Abstract
Escherichia coli O157:H7, a toxin-producing food and waterborne bacterial pathogen, has been linked to large outbreaks of gastrointestinal illness for more than two decades. E. coli O157 causes a wide range of clinical illness that varies by outbreak, although factors that contribute to variation in disease severity are poorly understood. Several recent outbreaks involving O157 contamination of fresh produce (e.g., spinach) were associated with more severe disease, as defined by higher hemolytic uremic syndrome and hospitalization frequencies, suggesting that increased virulence has evolved. To test this hypothesis, we developed a system that detects SNPs in 96 loci and applied it to >500 E. coli O157 clinical strains. Phylogenetic analyses identified 39 SNP genotypes that differ at 20% of SNP loci and are separated into nine distinct clades. Differences were observed between clades in the frequency and distribution of Shiga toxin genes and in the type of clinical disease reported. Patients with hemolytic uremic syndrome were significantly more likely to be infected with clade 8 strains, which have increased in frequency over the past 5 years. Genome sequencing of a spinach outbreak strain, a member of clade 8, also revealed substantial genomic differences. These findings suggest that an emergent subpopulation of the clade 8 lineage has acquired critical factors that contribute to more severe disease. The ability to detect and rapidly genotype O157 strains belonging to such lineages is important and will have a significant impact on both disease diagnosis and treatment guidelines.
Keywords: pathogens, polymorphisms, population genetics
Enterohemorrhagic Escherichia coli (EHEC) includes a diverse population of Shiga toxin-producing E. coli that causes outbreaks of food and waterborne disease (1–3). EHEC often resides in bovine reservoirs and is transmitted via many food vehicles including cooked meat, such as hamburger (4) and salami (5), and raw vegetables, such as lettuce (6, 7) and spinach (8). In North America, E. coli O157:H7 is the most common EHEC serotype contributing to >75,000 human infections (9) and 17 outbreaks (3) per year.
It is not clear why outbreaks of EHEC O157 vary dramatically in the severity of illness and the frequency of the most serious complication, hemolytic uremic syndrome (HUS) (10–12). The 1993 outbreak in western North America (4) and the large 1996 outbreak in Japan (13) had low rates of hospitalization and HUS (14, 15), whereas the 2006 North American spinach outbreak (8) had high rates of both hospitalization (>50%) and HUS (>10%). One hypothesis is that outbreak strains differ in virulence as a result of variation in the presence and expression of different Shiga toxin (Stx) gene combinations (16–19).
To assess the genetic diversity and variability in virulence among E. coli O157 strains, we developed a real-time PCR system for identifying synonymous and nonsynonymous mutations as SNPs (20–23). Although molecular subtyping methods, such as pulsed-field gel electrophoresis (PFGE), reveal extensive genomic diversity among O157 outbreaks, “DNA fingerprinting” data are not amenable to population genetic or phylogenetic analyses. PFGE analysis has demonstrated that differences between O157 strains result from discrete insertions or deletions that contribute to restriction site changes between strains rather than SNPs (24). Comparison of multiple O157 genomes has shown that bacteriophage variation is a major factor in generating genomic diversity (25) and presumably underlies most genomic variability detected by PFGE (24, 26). In contrast, the systematic analysis of SNPs, also useful for outbreak investigations, can resolve closely related bacterial genotypes, provide insights into the microevolutionary history of genome divergence (20, 27), and contribute to an epidemiologic assessment of associations between bacterial genotypes and disease. Here we genotyped >500 clinical strains of EHEC O157 based on 96 SNPs that separated strains into genetically distinct groups and sequenced the genome of the O157 strain implicated in the spinach outbreak. These data form a basis for addressing how EHEC O157 has diversified and evolved in genome content and for assessing intrinsic differences among O157 lineages with regard to clinical presentation and disease severity.
Results
SNP Genotyping and Diversity Among O157 Strains.
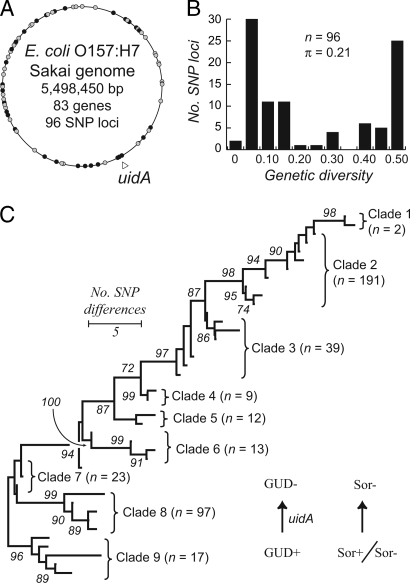
A total of 96 SNP loci were evaluated in 83 O157 genes (Fig. 1A); 68 sites were identified by comparative genome microarrays (23), 15 from housekeeping genes (28), four by comparisons between two O157 genomes (29, 30), and nine from three virulence genes (eae, espA, and fimA). Overall, 52 (54%) of the SNPs are nonsynonymous and 43 (45%) are synonymous substitutions (Fig. 1A). One SNP locus detects a guanosine (G) dinucleotide insertion that results in a frameshift in the uidA gene and produces a premature termination codon. This uidA SNP (Fig. 1A) was examined because the GG insertion is hypothesized to have occurred late in the emergence of E. coli O157:H7, and its early origin explains the absence of β-glucuronidase activity (i.e., GUD− phenotype) in most O157 strains (31).
Fig. 1.
Genetic relatedness of E. coli O157 among 403 O157 and closely related O55:H7 strains based on 96 SNPs. (A) The location of 83 genes within 96 SNP loci on the E. coli O157:H7 genomic map of the Sakai strain. Real-time PCR assays detected 52 loci with nonsynonymous polymorphisms (black circles), 43 loci with synonymous polymorphisms (white circles), and one locus (uidA-686) with a GG insertion (open triangle). (B) The distribution of nucleotide diversity (π) across 96 SNP loci. Diversity ranges from 0 for two monomorphic SNP loci to a maximum between 0.45 and 0.50 for 26 loci. The average π for the 96 loci is 0.212 ± 0.199. (C) Phylogenetic relationships among SNP genotypes (SGs) using the minimum evolution algorithm based on the distance matrix of pairwise differences between SGs. The consensus tree is shown with the percentages at the nodes of the >70% bootstrap confidence values based on 1,000 replicates. Both the GUD+ and Sor+, which occur in clade 9, are negative (GUD− and Sor−) in the derived clades 1–8.
Pairwise comparisons of the nucleotide profiles from 403 E. coli O157 and closely related strains from clinical sources worldwide distinguished 39 distinct SNP genotypes (SGs) [see supporting information (SI) Table 2]. Overall, the number of nucleotide differences between O157 SGs ranged from 1 to 57 with an average of 23.1 ± 1.6 across the 96 loci. The nucleotide diversity (π), a measure of the degree of polymorphism within the O157 population, is 0.212 ± 0.199, indicating that two strains selected at random differ on average at ≈20% of SNP loci (Fig. 1B). The minimum evolution (ME) algorithm, which infers that the theoretical tree is the smallest among all possible trees based on the sum of branch length estimates (32), revealed nine clusters among the 39 genotypes (Fig. 1C). Eight of the nine clusters are significant (multiple SGs grouped with >85% bootstrap support). The deepest node in the ME phylogeny occurs at 15 SNP locus differences and separates a lineage that includes ancestral O157 strains and close relatives with wild-type E. coli phenotypes (i.e., GUD+; sorbitol-positive, Sor+) from the evolutionarily derived lineages (GUD− and Sor−) (Fig. 1C).
Neighbor-net Resolves Clades.
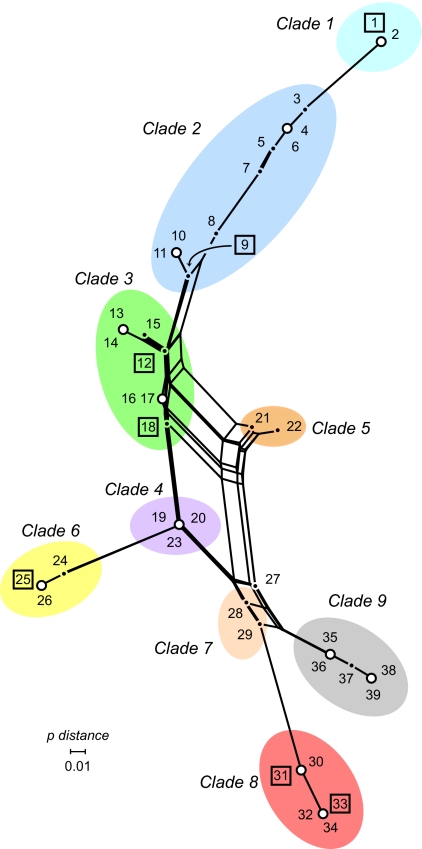
Subsequent analyses of the 39 SG profiles revealed phylogenetically informative loci, as defined by two variants found in two or more SGs. Among the 96 SNP loci, 71 sites had complete data, and, of these, there were 23 singletons and 48 parsimoniously informative (PI) sites. The 48 PI sites were used to construct a Neighbor-net tree (33) to determine whether the informative sites support conflicting phylogenies or a single tree (Fig. 2). In this analysis, the 39 SGs were resolved into 25 distinct nodes: 10 nodes contained two or more SGs with the same profiles across all 48 loci (Fig. 2). Clade 9 roots the phylogenetic network because it includes strains with wild-type E. coli phenotypes (e.g., GUD+ and Sor+), characteristics of the lineage most primitive to the derived EHEC O157 lineages (e.g., GUD− and Sor−) (31, 34). Rather than producing a unique bifurcating tree, the Neighbor-net reveals a central group of four clades (clade 3, 4, 5, and 7) connected by multiple paths. The presence of these parallel paths suggests that either recombination or recurrent mutation has contributed to the divergence of the central clades from the evolutionarily derived lineages. In contrast, clades 1, 2, 6, and 8 occur at the end of distinct branches with no evidence of conflicting phylogenetic signals, indicating that these lineages are diverging without evidence of recombination in background polymorphisms.
Fig. 2.
The phylogenetic network applied to 48 parsimoniously informative (PI) sites using the Neighbor-net algorithm for 528 E. coli O157 strains. The colored ellipses mark clades supported in the minimum evolution phylogeny. The numbers at the nodes denote the SNP genotypes (SGs) 1–39, and the white circle nodes contain two SGs that match at the 48 PI sites. The seven SGs found among multiple continents are marked with squares.
To further examine the distribution of O157 genotypes, we devised a minimum set of 32 SNP loci for resolving all 39 SGs and genotyped 135 additional O157 strains representing clinical sources, including five from well known outbreaks. In all, with the additional screening based on the minimal SNP set, 528 O157 strains were genotyped and classified into SGs and clades. Virtually all of the 528 strains were classified into one of nine clades, and >75% of strains belonged to one of four clades. The most common genotypes were SG-9 (n = 184; 35%) of clade 2 followed by SG-30 (n = 94; 18%) of clade 8; 20 of the 39 SGs were represented by only one or two strains (Fig. 3A and SI Table 2). In addition, seven SGs were found among O157 strains isolated from multiple continents and during different time periods (SI Table 2). Five of these seven SGs belonged to the four clades located at the end of long branches identified in the Neighbor-net analysis (Fig. 2) and may represent stable EHEC O157 lineages generated from the central clades. Strains N0436 (SG-15), N0303 (SG-11), and N0587 (SG-27), which were included in a prior study of O157 SNPs (23) because they had uncommon PFGE patterns via PulseNet, represented unique, single-strain SGs in this study as well. These SGs do not match other genotypes including SG-11 (N0303), which matches SG-10 at all 48 PI SNP loci.
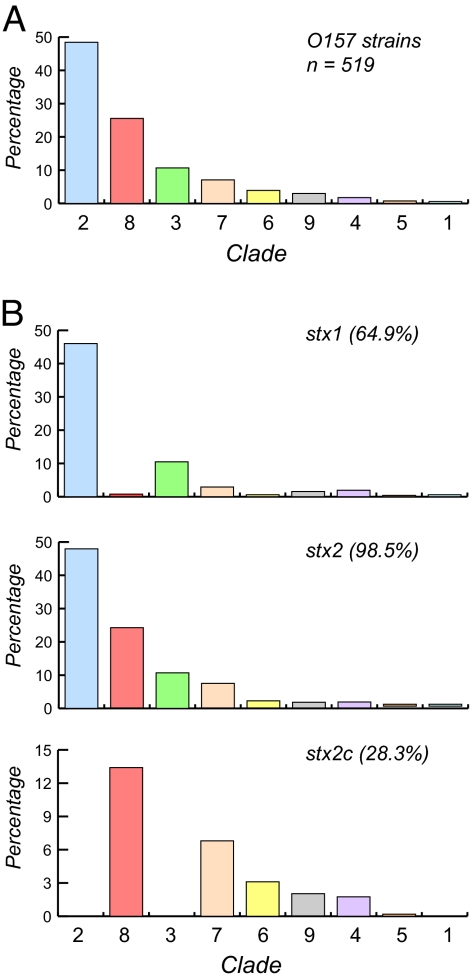
Fig. 3.
Distribution of Shiga toxin (Stx) genes in E. coli O157 clades. (A) The frequency of 528 O157 strains that were classified into one of nine clades based on SNP genotyping, ranked from left to right in the histogram by decreasing frequency. The four most common clades were clades 2 (47.6%), 8 (25.4%), 3 (10.6%), and 7 (7.3%). (B) Distribution of Shiga toxin variants (stx1, stx2, and stx2c) among 519 of the 528 O157 strains organized into nine clades. The percentage of PCR assay positive strains overall is given in parentheses.
Shiga Toxin Genes in Clades.
Because the production of Stx has been linked to virulence in O157 strains (35), we estimated the frequency of one or more of three Stx variants (stx1, stx2, and stx2c) by clade. Although stx1 was found in more than half (≈65%) of 519 of the 528 O157 strains tested, the distribution is highly nonrandom across clades (Fig. 3B). The stx1 gene was common in clade 2 strains (95.1% of all stx1-positive strains are in clade 2) but not clade 8 (3.7%). The stx2 gene was present in virtually all (98.5%) O157 strains evaluated (Fig. 3B), occurring most frequently in clade 2 (46.8% of 519 strains) and clade 8 (25.4%) strains. In total, 98.4% and 100% of clade 2 and clade 8 strains, respectively, were positive for stx2 (Fig. 3B).
The stx2c gene also has a nonrandom distribution and is concentrated in clades 4, 6, 7, and 8 (Fig. 3B) but is missing from clades 1, 2, and 3. Most noteworthy is that clade 8 strains were significantly more likely to have both the stx2 and stx2c genes when compared with the other stx2c-positive clades (P < 0.0001); 69 of the 79 O157 strains positive for both the stx2 and stx2c genes belonged to clade 8, but not all (57.6%) of the 128 clade 8 strains had stx2c.
Virulence Differences Between O157 Clades.
Clade 1 contains two SGs and includes the O157 genome strain Sakai (29) (SG-1), implicated in the 1996 Japanese outbreak (Table 1) linked to radish sprouts (13). Clade 2, the predominant lineage identified, contains nine SGs and includes strain 93-111 (SG-9) from the 1993 outbreak associated with contaminated hamburgers in western North America (4). Clade 3 consists of seven genotypes and includes the genome strain EDL-933 (30) (SG-12) from the first human O157 outbreak in 1982 linked to hamburgers sold at a chain of fast food restaurant outlets in Michigan and Oregon (36). Although these outbreaks representing clades 1, 2, and 3 affected ≈12,000 people combined, the rate of HUS and hospitalization was low for each (4, 14, 15, 36) compared with the average rates for 350 North American outbreaks (3) (Table 1). Clade 8, in contrast, consists of five SGs that include O157 strains from multistate outbreaks linked to contaminated spinach (37) and lettuce (7) (SG-30) in North America. These 2006 outbreaks caused reportable illnesses in >275 patients and resulted in remarkably high rates of hospitalization (average 63%) and HUS (average 13%), a rate that is three times greater than the average HUS rate for 350 outbreaks (Table 1).
Table 1.
SG and clade for several E. coli O157:H7 outbreak strains with hospitalization and HUS rates by outbreak
| Strain* | Year | SG | Clade | Outbreak | No. of cases | No. of hospitalizations (%) | No. of HUS (%) | Ref(s). |
|---|---|---|---|---|---|---|---|---|
| Sakai† | 1996 | 1 | 1 | Radish sprouts, Sakai, Japan | 5,000–12,680 | 398–425 (3–5) | 0–122 (0–3) | 13–15 |
| 93-111 | 1993 | 9 | 2 | Hamburger, northwest U.S. | 583 | 171 (29) | 41 (7) | 4 |
| EDL-933 | 1982 | 12 | 3 | Hamburger, Michigan and Oregon | 47 | 33 (70) | 0 (0) | 36 |
| TW14359 | 2006 | 30 | 8 | Spinach, western U.S. | 204 | 104 (51) | 31 (15) | 37 |
| TW14588 | 2006 | 30 | 8 | Lettuce, eastern U.S. | 71 | 53 (75) | 8 (11) | 7 |
| 350 O157 outbreaks in the U.S. (1982–2002) | 8,598 | 1,493 (17) | 354 (4) | 3 | ||||
*Sakai (RIMD-0509952) and EDL-933 have complete genome sequence available, and strain TW14359 has been sequenced by pyrosequencing (see text).
†The range is reported for the number of cases and frequency of HUS and hospitalization in the Sakai outbreak because the numbers vary in the literature.
Genome Sequencing of a Clade 8 Outbreak Strain.
To assess whether the high rates of severe disease associated with the spinach outbreak are attributable to intrinsic differences between the spinach outbreak strain (clade 8) and other previously sequenced strains (e.g., Sakai, clade 1; EDL-933, clade 3), we used massively parallel pyrosequencing (GS 20, 454; Life Sciences) to sequence the genome of a strain (TW14359) linked to the 2006 spinach outbreak. Contig alignment of the spinach outbreak strain to the O157 Sakai genome (29, 30) using MUMmer (38) revealed 5,061 (96.3%) significant matches to the 5,253 Sakai genes. The spinach strain genome was missing 192 Sakai genes, 26 of which are backbone genes and 166 of which are genes for prophage and prophage-like elements. For example, the Mu-like phage Sp18 that is integrated into the sorbose operon of the Sakai genome (25) is absent in the spinach strain genome. Alignment to the Sakai pO157 plasmid revealed that 111 of 112 pO157 genes are present in the spinach outbreak strain, suggesting that the plasmid is conserved in both pathogens.
Among the 4,103 shared backbone genes within the Sakai and spinach genomes, the average sequence identity is 99.8%, and, of the 958 shared island genes with Sakai, the average sequence identity is 97.96%. The average sequence identity for all shared genes (n = 5,061) is 99.25%. We then compared the conservation of backbone genes and identified 2,741 shared genes with <0.5% nucleotide divergence among all three O157 genomes (SI Fig. 5). Interestingly, the Sakai and EDL-933 genomes are more similar to each other in gene content and nucleotide sequence identity than to the clade 8 spinach outbreak strain, which carries additional genetic material including stx2c and the Stx2c lysogenic bacteriophage 2851 (39). This suggests that the spinach outbreak genome and, by inference, clade 8, has substantial time to diverge with respect to its genetic composition when compared with strains from other lineages.
Association Between Clades and Severe Disease.
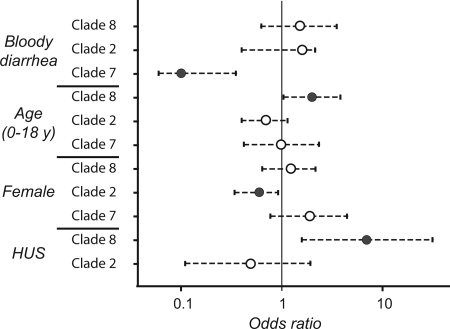
To determine whether the O157 infections caused by clade 8 pathogens differ with respect to clinical presentation, we examined epidemiological data for all laboratory-confirmed O157 cases (n = 333 patients) identified in Michigan since 2001 (40). There are significant associations between specific O157 clades and patient symptoms as well as disease severity via univariate (SI Table 3) and multivariate (SI Table 4) analyses. Patients infected with O157 strains of clade 8 were significantly more likely to be younger (ages 0–18), and, despite the small number (n = 11) of HUS cases identified, HUS patients were seven times more likely to be infected with clade 8 strains than patients with strains from clades 1–7 combined (Fig. 4). This HUS association could not be explained by the presence of stx2c in clade 8 strains, because only four of 11 HUS patients had stx2c-positive strains.
Fig. 4.
Odd ratios with 95% confidence intervals (dotted lines) highlighting the association between patient characteristics and infection with specific clades. Logistic regression models were adjusted for age, gender, bloody diarrhea, diarrhea, abdominal pain, chills, HUS, hospitalization, and body aches. Dark circles show significant associations.
Three HUS patients had infections caused by strains of clade 2, the most numerically dominant clade; however, patients with HUS were still more likely to have a clade 8 infection when compared with clade 2 (SI Tables 3 and 4). In this analysis, we also observed that clade 2 strains were more common in male patients, and clade 7 strains caused less severe disease, as measured by reporting frequencies of bloody diarrhea and other symptoms, although not all were significant (Fig. 4 and SI Tables 3 and 4).
Clade Frequencies over Time.
Because both the 2006 spinach and lettuce outbreaks were caused by members of the same SG within clade 8, we estimated the frequency of clade 8 over time in an epidemiologically relevant setting. There was a significant increase (Mantel–Haenszel χ2 = 32.5, df = 1, P < 0.0001) in the frequency of disease caused by clade 8 strains among all 444 O157 cases in Michigan (SI Fig. 6). Specifically, the frequency of clade 8 strains increased from 10% in 2002 to 46% in 2006 despite the steady decrease in all O157 cases identified via surveillance (40) since 2002 (SI Fig. 6).
Discussion
The population genetics and epidemiology of E. coli O157:H7 infections have changed dramatically since the first outbreaks of illness associated with contaminated ground beef occurred in the early 1980s (1). New routes of infection, including direct contact with animals, and survival in novel food vehicles, particularly fresh produce, have become major sources of new disease cases and have contributed to widespread epidemics (3). This changing epidemiology is also influenced by the genetic variation and “relentless evolution” (41) of the O157 pathogen population. As the population of EHEC O157 strains has increased in frequency and spread geographically, it has genetically diversified. Isolates of EHEC O157 from clinical and bovine sources have been shown to be genotypically diverse by different methods, including PFGE (26), octomer-based genome scanning (42), and multilocus variable number of tandem repeats analysis (43). Studies of prophage and prophage remnants in EHEC O157 strains have indicated that genotypic diversity is largely attributable to bacteriophage-related insertions, deletions, and duplications of variable sizes of DNA fragments (24, 25, 44).
Substantial variability in clinical presentation also has been observed among patients with EHEC O157 infections. This variation is even apparent among different O157 outbreaks, as some outbreaks have contributed to remarkably high frequencies of HUS and hospitalization relative to others (Table 1). Consequently, we hypothesize that there is extensive variation in virulence among distinct clades of O157. The genetic basis of virulence that contributes to variation by clade will require further investigation, as will assessing the ecological differences that contribute to variation in transmission rates and linkage to food and waterborne disease.
The evaluation of >500 O157 strains from clinical sources for up to 96 SNP loci highlights the degree of genetic variation among strains and identifies a specific O157 lineage (clade 8) that has increased in frequency (SI Fig. 6). This increase in clade 8 is surprising given that, at the same time, the overall national prevalence of EHEC O157 infections has been decreasing (45). Strains of the clade 8 lineage have caused two recent and unusually severe outbreaks linked to produce, are associated with HUS, and more frequently carry both the stx2 and stx2c genes. In concert, these results suggest that a more virulent subpopulation of EHEC O157 is increasing in its contribution to the overall disease burden associated with O157 infections. Although there are clear differences in the frequency and combination of stx genes among clades, the toxin–gene combination alone does not account for the variation in hospitalization and HUS rates by clade.
The observation that clade 8 strains more frequently have both the stx2 and stx2c genes implies that carriage of both the Stx2 and Stx2c phages contribute in part to the greater virulence of clade 8 strains. The Stx genes, encoded by lambda-like bacteriophages, can circulate among hundreds of different E. coli strains (46) and integrate into many sites in the O157 genome (25, 44). Previous studies have observed correlations between specific Stx genes and disease, particularly for stx2 and stx2c (18, 19), although it has not been suggested that having both variants together may increase virulence. Because not all clade 8 strains have both stx2 and stx2c, and none of the strains has only stx2c, the presence and presumable production of the Stx2c variant alone cannot be solely responsible for the enhanced virulence attributed to this lineage. This also is true for the production of Stx2, because it was detected in nearly every strain representing all nine clades. We cannot, however, rule out the possibility that stx2c is rapidly lost during infection, thereby inhibiting our ability to detect it in some strains. What accounts for the greater intrinsic virulence among clade 8 strains and other O157 genotypes has not been fully understood. There is a constellation of mobile genetic elements that contribute to the virulence of pathogenic E. coli (47), and it is possible that a novel combination of virulence factors has emerged in the clade 8 lineage. The extent to which other ancillary factors contribute to the full virulence of clade 8 strains requires further investigation.
Among the three most common clades (2, 7, and 8) examined, there are noteworthy differences in transmission and clinical disease characteristics (SI Table 3) in addition to the association between clade 8 and HUS. For example, patients infected with strains from both clades 2 and 8 reported bloody diarrhea more frequently when compared with patients with clade 7 infections. Furthermore, clades 7 and 8 were more common among female patients, and clade 8 was associated with disease in younger (<18 years) patients (Fig. 4). These observed differences among patients with O157 infections clearly reflect differences among the common clades that can result from variability in gene content or genetic variation in conserved, common genes. The sequence comparisons of the spinach outbreak genome (clade 8) with the two other complete genomes (clades 1 and 3) indicate that there has been sufficient evolution time for 5% mutational substitution (10% differences in sequence of 2,741 conserved genes). This is consistent with a study by Zhang et al. (23) that estimated the most recent ancestor for EHEC O157 strains in clades 1–8 (β-glucuronidase-negative, non-sorbitol-fermenting) to be ≈20 thousand years ago (based on the assumed rate of 4.7 × 10−9 per site per year).
To determine when specific clades first appeared in human disease and assess whether clade 8 strains have increased in frequency in strains recovered from outside of Michigan, we evaluated a subset of O157 strains isolated during different time periods. Through this screening, we identified clade 8 strains from clinical cases dating back to 1984 on multiple continents (SI Table 2), suggesting that clade 8 has not recently emerged. This result was confirmed by both the spinach outbreak genome (Fig. 4) and phylogenetic analyses (Fig. 1B), because clade 8 is more closely related to the evolutionarily ancestral O157 lineage (clade 9) than other lineages. In contrast to clade 8 strains from Michigan patients, the frequency of stx2c with or without stx2 did not increase in frequency over time, and stx2c was detected in a strain isolated in 1984, indicating that it, too, has not recently emerged.
It is clear that EHEC O157 is genetically diversified and comprises multiple detectable clades with substantial genomic, biological, and epidemiological variation. SNP genotyping has revealed the clades that reflect the genetic variability among pathogenic strains associated with clinical infection. These results support the hypothesis that the clade 8 lineage has recently acquired novel factors that contribute to enhanced virulence. Evolutionary changes in the clade 8 subpopulation could explain its emergence in several recent foodborne outbreaks; however, it is not clear why this virulent subpopulation is increasing in prevalence. Because humans are more an incidental host for EHEC O157, further investigation of the bovine reservoir (48, 49) and environment is critical, as is the evaluation of agricultural practices in areas where livestock and produce are farmed side by side. Identifying the underlying factors that lead to enhanced virulence and the successful transmission of EHEC O157 in contaminated food and water is imperative. Similarly, conducting large-scale molecular epidemiologic studies is necessary to assess the actual distribution of SGs, clades, and Stx variants in environmental reservoirs and broad geographic scales (50). The development and deployment of a rapid, inexpensive molecular test that can identify more virulent O157 subtypes also would be useful for clinical laboratories to identify patients with an increased likelihood of developing HUS.
Materials and Methods
Bacterial Strains.
A total of 528 EHEC O157 strains and close relatives were genotyped; 444 were from Michigan patients identified via surveillance by the Michigan Department of Community Health Bureau of Laboratories from 2001 to 2006 (40). Patients were confirmed to have O157-associated disease by culture, enzyme immunoassay, and real-time PCR for stx1 and stx2 (40). Strains with unique PFGE patterns or patterns present in two or fewer strains (n = 333) were included in the epidemiological analyses. The additional 94 strains were selected based on epidemiological data to provide a sample representing different geographic locations and collection dates.
SNP Loci and Real-Time PCR Assays.
The 96 SNP loci (SI Table 5) were identified from data generated by comparative genome sequencing microarrays (23), multilocus sequence typing (28), virulence gene sequencing, and in silico comparisons of the two O157 genomes (29, 30). Hairpin-shaped primers were designed by adding a 5′ tail complementary to the 3′ end of each linear primer (22) for each locus, and real-time PCR was used to identify the SNP. Six strains were duplicated to serve as internal controls; identical SNP profiles were observed.
To reduce the number of SNP assays, we used the SNPT program (21), which identified a subset of 32 loci to delineate all 39 SGs. Additional assays were performed to confirm certain SGs. The final set of 32 loci was obtained by substituting three SNP loci that resolved SNP types 35–39 and adding three different loci for classifying SGs 1–34.
Phylogenetic Analyses.
Distance between SGs was measured as the pairwise number of nucleotide difference. ME trees were used to infer the evolutionary relationships among the 39 SGs based on pairwise distance matrix with bootstrap replication for concatenated SNP data using MEGA3 (51). Bootstrap analysis of phylogenetic trees generated by the ME method were constructed by using MEGA3 (51), and bootstrap confidence levels (based on 1,000 replicate trees) were used to classify SGs into clades. A phylogenetic network based on the Neighbor-net algorithm (33) was applied to 48 PI sites by using the SplitsTree4 program (52).
Spinach Outbreak strain Genomic Analysis.
A culture isolated from a Michigan patient hospitalized in September 2006, linked by the PulseNet PFGE system (53) to the spinach outbreak pattern by the Michigan Department of Community Health and the Centers for Disease Control and Prevention, was sequenced. The Michigan State University Genomic Research Support Technical Facility used parallel pyrosequencing on the GS20 454 that included four standard sequencing runs and one paired end run. The final assembly had 201 large contigs (>500 nt) with ≈20 times coverage arranged into 79 scaffolds with a total of 5,307,096 nt, and 680 small contigs for a total of 213,699 nt (4% of the total assembled length). Contig alignments to published genomes [Sakai (29) and EDL-933 (30)] were conducted by MUMmer (38). Sakai/EDL-933 genes with at least one alignment of >90% nucleotide identity in the spinach genome were considered present in the spinach strain.
To evaluate the distribution of SNPs in the spinach genome, a strict set of comparison rules was applied. Conserved genes were included only if the alignment was 100% unique in both genomes (i.e., multicopied genes in either genome were excluded), the identity between the aligned regions was >90%, and the alignment region was >90% of the length of Sakai/EDL-933 genes. Insertions and deletions were excluded. A total of 2,741 genes that fit these criteria and occurred in all three genomes were compared by identify SNP differences. A map was plotted by GenomeViz (54).
Stx2c Detection.
Multiplex PCR was used to detect stx2c and the Stx2c-phage o and q genes (39) in 519 strains; stx data were missing for 19 strains, four of which were repeatedly stx-negative. The malate dehydrogenase (mdh) gene was used as a positive control. Strains were considered positive for stx2c if mdh (835 bp), stx2c (182 bp), o (533 bp), and q (321 bp) were present.
The multiplex PCR does not distinguish between stx2 and stx2c [both genes differ by only 3 aa in the B subunit (55)]; thus, we developed a RFLP-based method that amplifies a larger PCR product (1,152 bp) using primers stx2_F61 (5′-TATTCCCRGGARTTTAYGATAGA-3′) and stx2-2g_R1213 (5′-ATCCRGAGCCTGATKCACAG-3′). PCR conditions include a 10-min soak at 94°C and 35 cycles of 92°C or 1 min, 59°C for 30 sec, and 72°C for 1 min, followed by a 5-min soak at 72°C. Digestion with FokI at 37°C for 3 h yields banding patterns specific for stx2 (453 bp, 362 bp, 211 bp, and 126 bp) or stx2c (488 bp, 453 bp, and 211 bp). All bands from each pattern are visible in strains with both stx2 and stx2c.
Epidemiological Analyses.
We tested for differences in the frequency of clinical characteristics for Michigan patients using the likelihood χ2 test and described the distributions using odds ratios with 95% confidence intervals. Clade 9 was omitted from the analysis, as was one strain not part of a clade. To adjust for factors associated with infection by clade, we fit logistic regression models adjusting for age, gender, and symptoms. The final epidemiologic analysis was limited to 333 of the 444 Michigan patients, because only one strain from each outbreak or cluster was included.
Supplementary Material
Acknowledgments.
We thank C. Brenan, T. Morrison, and J. Hurley at Biotrove for the initial work with the OpenArray technology; K. Schaeffer, D. DeWitt, S. Tjugum-Holland, and K. Carr at Michigan State University; R. Madera, F. Pouch Downes, and others at the Michigan Department of Community Health; P. Tarr of Washington University; and the Wadsworth Center, New York State Department of Health, for supplying DNA from a 2006 lettuce outbreak. This project was supported in part by National Institutes of Health Contract N01-AI-30058 (to T.S.W.) and National Institutes of Health Grant AI049353 (to D.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The SNPs of E. coli O157 reported in this paper have been deposited in the STEC Center database, www.shigatox.net.
See Commentary on page 4535.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710834105/DC1.
References
- 1.Caprioli A, Morabito S, Brugere H, Oswald E. Enterohaemorrhagic Escherichia coli: Emerging issues on virulence and modes of transmission. Vet Res. 2005;36:289–311. doi: 10.1051/vetres:2005002. [DOI] [PubMed] [Google Scholar]
- 2.Mainil JG, Daube G. Verotoxigenic Escherichia coli from animals, humans and foods: Who's who? J Appl Microbiol. 2005;98:1332–1344. doi: 10.1111/j.1365-2672.2005.02653.x. [DOI] [PubMed] [Google Scholar]
- 3.Rangel JM, Sparling PH, Crowe C, Griffin PM, Swerdlow DL. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982–2002. Emerg Infect Dis. 2005;11:603–609. doi: 10.3201/eid1104.040739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CDC. Update: Multistate outbreak of Escherichia coli O157:H7 infections from hamburgers—western United States, 1992–1993. Morbid Mortal Wkly Rep. 1993;42:258–263. [PubMed] [Google Scholar]
- 5.CDC. Escherichia coli O157:H7 outbreak linked to commercially distributed dry-cured salami—Washington and California, 1994. Morbid Mortal Wkly Rep. 1995;44:157–160. [PubMed] [Google Scholar]
- 6.Hilborn ED, et al. A multistate outbreak of Escherichia coli O157:H7 infections associated with consumption of mesclun lettuce. Arch Intern Med. 1999;159:1758–1764. doi: 10.1001/archinte.159.15.1758. [DOI] [PubMed] [Google Scholar]
- 7.CDC. Multistate Outbreak of E. coli O157 Infections, November–December 2006. Atlanta: Centers for Disease Control and Prevention; 2006. [accessed 2007]. www.cdc.gov/ecoli/2006/december/121406.htm. [Google Scholar]
- 8.CDC. Ongoing multistate outbreak of Escherichia coli serotype O157:H7 infections associated with consumption of fresh spinach—United States, September 2006. Morbid Mortal Wkly Rep. 2006;55:1045–1046. [PubMed] [Google Scholar]
- 9.Mead PS, et al. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mead PS, Griffin PM. Escherichia coli O157:H7. Lancet. 1998;352:1207–1212. doi: 10.1016/S0140-6736(98)01267-7. [DOI] [PubMed] [Google Scholar]
- 11.Tarr PI, Gordon CA, Chandler WL. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet. 2005;365:1073–1086. doi: 10.1016/S0140-6736(05)71144-2. [DOI] [PubMed] [Google Scholar]
- 12.Reiss G, Kunz P, Koin D, Keeffe EB. Escherichia coli O157:H7 infection in nursing homes: Review of literature and report of recent outbreak. J Am Geriatr Soc. 2006;54:680–684. doi: 10.1111/j.1532-5415.2006.00682.x. [DOI] [PubMed] [Google Scholar]
- 13.Michino H, et al. Massive outbreak of Escherichia coli O157:H7 infection in schoolchildren in Sakai City, Japan, associated with consumption of white radish sprouts. Am J Epidemiol. 1999;150:787–796. doi: 10.1093/oxfordjournals.aje.a010082. [DOI] [PubMed] [Google Scholar]
- 14.Fukushima H, et al. Clinical experiences in Sakai City Hospital during the massive outbreak of enterohemorrhagic Escherichia coli O157 infections in Sakai City, 1996. Pediatr Int. 1999;41:213–217. doi: 10.1046/j.1442-200x.1999.4121041.x. [DOI] [PubMed] [Google Scholar]
- 15.Higami S, et al. Retrospective analysis of the relationship between HUS incidence and antibiotics among patients with Escherichia coli O157 enterocolitis in the Sakai outbreak. Kansenshogaku Zasshi. 1998;72:266–272. doi: 10.11150/kansenshogakuzasshi1970.72.266. (in Japanese) [DOI] [PubMed] [Google Scholar]
- 16.Ostroff SM, et al. Toxin genotypes and plasmid profiles as determinants of systemic sequelae in Escherichia coli O157:H7 infections. J Infect Dis. 1989;160:994–998. doi: 10.1093/infdis/160.6.994. [DOI] [PubMed] [Google Scholar]
- 17.Boerlin P, et al. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J Clin Microbiol. 1999;37:497–503. doi: 10.1128/jcm.37.3.497-503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jelacic JK, et al. Shiga toxin-producing Escherichia coli in Montana: Bacterial genotypes and clinical profiles. J Infect Dis. 2003;188:719–729. doi: 10.1086/376999. [DOI] [PubMed] [Google Scholar]
- 19.Persson S, Olsen KE, Ethelberg S, Scheutz F. Subtyping method for Escherichia coli shiga toxin (verocytotoxin) 2 variants and correlations to clinical manifestations. J Clin Microbiol. 2007;45:2020–2024. doi: 10.1128/JCM.02591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alland D, et al. Modeling bacterial evolution with comparative-genome-based marker systems: Application to Mycobacterium tuberculosis evolution and pathogenesis. J Bacteriol. 2003;185:3392–3399. doi: 10.1128/JB.185.11.3392-3399.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filliol I, et al. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: Insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J Bacteriol. 2006;188:759–772. doi: 10.1128/JB.188.2.759-772.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hazbon MH, Alland D. Hairpin primers for simplified single-nucleotide polymorphism analysis of Mycobacterium tuberculosis and other organisms. J Clin Microbiol. 2004;42:1236–1242. doi: 10.1128/JCM.42.3.1236-1242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, et al. Probing genomic diversity and evolution of Escherichia coli O157 by single nucleotide polymorphisms. Genome Res. 2006;16:757–767. doi: 10.1101/gr.4759706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kudva IT, et al. Strains of Escherichia coli O157:H7 differ primarily by insertions or deletions, not single-nucleotide polymorphisms. J Bacteriol. 2002;184:1873–1879. doi: 10.1128/JB.184.7.1873-1879.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohnishi M, et al. Genomic diversity of enterohemorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc Natl Acad Sci USA. 2002;99:17043–17048. doi: 10.1073/pnas.262441699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noller AC, et al. Multilocus sequence typing reveals a lack of diversity among Escherichia coli O157:H7 isolates that are distinct by pulsed-field gel electrophoresis. J Clin Microbiol. 2003;41:675–679. doi: 10.1128/JCM.41.2.675-679.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearson T, et al. Phylogenetic discovery bias in Bacillus anthracis using single-nucleotide polymorphisms from whole-genome sequencing. Proc Natl Acad Sci USA. 2004;101:13536–13541. doi: 10.1073/pnas.0403844101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hyma KE, et al. Evolutionary genetics of a new pathogenic Escherichia species: Escherichia albertii and related Shigella boydii strains. J Bacteriol. 2005;187:619–628. doi: 10.1128/JB.187.2.619-628.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayashi T, et al. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 30.Perna NT, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 31.Monday SR, Whittam TS, Feng PC. Genetic and evolutionary analysis of mutations in the gusA gene that cause the absence of beta-glucuronidase activity in Escherichia coli O157:H7. J Infect Dis. 2001;184:918–921. doi: 10.1086/323154. [DOI] [PubMed] [Google Scholar]
- 32.Rzhetsky A, Nei M. Theoretical foundation of the minimum-evolution method of phylogenetic inference. Mol Biol Evol. 1993;10:1073–1095. doi: 10.1093/oxfordjournals.molbev.a040056. [DOI] [PubMed] [Google Scholar]
- 33.Bryant D, Moulton V. Neighbor-net: An agglomerative method for the construction of phylogenetic networks. Mol Biol Evol. 2004;21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- 34.Feng P, Lampel KA, Karch H, Whittam TS. Genetic and phenotypic changes in the emergence of Escherichia coli O157:H7. J Infect Dis. 1998;177:1750–1753. doi: 10.1086/517438. [DOI] [PubMed] [Google Scholar]
- 35.Paton JC, Paton AW. Methods for detection of STEC in humans. An overview. Methods Mol Med. 2003;73:9–26. doi: 10.1385/1-59259-316-x:09. [DOI] [PubMed] [Google Scholar]
- 36.Riley LW, et al. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med. 1983;308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 37.FDA. Nationwide E. coli O157:H7 Outbreak: Questions & Answers. Rockville, MD: Food and Drug Administration; 2007. [accessed 2007]. www.cfsan.fda.gov/∼dms/spinacqa.html#howmany. [Google Scholar]
- 38.Delcher AL, Phillippy A, Carlton J, Salzberg SL. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strauch E, Schaudinn C, Beutin L. First-time isolation and characterization of a bacteriophage encoding the Shiga toxin 2c variant, which is globally spread in strains of Escherichia coli O157. Infect Immun. 2004;72:7030–7039. doi: 10.1128/IAI.72.12.7030-7039.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manning SD, et al. Surveillance for Shiga toxin-producing Escherichia coli (STEC) in Michigan, 2001–2005. Emerg Infect Dis. 2006;13:318–321. doi: 10.3201/eid1302.060813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robins-Browne RM. The relentless evolution of pathogenic Escherichia coli. Clin Infect Dis. 2005;41:793–794. doi: 10.1086/432725. [DOI] [PubMed] [Google Scholar]
- 42.Kim J, Nietfeldt J, Benson AK. Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc Natl Acad Sci USA. 1999;96:13288–13293. doi: 10.1073/pnas.96.23.13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noller AC, McEllistrem MC, Pacheco AG, Boxrud DJ, Harrison LH. Multilocus variable-number tandem repeat analysis distinguishes outbreak and sporadic Escherichia coli O157:H7 isolates. J Clin Microbiol. 2003;41:5389–5397. doi: 10.1128/JCM.41.12.5389-5397.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaikh N, Tarr PI. Escherichia coli O157:H7 Shiga toxin-encoding bacteriophages: Integrations, excisions, truncations, and evolutionary implications. J Bacteriol. 2003;185:3596–3605. doi: 10.1128/JB.185.12.3596-3605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.CDC. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 States, United States, 2005. Morbid Mortal Wkly Rep. 2006;55:392–395. [PubMed] [Google Scholar]
- 46.Schmidt H. Shiga-toxin-converting bacteriophages. Res Microbiol. 2001;152:687–695. doi: 10.1016/s0923-2508(01)01249-9. [DOI] [PubMed] [Google Scholar]
- 47.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 48.Besser TE, et al. Greater diversity of Shiga toxin-encoding bacteriophage insertion sites among Escherichia coli O157:H7 isolates from cattle than in those from humans. Appl Environ Microbiol. 2007;73:671–679. doi: 10.1128/AEM.01035-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steele M, et al. Identification of Escherichia coli O157:H7 genomic regions conserved in strains with a genotype associated with human infection. Appl Environ Microbiol. 2007;73:22–31. doi: 10.1128/AEM.00982-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim J, et al. Ancestral divergence, genome diversification, and phylogeographic variation in subpopulations of sorbitol-negative, beta-glucuronidase-negative enterohemorrhagic Escherichia coli O157. J Bacteriol. 2001;183:6885–6897. doi: 10.1128/JB.183.23.6885-6897.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinformatics. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 52.Huson DH. SplitsTree: Analyzing and visualizing evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 53.Swaminathan B, Barrett TJ, Hunter SB, Tauxe RV. PulseNet: The molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg Infect Dis. 2001;7:382–389. doi: 10.3201/eid0703.010303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghai R, Hain T, Chakraborty T. GenomeViz: Visualizing microbial genomes. BMC Bioinformatics. 2004;5:198. doi: 10.1186/1471-2105-5-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang W, Bielaszewska M, Friedrich AW, Kuczius T, Karch H. Transcriptional analysis of genes encoding Shiga toxin 2 and its variants in Escherichia coli. Appl Environ Microbiol. 2005;71:558–561. doi: 10.1128/AEM.71.1.558-561.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.