena gracilis are differentially expressed during light-induced chloroplast development. Mol Gen Genet. 1994;245:616–622. doi: 10.1007/BF00282224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.