Abstract
With the potential to give rise to all somatic cell types, human embryonic stem cells (hESC) have generated enormous interest as agents of cell replacement therapy. One potential limitation is their safety in vivo. Although several studies have focused on concerns over genomic stability ex vivo, few have analyzed epigenetic stability. Here, we use tools of the epigenetic phenomenon, X-chromosome inactivation (XCI), to investigate their epigenetic properties. Among 11 distinct hESC lines, we find a high degree of variability. We show that, like mouse ESC, hESC in principle have the capacity to recapitulate XCI when induced to differentiate in culture (class I lines). However, this capacity is seen in few hESC isolates. Many hESC lines have already undergone XCI (class II and III). Unexpectedly, there is a tendency to lose XIST RNA expression during culture (class III). Despite losing H3-K27 trimethylation, the inactive X of class III lines remains transcriptionally suppressed, as indicated by Cot-1 RNA exclusion. We conclude that hESC lines are subject to dynamic epigenetic reprogramming ex vivo. Given that XCI and cell differentiation are tightly linked, we consider implications for hESC pluripotency and differentiation potential.
Human embryonic stem cells (hESCs) (1) can be maintained in culture in a self-renewing state and differentiate into all three embryonic germ layers (2). Although they hold great promise for regenerative medicine, hESC-based cell therapy faces several challenges. Concerns have been raised regarding their genetic stability. For example, their tendency to gain chromosomes 12, 17, and X likens hESC to various germ cell tumors (3, 4). Although not yet fully investigated, concerns also exist over epigenetic stability (5, 6). Epigenetic changes make alterations to the nuclear program without changing the primary DNA sequence. Because epigenetic changes can substantially modify cellular behavior and are mitotically or meiotically heritable, investigation into the epigenetic properties of hESC is desirable prior to considering their use in vivo.
Here, we examine the epigenetic stability of hESC using markers of X-chromosome inactivation (XCI), a whole-chromosome silencing phenomenon that compensates for the female's extra X-chromosome dosage relative to the male's (7). We reasoned that, because XCI is one of the first measurable epigenetic changes in the early mammalian embryo and is coincident with differentiation into the three germ layers (8), it serves as an excellent tool to investigate the epigenetic behavior of hESC. In mouse embryonic stem cells (mESC), XCI is recapitulated when the cells are induced to differentiate ex vivo. Because hESC and mESC have similar derivation, hESC may also recapitulate XCI and enable the study of epigenetic change during early human development.
Early reports have not reached a consensus on the status of XCI in hESC. One report showed that, based on XIST expression levels, the female line, H9, is pre-XCI and can recapitulate XCI during differentiation (9). However, others have reported that XIST is already expressed in H9 and also in H7 and CyT25 (10–12). Inexplicably, this variability of the XCI marker occurs not only between distinct cell lines but also between sublines and subpassages of any one cell line (10, 11). Additionally, sampling of dozens of female hESC has revealed highly variable XIST expression levels (13). Thus, these studies indicate widely disparate XIST states and do not agree on whether hESC is fundamentally pre- or post-XCI. Below, we characterize 11 female hESC lines, both federally registered and nonregistered, and find that they can be subgrouped into three classes. We find that female hESC are pre-XCI and uncover a surprising explanation for the variability of XIST expression states.
Results and Discussion
Xist makes a noncoding RNA required to initiate silencing during XCI (reviewed in ref. 14). Before XCI and mESC differentiation, Xist is expressed at low levels. Upon cell differentiation and the onset of XCI, Xist RNA is transcriptionally induced and forms a “cloud” around the inactive X (Xi). To analyze the XCI status of hESC, we tested 11 female lines, four of which are NIH-approved [H7 and H9 (1), HES-2 and HES-3 (15)] and seven of which are not currently registered (HUES1, HUES5, HUES6, HUES9, HUES12, HUES14 and HUES15; ref. 16).
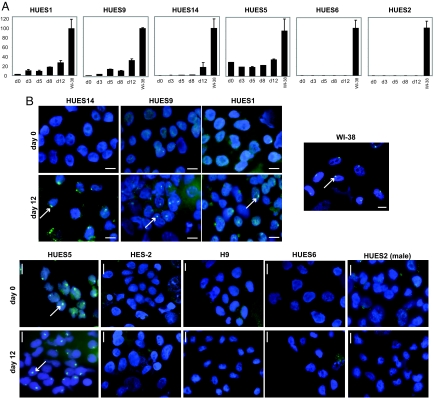
We first examined XIST expression by quantitative RT-PCR using primer pairs that detect XIST-specific splice patterns. HUES1, HUES9, and HUES14 all showed low level XIST on day 0 (d0) and increased XIST levels upon cell differentiation (Fig. 1A). RNA fluorescence in situ hybridization (FISH) verified these results (Fig. 1B). On d0, XIST RNA clouds occurred in <20% of HUES1, HUES9, and HUES14 cells; when placed in differentiation conditions, a significant percentage increase became evident (Fig. 1B and Table 1). Qualitatively, the XIST RNA domains resembled those in control female fibroblasts, WI-38. In contrast, the male HUES2 line did not express XIST at all. The presence of XIST+ female cells on d0 might be explained by spontaneous cellular differentiation, a frequent occurrence for mESC. In differentiated cultures, XIST induction was rarely seen in >60%. One potential explanation is incomplete differentiation, as often occurs for mESC. These data show that a majority of HUES1, HUES9, and HUES14 cells is pre-XCI on d0 and can reproduce XCI when placed under differentiation conditions. Thus, like mESC, hESC also have the ability to recapitulate XCI ex vivo.
Fig. 1.
RT-PCR and RNA FISH analysis of XIST expression during hESC differentiation. (A) Quantitative real-time RT-PCR for XIST expression on d0 (undifferentiated), d3, d5, d8, and d12 of differentiation compared with primary female fibroblasts (WI-38). XIST is normalized to GAPDH. (B) XIST RNA FISH in undifferentiated (d0) and differentiated (d12) hESC. Nuclei are stained with DAPI (blue). Arrows point to XIST clouds (green). (Scale bar, 10 μm.)
Table 1.
Summary of XCI and cell differentiation potential in hESC lines
| Class | hESC (passage) | XIST clouds |
Markers of silencing |
Cell differentiation quality |
|||
|---|---|---|---|---|---|---|---|
| d0 | d7–d16 | Cot-1 hole on one X | H3-K27me3 on X | EB character (SI Fig. 6C) | Lineage-specific markers | ||
| I | HUES1 (p24–29) | 20%, n = 726 | 38%, n = 172 | d0: 100% of XIST+ cells, n = 14 | XIST+ cells only | Fair | Present |
| d0: 34% of XIST−, n = 50 | |||||||
| HUES9 (p26–36) | 15%, n = 805 | 58%, n = 147 | d0: 100% of XIST+ cells, n = 31 | d13: 88% of XIST+, n = 81 | Robust | Present | |
| d0: 41% of XIST− cells, n = 58 | |||||||
| d9: 100% of XIST+ cells, n = 70 | |||||||
| d9: 42% of XIST− cells, n = 57 | |||||||
| HUES14 (p21–23) | 17%, n = 762 | 51%, n = 126 | d0: 35% of XIST− cells, n = 57 | XIST+ cells only | Robust | Not determined | |
| II | HUES5 | 70%, n = 157 | 72%, n = 149 | d0: 100% of XIST+ cells, n = 76 | d0: 92% of XIST+, n = 114 | Robust | Present |
| (p22; short-term culture) | d9: 100% of XIST+ cells, n = 75 | ||||||
| HUES5 | 24%, n = 123 | 30%, n = 119 | d0: 100% of XIST+ cells, n = 17 | ||||
| (p23; longer culture) | d0: 63% of XIST− cells, n = 49 | ||||||
| d20: 100% of XIST+ cells, n = 16 | |||||||
| HES-3 (p86–94) | 67%, n = 122 | 73%, n = 297 | Not determined | XIST+ cells only | Poor/Fair | Present | |
| III | HUES6 (p23–42) | 0%, n > 1,000 | 0%, n > 1,000 | d0: 84% of XIST− cells, n = 56 | 0%, n > 1,000 | Poor | Endoderm-poor |
| d9: 67% of XIST− cells, n = 61 | |||||||
| HUES12 (p25–28) | 0%, n > 1,000 | 0%, n >1,000 | Not determined | 0%, n > 1,000 | Fair/Robust | Present | |
| HUES15 (p35–40) | 0%, n > 1,000 | 0%, n >1,000 | d0: 84% of XIST− cells, n = 49 | 0%, n > 1,000 | Fair/Robust | Present | |
| d15: 73% of XIST− cells, n = 60 | |||||||
| H7 (p31–35) | 0%, n > 1,000 | 0%, n >1,000 | d0: 78% of XIST− cells, n = 41 | 0%, n > 1,000 | Poor/Fair | Present | |
| H9 (p35–47) | 0%, n > 1,000 | 0%, n >1,000 | d0: 78% of XIST− cells, n = 50 | 0%, n > 1,000 | Poor/Fair | Present | |
| HES-2 (p90–102) | 0%, n > 1,000 | 0%, n >1,000 | d0: 80% of XIST− cells, n = 52 | 0%, n > 1,000 | Poor/Fair | Present | |
| d15: 81% of XIST− cells, n = 52 | |||||||
| HUES2 (male) | 0%, n > 1,000 | 0%, n > 1,000 | 0%, n > 1,000 | 0%, n > 1,000 | Fair | Present | |
However, not all hESC lines possess this property. Our analyses showed that HUES5 and HES-3 expressed similar levels of XIST RNA before or after placement in differentiation conditions (Fig. 1A, Table 1, and data not shown). On d0, XIST was seen in 67% of HES-3 cells and 70% of HUES5 cells. After d7–16 in differentiation condition, 73% of HES-3 cells and 72% of HUES5 remained XIST+. Therefore, dosage compensation may have at least partially occurred in HUES5 and HES-3. By contrast, HUES6, HUES12, HUES15, HES-2, H7, and H9 revealed no detectable XIST at any time point (Fig. 1 and data not shown). Note that the XIST− states of H7 and H9 differ from previous reports of XIST+ states (9–12). By our analysis, 6 of 11 female lines may have lost the potential to carry out XCI.
The results demonstrate that hESC lines can be categorized into three epigenetic classes. Like mESC, class I hESC (HUES1, HUES9, HUES14) are pre-XCI with the capacity to recapitulate XCI upon differentiation. Class II lines (HUES5 and HES-3) exhibit consistently elevated XIST+ cells, suggesting the completion of XCI. Class III lines (HUES6, HUES12, HUES15, HES-2, H7, and H9) have lost the capacity to express XIST. Significantly, class II and III lines have no counterpart in the mESC system.
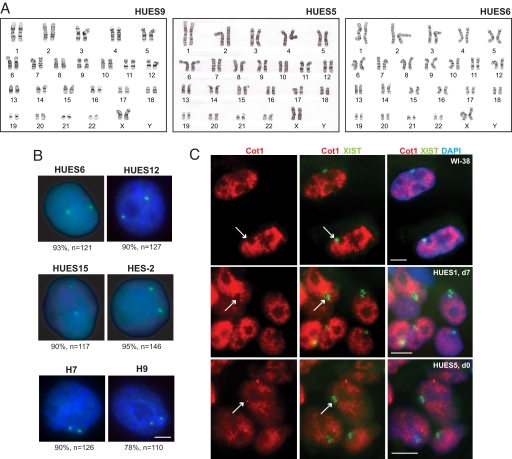
In principle, aberrant XIST expression may have several underlying causes. To rule out X-chromosome loss, which would obviate the need for dosage compensation, we performed X-chromosome painting [supporting information (SI) Fig. 5] and G-banding karyotype analysis (Fig. 2A). Both confirmed a 46XX karyotype. Therefore, the absence of XIST was not due to X-chromosome loss. Can it be explained by a deletion of the X-inactivation center (XIC)? DNA FISH using an XIST-containing PAC showed two signals for class III lines on d0; no significant change was seen after differentiation (Fig. 2B). Thus, XIC deletions also could not explain the absence of XIST. These data showed that, unlike female mESC, female hESC do not have the tendency to lose one X chromosome.
Fig. 2.
Karyotype analysis and Cot-1 RNA FISH. (A) Karyotype analysis by G-banding was performed at Cell Line Genetics on 20 metaphase spreads from d0 HUES9, HUES5, and HUES6. All are 46XX, although HUES9 and HUES5 appeared to carry a small but cytogenetically visible pericentric inversion on one chromosome 9. (B) Two XIC DNA FISH signals (green) are evident in a vast majority of nuclei in all class III lines (d0). Note: that after d14–d15 of differentiation, the frequency of nuclei with 2 XIC signals did not change significantly, e.g., 89% for HUES6 (n = 108), 89% for HUES15 (n = 104), 92% for H9 (n = 114), and 92% for HES-2 (n = 126). Percentage with XIC PAC signals and sample sizes (n) are shown. (Scale bar, 5 μm.) (C) RNA FISH shows that XIST RNA domains (green) exclude Cot-1 hybridization (red), indicating transcriptional silencing of the X chromosome in WI-38 fibroblasts and class I and II lines indicated. Arrows point to examples of Xs in Cot-1 holes. (Scale bar, 10 μm.)
In theory, although not known to occur naturally in mice, it is possible that XCI may be uncoupled from XIST: XIST expression may not necessarily indicate XCI, or the absence of XIST may not rule out XCI. To test this possibility, we carried out Cot-1 RNA FISH, a technique whereby nascent transcription from a chromosomal territory could be visualized by hybridization to highly repetitive elements (Cot-1 fraction) within prespliced hnRNA (17). As expected, we found that all XIST+ domains observed in class I and II excluded Cot-1 hybridization (Fig. 2C and Table 1): In class I cells, Cot-1 “holes” associated with XIST clouds increased between d0 and d9; in class II, XIST clouds were invariably associated with Cot-1 holes from d0 to d9. Thus, XIST RNA clouds correlated with XCI.
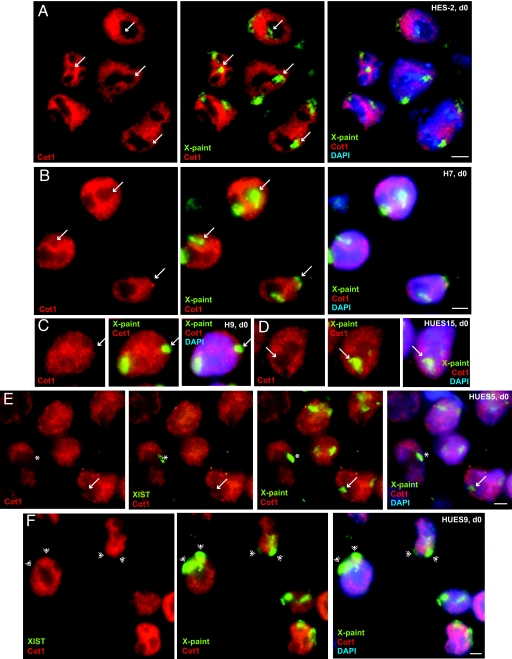
However, does the absence of XIST necessarily rule out XCI? To address this, we performed sequential RNA/DNA FISH on class III lines. We first performed Cot-1 RNA FISH on undenatured nuclei and then denatured the cells for X-chromosome painting to locate the two X chromosomes. Surprisingly, despite lacking XIST, one X excluded Cot-1 hybridization in a large majority of nuclei (Fig. 3, Table 1, and data not shown). For HES2, Cot-1 holes were already observed in 80% of nuclei on d0 and in 81% of nuclei on d15. Likewise, for HUES15, 73–84% excluded Cot-1 hybridization on one X on d0 and d15. For H7 and H9, Cot-1 holes were seen in 78% of d0 XIST− cells. These data led to the intriguing idea that class III cells might have undergone XCI but subsequently lost XIST expression.
Fig. 3.
Cot-1 RNA FISH reveals that XIST− nuclei of class II and III lines have Xi. (A–D) Combined RNA/DNA FISH reveals that class III lines have undergone XCI but lost XIST expression. Arrows point to Xi associated with a Cot-1 hole. (E) XIST− cells of p.23 HUES5 class II line also exhibit one Xi. Asterisk denotes XIST-coated Xi. (F) XIST− d0 cells of the class I line HUES9 largely do not exclude Cot-1 hybridization. Double arrowheads indicate Cot-1+ X chromosomes. (Scale bar, 5 μm.)
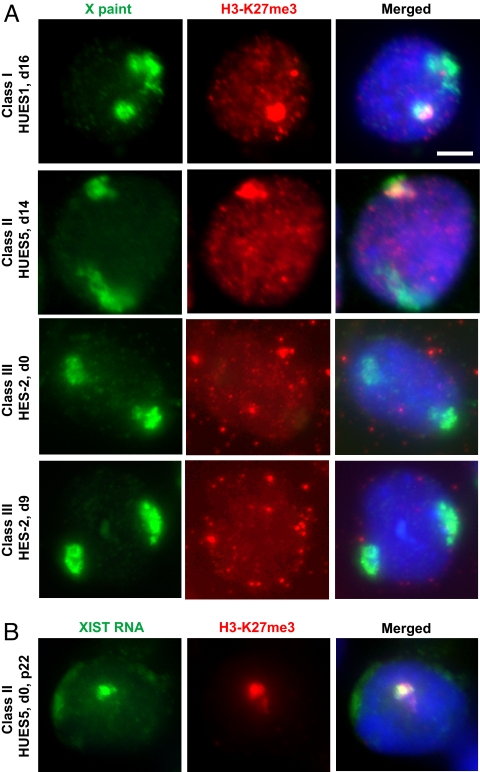
In the mouse, Xist up-regulation and XCI are accompanied by trimethylation of H3-K27 (H3-K27me3) on Xi (reviewed in ref. 14). In class I hESC, trimethylation also accompanied XCI (Fig. 4 and Table 1). In class II hESC, H3-K27me3 domains were observed in all XIST+ cells. In class III lines, H3-K27me3 domains were not observed at all. Thus, XIST clouds correlated perfectly with hypermethylation of H3-K27. Although a Cot-1 hole remained distinct (Fig. 3), loss of XIST in class III lines invariably led to H3-K27me3 depletion on the Xi. In mice, deleting Xist also causes H3-K27me3 depletion on Xi without losing the Cot-1-deficient domain (18).
Fig. 4.
The H3-K27me3 mark of the Xi is lost in XIST− cells. (A) ImmunoFISH combining immunostaining for H3-K27me3 (red) and X chromosome painting (green). (Scale bar, 5 μm.) (B) Immunostaining of H3-K27me3 (red) combined with XIST RNA FISH (green).
Our results raised the intriguing scenario that class II may differ from class III primarily in the degree of XIST loss. Indeed, our cell lines readily change from class II to class III. For example, a short-term culture of HUES5 at p.22 was XIST+ in 70% of cells on d0 (Table 1), but longer-term culture at p.23 reduced XIST+ frequency to 24% on d0 and 30% after differentiation. For class II, all XIST+ chromosomes resided within a Cot-1 hole, as expected. However, 63% of XIST− chromosomes at p.23 also excluded Cot-1 staining 63% (Fig. 3E). Therefore, a significant fraction of the later-passage class II cells was class III-like. By contrast, a majority of the XIST− chromosomes of class I lines did not exclude Cot-1 staining (Fig. 3F) and could up-regulate XIST upon differentiation (Table 1), indicating that class I lines, in contrast to class II lines, are largely pre-XCI on d0 but can recapitulate XCI during differentiation. Thus, class II cells may be an “intermediate” between the pre-XCI class I state and the XIST−, post-XCI class III state. Disturbingly, class I lines also readily transform into class III during culture (data not shown). On d0, some XIST− chromosomes of class I also exclude Cot-1 hybridization (Table 1 and data not shown). These data demonstrate the epigenetic fluidity of all hESC lines in culture.
Our results have several important implications. We believe that, like mESC, hESC in their most pristine condition are poised in the pre-XCI state but can recapitulate XCI when placed in differentiation conditions (class I). However, perhaps because optimal culture conditions have yet to be established for hESC, hESC may be prone to differentiate spontaneously and induce XCI (class II). Unlike mESC, hESC are prone to lose XIST expression once XCI is initiated ex vivo (class III). Although the molecular basis of this unusual epigenetic behavior is unknown, the data are clear in warning that an absence of XIST cannot be equated with an absence of XCI. Without XIST, the Xi loses H3-K27me3 but does not reactivate immediately, consistent with the stability of XCI during the maintenance phase, although partial reactivation can occur (18–20). These conclusions have implications for the interpretation of analyses based solely on XIST expression.
Because XCI in the mouse is tightly linked to differentiation (8, 21), our conclusions may have implications for hESC pluripotency. Regardless of class, all hESC lines expressed the OCT4, alkaline phosphatase, SSEA4, and TRA-1-81 pluripotency markers on d0, as determined by immunostaining and RT-PCR (SI Fig. 6A and data not shown). None of the lineage-specific markers (MAP2 for ectoderm, SMA for mesoderm, and FOXA2/AFP for endoderm) were expressed on d0. When placed in differentiation conditions, all hESC lines gradually lost expression of pluripotency markers (SI Fig. 6B and data not shown), but embryoid body (EB) quality and their ability to express lineage-specific differentiation markers varied somewhat between cell lines. Although class I and II cells showed fair to robust EB growth, class III lines exhibited a range of phenotypes: H7, H9, and HES-2 displayed scant EB outgrowth and HUES6 formed very poor EB (SI Fig. 6C and Table 1). Furthermore, whereas all lines after d15 showed robust staining with lineage-specific markers, the HUES6 showed poor staining for endodermal markers (SI Fig. 6D and Table 1). We conclude that hESC lines are not epigenetically or developmentally equivalent. However, we cannot rule out the possibility that anomalies unrelated to XCI may also cause aberrant differentiation.
In conclusion, our study provides a perspective on the epigenetic fluidity of hESC. A parallel report has reached similar conclusions (22). The “pristine” hESC is uncommitted to XCI and can recapitulate XCI during cell differentiation, but there is a strong tendency to initiate XCI ex vivo and lose XIST expression thereafter. Even lines categorized as class I show XIST clouds in 15–20% of nuclei, suggesting a pre-class II state. The tendency toward initiating XCI is further highlighted by the number of lines that fall into class III and the ease with which all lines eventually become class III. Thus, the three classes represent a progression of epigenotypes ex vivo, with class I being the closest approximation of the “original” pluripotent state, class II being a partially differentiated state, and class III being a state of further adaptation to culture conditions. These results provide a unifying explanation for the diverse patterns of XIST expression in the H9 and H7 sublines reported in refs. 9–12.
The potential for X-reactivation in hESC lines must be considered carefully when evaluating the safety of hESC for in vivo applications. Supernumerary active X chromosomes may confer a selective advantage in culture (3) and have been correlated with both male and female malignancies (23, 24). The possibility of suboptimal culture conditions being a cause of aberrant epigenetic reprogramming should be tested. Our present analysis, though performed in female hESC, has broader implications for both male and female lines, as indeed all hESC are isolated, cultured, and passaged under similar conditions.
Materials and Methods
Maintenance of hESC Lines.
All hESC protocols were approved by the Massachusetts General Hospital Office of Embryonic Stem Cell Research Oversight in conformity with federal regulations. H7, H9 (WiCell), HES-2, and HES-3 cell lines (ES Cell International) were cultured according to WiCell instructions. Undifferentiated cells were grown on ã-irradiated ICR (Taconic) MEF on gelatin-coated plates in DMEM-F12 (Invitrogen) supplemented with 20% knockout serum replacement (Invitrogen), 1% nonessential amino acids (Invitrogen), 1 mM glutamine (Invitrogen), 50 units/ml penicillin and 50 μg/ml streptomycin (Invitrogen) and 0.1 mM 2-mercaptoethanol (Sigma). Undifferentiated cultures were maintained in 10ng/ìl basic-FGF (R&D Systems). H7 and H9 cells were passaged by mechanical dissociation or by treatment with 0.05% trypsin-EDTA for 30 s. The HUES lines were maintained on MEFs in KO-DMEM (Invitrogen) supplemented with 10% serum replacement (Invitrogen), 10% plasmanate (Bayer), 1% nonessential amino acids, 2 mM glutamax-I, 50 units/ml penicillin and 50 μg/ml streptomycin, 0.055 mM 2-mercaptoethanol, and 10 ng/ìl basic-FGF. HUES cells were passaged by treatment with 0.05% trypsin-EDTA for 30 s. All lines were differentiated by EB suspension in low-attachment six-well plates (Corning) in media containing 10–20% FBS (Hyclone) in place of the serum replacement and plasmanate. After d5-d6, cells were plated onto Matrigel-coated 6-well plates and grown for 8–10 days.
RT-PCR.
Strand-specific RT-PCR was performed as described in ref. 25. PCR products were quantitated on the BioRad iCycler real-time PCR machine using SYBR green. Primer sequences are available on request.
RNA/DNA FISH and Immunofluorescence.
RNA and DNA FISH was carried out as described in ref. 25. Dispersed HESC were cytospun onto glass slides prior to fixation. StarFISH X chromosomal paints (Cambio) were hybridized according to manufacturer's instructions. DNA probes were generated to XIST exon 1 (GenBank U80460: 61251–69449) labeled with fluorescein-12-dUTP and Cot-1 labeled with Texas Red-5-dUTP (ChromaTide) using the Prime-It Fluor Labeling kit (Stratagene). XIST RNA exon 6 probes (GenBank U80460: 75081–78658) were transcribed by using T7 RNA polymerase (Roche) with Alexa Fluor 488–5-UTP. XIC PAC probe was labeled with fluorescein-12-dUTP by nick translation. In sequential RNA/DNA FISH, RNA FISH was performed first, 0.2-m z-section images were captured, their x–y coordinates were marked, and then the same slide was denatured for subsequent DNA FISH. Images were then overlaid by using DAPI nuclear staining as reference. H3-K27me3 immunoFISH was performed by using a rabbit polyclonal antibody (1:200 dilution, Upstate Biotechnology) and a goat anti-rabbit Alexa Fluor-555 secondary antibody (1:500). After fixing the immunofluorescence signals with 4% paraformaldeyde, X-chromosome, DNA FISH was performed. For XIST/H3-K27me3 RNA immunoFISH, RNA FISH was carried out first, the signals were fixed, and then immunostaining of H3-K27me3 was undertaken as described in ref. 18.
Immunostaining of Differentiation Markers.
See SI Text for details.
Giemsa Staining.
See SI Text for details.
Supplementary Material
Acknowledgments.
We are grateful to C. Cowan and D. Melton for providing the HUES lines and to S. Namekawa for microscopy advice. We thank all laboratory members for helpful discussion, and Z. Wang, T. Schlaeger, L. Daheron, and D. Datta for technical advice. Work on federally approved hESC lines was funded by a National Institutes of Health Grant R01-GM58839-S1; work on HUES lines was funded by a Harvard Stem Cell Institute grant (to J.T.L.). S.S.S. is partially funded by a predoctoral fellowship from the Gulbenkian Institute–Portugal (Fundação para a Ciência e Tecnologia SFRH/BD/9614/2002), S.M. by the Harvard MCB Graduate Program. J.T.L. is also an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712136105/DC1.
References
- 1.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Spagnoli FM, Hemmati-Brivanlou A. Guiding embryonic stem cells toward differentiation: Lessons from molecular embryology. Curr Opin Genet Dev. 2006;16:469–475. doi: 10.1016/j.gde.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Baker DE, et al. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol. 2007;25:207–215. doi: 10.1038/nbt1285. [DOI] [PubMed] [Google Scholar]
- 4.Maitra A, et al. Genomic alterations in cultured human embryonic stem cells. Nat Genet. 2005;37:1099–1103. doi: 10.1038/ng1631. [DOI] [PubMed] [Google Scholar]
- 5.Lagarkova MA, Volchkov PY, Lyakisheva AV, Philonenko ES, Kiselev SL. Diverse epigenetic profile of novel human embryonic stem cell lines. Cell Cycle. 2006;5:416–420. doi: 10.4161/cc.5.4.2440. [DOI] [PubMed] [Google Scholar]
- 6.Rugg-Gunn PJ, Ferguson-Smith AC, Pedersen RA. Epigenetic status of human embryonic stem cells. Nat Genet. 2005;37:585–587. doi: 10.1038/ng1556. [DOI] [PubMed] [Google Scholar]
- 7.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 8.Monk M, Harper MI. Sequential X chromosome inactivation coupled with cellular differentiation in early mouse embryos. Nature. 1979;281:311–313. doi: 10.1038/281311a0. [DOI] [PubMed] [Google Scholar]
- 9.Dhara SK, Benvenisty N. Gene trap as a tool for genome annotation and analysis of X chromosome inactivation in human embryonic stem cells. Nucleic Acids Res. 2004;32:3995–4002. doi: 10.1093/nar/gkh746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enver T, et al. Cellular differentiation hierarchies in normal and culture-adapted human embryonic stem cells. Hum Mol Genet. 2005;14:3129–3140. doi: 10.1093/hmg/ddi345. [DOI] [PubMed] [Google Scholar]
- 11.Hoffman LM, et al. X-inactivation status varies in human embryonic stem cell lines. Stem Cells. 2005;23:1468–1478. doi: 10.1634/stemcells.2004-0371. [DOI] [PubMed] [Google Scholar]
- 12.Sperger JM, et al. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc Natl Acad Sci USA. 2003;100:13350–13355. doi: 10.1073/pnas.2235735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adewumi O, et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- 14.Lucchesi JC, Kelly WG, Panning B. Chromatin remodeling in dosage compensation. Annu Rev Genet. 2005;39:615–651. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- 15.Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. Embryonic stem cell lines from human blastocysts: Somatic differentiation in vitro. Nat Biotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- 16.Cowan CA, et al. Derivation of embryonic stem-cell lines from human blastocysts. N Engl J Med. 2004;350:1353–1356. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- 17.Hall LL, Byron M, Sakai K, Carrel L, Willard HF, Lawrence JB. An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells. Proc Natl Acad Sci USA. 2002;99:8677–82. doi: 10.1073/pnas.132468999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang LF, Huynh KD, Lee JT. Perinucleolar targeting of the inactive X during S phase: Evidence for a role in the maintenance of silencing. Cell. 2007;129:693–706. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- 19.Brown CJ, Willard HF. The human X-inactivation centre is not required for maintenance of X-chromosome inactivation. Nature. 1994;368:154–156. doi: 10.1038/368154a0. [DOI] [PubMed] [Google Scholar]
- 20.Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol. 2001;153:773–783. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JT. Regulation of X-chromosome counting by Tsix and Xite sequences. Science. 2005;309:768–771. doi: 10.1126/science.1113673. [DOI] [PubMed] [Google Scholar]
- 22.Hall LL, et al. X-inactivation reveals epigenetic anomalies in most hESC but identifies sublines that initiate as expected. J Cell Physiol. 2008 doi: 10.1002/jcp.21411. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawakami T, et al. The roles of supernumerical X chromosomes and XIST expression in testicular germ cell tumors. J Urol. 2003;169:1546–1552. doi: 10.1097/01.ju.0000044927.23323.5a. [DOI] [PubMed] [Google Scholar]
- 24.Sirchia SM, et al. Loss of the inactive X chromosome and replication of the active X in BRCA1-defective and wild-type breast cancer cells. Cancer Res. 2005;65:2139–2146. doi: 10.1158/0008-5472.CAN-04-3465. [DOI] [PubMed] [Google Scholar]
- 25.Lee JT, Lu N. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell. 1999;99:47–57. doi: 10.1016/s0092-8674(00)80061-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.