Abstract
Cell adaptation to changes in oxygen (O2) availability is controlled by two subfamilies of O2-dependent enzymes: the hypoxia inducible factor (HIF)–prolyl and asparaginyl hydroxylases [prolyl hydroxylases domain (PHDs) and factor inhibiting HIF (FIH)]. These oxygen sensors regulate the activity of the HIF, a transcriptional complex central in O2 homeostasis. In well oxygenated cells, PHDs hydroxylate the HIFα subunits, thereby targeting them for proteasomal degradation. In contrast, acute hypoxia inhibits PHDs, leading to HIFα stabilisation. However, here we show that chronic hypoxia induces HIF1/2α“desensitization” in cellulo and in mice. At the basis of this general adaptative mechanism, we demonstrate that chronic hypoxia not only increases the pool of PHDs but also overactivates the three PHD isoforms. This overactivation appears to be mediated by an increase in intracellular O2 availability consequent to the inhibition of mitochondrial respiration. By using in cellulo and in vivo siRNA, we found that the PHDs are the key enzymes triggering HIFα desensitization, a feedback mechanism required to protect cells against necrotic cell death and thus to adapt them across a chronic hypoxia. Hence, PHDs serve as dual enzymes, for which inactivation and later overactivation is necessary for cell survival in acute or chronic hypoxia, respectively.
Keywords: cell survival, oxygen sensing
The transcriptional complex hypoxia inducible factor (HIF) plays a central role in the maintenance of oxygen (O2) homeostasis, which is essential for cell survival (1). HIF is tightly regulated in an O2-dependent manner by hydroxylation of one of the three HIFα subunits (HIF1α, HIF2α, and HIF3α) (2, 3). In well oxygenated cells (normoxia), the hydroxylation of two proline residues (P402 and P564 in human HIF1α) by the HIF-prolyl hydroxylases [prolyl hydroxylases domains (PHDs)] allows the specific recognition and polyubiquitination by the von Hippel-Lindau protein (pVHL) E3–ligase complex, leading to proteasomal degradation (4). Moreover, the hydroxylation of an asparagine residue (N803 in human HIF1α) by the factor inhibiting HIF (FIH) prevents binding of the coactivator p300/CBP and hence blocks HIF transcriptional activity (5). In contrast, restricted O2 availability, by relaxing HIFα hydroxylation, results in HIFα stabilization and activation of the HIF transcriptional complex. Like FIH, the PHDs belong to the super family of iron- and 2-oxoglutarate-dependent dioxygenases, which, by using O2 as a cosubstrate, provide the molecular basis for their O2-sensing function (6). In mammalian cells, three PHDs isoforms have been identified (PHD1, PHD2, and PHD3) and shown to hydroxylate HIF1α in cellulo depending on their relative abundance (7). Nevertheless, we report that PHD2 has a dominant role, as it is the rate-limiting enzyme that sets the low steady-state level of HIF1α in normoxia (8).
In line with our previous work, we sought to look for HIFα regulation during long-term hypoxia. Contrary to acute hypoxia, we observed that chronic hypoxia is not able to accumulate HIF1α nor HIF2α in any of the cell systems analyzed so far. HIFα proteins are degraded because of hydroxylation, ubiquitination, and their targeting through the proteasome, suggesting that upon long-term hypoxia PHDs are active despite the hypoxic conditions. Here, we highlight an unexpected overactivation of the three PHD isoforms during chronic hypoxic stress. By using a respiratory deficient cell line, we show that chronic hypoxia enhances O2 availability for PHDs. Because hypoxia also increases the pool of PHD proteins, both events converge to overactivate PHDs and consequently to reduce the HIFα levels that we observed upon chronic hypoxia. Moreover, overactivation of PHDs enzymes was also measured in mice exposed to prolonged hypoxia, and we confirmed their contribution to HIFα“desensitization” by using the siRNA approach in vivo. Finally, in contrast to the rapid HIFα stabilization required for cell adaptation in acute hypoxia, here we show that HIFα desensitization during chronic hypoxic stress is essential for cell survival.
Results
HIFα Proteins Decline During Chronic Hypoxia.
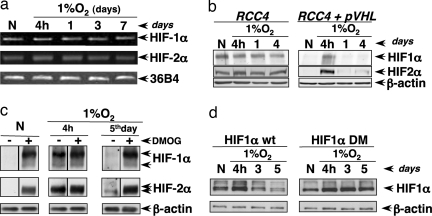
We previously showed that, in normoxia, silencing of PHD2 promotes HIF1α stabilization. However, its prolonged silencing is no more sufficient to stabilize HIF1α (8). Hence, we sought to determine HIFα regulation during chronic hypoxia. As shown in Fig. 1a, hypoxia at 1%O2 induced HIF1 and HIF2α protein accumulation after 4 h followed by a strong desensitization (loss of the protein) after 24 h to 7 days. We observed that HIF1/2α desensitization still occurred at 3% O2, whereas this phenomenon was strongly attenuated in drastic hypoxia (0.1% O2; Fig. 1b). To further analyze whether long-term induced HIFα desensitization was inherent to any cellular context, we investigated a battery of human cells. First, we tested immortalized cell lines such as CAL51 (breast cancer), HT1080 (fibrosarcoma), and LS174 (colon cancer). Second, we assessed primary cultures of human dermal fibroblasts (NHFs) and human epidermal keratinocyts (NHKs). In a way similar to HeLa cells (cervix adenocarcinoma), HIFα protein level decreased from acute (4 h) to chronic hypoxia (3–7 days) at 1% O2 in all of the cell systems tested so far (Fig. 1c and data not shown).
Fig. 1.
Levels of HIFα proteins decline during chronic hypoxia. Cells were incubated in hypoxic conditions for different periods of time, and the levels of HIF1α, HIF2α, and β-actin (loading control) were analyzed by Western blotting. (a) HeLa cells at 1% O2. (b) HeLa cells at 3% and 0.1% O2. (c) HT1080 and NHF cells at 1% O2.
Chronic Hypoxia Induces HIFα Protein Degradation Depending on the Hydroxylation–pVHL–Proteasome Pathway.
Because the HIFα proteins decrease might be caused by modification of mRNA and/or protein stability, we analyzed HIFα mRNA levels in cells exposed to acute or chronic hypoxia. As shown in Fig. 2a, the mRNA levels did not change during prolonged hypoxia in HeLa cells. Therefore, we tested an eventual increase in protein degradation rate. By Western blot analysis, we observed that proteasome inhibition induced a massive accumulation of HIF1/2α proteins in cells exposed for 5 days to hypoxia as happened in control cells (data not shown). Based on the bona fide HIFα degradation pathway, we tested whether during chronic hypoxia pVHL was the E3 ligase targeting both isoforms for degradation. We exposed a pVHL-deficient cell line, RCC4, under hypoxia at 1% O2 for 4 h to 4 days and looked for the HIFα proteins. In these cells, HIFα level never declined, whereas in the stable clone RCC4-pVHL (RCC4 in which pVHL had been reintroduced) HIFα dramatically decreased after 1 day at 1% O2 (Fig. 2b). Finally, we investigated HIFα protein hydroxylation as the signal allowing pVHL binding, and we observed that the addition of the hydroxylase inhibitor, dimethyloxallyl glycine (DMOG), restored HIFα accumulation across hypoxic stress (Fig. 2c). Accordingly, the HIF1α construct mutated on the two proline residues (Pro402 and Pro564) remained stable upon chronic hypoxia (Fig. 2d). Therefore, during chronic hypoxia, HIFα levels decline because of increased protein instability. Indeed, HIFα proteins appear to be hydroxylated, ubiquitinated, and targeted through the proteasome, suggesting that PHDs are “active” despite the hypoxic conditions.
Fig. 2.
Chronic hypoxia promotes HIFα protein degradation depending on the hydroxylation–pVHL–proteasome pathway. (a) Hypoxic kinetics in HeLa cells at 1% O2. HIF-1α, HIF-2α, and 36B4 mRNA levels were analyzed by RT-PCR. (b) RCC4 cells (pVHL-deficient) or the stable clone RCC4-pVHL (which overexpresses pVHL) were incubated in normoxia (20% O2) or hypoxia (1% O2) for different periods of time. (c) HeLa cells were incubated in normoxic (20% O2) or hypoxic conditions (1% O2) for 4 h or 5 days, and 4 h before the lysis, cells were incubated in the presence or absence of DMOG (1 mM). The levels of HIF1α, HIF2α, and β-actin (loading control) were analyzed by Western blotting. (d) 293 cells were incubated in normoxia (20% O2) or hypoxia (1% O2) for different periods of time, and 48 h before the lysis, cells were transfected with the HIF1α WT or DM constructs. The levels of HIF1α were analyzed by Western blotting using the Myc-tag antibody.
Chronic Hypoxia Overactivates the Three PHDs.
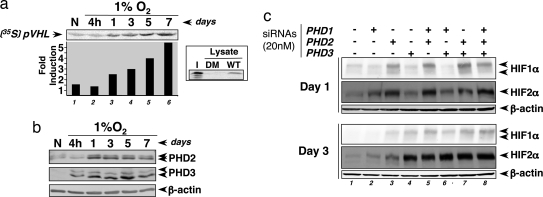
To evaluate PHD activity during chronic hypoxia, we performed in vitro pVHL capture assays. HeLa cells were exposed to hypoxia at 1% O2 for 4 h up to 7 days. GST-HIF1α constructs were incubated with the cell lysates and thereafter with the radio-labeled pVHL protein. Because pVHL binds to HIFα only when the relevant proline residues have been previously hydroxylated by the PHDs, the binding of in vitro-translated [35S]pVHL to GST-HIF1α is accepted as a reliable measure of PHD activity (Fig. 3a Inset). Compared with normoxia, as soon as 24 h of 1% O2, PHDs appeared overactivated (Fig. 3a, lane 3). Furthermore, from day 1 to day 7, the assay highlighted a stronger and progressive activation of the PHDs (Fig. 3a). PHD activation was indeed observed with the recombinant GST-HIF1α P402A mutant constructs but not with P564A or P402/564A, meaning that P564 hydroxylation is absolutely required and limits HIF1α hydroxylation (data not shown). Because PHD activity could be caused by an increase in the pool of these proteins, we quantified the level of the hypoxia up-regulated PHD2 and PHD3 isoforms. As expected, both proteins are induced under hypoxia but their maximal induction were reached after 24 h and the proteins were no longer increased in subsequent days (Fig. 3b). This result strongly suggests that in addition to an increase in the amount of PHD proteins, chronic hypoxia overactivates these enzymes.
Fig. 3.
Chronic hypoxia overactivates the three PHDs. (a) Autoradiogram showing PHD activity during hypoxic kinetics in HeLa cells at 1% O2. (Inset) [35S]pVHL input (I) and pVHL binding using the P402/564A mutant (DM) or the WT GST-HIF1α constructs. (b) Western blot analysis showing PHD2 and PHD3 protein levels during hypoxic kinetics. (c) To evaluate the specific contribution of each PHD isoform, 48 h before extraction, cells were transfected with siRNAs as indicated. The levels of HIF1α, HIF2α, and β-actin were analyzed by Western blotting.
We investigated the specific contribution of each PHD isoform on the HIFα degradation pathway by using the siRNA approach. During hypoxic kinetics at 1% O2 and at two different points (on days 1 and 3 according to our PHD activity assay), PHDs isoforms were invalidated alone or in combination. Fig. 3c shows that at day 1 and consistent with our previous work (8), only PHD2 silencing leads to HIF1α stabilization (lane 3). For HIF2α, in addition to PHD2, we observed a slight contribution of PHD1 (Fig. 3c, lanes 2 and 3). Surprisingly, at day 3, not only PHD2 but also PHD3 silencing rescued HIFα protein accumulation (Fig. 3c, lanes 3 and 4). Moreover, cosilencing of PHDs suggests that, in combination with the other isoforms, PHD1 acts on HIFα regulation (Fig. 3c, lanes 5, 6, and 8). We conclude that, in contrast to normoxia, during chronic hypoxia, the three PHDs contribute to HIFα degradation.
Chronic Hypoxia Increases O2 Availability for PHDs.
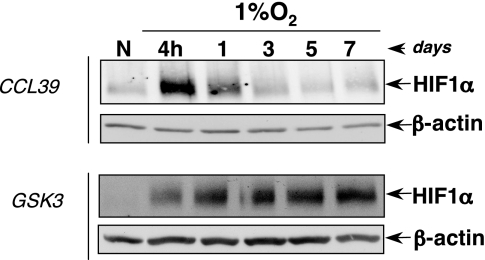
Hagen et al. (9) showed that NO and other chemical inhibitors of mitochondrial respiration prevent hypoxia-induced HIF1α stabilization. Indeed, because mitochondrial respiration pumps most of the intracellular O2, its inhibition increases intracellular O2 availability. Moreover, PDK1 (pyruvate dehydrogenase kinase), which is a HIF1-dependent gene product, has been recently reported as a natural inhibitor of mitochondrial activity in hypoxia (10, 11). Based on these results and because we observed that HIFα desensitization did not occur in drastic hypoxic conditions (0.1% O2; Fig. 1b), we assumed that chronic hypoxia by inhibiting mitochondrial activity increases intracellular O2 availability. We further decided to use GSK3 cells, a respiratory-deficient cell line (12). Indeed, we hypothesized that in GSK3 cells, in which O2 cannot be redistributed because of the constitutive inhibition of mitochondrial inhibition, we would prevent PHDs reactivation and thus chronic hypoxia-induced HIFα desensitization. According to our hypothesis, we observed that HIF1α protein, which accumulates after acute hypoxia, was not degraded during chronic hypoxia in GSK3 cells unlike in the control cells (Fig. 4). Therefore, we conclude that chronic hypoxia, by inhibiting mitochondrial respiration, progressively increases O2 availability for PHDs, leading to their overactivation and thus triggering HIFα desensitization.
Fig. 4.
Intracellular O2 redistribution is required for PHD overactivation. Hypoxic kinetics in CCL39 and GSK3 cells at 1% O2 are shown. HIF1α and β-actin levels were analyzed by Western blotting.
Chronic Hypoxia-Induced HIFα Desensitization Protects Cells from Necrosis.
To elucidate the physiological relevance of the intracellular O2 redistribution during chronic hypoxia, we exposed GSK3 cells to acute or chronic 1% O2 hypoxia and measured cell death by determining the percentage of permeabilized (trypan blue-positive) cells. As shown in Fig. 5a Left, in chronic hypoxia the GSK3 cells (deficient for HIFα desensitization) showed a drastic increase in cell death compared with the control cells fully competent to degrade HIFα. Indeed, almost 100% of GSK3 cells died after 7 days, suggesting that HIFα degradation is essential for cell survival in chronic hypoxia.
Fig. 5.
Chronic hypoxia-induced HIFα desensitization protects cells from necrosis. Cell death was measured in cells incubated in hypoxia at 1% O2 for different periods of time by a trypan blue incorporation assay. (a) CCL39 and GSK3 cells (Left) or HeLa cells transfected with siRNA targeting the three PHDs isoforms or an irrelevant sequence (Right) for 5 days and incubated either in normoxia (N) or upon hypoxia for different periods of time (4 h, 3 days, or 5 days). (b) GSK3 cells (Left) and HeLa cells transfected as described in a (Right) in which HIFα expression has been silenced. The siHIFα transfections started 16 h after the cells were incubated upon hypoxia and were carried on every day until the end of the experiment for days 3 and 5. In the case of the N and H (4 h) conditions, siHIFα were transfected during the last 24 h.
To support our hypothesis and confirm the role of PHDs, we performed hypoxic kinetics in HeLa cells and forced sustained HIF1/2α expression by silencing the PHDs. As we expected, PHDs silencing-mediated HIFα stabilization during chronic hypoxia led to massive cell death (Fig. 5a Right). To definitively prove that HIFα subunits mediate this process, we cosilenced the PHDs and HIFα subunits (HIF1α and HIF2α) at the same time upon chronic hypoxia. Interestingly, silencing of HIFα subunits completely blocks PHD silencing-mediated cell death (Fig. 5b Right). Similar results were obtained when we silenced HIF1α expression in GSK3 cells upon chronic hypoxia (Fig. 5b Left). Therefore, we conclude that HIFα desensitization induced by chronic hypoxia-mediated PHD overactivation is required to prevent cells from cell death.
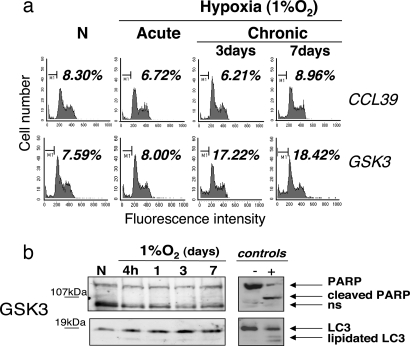
To characterize this cell death, we analyzed by flow cytometry the proportion of apoptotic cells represented by the sub-G1 population stained for propidium iodide (PI). We measured a slight and nonsignificative increase in the number of apoptotic cells in GSK3 cells or PHD-invalidated cells after chronic hypoxia (Fig. 6 and data not shown). Furthermore, Western blot analysis showed no cleavage of the apoptotic marker, poly(ADP-ribose)polymerase (PARP), and LC3 lipidation was not involved in the autophagic process during the chronic hypoxia (Fig. 6 and data not shown). Taken together, we conclude that during chronic hypoxia, HIFα desensitization is required to prevent cells from a massive cell death that is largely independent of the apoptotic and/or autophagic pathways. Moreover, as a feedback survival mechanism, chronic hypoxia-induced PHD overactivation, by ensuring HIFα desensitization, escapes cells from necrosis.
Fig. 6.
Chronic hypoxia-induced cell death is independent of the apoptotic and/or autophagic pathways. Cells were incubated in hypoxia (1% O2) for different periods of time. (a) Measurement by flow cytometry of the sub-G1 PI-positive population in CCL39 and GSK3 cells. (b) PARP and LC3 proteins were analyzed by Western blotting on GSK3 extracts. Staurosporin (1 μM) was used as an apoptotic cell death activator. Nutrient-starved cells were used as a positive control for autophagy.
Mice Adapt to Chronic Hypoxia by Overactivating PHDs to Desensitize HIFα.
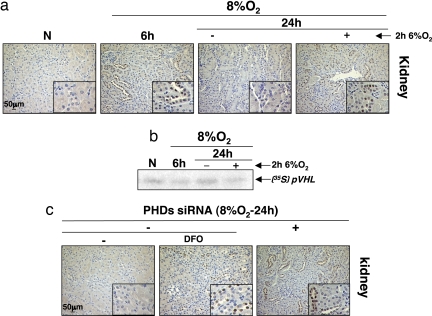
To confirm our results in vivo, we exposed mice to hypoxia at 8% O2 for 6 h (acute) and 24 h (chronic). HIF1α protein level in the kidney (Fig. 7a), the brain, and thymus (data not shown) of mice was then analyzed by immunohistochemistry. We observed an HIF1α increase in the tissues of mice exposed to 6 h hypoxia but none in the 24 h group. Nevertheless, when these mice were exposed for an additional 2 h at more drastic hypoxia (6% O2), we observed a reaccumulation of HIF1α protein (Fig. 7a). Therefore, we hypothesized that mice, as cells, “adapt” to chronic hypoxia by increasing the rate of HIF1α degradation. To test whether the HIF1α desensitization we observed in mice tissues was caused by PHD overactivation, we performed a pVHL capture assay. Fig. 7b shows that after 6 h of hypoxia PHDs are less active compared with the control group. However, after chronic hypoxia of 24 h, the assay reveals a reactivation of the hydroxylases. Moreover, the autoradiogram shows less pVHL binding, reflecting a new decrease in the activity of the PHDs, in mice subjected to an additional and more severe hypoxic exposure (2 h at 6% O2). These results showed a perfect correlation between the expression of HIF1α and the inhibition of PHDs activity in vivo. Finally, to confirm the implication of PHDs in this mechanism, we used the siRNA approach in vivo. Mice were injected with siRNAs targeting the three PHDs or an irrelevant sequence before the hypoxic exposure, and HIF1α protein expression was analyzed (Fig. 7c). As previously observed in the control group, we did not detect HIF1α protein after 24 h at 8% O2, whereas in mice injected with siRNAs silencing the three PHDs isoforms, we observed a clear stabilization of HIF1α protein similar to the desferrioxamine (DFO)-treated animals (Fig. 7c). These data reinforced our in cellulo experiments, showing that mice, which accumulate HIFα protein during acute hypoxia, also adapt to chronic hypoxia by overactivating PHDs to desensitize HIFα.
Fig. 7.
Mice adapt to chronic hypoxia by activating PHDs to desensitize HIFα. (a) BL6 mice were exposed or not to hypoxia at 8%O2 for 6 or 24 h or to 8%O2+2h at 6%O2 for 24 h. HIF-1α staining in kidney was analyzed by immunohistochemistry. (b) pVHL capture assay was performed with brain extracts from mice exposed to the experimental conditions described in a. (c) BL6 mice were injected for 7 days with siRNAs targeting the three PHD isoforms or an irrelevant sequence before being exposed to hypoxia at 8% O2 for 24 h. Control mice were also injected with DFO (1,200 mg/kg) as a positive control. The HIF1α protein level in kidney was analyzed by immunohistochemistry.
Discussion
A perfect balance between O2 delivery and demand in tissues (O2 homeostasis) is essential for survival. When O2 balance is disrupted, organisms have developed adaptative mechanisms to restore the equilibrium and go out of danger. The transcription factor HIF is at the heart of these mechanisms implicated in several physiopathological processes such as cancer or ischemia.
HIFα subunits are tightly regulated by O2. When O2 is restricted, the high turnover rate of the HIFα subunits allows their rapid stabilization and activation. Nevertheless, we show here that chronic hypoxic stress desensitizes HIFα proteins. Our results agree with previous reports showing that HIF1α proteins vanish with prolonged hypoxia (13) and extend this regulatory pathway to HIF2α. Indeed, in contrast with previous data (14, 15), HIF2α behavior appears very close to that of HIF1α. Furthermore, we demonstrate that HIF1/2α decline is observed in a large range of O2 levels and cell types (Fig. 1 and data not shown), supporting HIFα desensitization during chronic hypoxia as a general adaptative mechanism.
Analysis of the HIFα mRNAs did not show any variation in acute or chronic hypoxia, ruling out the putative contribution of the antisense HIF1α previously described (16). However, we show that the bona fide hydroxylation–ubiquitination–proteasome pathway mediates HIFα protein decrease as recently reported by Stiehl et al. (13). Furthermore, we demonstrate an unexpected and gradual PHD overactivation across chronic hypoxia despite low global O2 availability (Fig. 3a). P564 remains the first hydroxylated residue, and mutation on this proline results in the complete loss of P402 hydroxylation (data not shown). Moreover, siRNA experiments reveal that, in contrast to normoxia, the three PHD isoforms contribute to HIFα degradation and hence to the HIFα desensitization mechanism set up during chronic hypoxia (Fig. 3c). However, these results point out differences in the affinity of each PHD isoform for HIF1α and/or HIF2α that need further work to be clarified.
Accumulation of PHD2 and PHD3, could account, at least partially, for the increase in the overall PHD activity. However, PHD2 and PHD3 analysis revealed no additional increase in the pool of these proteins after 1 day of hypoxia (Fig. 3b), suggesting that additional mechanisms cooperate to provide the gradual PHD overactivation that we demonstrate. Here, we tried to elucidate the underlying regulatory pathways.
In addition to O2, these enzymes need 2-oxoglutarate and ferrous iron for full activity (17, 18). Gerald et al. (19) showed that ROS generation, by interfering with Fe(II) availability, can regulate PHD activity and hence HIFα stability. Thus, we measured ROS production across hypoxic kinetics. However, the quantification of ROS production by using the cell-permeant molecule CM-H2DCFDA did not show any variation (data not shown).
Hagen et al. (9) reported that inhibition of mitochondrial respiration by NO or chemical inhibitors leads to intracellular O2 redistribution (measured by using the Renilla luciferase) and prevents hypoxia-induced HIF1α stabilization. Furthermore, they showed that mitochondrial inhibition has no effect on HIF1α stability in drastic hypoxia as we report here for HIF1/2α desensitization (Fig. 1b). More recently, HIF-dependent up-regulation of PDK1 has been shown to mediate a metabolic switch required for cellular adaptation to hypoxia (10, 11). By inducing this gene, pyruvate entry into the tricarboxylic acid cycle is reduced and mitochondrial function is down-regulated. As elegantly demonstrated by Papandreou et al. (11), this down-regulation causes a drop in oxygen consumption that results in increased O2 availability and reduces the toxicity of the hypoxic cytotoxin tirapazamine (11). We speculated that in cells, which cannot redistribute intracellular O2, we would prevent PHDs overactivation and in this way HIFα desensitization during chronic hypoxia. We confirmed our hypothesis by analyzing HIF1α level in respiratory-deficient cells (GSK3). Indeed, as we presumed, in this mutant because of the basal reduced O2 consumption based on the constitutive inhibition of mitochondrial respiration, the absence of O2 redistribution totally prevented HIF1α desensitization. These results support the notion that mitochondrial inhibition goes beyond the metabolic switch required for adaptation to hypoxia. Furthermore, these data minimize the impact of an increase in the pool of PHD2 and PHD3 proteins in the HIFα desensitization process, at least in this particular model, and support the need for additional activating mechanisms.
Chronic hypoxia-induced inhibition of mitochondrial respiration leads to an increase in O2 availability within the cell that appears to be significant and accounts for PHD overactivation even if in vitro experiments revealed Km values for the three PHDs close to atmospheric O2 level (20). It seems that upon long hypoxia even a subtle increase in O2 availability allows PHDs to escape from the hypoxic setpoint. Nevertheless, we did not observe comparable O2 dependence between the three PHDs, as PHD3 seems to need longer exposure, suggesting that isoform-specific regulation depends on additional cofactors to assure optimal O2-sensing properties.
It would be also interesting to evaluate the status of FIH, which controls HIF transcriptional activity. As we have shown for the PHDs, we could speculate that FIH (even if it shows in vitro lower Km values for O2 than the PHDs) will respond to the chronic hypoxia-induced increase in O2 availability and hence cooperate with PHDs to attenuate the hypoxic cascade.
Interestingly we show here that HIFα desensitization is absolutely required for cell survival under chronic hypoxia. Indeed, HIFα desensitized cells are alive, whereas ≈100% of the GSK3 cells die when HIF1α expression is maintained across hypoxic stress (Fig. 5). HIF as a survival factor during the first hours of hypoxia appears as a death-promoting factor when the hypoxia is prolonged in cellulo and in vivo. Furthermore, we show that forced expression of HIFα upon chronic hypoxia induces cell death that is mostly independent of the apoptotic and/or autophagic pathways. Indeed, preventing HIFα desensitization leads to massive cell death by necrosis.
In summary, we show that PHD overactivation during chronic hypoxia appears as a critical adaptative mechanism for triggering HIFα desensitization and protecting cells from necrotic cell death. Therefore, PHDs serve as dual enzymes for cell survival. Upon acute hypoxia, PHD inactivation, by ensuring HIFα stabilization and HIF activation, assures O2 homeostasis. Upon chronic hypoxia, once the adaptative mechanisms are set up and a new O2 homeostasis established, PHD overactivation, despite reduced O2 availability, guarantees survival. Supporting these data, we show in vivo that mice adapt to acute hypoxia by inhibiting PHDs and thereby accumulating HIF1α, whereas during chronic hypoxia PHDs become overactivated and thus HIF1α is degraded. Moreover, HIF1α desensitization is indeed impaired by PHD silencing or by disrupting the established equilibrium by reducing once again O2 availability.
Necrosis results in cell lysis and inflammation. The inflammatory response stimulates proliferation and angiogenesis but also promotes the immune response. Thus, understanding how necrosis favors or, in contrast, suppresses tumor progression, wound healing, or ischemic diseases may yield insights into the suitable use of PHDs as an attractive therapeutic strategy for those pathologies in which hypoxia is implicated.
Methods
Cell Culture.
HeLa, HT1080, NHF, LS174, CAL51, 293, CCL39, and GSK3 (12) cells were grown in DMEM with 7.5% FBS. RCC4 cells and the stable clone RCC4+pVHL we generated were grown in DMEM/Ham's F10 (1:1) medium with 10% FBS. NHK cells were grown in a keratinocyt serum-free medium (Invitrogen) supplemented in l-glutamine, recombinant EGF (0.2 μg/ml) and bovine pituitary extracts (30 μg/ml).
Hypoxic conditions were performed by incubation of cells in a “bug box” anaerobic workstation (Ruskin Technologies). The oxygen concentrations were fixed with a gas mixture containing 5% carbon dioxide and balance nitrogen. For hypoxic kinetics, cells were split sparsely (to reach 60% of confluency at the end of the experiment) 3 days before lysis, outside or inside the bug box depending on the timing, and cell medium was changed every day. Cell extraction was performed inside the bug box to avoid reoxygenation.
Transfections were carried out as described (8). siRNAs were purchased from Eurogentec. Sequences of the irrelevant siRNA and the human HIF1α and PHDs have been reported (8). The same regions were targeted by the mouse siPHDs sequences. siRNA sequence targeting HIF2α (GenBank accession no. NM_001430) corresponds to regions 1590–1609 relative to the start codon. The HIF1α expression vector mutated on Pro402 and Pro564 [HIF1α double mutant (DM)] was generated by mutagenesis using the QuikChange Site-Directed Mutagenesis kit (Stratagene).
Animal Studies.
C57BL/6 mice (8-week-old females) were subjected to systemic hypoxia in the anaerobic workstation at 25°C and in an humidified atmosphere (95%) supplied with the gas mixture allowing it to adjust to the hypoxic environment by gradually decreasing the O2 level from 20.9% to 8% and 92% N2 during an adaptation time of 2 h 30 min. The mice were provided with food and water ad libitum. For the siRNA experiment, mice were i.p.-injected for 7 days every day with 3 μg of each siRNA by using jetPEI (Polyplus Transfection). The animal study protocols were conducted according to institutional guidelines for animal use. Three mice were used for each experimental condition and experiments were reproduced twice. Mice were killed inside the bug box. Half of the dissected organs were frozen and stored at −80°C (to measure PHDs activity), and the rest were fixed in 4% formaldehyde.
RT-PCR, Western Blotting, and Immunohistochemistry.
RNA was purified with the RNeasy Mini Kit (Qiagen). The oligonucleotide sequences for RT-PCR are available on request.
For Western blot analysis, cells were lysed in Laemmli buffer. Protein concentration was determined with Lowry assay, and 50 μg of cell extracts was resolved by SDS/PAGE and electrophoretically transferred onto a PVDF (Millipore) or nitrocellulose membrane (Amersham Biosciences). Immunoreactive bands were visualized with the Amersham Biosciences ECL system.
HIF1α immunohistochemistry was performed as described (21). Immunohistochemical studies were analyzed with a Leica Zeiss inverted microscope (Axiovert 200 M, ×40 objective) equipped with a CoolSnap HQ cooled CCD camera (Roeper Scientifique).
Antibodies.
The anti-HIF1α has been characterized (22). The Ab to PHD2 (antiserum 923) was raised in rabbits immunised against the first 15 N-terminal amino acids of human PHD2. Antibodies recognizing β-actin (Sigma), PHD3 (Interchim and Novus Biologicals), HIF2α (Novus Biologicals), Myc (9B11; Cell Signaling), PARP (Biomol), and LC3 (MBL) were used.
pVHL Capture Assay.
GST-HIF1α WT was obtained by subcloning the 0.714-kb BamHI/XmaI PCR fragment of the pcDNA3-HA-HIF1α plasmid (corresponding to the 1032–1746 nucleotides of the human HIF1α sequence) into the BamHI/SmaI site of PGEX-6P-3. GST-HIF1α P402A, P564A, and P402AP564A mutants were obtained by mutagenesis of the GST-HIF1α construct with the QuikChange Site-Directed Mutagenesis Kit (Stratagene).
pVHL was in vitro-translated by using a TNT Transcription/Translation Kit (Promega) supplemented with Redivue l- [35S]methionine (Amersham Biosciences) to produce [35S]pVHL. The protocol used to cell extracts preparation and hydroxylation reaction has been described (23). It is important to note that cell lysis and hydroxylation assay of hypoxic extracts were performed inside the bug box. The same procedure was applied for tissue hydroxylation assay.
Cell Death and Apoptosis.
Cell death was analyzed by using the trypan blue staining procedure (24).
For PI staining, total cell pellets were washed once with PBS and finally fixed in ice-cold 70% ethanol. The proportion of apoptotic cells was determined as described (25).
All experiments were repeated at least three times (excepted for the mice studies) with similar results. Representative immunostainings, blots, and autoradiograms are shown.
Acknowledgments.
We thank Dr. R. Buscà (Faculty of Medicine, University of Nice) for providing NHK and NHF cells and critically reading the manuscript, Dr. A. O. Hueber and her team for help with the death experiments, and Drs. P. Lenormand and E. Vial for their comments. Financial support was from the Centre National de la Recherche Scientifique, Ministère de l'Education, de la Recherche et de la Technologie, Ligue Nationale Contre le Cancer (Equipe Labellisée), Cancéropôle Provence-Alpes-Cote d'Azur, and Association pour la Recherche sur le Cancer (subvention 3693). E.B. is supported by Ministerio de Educación y Ciencia Grant SAF2007-64597, ETORTEK Research Program 05-07, and the Bizkaia Xede Program from Bizkaia County.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Semenza GL. Curr Opin Genet Dev. 1998;8:588–594. doi: 10.1016/s0959-437x(98)80016-6. [DOI] [PubMed] [Google Scholar]
- 2.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 3.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, et al. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 4.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 5.Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berra E, Ginouves A, Pouyssegur J. EMBO Rep. 2006;7:41–45. doi: 10.1038/sj.embor.7400598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Appelhoff RJ, Tian YM, Raval RR, Turley H, Harris AL, Pugh CW, Ratcliffe PJ, Gleadle JM. J Biol Chem. 2004;279:38458–38465. doi: 10.1074/jbc.M406026200. [DOI] [PubMed] [Google Scholar]
- 8.Berra E, Benizri E, Ginouves A, Volmat V, Roux D, Pouyssegur J. EMBO J. 2003;22:4082–4090. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagen T, Taylor CT, Lam F, Moncada S. Science. 2003;302:1975–1978. doi: 10.1126/science.1088805. [DOI] [PubMed] [Google Scholar]
- 10.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Pouyssegur J, Franchi A, Pages G. Novartis Found Symp. 2001;240:186–196. doi: 10.1002/0470868716.ch13. discussion 196–188. [DOI] [PubMed] [Google Scholar]
- 13.Stiehl DP, Wirthner R, Koditz J, Spielmann P, Camenisch G, Wenger RH. J Biol Chem. 2006;281:23482–23491. doi: 10.1074/jbc.M601719200. [DOI] [PubMed] [Google Scholar]
- 14.Holmquist-Mengelbier L, Fredlund E, Lofstedt T, Noguera R, Navarro S, Nilsson H, Pietras A, Vallon-Christersson J, Borg A, Gradin K, et al. Cancer Cell. 2006;10:413–423. doi: 10.1016/j.ccr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 15.Uchida T, Rossignol F, Matthay MA, Mounier R, Couette S, Clottes E, Clerici C. J Biol Chem. 2004;279:14871–14878. doi: 10.1074/jbc.M400461200. [DOI] [PubMed] [Google Scholar]
- 16.Thrash-Bingham CA, Tartof KD. J Natl Cancer Inst. 1999;91:143–151. doi: 10.1093/jnci/91.2.143. [DOI] [PubMed] [Google Scholar]
- 17.Schofield CJ, Ratcliffe PJ. Nat Rev Mol Cell Biol. 2004;5:343–354. doi: 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- 18.Schofield CJ, Zhang Z. Curr Opin Struct Biol. 1999;9:722–731. doi: 10.1016/s0959-440x(99)00036-6. [DOI] [PubMed] [Google Scholar]
- 19.Gerald D, Berra E, Frapart YM, Chan DA, Giaccia AJ, Mansuy D, Pouyssegur J, Yaniv M, Mechta-Grigoriou F. Cell. 2004;118:781–794. doi: 10.1016/j.cell.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 20.Tuckerman JR, Zhao Y, Hewitson KS, Tian YM, Pugh CW, Ratcliffe PJ, Mole DR. FEBS Lett. 2004;576:145–150. doi: 10.1016/j.febslet.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 21.Trastour C, Benizri E, Ettore F, Ramaioli A, Chamorey E, Pouyssegur J, Berra E. Int J Cancer. 2007;120:1443–1450. doi: 10.1002/ijc.22436. [DOI] [PubMed] [Google Scholar]
- 22.Richard DE, Berra E, Gothie E, Roux D, Pouyssegur J. J Biol Chem. 1999;274:32631–32637. doi: 10.1074/jbc.274.46.32631. [DOI] [PubMed] [Google Scholar]
- 23.Huang J, Zhao Q, Mooney SM, Lee FS. J Biol Chem. 2002;277:39792–39800. doi: 10.1074/jbc.M206955200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forcet C, Ye X, Granger L, Corset V, Shin H, Bredesen DE, Mehlen P. Proc Natl Acad Sci USA. 2001;98:3416–3421. doi: 10.1073/pnas.051378298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cahuzac N, Baum W, Kirkin V, Conchonaud F, Wawrezinieck L, Marguet D, Janssen O, Zornig M, Hueber AO. Blood. 2006;107:2384–2391. doi: 10.1182/blood-2005-07-2883. [DOI] [PubMed] [Google Scholar]