Abstract
Organelle transport to the periphery of the cell involves coordinated transport between the processive motors kinesin and myosin V. Long-range transport takes place on microtubule tracks, whereas final delivery involves shorter actin-based movements. The concept that motors only function on their appropriate track required further investigation with the recent observation that myosin V undergoes a diffusional search on microtubules. Here we show, using single-molecule techniques, that a functional consequence of myosin V's diffusion on microtubules is a significant enhancement of the processive run length of kinesin when both motors are present on the same cargo. The degree of run length enhancement correlated with the net positive charge in loop 2 of myosin V. On actin, myosin V also undergoes longer processive runs when kinesin is present on the same cargo. The process that causes run length enhancement on both cytoskeletal tracks is electrostatic. We propose that one motor acts as a tether for the other and prevents its diffusion away from the track, thus allowing more steps to be taken before dissociation. The resulting run length enhancement likely contributes to the successful delivery of cargo in the cell.
Keywords: actin, microtubule, molecular motor, Qdot, electrostatic
Movement of membrane-bound organelles from the center of the cell to its periphery and back involves the interplay between processive molecular motors on both microtubule (MT) and actin tracks. In amphibian melanophores, where pigment granules disperse and aggregate to cause skin color changes, long-range transport toward the plus-end of microtubules is carried out by kinesin-2, whereas myosin V (myoV) takes over in the actin-rich cell cortex to deliver melanosomes to the cell periphery during dispersion (1). This process requires switching between cytoskeletal tracks. We have shown, at the single-molecule level, that myoV could effectively navigate actin–actin intersections that normally exist within cells, either by executing a turn, or by stepping over the crossing actin filament (2). The frequency of these two events appeared to be related to myoV's inherent flexibility and the availability of actin-binding sites on the intersecting filaments that were within reach of the leading head. Surprisingly, myoV could also undergo a diffusional search on MTs, resulting from an electrostatic interaction between the myoV head and the negatively charged tubulin E-hook. We hypothesized that this diffusive process helps myoV find cargo that is undergoing MT-based movement, and/or facilitates the binding of myoV to kinesin if the two motors directly interact (3).
Can myoV's interaction with the MT affect kinesin's processivity when both motors are present on the same cargo? Here we show that this interaction enhances the processivity of kinesin-based cargo transport. We also provide evidence that the structural element on myoV that acts as an electrostatic tether is loop 2, a charged surface loop that also influences myoV's processive run length on actin (4). On actin, kinesin also enhances the run length of myoV. We propose that both motors act as tethers for each other when bound to the same cargo, preventing dissociation from the track when a run would normally terminate. In this case, the tether is a motor that interacts with the heterologous track. Binding partners such as dynactin have been shown to increase the processivity of dynein and kinesin-2 (5–7), but this is the first example of two motor proteins enhancing each other's processivity.
Results
Myosin V Enhances Kinesin Processivity.
A model in vitro system was used to investigate functional interactions between kinesin and myoV when both motors are present on the same cargo. MTs and/or actin were adhered to a glass coverslip. Quantum dots (Qdots), which are readily visualized in an objective-type total internal reflectance microscope, acted as “cargo” to which the motors were bound (Fig. 1A). Full-length kinesin adopts an inhibited, folded conformation at low salt in solution, but is activated by binding to carboxylated-Qdots, presumably by abolishing the motor domain-tail interactions that stabilize the inhibited state (8–10). To ensure that Qdot movement on MTs was being driven by a single kinesin motor, the Qdot to kinesin molar ratio was 16:1, where the characteristic run length was 1.7 ± 0.1 μm (Table 1). At this molar ratio, statistically ≈95% of moving Qdots should have only a single motor bound (4). Full-length myoV is also in a folded inhibited conformation at low salt that is not capable of processive motion (11). Full-length myoV was also activated when attached to carboxylated Qdots, and moved processively on actin filaments (data not shown) and diffused on MTs (Fig. 1B) with a diffusion coefficient of 0.18 ± 0.16 (SEM) μm2/s (n = 30), similar to that described in ref. 2 for myoV-HMM.
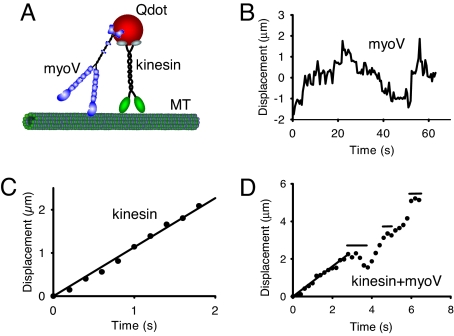
Fig. 1.
Motor movement on microtubules. (A) Cartoon of the experimental setup. Microtubules (MT) are adhered to a glass coverslip. Full-length constructs of myoV, kinesin, or both motors are attached to carboxylated Qdots (see Methods), and visualized by in an objective-type total internal reflectance microscope. Experiments with constitutively active constructs of myoV and kinesin, attached to streptavidin-Qdots via a C-terminal biotin tag, were also performed. (B) One-dimensional diffusion of myoV on microtubules with no net displacement. (C) Smooth movement routinely observed when kinesin alone is attached to the Qdot. Data obtained at the onset of movement when the Qdot appeared on the microtubule until its disappearance when kinesin terminated its run. The velocity for this kinesin molecule was 1.13 μm/s, estimated from the linear regression. Images were captured at 5 frames/s. (D) New behavior seen when both kinesin and myoV are attached to a Qdot (data are from SI Movie 1). For kinesin/myoV-labeled Qdots, numerous runs were characterized by periods of smooth directed movement, as highlighted by the regression through the first 3 s, interspersed with diffusive searches identified by horizontal lines. The periods of smooth movement were characteristic of the movement seen with Qdots labeled only with kinesin (as in C), having velocities (0.80 μm/s for the first 3 s of this run based on regression) comparable to that of kinesin alone (see Table 1). Data obtained at the onset of movement when the Qdot appeared on the microtubule until its disappearance when the run terminated. Note that the scales are different when comparing the Qdot movement here which has both kinesin and myoV attached to that of a Qdot labeled solely with kinesin in C. Image capture rate of 5 frames/s.
Table 1.
Processive run length and velocity of different combinations of motors and tracks
| Kinesin | myoV | Track | Run length, μm | Velocity, μm/s | n | Type of processive events analyzed |
|---|---|---|---|---|---|---|
| + | − | MT | 1.7 ± 0.1 | 0.88 ± 0.20 | 61 | All runs |
| + | + | MT | 3.7 ± 0.3* | 0.73 ± 0.30* | 74 | All runs |
| + | + | MT | 7.1 ± 1.7* | 0.55 ± 0.15* | 21 | Only runs that had obvious diffusive events interspersed |
| + | + | MT | 1.8 ± 0.2 | 0.84 ± 0.13 | 54 | Analysis of run segments between diffusive events |
| − | + | Actin | 0.76 ± 0.05 | 0.46 ± 0.15 | 208 | All runs |
| + | + | Actin | 1.09 ± 0.05# | 0.42 ± 0.15 | 299 | All runs |
Run length values are mean and standard errors from exponential fits to the data (see Materials and Methods). Velocity is the mean and standard deviation. n is the number of processive runs. Statistical significance at P < 0.05 was determined using the Kolmogorov–Smirnov Test for run length comparisons and the Student's t-Test for velocity comparisons to kinesin on MT (*) and to myoV on actin (#). On MT tracks, the ratio of Qdot:myoV:kinesin was 16:1:32. On actin tracks, data for two ratios of Qdot:kinesin:myoV with the same run lengths were pooled.
To obtain Qdots with both motors bound, Qdots and kinesin were first mixed in a 16:1 molar ratio, followed by addition of excess myoV (see Methods). When both actin filaments and MTs were adhered to a glass surface, these Qdots were observed to switch between and travel along these two cytoskeletal tracks, confirming that both motors were present on the same cargo (data not shown). Strikingly, the average run length of kinesin in the presence of myoV was increased to 3.7 ± 0.3 μm, a 2.2-fold increase over the run length of kinesin alone (Fig. 2A and Table 1). Processivity was enhanced to a lesser extent as the salt concentration was increased, indicating that the enhancement process was electrostatic in nature [see supporting information (SI) Table 2]. In the presence of myoV, the average velocity was decreased to 83% the value obtained with kinesin alone (Fig. 2B and Table 1).
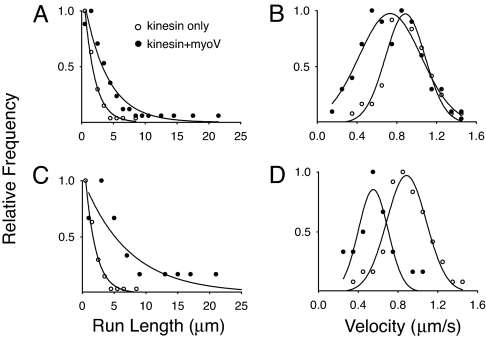
Fig. 2.
Run length (A and C) and velocity (B and D) distributions on microtubules. (A and B) Data obtained when Qdots were incubated with both kinesin and myoV (filled circles), or with kinesin alone (open circles). (C and D) Analysis of the subpopulation of runs (22% of total) that included obvious diffusive events or pauses. The run length and velocity of the entire run (filled circles), are compared with the run length and velocity of kinesin alone (open circles, as in A and B). The relative frequencies were determined by using the total number of runs for a given experimental condition as reported in Table 1.
An alternative attachment strategy to the Qdot was used with constitutively active truncated constructs of both kinesin (kinesin-ΔC) and myoV (myoV-HMM). Both constructs contained a C-terminal biotin tag for attachment to streptavidin-Qdots (SA-Qdots) (see Methods). This approach removes the possibility that the globular tail of myoV could bind to microtubules (12) or to kinesin (3). The run length of kinesin-ΔC in the presence of myoV-HMM was 1.8-fold higher (2.8 ± 0.2 μm) than kinesin-ΔC alone (1.6 ± 0.1 μm) at a molar ratio of Qdot:kinesin:myoV = 16:1:32 (SI Table 3), similar to the degree of enhancement observed with the full-length constructs.
Myosin V Causes Diffusive Events and Pauses During Processive Runs by Kinesin.
Closer inspection of the data revealed that a subpopulation of Qdots (22%) showed occasional diffusive events and pauses interspersed during a processive run (Fig. 1D and SI Movie 1), behaviors not observed with kinesin alone (Fig. 1C). Simultaneous visualization of both the MT track and the Qdot showed that the run interruptions occurred on single MTs, and were not due to MT–MT intersections or to MT bundles acting as roadblocks or opportunities for switching between tracks. Processive runs that showed clear interruptions or diffusive events had characteristic run lengths of 7.1 ± 1.7 μm, 4.2-fold longer than that of kinesin alone (Fig. 2C and Table 1). The run length of individual segments between pauses and diffusional events were not statistically different (P > 0.05) than those observed with kinesin alone (Table 1). On average, there were two to three apparent interruptions per run, suggesting that the total distance traveled was the sum of three or more individual runs. As many as six diffusive searches/pauses were observed within a single run, and the maximum distance traveled by a Qdot was 22 μm. The observation of diffusive events during Qdot transport by kinesin strongly suggests that myoV is involved in prolonging kinesin's effective run length.
The average velocity over such runs was 63% that of kinesin alone (Fig. 2D and Table 1), because the diffusive events increase the time spent on the MT without contributing to net movement. Although the diffusion coefficient of a Qdot with both motors bound (0.11 ± 0.12 μm2/s, n = 28) was similar to that with only myoV bound, the average lifetime of the diffusive events was 4.5 ± 0.5 s (n = 42), 11 times shorter than the diffusive events of myoV alone on MTs (50 ± 8 s, n = 43) (2), suggesting that kinesin ends the diffusive event as it begins another processive run. The velocity during segments of directed movement was virtually identical to that of kinesin alone (Table 1), implying that myoV's interaction with the MT does not exert a significant drag force. Alternatively, spatial constraints allow only one motor to interact with the MT at any point in time, which may explain the clear distinction between processive and diffusive events for this subpopulation of runs.
Net Positive Charge in Loop 2 of Myosin V Affects the Degree of Run Length Enhancement.
We previously proposed that the positively charged loop 2 of myoV is the structural element that electrostatically interacts with the negatively charged E-hook of tubulin (2). To test whether loop 2 also serves as a tether to enhance kinesin's processivity, we assessed the effect of two previously characterized myoV loop 2 charge mutants on kinesin run length (4). Net positive charge in loop 2 was increased by three in the mutant “Plus8,” and decreased by three in the mutant “AAA” compared with the wild-type control (WT) (see Methods). The myoV-HMM constructs were attached via a biotin tag at their C terminus to an SA-Qdot. The relative binding affinity of myoV-HMM for the MT was assessed by counting the number of Qdots bound per μm of MT (>400 μm MT per construct). Plus8 had a higher affinity for the MT than WT (≈1.4-fold), whereas the AAA mutant had a lower affinity (one-third that of WT). The diffusion constants also varied with the net positive charge of loop 2 (Plus8, 0.17 μm2/s; WT, 0.24 μm2/s; and AAA, 0.34 μm2/s). The lifetime of MT-myoV association was longer for the Plus8 mutant (57 ± 6 s) than for AAA (38 ± 5 s). Importantly, the processive run length of kinesin-ΔC was enhanced 94% by Plus8, 75% by WT, and only 25% by AAA (see SI Table 3). The enhancement of kinesin's run length is clearly dependent on the net positive charge in loop 2 of myoV, strongly suggesting that this structural element is responsible for the electrostatic interaction between myoV and the MT.
Kinesin Enhances Myosin V Processivity.
Does kinesin return the favor and enhance the processivity of myoV on actin? MyoV-HMM and kinesin-ΔC were attached to SA-Qdots via a C-terminal biotin tag (Fig. 3A). The characteristic run length of myoV-HMM alone was 0.76 ± 0.05 μm (Table 1), in good agreement with results obtained by directly visualizing single molecules of myoV-HMM-YFP (11), thus ensuring that we were observing single motor transport. When SA-Qdots were first mixed with myoV-HMM, and then with kinesin-ΔC (see Methods), the run length of the complex increased significantly (P < 0.05) to 1.1 ± 0.05 μm, a 1.4-fold increase over the run length of myoV-HMM alone (Fig. 3B and Table 1). Control experiments with nonbiotinated kinesin showed no increase in run length, suggesting that kinesin needs to be bound to the same SA-Qdot as myoV-HMM to exert its effect on processivity. Additional controls with biotinated BSA showed no increase in run length, suggesting that not all proteins can produce run length enhancement. As the salt concentration was increased, the kinesin-dependent enhancement of the myoV-HMM run length decreased (Fig. 3B), suggesting that kinesin interacts electrostatically with the actin filament to prolong the myoV-HMM run length. Unlike run length, the velocity of myoV-HMM was almost unchanged in the presence of kinesin, independent of ionic strength (Table 1 and SI Table 2). The movement trajectories of myoV-HMM in the presence of kinesin are smooth, and show no signs of stops and starts, indistinguishable from those with myoV-HMM alone (data not shown).
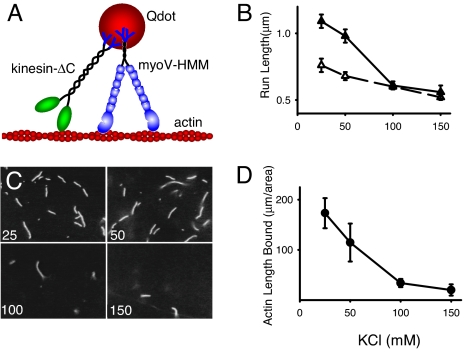
Fig. 3.
Processive movement on actin filaments. (A) Cartoon of the experimental setup. Actin filaments are adhered to a glass coverslip. Constitutively active constructs of myoV-HMM and kinesin-ΔC, both of which contain a biotinated tag at their C terminus, are attached to streptavidin-Qdots (see Methods), and visualized by in an objective-type total internal reflectance microscope. (B) Run lengths as a function of KCl concentration for myoV-HMM alone (open triangles) and for myoV in the presence of kinesin-ΔC (filled triangles). Run lengths at both 25 and 50 mM KCl in the presence of kinesin were statistically different (P < 0.05) from those in the absence of kinesin, using the Kolmogorov-Smirnov Test. (C) Raw images obtained of actin bound to a kinesin-coated glass coverslip at varying KCl concentration (concentration in mM KCl in lower left corner). (D) Average total length of actin bound per 8,700 μm2 of a kinesin-coated glass coverslip, as a function of KCl concentration.
If kinesin must be attached to the Qdot to enhance myoV-HMM processivity, then presumably this enhancement is brought about by kinesin interacting with the actin filament. Even though we did not observe kinesin bound to SA-Qdots diffusing on actin, we could demonstrate a weak electrostatic interaction between kinesin and actin by inverting the assay, an approach used previously to demonstrate an interaction between actin and myosin with weak-binding affinity (13). Multiple kinesins were adhered to a glass coverslip, and the amount of actin that bound to the surface quantified (see Methods). When the surface was blocked with BSA, actin filaments did not bind, but with kinesin on the surface, actin filaments bound in a salt-dependent manner, the extent of which paralleled the enhancement of myoV-HMM run length (Fig. 3 B–D). This result suggests that a weak electrostatic interaction between kinesin and actin is responsible for the increase of myoV run length.
Effect of Kinesin on the Processivity of Myosin V Mutants.
If myoV run length enhancement is due to the nonspecific, electrostatic interaction between kinesin-ΔC and actin filaments, then this enhancement should be observed with other processive myosin motors. We tested two myosin V mutants, one which is less processive than WT myoV-HMM (AAA), and one that is more processive than WT (Plus8) (4). As expected, the characteristic run length of the AAA mutant showed a 1.6-fold increase in the presence of kinesin (SI Table 4), whereas velocity was unaffected, similar to that of WT. Interestingly, the run length of Plus8 was unchanged in the presence of kinesin (SI Table 4). The run length of Plus 8 alone, which is greater than that of WT, may reflect an already optimized rebinding of the lead head, which is not helped further by the tethering kinesin. The mechanism by which kinesin enhances myosin V's processivity on actin may therefore differ somewhat from the joining together of multiple runs proposed for the enhancement of kinesin run length by myoV.
Discussion
Here we show that having both myosin V and kinesin present on the same cargo provides a distinct biological advantage, namely a significant increase in processive run lengths. This is the first observation of two different classes of motor proteins enhancing each others' processivity by acting as partners and taking turns being an electrostatic tether on a heterologous track (Fig. 4). As a tether, the passive motor prevents diffusion of the cargo away from the track, thus allowing the active, processive motor (either myoV or kinesin) to continue moving forward instead of dissociating from its track. We also provide evidence that the electrostatic interaction between myoV and MTs is mediated by positively charged residues in loop 2 of myoV and the negatively charged E-hook of tubulin (2).
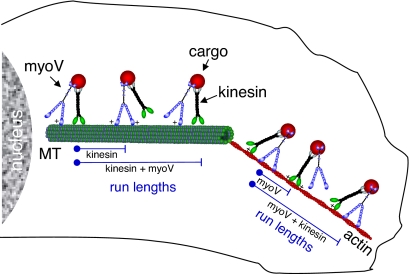
Fig. 4.
Model illustrating how myoV and kinesin act as molecular tethers for each other to enhance processivity inside the cell. MyoV and kinesin are bound to the same cargo. Through an electrostatic interaction (+) between myoV and the microtubule (MT), the run length for kinesin is extended beyond the point at which kinesin alone would have terminated its run. This effective tether contributes to an enhancement of kinesin's run length. The reciprocal mechanism is illustrated on actin with kinesin serving as the tether for myoV.
For kinesin, the tradeoff for enhanced run length was a decreased speed over the length of the run (Fig. 2 B and D). The effectively slower speed for kinesin in the presence of myoV was accounted for by the extra time spent during the diffusive events of myoV, which generate no net movement (Fig. 1 B and D). The myoV-dependent effect on kinesin's velocity may in part explain why the speed of melanosome movement by kinesin on MTs in vivo increased when myoV function was blocked (1). In contrast, for the reciprocal partnership, kinesin's interaction with actin did not affect myoV's speed of transport but did enhance its run length, suggesting that the underlying mechanism may be different. Based on the force:velocity relationship for myoV and for kinesin (14, 15), the electrostatic interaction between the tethering motor and its heterologous track must be relatively weak and does not create an effective drag force >1 pN, which would be required to slow the movement of the active motor.
Alternative explanations for the run length enhancement were considered. In principle, the two motors could interact directly (3), modulating the attachment and detachment kinetics of the active motor so that it becomes more processive. However, we have not been able to demonstrate a direct interaction between the two full-length proteins in the present assay (SI Text). Alternatively, the increase in size of the complex when two motors bind a Qdot could reduce its free diffusion, allowing the motor to reattach to the track before the motor–cargo complex can escape. This would result in higher processivity as proposed for single-headed myosin VI when attached to a 200-nm polystyrene bead (16). The ionic strength dependence of the process argues against this potential explanation.
Although kinesin presumably interacts electrostatically with actin, the significantly lower surface charge density presented by the smaller 7-nm-diameter actin compared with the larger 25-nm MT may explain why a single kinesin does not diffuse along an actin filament. However, we do show that multiple kinesins can effectively capture an actin filament through an interaction that is ionic strength dependent (Fig. 3 C and D). This implies that during a processive run, transient electrostatic interactions between a positively charged kinesin domain and negatively charged patches in subdomain-1 of actin (17) hold myoV close to the actin filament, reducing the rate of dissociation. This effect was more prominent for motors that have a weaker initial interaction with actin than for those motors that already interact strongly with actin, such as a loop 2 mutant engineered to have increased positive charge. Additionally, because the interaction between kinesin and actin is weaker than the interaction between myoV and the MT, the run length enhancement is less on an actin track than on a MT.
This concept is attractive because any cargo that myoV or kinesin carries that has a charged domain can in principle increase processivity, even without a specific interaction with the track. For example, the diffusive search on MTs is not specific to myoV, because this also occurs with a smooth muscle myosin head (2). Run length enhancement of kinesin was also observed when smooth muscle HMM was attached to the same Qdot (SI Table 3). In fact, molecular motors that have relatively low processivity in vitro may have much higher processivity in vivo when carrying a suitable cargo and/or partnering with another class of motor, or with the same motor in an inhibited state.
The potential for the negatively charged E-hook at the C terminus of tubulin to act as a highly charged domain that contributes to either one dimensional diffusive searches (2, 18–20) or processivity for various MT-based motors has been demonstrated previously (21, 22). Non-motor proteins have also been shown to act as tethers, most notably dynactin, which enhances the processivity of both dynein (7) and kinesin-2 (5, 23). Given the large and still growing number of proteins, or protein domains, that act as electrostatic tethers, it is likely that such a common mechanism will be pertinent to in vivo function involving processive motion or protein targeting (reviewed in refs. 6 and 24). Here we add a twist to the story by showing that two motors can assist each other by acting as a tether when present on the heterologous cytoskeletal track.
Because these tethers act via charge interactions, do they exist within the cell? Although high salt concentration was used to weaken the electrostatic attraction, we and others (22) have shown that the use of acetate instead of chloride, which is a closer mimic of the physiologic anionic environment (25), allows enhanced processivity to persist at higher ionic strengths (SI Table 2). The extent to which the high internal concentration of macromolecules and the constraints of cell architecture can influence these interactions is also not yet fully understood (26).
It is reasonable that both myoV and kinesin can bind to the same cargo in vivo. Frog melanosomes have been estimated to bind >60 myoV motors, and switching of melanosomes and endocytic vesicles from MT to actin tracks has been observed, implying that both motors are present (1, 27, 28). Our observation of kinesin and myoV enhancing each others' function via an electrostatic tethering mechanism may well be one of several ways to ensure successful delivery of cargo from nucleus to cell periphery. Multiple motors of the same type can also increase processive run length (29), and so depending on the number and type of motors recruited to a particular organelle, and their spatial distribution on that organelle, various mechanisms could alternately contribute to continuing a processive run. These features that enhance processivity may be required within the cell to counteract the crowded and more viscous intracellular milieu, as well as retarding forces that might be acting on the organelle.
Materials and Methods
Protein Expression and Purification.
The full-length murine myoV was the construct used in (30). MyoV-HMM was truncated at amino acid 1098 and contained a biotin tag at the C terminus, followed by a FLAG tag to facilitate purification. The biotin tag is an 88 aa sequence segment from the Escherichia coli biotin carboxyl carrier protein (31), which is biotinated at a single lysine during expression in Sf9 cells. This tag was used for attachment to streptavidin-conjugated Qdots. The myoV constructs were coexpressed in Sf9 cells with calmodulin and purified as described in ref. 30. The loop 2 charge mutants used here were characterized as described in ref. 4. For the AAA mutant, three lysines in the C-terminal region of loop 2 were changed to alanines (K629A, K632A, and K633A). For Plus8, two threonines were changed to lysines and a glutamic acid was changed to glutamine (T605K, T627K, and E630Q). Like WT, the loop 2 mutants contain a C-terminal biotin tag. The Drosophila melanogaster kinesin heavy chain (GenBank accession number AF053733) and light chain gene (GenBank accession number AF055298) (both generously provided by Will Hancock, Pennsylvania State University, State College, PA) were used as PCR templates for cloning each separately into the baculovirus transfer vector pAcSG2 (BD Biosciences). The full-length kinesin heavy chain gene was cloned with a C terminus hexa-HIS tag, as was the kinesin light chain. A truncated version of the kinesin heavy chain (kinesin-ΔC), ending at Ala910, was cloned with a biotin-tag for attachment to streptavidin Qdots, followed by a FLAG epitope at its C terminus to facilitate purification. Recombinant baculovirus was prepared by standard protocols (32).
For full-length kinesin, Sf9 cells were infected with recombinant baculovirus coding for HIS-tagged kinesin heavy chain, and grown in suspension for 72 h. Cells were sonicated in buffer containing 10 mM sodium phosphate, pH 7.5, 0.3 M NaCl, 0.5% glycerol, 7% sucrose, 2 mM 2-mercaptoethanol, 0.5 mM AEBSF, 5 μg/ml leupeptin, and 5 mM benzamidine. The cell lysate was clarified at 200,000 × g for 30 min, and the supernatant applied to a HIS-Select nickel affinity column (Sigma-Aldrich) at a flow rate of 0.5 ml/min. The resin was washed first with buffer A (10 mM sodium phosphate, 10 mM imidazole, pH 7.5, 0.3 M NaCl, 0.5 mM AEBSF, 5 μg/ml leupeptin and 5 mM benzamidine), and then with buffer A containing 30 mM imidazole. Kinesin was eluted from the column with 10 mM sodium phosphate, 200 mM imidazole, pH 7.5, 0.3 M NaCl, and 1 μg/ml leupeptin. The fractions of interest were combined and concentrated by using an Amicon centrifugal filter device (Millipore), and dialyzed in 10 mM Hepes, pH 7.3, 200 mM NaCl, 50% glycerol, 1 mM DTT, 10 μM MgATP and 1 μg/ml leupeptin for storage at −20°C.
For the constitutively active truncated kinesin, Sf9 cells were coinfected with recombinant baculovirus coding for kinesin-ΔC heavy chain with biotin and FLAG tag at the C terminus and HIS-tagged light chain, and grown in suspension for 72 h. Cells were sonicated in buffer containing 10 mM imidazole, pH 7.0, 0.3 M NaCl, 1 mM EGTA, 5 mM MgCl2, 7% sucrose, 2 mM 2-mercaptoethanol, 0.5 mM AEBSF, 5 μg/ml leupeptin, and 5 mM benzamidine. The cell lysate was clarified at 200,000 × g for 30 min, and the supernatant was applied to a FLAG affinity resin column (Sigma-Aldrich) at a flow rate of 0.5 ml/min. The resin was washed with buffer containing 10 mM imidazole, pH 7.0, 0.3 M NaCl, 1 mM EGTA, 1 mM NaN3, 1 mM DTT and 1 μg/ml leupeptin. Kinesin was eluted from the column by using a 0.1 mg/ml solution of Flag peptide in wash buffer. The fractions of interest were combined and concentrated with an Amicon centrifugal filter device (Millipore) and dialyzed in 10 mM Hepes, pH 7.3, 200 mM NaCl, 50% glycerol, 1 mM DTT, 10 μM MgATP and 1 μg/ml leupeptin for storage at −20°C.
Microtubule Preparation.
Rhodamine-labeled and unlabeled tubulin (Cytoskeleton) were mixed at 1:5 ratio, and polymerized as described by the vendor. To prepare biotinated microtubules, Rhodamine-labeled tubulin, unlabeled tubulin and biotinated tubulin were mixed at a molar ratio to 1:4:1. Microtubules were stabilized with 5 μM taxol.
Buffers.
Buffers used in the subsequent sections are defined as: Buffer A (25 mM imidazole, pH 7.4, 4 mM MgCl2, 1 mM EGTA, 25 mM KCl, 10 mM DTT); Buffer B (25 mM imidazole, pH 7.4, 4 mM MgCl2, 1 mM EGTA, 0.3 M KCl, 10 mM DTT); Buffer C (Buffer A plus 1 mg/ml BSA, 1 mM MgATP, and an oxygen scavenging system composed of 0.1 mg/ml glucose oxidase, 0.02–0.18 mg/ml catalase, and 3.0 mg/ml glucose).
Protein Attachment to Qdots.
Carboxylated Qdots, emitting at 655 nm (Invitrogen-Molecular Probe, Eugene, OR), were attached to kinesin by incubating a 16:1 or 8:1 mixture of Qdots/kinesin (kinesin concentration was 12–22 nM) for 10 min at room temperature in Buffer B with oxygen scavengers and 0.1 mg/ml BSA. A similar binding strategy between carboxylated beads and motors was used previously (15, 33–35). To attach both motors to a Qdot, myoV was added to the preformed Qdot-kinesin complex for 10 min at room temperature to generate molar ratios of Qdot/kinesin/myoV = 16:1:32 or 8:1:16. Excess myoV was added to ensure that most kinesin–Qdots contained at least one myoV molecule. The mixture was further diluted to a final kinesin concentration of 0.1–0.2 nM before infusion into the flow cell.
To form the myoV–streptavidin Qdot complex, biotinated myoV-HMM (WT or mutants) (12 nM) was incubated with streptavidin 655 Qdot (200 nM) for 10 min at room temperature in Buffer B. To attach both motors, kinesin was added to the preformed myoV-streptavidin Qdot complex and incubated for 5 min at room temperature (ratio of 66.7 nM Qdots:4.2 nM myoV-HMM:133 nM kinesin for most experiments). The mixture was further diluted to a final myoV concentration of 0.2 nM before infusion into the flow cell.
Processivity Assay.
For experiments in which the cytoskeletal track was MTs, the following solutions were introduced into a 10 μl flow-cell. N-ethyl maleimide (NEM)-modified myosin (0.5 mg/ml) in Buffer B for 2 min, wash with Buffer A, incubate with 1 mg/ml BSA in Buffer A for 2 min, followed by a wash with Buffer A. Fluorescent microtubules (0.6 μM) (1 mM MgCl2, 80 mM Pipes (piperazine-1,4-bis(2-ethanesulfonic acid), pH 6.9, 1 mM EGTA, and 5 μM taxol) were infused into the flow-cell chamber and incubated for 2 min, followed by a wash with room temperature Buffer A. Qdots bound to kinesin alone or to both motors were then added to the flow cell at 0.1–0.2 nM kinesin in Buffer C. MyoV concentration in the flow cell varied from 0.8–1.6 nM. Alternatively, MTs were adhered via a biotin-avidin linkage because MTs were not tightly bound to NEM or the glass surface at 100 mM KCl or 100 mM potassium acetate. Therefore, biotinated BSA (2 mg/ml) in Buffer A was infused and incubated for 2 min, washed with Buffer A, incubated with 2 mg/ml neutravidin (Buffer A) for 2 min, followed by a wash with Buffer A. Fluorescent biotinated MTs were flowed in and incubated for 2 min, followed by a wash with Buffer A. Subsequent steps are as described above.
For experiments in which the cytoskeletal track was actin, flow cells were first incubated with 0.1 mg/ml NEM-modified myosin in Buffer B for 2 min, rinsed with Buffer A, incubated with 0.5 μM TRITC-phalloidin labeled actin filaments in Buffer A for 2 min, rinsed with Buffer A, and then incubated with 5 mg/ml BSA in Buffer A for 2 min. The Qdot-motor complexes were diluted with Buffer C (plus 0.1 mg/ml CaMΔall; ref. 30), to a final myoV concentration of 0.2 nM before infusion into the flow cell.
Data Acquisition, Image and Data Analysis.
Single motor processivity assays were performed at room temperature (25 ± 1°C) on a Nikon TE2000-U microscope equipped with a PlanApo objective lens (100×; numerical aperture, 1.45) for through-the-objective TIRF microscopy. A software-controlled filter wheel was used to switch between Qdot and microtubule or actin emission filters at 2 or 5 frames/s. Qdots were excited with the 488-nm argon laser line. Single-color images were obtained by using a DVC-1412:GenIV Intensified high-resolution 12-bit digital camera (DVC Company, Austin, TX) at 166-ms integration time. Images were processed using QED In Vivo software (Media Cybernetics, Silver Spring, MD). Typically, 300 or 500 images per color were recorded for a total of 150 or 100 s, respectively.
Microtubule-based movement, i.e., run length and velocity, were measured by using a manual object tracking plugin, MTrack J, for Image J 1.34s digital image processing software (National Institutes of Health, Bethesda). Using this program, we set a 15 × 15 pixel window (55 nm/pixel) over the Qdot image after which the program determined the “bright centroid” based on image intensity with 20 nm resolution. Run lengths >0.3 μm were recorded, whereas velocities were calculated as the run length/time of run. For actin-based movement, the image sequence was processed as described in ref. 11. Run length distribution histograms were fit with p(x) = Ae(−x/λ), to determine the characteristic run length λ, where p(x) is the relative frequency of the motor traveling a distance x along a track and A is a constant. Statistical comparison of run lengths were performed by using the Kolmogorov–Smirnov Test, a nonparametric distribution and bin width independent statistical measure (36).
Diffusion constants and lifetimes for myoV-based diffusional search components on microtubules were measured as described in ref. 2.
Kinesin Binding to Actin.
Kinesin (20 μl of 0.1 mg/ml) in buffer A was flowed into a nitrocellulose coated cell for 1 min, and then rinsed with buffer C. Twenty microliters of 0.5 μM rhodamine—phalloidin-labeled actin was flowed in and allowed to bind for 1 min. Unbound actin was removed by washing twice with buffer A containing different KCl concentrations. The actin was visualized on an inverted microscope (Zeiss Axiovert 10) equipped for epifluorescence with a 100-watt mercury lamp and rhodamine filter set. A high numerical aperature objective (63X Zeiss planapochromat, N.A. 1.4) and image intensified CCD camera (Hamamatsu C2400–80) were used to record actin images on video tape (Panasonic S-VHS AG-7300 VCR). For each salt concentration, at least six 108 × 81 μm2 fields were recorded. The images were then digitized and imported into ImageJ (NIH). The lengths of actin filaments in an individual field were manually measured in ImageJ and added to get the total actin length. Data from all fields of view were then averaged for each salt concentration.
Supplementary Material
Acknowledgments.
We thank Sam Walcott for his modeling efforts, Neil Kad, Ned Debold, Alex Hodges and Susan Lowey for helpful discussions, Guy Kennedy (UVM Instrumentation and Model Facility) for his optomechanical expertise, Samantha Beck for technical assistance, and Elena Krementsova for expression of myoV constructs. This work was supported by National Institutes of Health Grants GM078097 (to K.M.T.) and HL059408 and HL085489 (to D.M.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711531105/DC1.
References
- 1.Gross SP, Tuma MC, Deacon SW, Serpinskaya AS, Reilein AR, Gelfand VI. Interactions and regulation of molecular motors in Xenopus melanophores. J Cell Biol. 2002;156:855–865. doi: 10.1083/jcb.200105055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali MY, Krementsova EB, Kennedy GG, Mahaffy R, Pollard TD, Trybus KM, Warshaw DM. Myosin Va maneuvers through actin intersections and diffuses along microtubules. Proc Natl Acad Sci USA. 2007;104:4332–4336. doi: 10.1073/pnas.0611471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang JD, Brady ST, Richards BW, Stenolen D, Resau JH, Copeland NG, Jenkins NA. Direct interaction of microtubule- and actin-based transport motors. Nature. 1999;397:267–270. doi: 10.1038/16722. [DOI] [PubMed] [Google Scholar]
- 4.Hodges AR, Krementsova EB, Trybus KM. Engineering the processive run length of Myosin V. J Biol Chem. 2007;282:27192–27197. doi: 10.1074/jbc.M703968200. [DOI] [PubMed] [Google Scholar]
- 5.Berezuk MA, Schroer TA. Dynactin enhances the processivity of kinesin-2. Traffic. 2007;8:124–129. doi: 10.1111/j.1600-0854.2006.00517.x. [DOI] [PubMed] [Google Scholar]
- 6.Kincaid MM, King SJ. Motors and their tethers: the role of secondary binding sites in processive motility. Cell Cycle. 2006;5:2733–2737. doi: 10.4161/cc.5.23.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King SJ, Schroer TA. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat Cell Biol. 2000;2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- 8.Coy DL, Hancock WO, Wagenbach M, Howard J. Kinesin's tail domain is an inhibitory regulator of the motor domain. Nat Cell Biol. 1999;1:288–292. doi: 10.1038/13001. [DOI] [PubMed] [Google Scholar]
- 9.Friedman DS, Vale RD. Single-molecule analysis of kinesin motility reveals regulation by the cargo-binding tail domain. Nat Cell Biol. 1999;1:293–297. doi: 10.1038/13008. [DOI] [PubMed] [Google Scholar]
- 10.Hackney DD, Stock MF. Kinesin's IAK tail domain inhibits initial microtubule-stimulated ADP release. Nat Cell Biol. 2000;2:257–260. doi: 10.1038/35010525. [DOI] [PubMed] [Google Scholar]
- 11.Lu H, Krementsova EB, Trybus KM. Regulation of myosin V processivity by calcium at the single molecule level. J Biol Chem. 2006;281:31987–31994. doi: 10.1074/jbc.M605181200. [DOI] [PubMed] [Google Scholar]
- 12.Cao TT, Chang W, Masters SE, Mooseker MS. Myosin-Va binds to and mechanochemically couples microtubules to actin filaments. Mol Biol Cell. 2004;15:151–161. doi: 10.1091/mbc.E03-07-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warshaw DM, Desrosiers JM, Work SS, Trybus KM. Smooth muscle myosin cross-bridge interactions modulate actin filament sliding velocity in vitro. J Cell Biol. 1990;111:453–463. doi: 10.1083/jcb.111.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uemura S, Higuchi H, Olivares AO, De La Cruz EM, Ishiwata S. Mechanochemical coupling of two substeps in a single myosin V motor. Nat Struct Mol Biol. 2004;11:877–883. doi: 10.1038/nsmb806. [DOI] [PubMed] [Google Scholar]
- 15.Svoboda K, Block SM. Force and velocity measured for single kinesin molecules. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 16.Iwaki M, Tanaka H, Iwane AH, Katayama E, Ikebe M, Yanagida T. Cargo-binding makes a wild-type single-headed myosin-VI move processively. Biophys J. 2006;90:3643–3652. doi: 10.1529/biophysj.105.075721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong WW, Doyle TC, Reisler E. Nonspecific weak actomyosin interactions: relocation of charged residues in subdomain 1 of actin does not alter actomyosin function. Biochemistry. 1999;38:1365–1370. doi: 10.1021/bi982467g. [DOI] [PubMed] [Google Scholar]
- 18.Vale RD, Soll DR, Gibbons IR. One-dimensional diffusion of microtubules bound to flagellar dynein. Cell. 1989;59:915–925. doi: 10.1016/0092-8674(89)90614-4. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z, Sheetz MP. One-dimensional diffusion on microtubules of particles coated with cytoplasmic dynein and immunoglobulins. Cell Struct Funct. 1999;24:373–383. doi: 10.1247/csf.24.373. [DOI] [PubMed] [Google Scholar]
- 20.Helenius J, Brouhard G, Kalaidzidis Y, Diez S, Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–119. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- 21.Lakamper S, Meyhofer E. The E-hook of tubulin interacts with kinesin's head to increase processivity and speed. Biophys J. 2005;89:3223–3234. doi: 10.1529/biophysj.104.057505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thorn KS, Ubersax JA, Vale RD. Engineering the processive run length of the kinesin motor. J Cell Biol. 2000;151:1093–1100. doi: 10.1083/jcb.151.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ross JL, Wallace K, Shuman H, Goldman YE, Holzbaur EL. Processive bidirectional motion of dynein-dynactin complexes in vitro. Nat Cell Biol. 2006;8:562–570. doi: 10.1038/ncb1421. [DOI] [PubMed] [Google Scholar]
- 24.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burton RF. The composition of animal cells: solutes contributing to osmotic pressure and charge balance. Comp Biochem Physiol B. 1983;76:663–671. doi: 10.1016/0305-0491(83)90375-9. [DOI] [PubMed] [Google Scholar]
- 26.Minton AP. How can biochemical reactions within cells differ from those in test tubes? J Cell Sci. 2006;119:2863–2869. doi: 10.1242/jcs.03063. [DOI] [PubMed] [Google Scholar]
- 27.Kural C, Serpinskaya AS, Chou YH, Goldman RD, Gelfand VI, Selvin PR. Tracking melanosomes inside a cell to study molecular motors and their interaction. Proc Natl Acad Sci USA. 2007;104:5378–5382. doi: 10.1073/pnas.0700145104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe TM, Higuchi H. Stepwise Movements in Vesicle Transport of HER2 by Motor Proteins in Living Cells. Biophys J. 2007;92:4109–4120. doi: 10.1529/biophysj.106.094649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vershinin M, Carter BC, Razafsky DS, King SJ, Gross SP. Multiple-motor based transport and its regulation by Tau. Proc Natl Acad Sci USA. 2007;104:87–92. doi: 10.1073/pnas.0607919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krementsov DN, Krementsova EB, Trybus KM. Myosin V: Regulation by calcium, calmodulin, and the tail domain. J Cell Biol. 2004;164:877–886. doi: 10.1083/jcb.200310065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cronan JE., Jr Biotination of proteins in vivo. A post-translational modification to label, purify, and study proteins. J Biol Chem. 1990;265:10327–10333. [PubMed] [Google Scholar]
- 32.O'Reilly DR, Miller LK, Luckow VA. Baculovirus expression vectors, A laboratory manual. New York: W. H. Freeman; 1992. [Google Scholar]
- 33.Higuchi H, Muto E, Inoue Y, Yanagida T. Kinetics of force generation by single kinesin molecules activated by laser photolysis of caged ATP. Proc Natl Acad Sci USA. 1997;94:4395–4400. doi: 10.1073/pnas.94.9.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mehta AD, Rock RS, Rief M, Spudich JA, Mooseker MS, Cheney RE. Myosin-V is a processive actin-based motor. Nature. 1999;400:590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- 35.Ali MY, Uemura S, Adachi K, Itoh H, Kinosita K, Jr, Ishiwata S. Myosin V is a left-handed spiral motor on the right-handed actin helix. Nat Struct Biol. 2002;9:464–467. doi: 10.1038/nsb803. [DOI] [PubMed] [Google Scholar]
- 36.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical Recipes in C: The Art of Scientific Computing. Cambridge, UK: Cambridge Univ Press; 1992. pp. 623–626. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.