Abstract
The hedgehog (Hh) signaling pathway regulates the development of many organs in mammals, and activation of this pathway is widely observed in human cancers. Although it is known that Hh signaling activates the expression of genes involved in cell growth, the precise role of the Hh pathway in cancer development is still unclear. Here, we show that constitutively activated mutants of Smoothened (Smo), a transducer of the Hh signaling pathway, inhibit the accumulation of the tumor suppressor protein p53. This inhibition was also observed in the presence of Hh ligand or with the overexpression of the transcription factors Gli1 and Gli2, downstream effectors of Smo, indicating that this inhibition is specific for the Hh pathway. We also report that Smo mutants augment p53 binding to the E3 ubiquitin-protein ligase Mdm2 and promote p53 ubiquitination. Furthermore, Hh signaling induced the phosphorylation of human Mdm2 protein on serines 166 and 186, which are activating phosphorylation sites of Mdm2. Smo mutants enhanced the proliferation of mouse embryonic fibroblasts (MEFs) while inducing a DNA-damage response. Moreover, Smo partially inhibited p53-dependent apoptosis and cell growth inhibition in oncogene-expressing MEFs. We also found that accumulation of p53 is inhibited by Hh signaling in several human cancer cell lines. Therefore, the Hh pathway may be a powerful accelerator of oncogenesis by activating cell proliferation and inhibiting the p53-mediated anti-cancer barrier induced by oncogenic stress.
Keywords: oncogenesis, ubiquitination, cell growth, apoptosis
The hedgehog (Hh) signaling pathway directs tissue patterning during embryonic development and stem cell maintenance (1–3). By binding to its receptor, Patched (Ptch), Hh is able to alleviate Ptch-mediated suppression of Smoothened (Smo). Activated Smo subsequently induces the activation of Gli family transcription factors by causing the dissociation of cytoplasmic inhibitory proteins, including Suppressor of Fused (SuFu), resulting in the transcriptional activation of Gli target genes (3). Accumulating evidence has shown that uncontrolled activation of the Hh signaling pathway results in distinct cancers. Constitutive activation of this pathway in human cancers is caused by genetic mutations of its molecular components, such as loss-of-function mutations in negative pathway regulators, for example Ptch and SuFu, and gain-of-function mutations in Smo (1, 4). These genetic alterations are mainly observed in basal-cell carcinomas (BCCs) and medulloblastomas. Recently, it was shown that Hh signaling activation due to the overproduction of the Hh ligands Sonic hedgehog (Shh) and Indian hedgehog (Ihh) is widely observed in human cancers, including cancers of the esophagus, stomach, pancreas, and lungs, suggesting a role for Hh signaling in oncogenesis (5–8).
The p53 tumor suppressor gene is the most common target of genetic alterations in human cancers (9). The p53 protein is highly unstable and is constitutively degraded by the ubiquitin-proteasome system (10). In response to DNA damage, p53 is covalently modified and stabilized, resulting in its accumulation (11). Recently, it has been shown that activation of oncogenes evokes a DNA damage response resulting in p53 activation. During the early tumorigenic stages, aberrant oncogene activation can give rise to “oncogenic stress,” which evokes a p53-mediated counterresponse to eliminate hazardous cells (12, 13) through the induction of apoptosis or senescence (14, 15). Therefore, during oncogenesis, p53 inhibition is crucial for the survival and maintenance of such oncogene-expressing cells.
In the present study, we determined that activation of Hh signaling inhibits the accumulation of p53 at the basal level and in response to DNA damage. We also found that Hh signaling promotes p53 ubiquitination mediated by Mdm2. Moreover, Hh signaling inhibited p53-dependent apoptosis and growth arrest in oncogene-expressing mouse embryonic fibroblasts (MEFs). These results suggest that Hh signaling accelerates oncogenesis through activation of cell proliferation and inhibition of p53-mediated tumor suppression.
Results
Hh Signaling Down-Regulates p53.
Previously, two activating somatic missense mutations of Smo, SMO-M1 (Arg-562 to Gln) and SMO-M2 (Trp-535 to Leu), were identified in sporadic BCCs (16). Interestingly, it was shown that the Smo mutants act in conjunction with adenovirus E1A to transform rat embryonic fibroblasts (16). Conversely, it has been shown that E1A strongly sensitizes primary fibroblasts to apoptosis in response to DNA damage and that this apoptosis is largely dependent on p53 (17, 18). These observations raise the possibility that Smo mutants inhibit p53 activity. To test this hypothesis, cells were cotransfected with expression vectors for SMO-M1 or SMO-M2, a p53-expression vector and a p53-responsive luciferase reporter plasmid. As shown in Fig. 1a, SMO-M1 and SMO-M2 efficiently inhibited p53-mediated transcription in p53-null human Saos-2 cells. The same effect was also observed in murine C3H10T1/2 cells (Fig. 1b), which are widely used to analyze Hh signaling (19); moreover, the endogenous WT p53 of C3H10T1/2 cells was inhibited by WT Smo. Overexpression of SMO-M1 and SMO-M2 markedly decreased the amount of ectopically expressed p53 (Fig. 1c) and endogenous p53 (Fig. 1d). The amount of endogenous p53 also decreased after overexpression of WT Smo (Fig. 1d). Furthermore, Shh decreased the expression level of endogenous p53 (Fig. 1e).
Fig. 1.
Hh signaling inhibits p53 activity and down-regulates p53. (a and b) A WT (SMO-WT) or mutant (SMO-M1 or SMO-M2) Smo expression plasmid was transfected into Saos-2 cells (a) or C3H10T1/2 cells (b), with (+p53) or without the p53 expression plasmid and with the RGC-luc reporter plasmid containing synthetic p53-binding sequences (39). Luciferase activity was measured 48 h after transfection. The histogram shows the means of three independent experiments, and error bars show standard deviations (SDs). (c) p53 (+p53) and gD-tagged Smo expression plasmids (SMO-WT, SMO-M1, and SMO-M2) were transfected into C3H10T1/2 cells, and the amount of p53 was determined by immunoblotting. The Smo protein level was determined by using an anti-gD antibody. (d) The Smo expression plasmid was transfected into C3H10T1/2 cells. Forty-eight hours after transfection, p53 was detected by immunoprecipitation (IP), using anti-p53 (FL-393) antibody, followed by immunoblotting with a different anti-p53 (Pab246) antibody. (e) C3H10T1/2 cells were treated with the indicated amounts of mouse sonic hedgehog N-terminal peptide (Shh-N; R&D Systems) for 48 h. The amount of p53 was determined as described in d.
Hh Signaling Promotes p53 Ubiquitination and Degradation.
The expression of p53 mRNA was not suppressed by Smo mutants (Fig. 2a), suggesting that Hh signaling inhibits p53 accumulation. Indeed, the Smo mutants-mediated decreases in p53 expression level were significantly reversed in the presence of the proteasome inhibitor MG132 (Fig. 2b). Moreover, expression of Smo mutants augmented the ubiquitination of p53 (Fig. 2c), suggesting that Hh signaling accelerates p53 degradation through the ubiquitin-proteasome system. Shh also enhanced ubiquitination of exogenously expressed p53 (Fig. 2d) and endogenous p53 (Fig. 2e). p53 ubiquitination has been shown to be mediated by multiple E3 ubiquitin ligases, including Mdm2, Pirh2 (20), and COP1 (21). However, Smo mutant expression did not increase the mRNA levels of these ligases (Fig. 2a). We then analyzed the role of Mdm2 in the p53-related effects of the Hh pathway. Mdm2 associates with p53, resulting in the ubiquitination and cytoplasmic export of p53 from the nucleus (11). As shown in Fig. 2 f and g, expression of Smo mutants induced the cytoplasmic translocation of p53 from the nucleus. Moreover, the binding of p53 to Mdm2 was augmented in the presence of Smo mutants (Fig. 2h), offering a molecular explanation for its elevated ubiquitination. Indeed, enhanced p53 ubiquitination induced by Smo mutants was clearly suppressed by the RNAi-mediated suppression of Mdm2, using an shRNA expression system (Fig. 2i). Moreover, the Mdm2 antagonist Nutlin-3 inhibited the Smo mutant-mediated decrease in p53 expression (Fig. 2j), suggesting that Hh signaling activates the Mdm2-mediated degradation of p53.
Fig. 2.
Hh signaling promotes p53 ubiquitination and activates Mdm2. (a) RNA blotting, using total RNA from MEFs carrying the control vector and expressing Smo mutants, was carried out by using p53, Mdm2, Pirh2, and COP1 cDNA probes. 28s rRNA was examined as a loading control. (b) p53 and Smo expression plasmids were transfected into C3H10T1/2 cells, which were treated with 50 μM MG132 for 4 h before collection. Forty-eight hours after transfection, the amount of p53 was determined by immunoblotting. (c) C3H10T1/2 cells were transfected with expression plasmds for Flag-tagged ubiquitin, HA-p53, and the Smo mutant and treated with 50 μM MG132 for 4 h before harvesting. Twenty-four hours after transfection, ubiquitinated p53 was detected by immunoprecipitation with anti-HA antibody and immunoblotting with anti-Flag or anti-p53 antibody. Mono-ubiquitinated (Ub) and polyubiqutinated (Ub) p53 are indicated. (d) HA-tagged p53 expression plasmid was transfected into C3H10T1/2 cells, and the cells were incubated with 5 μg/ml Shh-N for 24 h. Ubiquitinated p53 was detected by immunoprecipitation with anti-HA antibody and immunobloting with anti-p53 (FL-393) antibody. (e) C3H10T1/2 cells were incubated with indicated amount of Shh-N for 48h. Ubiquitinated p53 was detected by immunoprecipitation with anti-p53 polyclonal antibody and immunobloting with anti-p53 monoclonal antibody. (f) The p53 expression vector was transfected into C3H10T1/2 cells with or without Smo mutants. p53 (red) was detected by using anti-p53 (FL-393) antibody; Smo (green) was visualized by using anti-gD antibody. The nucleus (blue) was stained with DAPI. (g) Quantification of p53 staining was performed as described in f. The staining pattern for p53 in cells expressing p53 and Smo mutants was scored for 100 cells in three separate experiments. The graph indicates the percentages of cells with the indicated p53 localization patterns. Bars indicate the SD of three independent experiments. (h) SMO-M1 or -M2 expression plasmid was transfected into C3H10T1/2 cells, and the cells were then treated with 50 μM MG132 for 4 h before harvest. Forty-eight hours after transfection, cell lysates were subjected to immunoprecipitation with anti-p53 antibody (FL-393) and immunoblotting with anti-p53 (Pab246) or anti-Mdm2 (2A10) antibody. (i) HA-tagged p53, mutant Smo, and Flag-tagged ubiquitin expression plasmids were transfected into GFP siRNA (Control)- and Mdm2 siRNA-expressing C3H10T1/2 cells. Twenty-four hours after transfection, detection of ubiquitination of p53 was performed as described in c. (j) Mutant Smo expression vectors were transfected into C3H10T1/2 cells. Cells were incubated with 15 μM Nutlin-3 (Calbiochem) for 16 h. p53 was detected by immunoprecipitation (IP), using anti-p53 (FL-393) antibody, followed by immunoblotting (IB) with a different anti-p53 (Pab246) antibody.
Hh Signaling Induces Activating Phosphorylation of Mdm2.
The phosphorylation of two serine residues, S166 and S186, of human Mdm2 (Hdm2) was enhanced by the expression of Smo mutants (Fig. 3a). S166 and S186 are known to be phosphorylated by Akt, which activates the Mdm2-mediated degradation of p53 (22, 23). Recently, it was reported that Smo activates phosphoinositide 3-kinase (PI3K) and Akt, both of which belong to an essential Shh signaling pathway (24). As shown in Fig. 3b, Shh slightly enhanced the phosphorylation of Akt (S473) and decreased p53 expression; however, the Shh-mediated reduction in p53 expression was suppressed by the PI3K inhibitor LY294002. In the Hh signaling pathway, the activation of Smo results in the activation of Gli family transcription factors (25). It was also shown that the PI3K/Akt pathway activates Glis (24). In the context of cancer, Gli1 was originally identified to be amplified in human gliomas; the ectopic expression of Gli1 or Gli2 in the skin results in tumor formation in mice (26, 27). Interestingly, forced expression of Gli1 or Gli2 decreased endogenous p53 expression, and this effect was suppressed by MG132 (Fig. 3 c and d), suggesting that Gli1 and Gli2 induce ubiquitin-mediated p53 degradation. By contrast, forced expression of Gli3 had no effect (Fig. 3e). The involvement of Gli in this system was also confirmed by inhibition of the SMO-M2-mediated decrease in p53 expression induced by SuFu, a negative regulator of Gli activation by Hh signaling (Fig. 3f). Moreover, phosphorylation of S166 and S186 was also enhanced in the presence of Shh, but inhibited by the protein synthesis blocker cycloheximide (Fig. 3g). Therefore, these results suggest that Hh signaling augments p53 protein degradation by direct phosphorylation of Mdm2 through a Gli-induced factor(s).
Fig. 3.
Hh signaling induces Hdm2 activation and p53 destabilization. (a) Flag-tagged Hdm2 and Smo expression plasmids were transfected into C3H10T1/2 cells. The cells were subjected to serum starvation for 6 h before collection, and Hdm2 phosphorylation was detected by immunoprecipitation with anti-Flag antibody and immunoblotting with anti-phospho-Hdm2 (Ser-166 or Ser-186) antibody. (b) C3H10T1/2 cells were treated with 3 μg/ml Shh-N for 48 h. The amount of p53 was determined as described above. Endogenous Akt phosphorylation was detected by immunoblotting, using anti-phospho-Akt (Ser-473) antibody. (c–e) HA-tagged Gli1 (c), Myc-tagged Gli2 (d), or HA-tagged Gli3 (e) expression plasmid was transfected into C3H10T1/2 cells. MG132 (50 μM) was treated for 4 h before harvest (a and d). The amount of p53 protein was determined as described in Fig. 1d. Gli1 and Gli2 expression was determined by immunoblotting, using antibodies against each tag. (f) p53 and SMO-M2 expression plasmids were transfected into C3H10T1/2 cells with or without the Myc-tagged SuFu expression plasmid (40). Forty-eight hours after transfection, the amount of p53 protein was determined by immunoblotting. The expression of SuFu was also determined by using anti-Myc-tag antibody. (g) The Flag-tagged Hdm2 expression vector was transfected into C3H10T1/2 cells, and the cells were incubated with 1 μg/ml Shh-N for 48 h. To inhibit protein synthesis, 80 μg/ml cycloheximide (CHX) was added to the culture, which was then incubated for 2 h before harvest. Detection of phosphorylated Hdm2 was performed as described in a.
Hh Signaling Inhibits p53-Mediated Apoptosis and Cell Growth Inhibition.
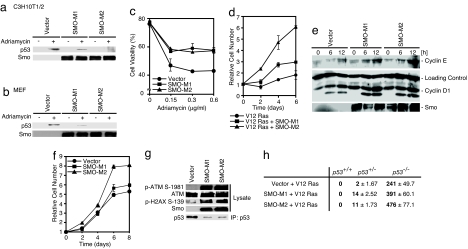
To explore the contribution of the Hh signaling pathway to oncogenesis, we analyzed whether the DNA damage-induced accumulation of p53 is inhibited by mutant Smo. As shown in Fig. 4a, p53 induction in response to adriamycin was attenuated by the expression of Smo mutants in C3H10T1/2 cells and MEFs (Fig. 4b). Apoptotic elimination of oncogene-expressing cells may be an important function of p53 in tumor suppression (17, 18). Therefore, we next evaluated the effect of mutant Smo on E1A-expressing MEFs treated with adriamycin. Indeed, SMO-M1 and SMO-M2 expression in E1A-expressing MEFs led to the attenuation of adriamycin-induced apoptosis (Fig. 4c). It is also known that oncogenic ras induces p53-dependent premature cell senescence (18). As shown in Fig. 4d, oncogenic ras (V12 Ras)-induced growth suppression was also inhibited by SMO-M1 and SMO-M2 expression.
Fig. 4.
Smo mutants activate cell growth, evoke DNA-damage-induced responses, and suppress p53 expression. (a and b) Mutant Smo expression plasmid or control plasmid (Vector) was transfected into C3H10T1/2 cells (a). Forty eight hours after transfection, the cells were incubated with 0.6 μg/ml adriamycin for 4 h. MEFs expressing SMO-M1 or SMO-M2 via recombinant retrovirus were also treated with adriamycin b. The amount of p53 was determined as described in Fig. 1c. (c) MEFs expressing E1A via retrovirus were also infected with control retrovirus (Vector) or Smo mutant (SMO-M1 or SMO-M2)-expressing retrovirus. Cells were incubated with adriamycin for 24 h and then subjected to a cell viability assay. Results are shown as means ± SD of triplicate experiments. (d) Growth curves for V12 Ras- and mutant Smo-expressing MEFs via retrovirus are shown. Bars indicate the SD from triplicate experiments. (e) The cell cycle of SMO-M1- or SMO-M2-expressing MEFs via retrovirus was arrested at the G1-phase by serum starvation, then the cells were cultured with 10% FBS to continue the cell cycle. At the indicated time points, the cells were harvested, and cyclins D1 and E were detected by immunoblotting. (f) The growth curves for control retrovirus-carrying (Vector), and SMO-M1- or SMO-M2-expressing retrovirus-carrying MEFs are shown. Bars indicate the SD from triplicate experiments. (g) Mutant Smo was stably expressed via transfection of recombinant retrovirus into MEFs. DNA-damage responses were determined by immunoblotting with anti-phospho-ATM (Ser-1981) and anti-phospho-histone H2AX (Ser-139) antibodies. The expression levels of ATM and Smo mutants were also determined by immunoblotting. The amount of p53 was determined as described in Fig. 1d. (h) Mutant Smo and V12 Ras were expressed via recombinant retrovirus in MEFs. A quantitative colony formation assay was performed by plating mutant Smo- and V12Ras-expressing cells at a density of 1 × 104 cells per 35-mm dish and incubating them for 14 days. Surviving colonies were counted and represented as the means ± SD of three independent dishes.
Hh signaling activates the expression of genes involved in cell proliferation, such as cyclins D and E (28). Indeed, Smo mutants enhanced cyclin D1 and E expression in MEFs (Fig. 4e), and also increased the rate of proliferation of these cells (Fig. 4f). Recently, it was suggested that deregulated expression of oncogenes might evoke a DNA-damage response, resulting in p53 activation (12, 13). Interestingly, Smo mutants induced a DNA-damage response, evidenced by the activation of ATM and the phosphorylation of histone H2AX (Fig. 4g). These results suggest that Hh signaling activates cell proliferation and enhances DNA damage responses; however, at the same time, p53-mediated tumor suppression was inhibited through the activation of Mdm2. We also analyzed cell transformation, using V12 Ras and Smo mutants. Although V12 Ras and Smo mutant-expressing WT MEFs did not show anchorage-independent colony formation in soft agar, Smo mutants enhanced the V12 Ras-induced transformation of p53 heterozygote (p53+/−) MEFs (Fig. 4h), suggesting that Hh signaling inhibits p53-mediated tumor suppression. In addition, Smo mutants enhanced colony formation by V12 Ras-expressing p53-null (p53−/−) MEFs, suggesting that some Smo mutant-mediated oncogenic function, for example, activation of cyclins D1 and E, enhances V12 Ras-induced cell transformation (Fig. 4h).
Inhibition of Hh Signaling Recovers p53 Expression and Function in Breast Cancer Cell Lines.
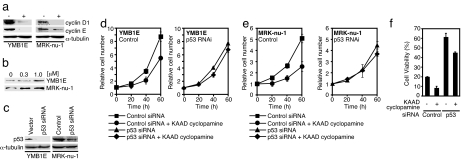
We next analyzed two human breast cancer cell lines containing WT p53 (29). As shown in Fig. 5a, KAAD cyclopamine (30), a specific inhibitor of the Hh pathway, inhibited the expression of cyclins D1 and E. Moreover, KAAD cyclopamine treatment resulted in the accumulation of endogenous WT p53 (Fig. 5b), suggesting that Hh signaling inhibited the production of p53. The growth of these two cell lines was efficiently inhibited by KAAD cyclopamine; however, this growth inhibition disappeared subsequent to p53 expression knockdown by RNAi, using an shRNA expression system (Fig. 5 c–e). Furthermore, the DNA damage-induced apoptosis of YMB1E cells was accelerated by KAAD cyclopamine; however, this acceleration was clearly inhibited by p53 expression knockdown (Fig. 5f). The acceleration of apoptosis by KAAD cyclopamine was not observed in MRK-nu-1 cells (data not shown), and the reason for this differential response is presently unknown. However, these results suggest that constitutive Hh signaling inhibits p53 accumulation in the tested human cancer cells.
Fig. 5.
KAAD cyclopamine treatment restores p53 expression in breast cancer cell lines. (a) Cells were treated with 1 μM KAAD cyclopamine (Calbiochem) for 72 h and collected, and the levels of cyclins D1 and E were determined by immunoblotting. (b) The cells were treated with KAAD cyclopamine at the indicated doses for 48 h and collected, and the p53 level was determined by immunoblotting. (c) p53 siRNA was expressed (41) by recombinant retrovirus. p53 level was determined by immunoblotting. (d and e) YMB1E (2 × 105) (d) and MRK-nu-1 (e) cells were incubated with 1 μM KAAD cyclopamine for the indicated lengths of time, and viable cells were counted. Results are shown as means ± SD from triplicate experiments. (f) YMB1E cells were incubated with 1 μM KAAD cyclopamine and 0.6 μg/ml adriamycin for 48 h and subjected to a cell viability assay. Results are shown as means ± SD from triplicate experiments.
Discussion
In this study, we found that Hh signaling inhibits p53-mediated tumor suppression by activating Mdm2. p53 functions as a critical tumor suppressor by activating the expression of genes that induce cell growth inhibition, apoptosis, and DNA repair (17, 31, 32). Recent studies have clearly shown that oncogene activation evokes a DNA-damage response, which results in p53 activation (12, 13). Moreover, many studies have shown that p53 induces apoptosis or replicative senescence in oncogene-expressing cells (17, 18, 31), suggesting that the elimination of such cells is important for tumor suppression. Therefore, it is possible that escape from such a p53-mediated tumor surveillance system is important during the early stages of oncogenesis. In this context, Hh signaling is an interesting example with which to consider oncogenesis. Hh signaling induces cell growth by activating the expression of cyclins D1 and E (Fig. 4 e and f). At the same time, Hh signaling evokes a DNA-damage-induced signal (Fig. 4g). This DNA-damage-induced signal may be caused by oncogenic stress (12, 13, 31, 32); however, at the same time, Hh signaling down-regulates p53 by activating Mdm2 (Figs. 1–3). As a result, an important function of p53 in tumor suppression, the induction of apoptosis or senescence in oncogene-expressing cells, is partially inhibited (Fig. 4 c and d). These results suggest that Hh signaling functions as an accelerator of oncogenesis, activating cell proliferation and, at the same time, inhibiting the p53-mediated anti-cancer barrier induced by oncogenic stress.
In this study, we showed that Hh signaling activates phosphorylation of Mdm2 at serines 166 and 186, which were identified as sites of activating phosphorylation by Akt (Fig. 3a and refs. 22 and 23). It was recently reported that Hh signaling activates PI3K and Akt (24). Indeed, phosphorylation of these sites, induced by Hh signaling, is reduced by the PI3K inhibitor (Fig. 3b). These results indicate that induction of the PI3K/Akt pathway by Hh signaling is involved in the activation of Mdm2. However, we also found that the forced expression of Gli1 or Gli2 reduced the amount of p53 in cells (Fig. 3 c and d). This result was also supported by the findings that the forced expression of SuFu, a negative regulator of Gli activation in the Hh pathway, restored the Smo mutant-meditated reduction in the amount of p53 in cells (Fig. 3f), and that the phosphorylation of Mdm2 at serines 166 and 186 was inhibited by cycloheximide (Fig. 3g), suggesting that de novo protein synthesis is important for these phosphorylation events. These results strongly suggest that a Gli-induced factor(s) underlies this phenomenon. At present, the kinase(s) directly induced or indirectly activated by Hh signaling remain to be identified. However, when considering oncogenesis, it is interesting that the oncogenic transcription factor Gli also decreased the amount of p53 in cells. Hdm2 and Gli1 genes are relatively close to each other on the same human chromosome, and they are sometimes coamplified in cancers (33), suggesting that a combination of excess Gli1 together with excess Mdm2 is sufficient to efficiently inactivate p53.
Hh signaling has been implicated in tissue repair and the maintenance of stem or progenitor cells in adult tissues, including skin, blood, gut, prostate, muscle, and the nervous system (1–3). It is also well known that chronic tissue injury caused by inflammation is associated with tumorigenesis (1). Moreover, recently, it was also shown that Hh signaling maintains a tumor stem cell compartment (34). Therefore, it is possible that, in the early stage of tumorigenesis, Hh signaling promotes cell growth and partially inhibits p53 function, thereby enabling these cells to survive and grow. This model is also supported by a recent study that demonstrated cooperation of Hedgehog and Ras signaling during the earliest stages of pancreatic ductal adenocarcinoma formation in an animal model (35). By contrast, the effect of Hh signaling in the inhibition of p53 is not complete, but partial (see Fig. 1 d and e and Fig. 4 a and b). This partial effect is seen in the result showing that Smo mutants enhanced activated Ras-induced transformation in p53 heterozygote (p53+/−) MEFs, but not in WT MEFs (Fig. 4h), suggesting that activation of Hh signaling only is not sufficient for the inhibition of p53-mediated tumor suppression. This issue is also supported because mutations in both Ptch (or Smo) and p53 are often observed together in BCC tumors (36, 37). We also showed that, in human breast cancer cell lines, the levels of p53 are decreased by activated Hh signaling and that inhibition of Hh signaling by KAAD cyclopamine results in accumulation of p53 (Fig. 5b). Therefore, at least in some types of cancer, activation of the Hh signaling pathway partially abrogates the tumor suppressive effects of p53. Altogether, our observations imply that constitutive Hh pathway activation functions as an accelerator of oncogenesis and simultaneously induces cell proliferation by overriding p53-mediated tumor suppression.
Materials and Methods
Immunoprecipitation and Immunoblotting.
To detect the amount of endogenous or transiently produced p53 protein, cells were lysed in RIPA buffer [1% Nonidet P-40, 0.5% sodium-deoxycholate, 0.1% SDS, 10 mM Tris·HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, and a protease inhibitor mixture (Nacalai)]. To detect binding between endogenous p53 and Mdm2, or phosphorylation of Mdm2 and Akt, cells were lysed in lysis buffer [0.5% Nonidet P-40, 150 mM NaCl, 10 mM Tris·HCl (pH 8.0), 1 mM EDTA, 1 mM NaF, 1 mM orthovanadate 0.1 mM DTT, and a protease inhibitor mixture]. Immunoprecipitation was carried out as described in ref. 38. Total cell lysate protein was quantified by using a protein assay kit (Bio-Rad), and an equal amount of protein was used for immunoprecipitation and immunoblotting. Additional materials and methods are provided in supporting information (SI) Materials and Methods.
Supplementary Material
Acknowledgments.
We thank F. J. de Sauvage (Genetech, San Francisco, CA) for pRK-7-gD-SMO-WT, -M1, -M2, and pcDNA3-Myc-Gli2 plasmids and the anti-gD antibody; B. Vogelstein (Johns Hopkins School of Medicine, Baltimore, MD) for human p53, and Gli1 and Gli3 cDNAs; Y. Gotoh (Institute of Molecular and Cellular Biosciences, University of Tokyo, Tokyo, Japan) for the HA-tagged human p53 and dominant negative Akt expression vectors; M. Nakafuku (Cincinnati Children's Hospital, Cincinnati, OH) for the Myc-tagged mouse SuFu expression vector; and F. Tokunaga (Osaka City University Graduate School of Medicine, Osaka, Japan) for the Flag-tagged ubiquitin expression vector. This work was supported by Grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712216105/DC1.
References
- 1.Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432:324–331. doi: 10.1038/nature03100. [DOI] [PubMed] [Google Scholar]
- 2.Ingham PW, McMahon AP. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 3.Ingham PW, Placzek M. Orchestrating ontogenesis: Variations on a theme by sonic hedgehog. Nat Rev Genet. 2006;7:841–850. doi: 10.1038/nrg1969. [DOI] [PubMed] [Google Scholar]
- 4.Pasca di Magliano M, Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nat Rev Cancer. 2003;3:903–911. doi: 10.1038/nrc1229. [DOI] [PubMed] [Google Scholar]
- 5.Berman DM, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 6.Karhadkar SS, et al. Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature. 2004;431:707–712. doi: 10.1038/nature02962. [DOI] [PubMed] [Google Scholar]
- 7.Thayer SP, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watkins DN, et al. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422:313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 9.Soussi T, Ishioka C, Claustres M, Beroud C. Locus-specific mutation databases: Pitfalls and good practice based on the p53 experience. Nat Rev Cancer. 2006;6:83–90. doi: 10.1038/nrc1783. [DOI] [PubMed] [Google Scholar]
- 10.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 11.Michael D, Oren M. The p53-Mdm2 module and the ubiquitin system. Semin Cancer Biol. 2003;13:49–58. doi: 10.1016/s1044-579x(02)00099-8. [DOI] [PubMed] [Google Scholar]
- 12.Bartkova J, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 13.Di Micco R, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 14.Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 15.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 16.Xie J, et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391:90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- 17.Vousden KH, Lu X. Live or let die: The cell's response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 18.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 19.Murone M, Rosenthal A, de Sauvage FJ. Sonic hedgehog signaling by the patched-smoothened receptor complex. Curr Biol. 1999;9:76–84. doi: 10.1016/s0960-9822(99)80018-9. [DOI] [PubMed] [Google Scholar]
- 20.Leng RP, et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 21.Dornan D, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 22.Zhou BP, et al. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol. 2001;3:973–982. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- 23.Ogawara Y, et al. Akt enhances Mdm2-mediated ubiquitination and degradation of p53. J Biol Chem. 2002;277:21843–21850. doi: 10.1074/jbc.M109745200. [DOI] [PubMed] [Google Scholar]
- 24.Riobo NA, Lu K, Ai X, Haines GM, Emerson CP., Jr Phosphoinositide 3-kinase and Akt are essential for Sonic Hedgehog signaling. Proc Natl Acad Sci USA. 2006;103:4505–4510. doi: 10.1073/pnas.0504337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koebernick K, Pieler T. Gli-type zinc finger proteins as bipotential transducers of Hedgehog signaling. Differentiation. 2002;70:69–76. doi: 10.1046/j.1432-0436.2002.700201.x. [DOI] [PubMed] [Google Scholar]
- 26.Grachtchouk M, et al. Basal cell carcinomas in mice overexpressing Gli2 in skin. Nat Genet. 2000;24:216–217. doi: 10.1038/73417. [DOI] [PubMed] [Google Scholar]
- 27.Nilsson M, et al. Induction of basal cell carcinomas and trichoepitheliomas in mice overexpressing GLI-1. Proc Natl Acad Sci USA. 2000;97:3438–3443. doi: 10.1073/pnas.050467397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duman-Scheel M, Weng L, Xin S, Du W. Hedgehog regulates cell growth and proliferation by inducing Cyclin D and Cyclin E. Nature. 2002;417:299–304. doi: 10.1038/417299a. [DOI] [PubMed] [Google Scholar]
- 29.Jia LQ, et al. Screening the p53 status of human cell lines using a yeast functional assay. Mol Carcinog. 1997;19:243–253. doi: 10.1002/(sici)1098-2744(199708)19:4<243::aid-mc5>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 30.Taipale J, et al. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 31.Oren M. Decision making by p53: Life, death and cancer. Cell Death Differ. 2003;10:431–442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- 32.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 33.Gordon AT, et al. A novel and consistent amplicon at 13q31 associated with alveolar rhabdomyosarcoma. Genes Chromosomes Cancer. 2000;28:220–226. [PubMed] [Google Scholar]
- 34.Peacock CD, et al. Hedgehog signaling maintains a tumor stem cell compartment in multiple myeloma. Proc Natl Acad Sci USA. 2007;104:4048–4053. doi: 10.1073/pnas.0611682104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pasca di Magliano M, et al. Hedgehog/Ras interactions regulate early stages of pancreatic cancer. Genes Dev. 2006;20:3161–3173. doi: 10.1101/gad.1470806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim MY, Park HJ, Baek SC, Byun DG, Houh D. Mutations of the p53 and PTCH gene in basal cell carcinomas: UV mutation signature and strand bias. J Dermatol Sci. 2002;29:1–9. doi: 10.1016/s0923-1811(01)00170-0. [DOI] [PubMed] [Google Scholar]
- 37.Reifenberger J, et al. Somatic mutations in the PTCH, SMOH, SUFUH and TP53 genes in sporadic basal cell carcinomas. Br J Dermatol. 2005;152:43–51. doi: 10.1111/j.1365-2133.2005.06353.x. [DOI] [PubMed] [Google Scholar]
- 38.Shinozaki T, Nota A, Taya Y, Okamoto K. Functional role of Mdm2 phosphorylation by ATR in attenuation of p53 nuclear export. Oncogene. 2003;22:8870–8880. doi: 10.1038/sj.onc.1207176. [DOI] [PubMed] [Google Scholar]
- 39.Haupt Y, Rowan S, Shaulian E, Vousden KH, Oren M. Induction of apoptosis in HeLa cells by trans-activation-deficient p53. Genes Dev. 1995;9:2170–2183. doi: 10.1101/gad.9.17.2170. [DOI] [PubMed] [Google Scholar]
- 40.Ding Q, et al. Mouse suppressor of fused is a negative regulator of sonic hedgehog signaling and alters the subcellular distribution of Gli1. Curr Biol. 1999;9:1119–1122. doi: 10.1016/s0960-9822(99)80482-5. [DOI] [PubMed] [Google Scholar]
- 41.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.