Abstract
Recent research has shown that genetic drift may have produced many cranial differences between Neandertals and modern humans. If this is the case, then it should be possible to estimate population genetic parameters from Neandertal and modern human cranial measurements in a manner analogous to how estimates are made from DNA sequences. Building on previous work in evolutionary quantitative genetics and on microsatellites, we present a divergence time estimator for neutrally evolving morphological measurements. We then apply this estimator to 37 standard cranial measurements collected on 2,524 modern humans from 30 globally distributed populations and 20 Neandertal specimens. We calculate that the lineages leading to Neandertals and modern humans split ≈311,000 (95% C.I.: 182,000 to 466,000) or 435,000 (95% C.I.: 308,000 to 592,000) years ago, depending on assumptions about changes in within-population variation. These dates are quite similar to those recently derived from ancient Neandertal and extant human DNA sequences. Close correspondence between cranial and DNA-sequence results implies that both datasets largely, although not necessarily exclusively, reflect neutral divergence, causing them to track population history or phylogeny rather than the action of diversifying natural selection. The cranial dataset covers only aspects of cranial anatomy that can be readily quantified with standard osteometric tools, so future research will be needed to determine whether these results are representative. Nonetheless, for the measurements we consider here, we find no conflict between molecules and morphology.
Keywords: craniometrics, evolutionary quantitative genetics, microsatellites, population genetics, human evolution
Lately, evidence has been accumulating for the importance of neutral evolution in producing cranial differences among human populations and between Neandertals‖ and modern humans. Under neutral evolution, genetic drift provides the mechanism, and mutation provides the raw material for divergence between groups, with natural selection being relegated to a role of slowing down divergence (1–3). Lynch (4) and Relethford (5) provided some of the first evidence of the importance of neutral evolution for understanding human cranial differences. Lynch (4) found that cranial distances among human populations corresponded well with relative and absolute rates of divergence predicted by neutral evolution. Relethford (5) showed that estimates of FST (a measure of among-population differentiation) from human cranial measurements were similar to those from presumably neutral genetic loci. Further work established that cranial and molecular distances among human populations tend to be significantly associated with each other (6–9), both cranial and molecular distances are correlated with geographic distances among globally distributed human populations (10, 11), cranial measurements appear to fit neutral expectations as well as microsatellites for humans (12), and statistical tests fail to detect deviations from neutrality for cranial differences between Neandertals and modern humans, even though the tests look reasonably powerful (12). Furthermore, within-population cranial diversity seems to decrease with geographic distance from subSaharan Africa (13). This decrease may mirror a decrease in molecular diversity (14–17), but the strength of the cranial relationship is much weaker.
If neutral evolution is responsible for cranial differences among human populations and between Neandertals and modern humans, then it should be possible to estimate population genetic parameters from cranial measurements in a manner analogous to how estimates are made from DNA sequences. Building on previous work in evolutionary quantitative genetics (18–21) and on microsatellites (22–24), we present a divergence time estimator for neutrally evolving morphological measurements. We then apply this estimator to cranial measurements to infer when Neandertals and modern humans diverged, and we compare our results with those from ancient Neandertal and extant human DNA sequences.
Morphological Divergence Time Estimator
Divergence Time.
To estimate the divergence time of two populations (or species), we adapt the TD estimator that was originally developed for microsatellites (23). Microsatellites are rapidly evolving blocks of DNA for which a simple DNA sequence is repeated multiple times, and individuals vary in their number of repeats. So, like morphological measurements (metric characteristics), microsatellites are quantitative characters that change by lengthening and shortening (24–26), making the adaptation fairly straightforward. We call the new estimator PTD for phenotypic TD and to distinguish it from TD for microsatellites. By divergence time, we mean when the two populations last shared a randomly mating common ancestor.
We define PTD as follows. For two daughter populations at mutation drift equilibrium (balance between the addition of variation by mutation and the removal of variation by genetic drift), the between-population variance for a measurement is expected to increase at the rate of 2 Vm per generation (20, 21), where Vm is the average amount of new additive genetic variance introduced by mutation per zygote per generation in each population. This result holds for many different underlying genetic models (20) as long as the divergence is by genetic drift rather than by natural selection. Let x1 and x2 be measurement means for populations nos. 1 and 2, respectively. An estimator for the time in generations when the two populations diverged, under the assumption of mutation drift equilibrium, is:
Note that (x1−x2)2 equals twice the between-population variance, which is the quantity most commonly used in the evolutionary quantitative genetics literature (21). Also note that the numerator in Eq. 1 is mathematically analogous to the expression for (δμ)2, a genetic distance for microsatellites (22, 23), if a measurement is equated to a single microsatellite. The denominator in Eq. 1, or the expected rate of increase, is four times the mutation parameter, Vm, whereas for (δμ)2, it is twice the mutation parameter (22, 23). This is because of differences in the definitions of the mutation parameters. By necessity, all within-population and mutational variances in the quantitative genetics literature are zygotic, whereas they are gametic for microsatellites. Gametic variances include differences both among individuals and among the paired chromosomes of a given individual; zygotic variances only include among-individual differences. At Hardy–Weinberg equilibrium, a gametic variance should be twice a zygotic variance (27), hence the difference by a factor of two between the quantitative genetic and microsatellite formulas.
Following results for microsatellites (23), if the assumption of mutation drift equilibrium is relaxed, three additional variance terms must be added to the numerator (with modifications to account for zygotic vs. gametic variances as before). These terms are measures of the degree of departure from mutation drift equilibrium. Departures could happen with population expansions or migration between populations. Let V1 and V2 be the additive genetic variances for the measurements for populations nos. 1 and 2, respectively. Let V0 be the additive genetic variance in the ancestral population before subdivision into two daughter populations. Under a more general model, an estimator for the time in generations when the two populations diverged is:
Let h2 be the narrow-sense heritability for the measurement in both populations and σ12 and σ22 be the phenotypic variances for populations nos. 1 and 2, respectively. Following Falconer (28),
Let m be a mutation parameter and σP2 be the pooled within population phenotypic variance. Following Lynch (29),
Combining Eqs. 3, 4, and 5 with Eq. 2 results in
Setting V0 = 0 provides a maximum estimate of divergence time. Using V0 = (V1 + V2)/2 reduces Eq. 6 to a mutation-drift-equilibrium model (Eq. 1), which provides a minimum estimate unless there was a decline in population size (23).
The rationale behind the three additional variance terms can be illustrated with an example. Imagine two daughter populations with very small effective population sizes that are at mutation drift equilibrium. Because of their small sizes, these populations will be very homogeneous. If they expand rapidly to large effective sizes, at first they will remain homogeneous, but over time, mutations will continue to introduce variation until a new equilibrium is reached. In the initial generations after the split, the morphological divergence will be very slow, because genetic drift will be very weak. This is because in a large population, the chance effects of sampling are small, and if the population is homogeneous, it does not matter much which individuals give rise to the next generation. As the populations become less homogeneous with the addition of mutational variance over time, the rate of divergence will increase until the mutation-drift-equilibrium rate is reached. Because the divergence is slow initially, assuming that the populations were always at mutation, drift equilibrium will underestimate the actual divergence time. The three additional variance terms correct for this bias.
Effective Population Size.
If the amount of additive genetic variance introduced by mutation per generation, Vm, is known, then it should be possible to estimate the effective population size of a particular group (populations or species) from the amount of additive genetic variation found within that group. Let Ne be the effective size of a population. Following Lynch and Hill (20), at mutation drift equilibrium, the expected amount of additive genetic variance, V, found within the population is
Eq. 7 has been tested empirically, and it appears to hold reasonably well, at least on average, in inbreeding experiments on Drosophila melanogaster (30). Substituting V for V1 and σ for σ1 in Eq. 3, combining Eqs. 3 and 5 with Eq. 7, and solving for N results in an estimator for the effective size of the population given by
Statistical Analyses
Data.
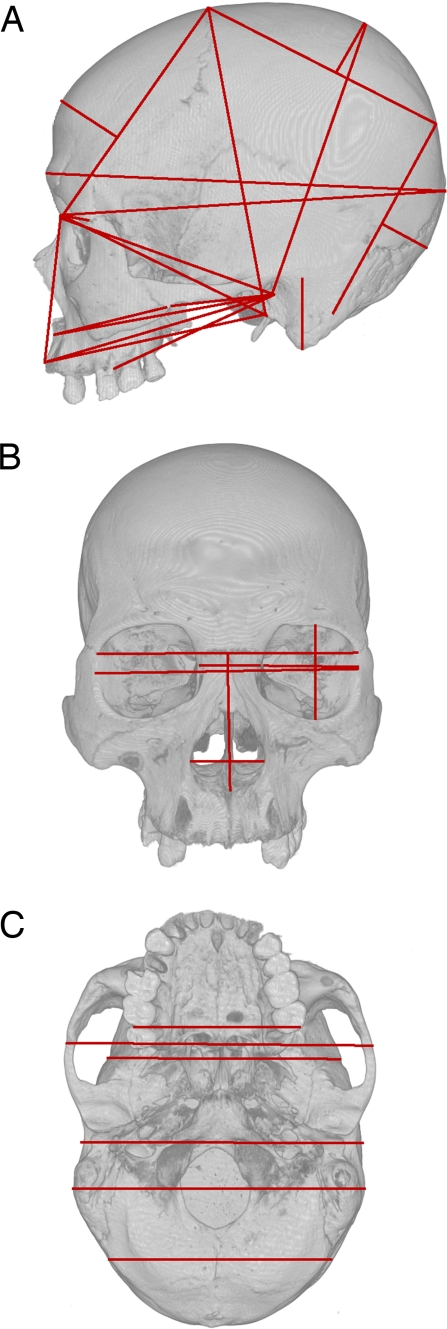
Our analyses are based on 37 standard cranial measurements (Fig. 1) collected on 2,524 modern humans from 30 globally distributed populations and 20 Neandertal specimens. More details about the sample and measurements can be found elsewhere (12, 31–33). We have previously shown that these 37 measurements appear to be evolving neutrally in Neandertals and modern humans (12), making them candidates for use in estimating divergence times.
Fig. 1.
The approximate locations of the cranial measurements used in the analyses are superimposed as red lines on lateral (A), anterior (B), and inferior (C) views of a human cranium. Note that when the endpoints of a measurement are not visible, the line is projected into a plane situated in front of the cranium.
Treatment of Multiple Measurements.
Divergence time can be estimated from multiple measurements as PTD, the mean of PTD estimates for individual measurements. The expected value for PTD for each measurement is the divergence time, so the expected value for PTD is also the divergence time, regardless of whether the measurements are correlated with each other. If the measurements are completely independent (uncorrelated within and between groups), bootstrapping (34) can be used to estimate the variance of PTD, but this procedure will underestimate Var {PTD} if the measurements are not independent.
It is always possible, however, to find a basis for which the measurements are uncorrelated within groups as the eigenvectors of the pooled within group variance–covariance matrix. There are as many eigenvectors as original measurements. Each eigenvector (sometimes called a principal component) represents a new measurement that is a linear combination of the original measurements and is uncorrelated with the other eigenvectors within groups. For neutral divergence, the elements of the between-group variance–covariance matrix are expected to be proportional to the elements of the within-group variance–covariance matrix (35–37), so the eigenvectors should be approximately uncorrelated between groups as well. Eq. 8 for effective population size can be extended to multiple measurements in a similar manner.
Calibration.
As with divergence time estimates from molecular data, to estimate an “unknown” divergence time, we must first calibrate certain parameters using “known” reference points. In our case, we need to calibrate m and h2. We pick two “known” reference points. The first is PTD under a mutation-drift-equilibrium model for the split between subSaharan African and other human populations. This divergence is the oldest among human populations, so it is the most appropriate calibration point for estimating even older divergence times. Zhivotovsky (23) estimated TD as ≈30,000 years ago based on 131 autosomal and four Y-chromosomal microsatellites and from comparisons with other studies. This divergence time estimate is almost certainly too recent, because the mutation-drift-equilibrium model assumes that there was no postdivergence gene flow between subSaharan African and other human populations, and that human populations have not expanded in size recently. However, by calibrating to a divergence time based on a mutation-drift-equilibrium model, we are just assuming that the morphological and microsatellite estimates should match up under the same model, not that this is the most realistic model to use to infer the actual divergence time. In other words, because the mutation-drift-equilibrium model is expected to underestimate the actual divergence time between subSaharan African and other human populations by the same amount for morphology and microsatellites, using 30,000 years ago as the calibration point will not result in an underestimate of the divergence time between Neandertals and modern humans.
The second reference point is the effective population size, PNe, under a mutation–drift–equilibrium model for subSaharan African human populations. Zhivotovsky and colleagues (17) estimated Ne from 271 microsatellites using an equation equivalent to our Eq. 7 as ≈2,700 individuals. Once again, we are just assuming that the morphological and microsatellite estimates should match up under the same model, not that this is the most realistic model to use to infer the actual effective population size. Using V0 = (V1 + V2)/2 in Eq. 8 for mutation drift equilibrium and setting PTD = TD in Eq. 6 and PNe = Ne in Eq. 8 produces two equations that can be solved for h2 and m, the two remaining unknown parameters. Following Zhivotovsky (23), we use a generation length of 25 years for all our calculations.
Confidence Limits.
We use bootstrapping (34) to calculate approximate 95% confidence limits for PTD. We resampled with replacement 10,000 times from the PTD estimates for the individual eigenvectors and calculated an estimate of PTD for each resample, producing a distribution of PTD estimates. The lower and upper confidence limits are the 2.5% and 97.5% quantiles respectively of this distribution. These confidence limits account for evolutionary stochasticity in the amount of mutational variance actually introduced per generation, but they do not account for uncertainty in the calibration. Because uncertainty in the calibration is difficult to quantify accurately, it is not usually included in confidence limits for divergence times estimated from DNA sequences. We follow this practice here, but it is important to point out that this additional uncertainly can often be quite large.
Calculations.
We wrote scripts in MATLAB (Mathworks) to perform all of the calculations, making use of the statistical toolbox and Richard E. Strauss' MATLAB function package.
Results
Calibration.
From our calibration, we estimate h2 = 0.37. Studies of human cranial measurements commonly use h2 = 0.55 (5, 6, 38, 39), but this value actually derives from soft-tissue head measurements, which appear to have higher heritabilities than skeletal measurements (40). In a recent study of skeletal measurements collected on a pedigreed human sample, Carson (40) estimated heritabilities for 21 of the 37 measurements we use here with mean h2 = 0.36. The sample sizes were small for three measurements, potentially making the h2 estimates unreliable (40). For the remaining 18 measurements, mean h2 = 0.31. Both estimates are quite close to our calibration-based estimate, especially considering the considerable uncertainty in h2 estimates. Additionally, we do not expect an exact match, because the populations are different, and our calibration-based estimate reflects average h2 over hundreds to thousands of generations (evolutionary time) rather than over just a few generations.
From our calibration, we estimate m = 1.20 × 10−4. Experimental studies of a number of different taxa for a variety of traits (21, 29, 41) suggest a rough range of 10−4 to 10−2 for m. Our calibration-based estimate is at the lower end of the range, which is expected, because most experimental values for m are overestimates. Experimental estimates consider all mutations, not just neutral ones, and over evolutionary time stabilizing selection will remove some mutations from the population. The overestimation may sometimes be substantial, because the experimental populations have low effective sizes, which would weaken stabilizing selection (1). A similar overestimation problem is documented for molecular evolution where mutation rates estimated from pedigrees are much higher than those estimated from phylogenetic calibration points (42, 43).
Neandertal and Modern Human Divergence.
We estimate that Neandertals and modern humans diverged ≈311,000 years ago (95% C.I.: 182,000–466,000) assuming mutation drift equilibrium or 435,000 years ago (95% C.I.: 308,000–592,000) assuming V0 = 0. For both estimates, we added 25,000 years to account for the fact (averaging dates) that Neandertals lived ≈50,000 years ago. When we compare Neandertals with only male recent humans, the point estimates and C.I.s decrease by <10,000 years, so our estimates would not be strongly biased, even if the entire Neandertal sample were male.
It is difficult to decide which V0 model is most appropriate. The V0 = 0 result is probably an overestimate for at least two reasons. (i) Because the Neandertal sample is too small to accurately estimate within-population variation in Neandertals, we used the human value for both V1 and V2. If Neandertals were actually less variable than present-day human populations, the V0 = 0 result would be an overestimate (i.e., the Neandertal lineage would maximally deviate from mutation drift equilibrium less than the modern human lineage). (ii) The additive genetic variance in the last common ancestor of Neandertals and modern humans must have been greater than zero, making the V0 = 0 result an overestimate. In contrast, the mutation-drift equilibrium, V0 = (V1 + V2)/2, result could be an underestimate for at least two reasons. (i) Human populations have grown in size recently, which would make the mutation-drift-equilibrium result an underestimate as long as this growth in census size corresponds to growth in effective size. (ii) Postdivergence gene flow between Neandertals and modern humans would make the mutation-drift-equilibrium result an underestimate (23). Given this uncertainty, the mean of the two estimates, 373,000 years ago, seems to be a reasonable point estimate.
Discussion
If we consider the maximum extent of the 95% confidence limits for both the mutation-drift equilibrium and the V0 = 0 estimates, then the Neandertal and modern human lineages split between 182,000 and 592,000 years ago. Although this range is quite large, it still allows for some observations with respect to the human fossil record. First, even the lower limit is within the Middle Pleistocene, suggesting a relatively deep divergence of Neandertals and modern humans, which is consistent with the presence of derived Neandertal features on Middle Pleistocene fossils from Europe (44–47). Second, recent dates suggesting that the Sima de los Huesos site is >530,000 years old (48) would put the fossils from this site, which appear to have multiple derived Neandertal features (46), at or potentially before the split of the Neandertal and modern human lineages. Third, although the ≈800,000-year-old Atapuerca-TD6 humans (49) could be ancestral to Neandertals and modern humans, their morphology may not be representative of the source population that actually gave rise to Neandertals and modern humans, because they date from, at minimum, ≈200,000 years before the split time.
Our PTD results are estimates of when the ancestral Neandertal and modern human populations last shared a randomly mating common ancestor (split time), whereas most molecular estimates are DNA sequence coalescence times. As long as the mutation rate is correct, coalescence times will equal or predate the split time by an amount that depends, on average, on ancestral effective population size (50). For example, if the ancestral population had a constant effective size of 2,500 individuals, an autosomal coalescence time based on one Neandertal and one extant human sequence would be expected to predate the split time by 125,000 years, assuming a generation length of 25 years. Point estimates for the coalescence of ancient Neandertal and extant human sequences for both mitochondrial and nuclear DNA range from ≈300,000 to 800,000 years ago (42, 51–55). Additionally, Noonan and colleagues (53) estimated that the Neandertal and modern human lineages split ≈370,000 years ago based on comparisons at >36,000 autosomal DNA sites. This split time is very similar to 373,000 years ago, the average of our mutation-drift-equilibrium and V0 = 0 point estimates.
The divergence time estimates could change somewhat if the separation between Neandertals and modern humans was actually more complicated than a simple splitting of populations. Additionally, although the ancestors of Neandertals and modern humans may have split ≈400,000 years ago, modern humans are sometimes thought to have originated with a subsequent speciation event ≈150,000 years ago. It is unclear what this view implies demographically, but it could be taken to mean that the emergence of modern humans involved a bottleneck, which is a sharp reduction followed by a rapid expansion in population size. If such a bottleneck occurred, the mutation-drift-equilibrium result would likely be an underestimate, and the V0 = 0 estimate would be closer to the actual split time.
Regardless of these potential complications, the close correspondence between the cranial and DNA-sequence estimates implies that both datasets largely, although not necessarily exclusively, reflect neutral divergence, causing them to track population history or phylogeny rather than the action of diversifying natural selection. The overall pattern for the measurements considered here appears to be neutral divergence, but as is the case among human populations (6–8, 38), a few measurements could still have diverged by diversifying natural selection. Additionally, future research will be needed to determine if this pattern is representative of Neandertal and modern human cranial divergence in general, or whether it applies only to the measurements included in our study. In particular, more detailed or internal cranial structures and other aspects of cranial anatomy that are difficult to quantify with standard osteometric tools many have been shaped by diversifying natural selection.
Brain size relative to body size appears to have increased in both the Neandertal and the modern human lineages (56, 57). These parallel trajectories may indicate that directional natural selection was acting on both lineages independently, resulting in differently shaped but similarly sized brain cases (58). To the extent that the measurements considered here reflect these changes, our results would imply that although natural selection may have produced the similarities in size, genetic drift lead to the differences in shape. We are not arguing that natural selection had no effect, just that it appears not to have played a dominant role in producing differences.
One misconception about neutral evolution is that it is slow, because it is driven by genetic drift rather than by natural selection. However, if stabilizing natural selection is more prevalent than directional natural selection, then neutral evolution will actually be comparatively fast. Lynch (59) estimated that human cranial evolution was rapid relative to morphological evolution in other mammals, but the absolute rate was consistent with neutral evolution. It follows from these results and ours that Neandertal and modern human crania may have been released from the constraints of stabilizing natural selection that limit the rates of morphological evolution in other mammals.
Perhaps the most significant implication of our results is that there is no conflict between molecules and morphology. This contrasts with a common characterization of debates about the origins of modern humans as molecules vs. morphology (60). In fact, at least for the measurements considered here, there is a close quantitative correspondence between the amount of cranial divergence and the amount of DNA sequence divergence between Neandertals and modern humans.
Acknowledgments.
We thank D. DeGusta, R. Fournier, B. Henn, J.-J. Hublin, K. A. Horsburg, R. Klein, J. Mountain, N. Rosenberg, T. Steele, M. Stoneking, and two anonymous reviewers for valuable comments on drafts or feedback; the late W. W. Howells for making his raw data publicly available; the institutes that made their fossil material available for study; M. v. Harling for the CT scans used to create Fig. 1; and the Max Planck Society for financial support.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Because of a spelling change in German, there are two acceptable spellings for the name of the group. “Neanderthal” rather than “Neandertal” is the preferred spelling of C.B.S.
References
- 1.Ohta T. The current significance and standing of neutral and nearly neutral theories. BioEssays. 1996;18:673–677. doi: 10.1002/bies.950180811. [DOI] [PubMed] [Google Scholar]
- 2.Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- 3.Kimura M. The neutral theory of molecular evolution and the world view of the neutralists. Genome. 1989;31:24–31. doi: 10.1139/g89-009. [DOI] [PubMed] [Google Scholar]
- 4.Lynch M. Phylogenetic hypotheses under the assumption of neutral quantitative-genetic variation. Evolution (Lawrence, Kans) 1989;43:1–17. doi: 10.1111/j.1558-5646.1989.tb04203.x. [DOI] [PubMed] [Google Scholar]
- 5.Relethford JH. Craniometric variation among modern human populations. Am J Phys Anthropol. 1994;95:53–62. doi: 10.1002/ajpa.1330950105. [DOI] [PubMed] [Google Scholar]
- 6.Roseman CC. Detection of interregionally diversifying natural selection on modern human cranial form by using matched molecular and morphometric data. Proc Natl Acad Sci USA. 2004;101:12824–12829. doi: 10.1073/pnas.0402637101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harvati K, Weaver TD. Human cranial anatomy and the differential preservation of population history and climate signatures. Anat Rec. 2006;288A:1225–1233. doi: 10.1002/ar.a.20395. [DOI] [PubMed] [Google Scholar]
- 8.Harvati K, Weaver TD. In: Neanderthals Revisited: New Approaches and Perspectives. Harvati K, Harrison T, editors. New York: Springer; 2006. pp. 239–254. [Google Scholar]
- 9.Gonzáles-Jose R, Van der Molen S, Gonzales-Perez E, Hernandez M. Patterns of phenotypic covariation and correlation in modern humans as viewed from morphological integration. Am J Phys Anthropol. 2004;123:69–77. doi: 10.1002/ajpa.10302. [DOI] [PubMed] [Google Scholar]
- 10.Relethford JH. Boas and beyond: migration and craniometric variation. Am J Hum Biol. 2004;16:379–386. doi: 10.1002/ajhb.20045. [DOI] [PubMed] [Google Scholar]
- 11.Relethford JH. Global patterns of isolation by distance based on genetic and morphological data. Hum Biol. 2004;76:499–513. doi: 10.1353/hub.2004.0060. [DOI] [PubMed] [Google Scholar]
- 12.Weaver TD, Roseman CC, Stringer CB. Were Neandertal and modern human cranial differences produced by natural selection or genetic drift? J Hum Evol. 2007;53:135–145. doi: 10.1016/j.jhevol.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 13.Manica A, Amos W, Balloux F, Hanihara T. The effect of ancient population bottlenecks on human phenotypic variation. Nature. 2007;448:346–349. doi: 10.1038/nature05951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conrad DF, et al. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat Genet. 2006;38:1251–1260. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- 15.Prugnolle F, Manica A, Balloux F. Geography predicts neutral genetic diversity of human populations. Curr Biol. 2005;15:R159–R160. doi: 10.1016/j.cub.2005.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramachandran S, et al. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci USA. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhivotovsky LA, Rosenberg NA, Feldman MW. Features of evolution and expansion of modern humans, inferred from genomewide microsatellite markers. Am J Hum Genet. 2003;72:1171–1186. doi: 10.1086/375120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lande R. Natural selection and random genetic drift in phenotypic evolution. Evolution (Lawrence, Kans) 1976;30:314–334. doi: 10.1111/j.1558-5646.1976.tb00911.x. [DOI] [PubMed] [Google Scholar]
- 19.Lande R. Statistical tests for natural selection on quantitative characters. Evolution (Lawrence, Kans) 1977;31:442–444. doi: 10.1111/j.1558-5646.1977.tb01025.x. [DOI] [PubMed] [Google Scholar]
- 20.Lynch M, Hill WG. Phenotypic evolution by neutral mutation. Evolution (Lawrence, Kans) 1986;40:915–935. doi: 10.1111/j.1558-5646.1986.tb00561.x. [DOI] [PubMed] [Google Scholar]
- 21.Turelli M, Gillespie JH, Lande R. Rate tests for selection on quantitative characters during macroevolution and microevolution. Evolution (Lawrence, Kans) 1988;42:1085–1089. doi: 10.1111/j.1558-5646.1988.tb02526.x. [DOI] [PubMed] [Google Scholar]
- 22.Goldstein DB, Ruiz Linares A, Cavalli-Sforza LL, Feldman MW. Genetic absolute dating based on microsatellites and the origin of modern humans. Proc Natl Acad Sci USA. 1995;92:6723–6727. doi: 10.1073/pnas.92.15.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhivotovsky LA. Estimating divergence time with the use of microsatellite genetic distances: impacts of population growth and gene flow. Mol Biol Evol. 2001;18:700–709. doi: 10.1093/oxfordjournals.molbev.a003852. [DOI] [PubMed] [Google Scholar]
- 24.Zhivotovsky LA, Feldman MW. Microsatellite variability and genetic distances. Proc Natl Acad Sci USA. 1995;92:11549–11552. doi: 10.1073/pnas.92.25.11549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldstein DB, Linares AR, Cavalli-Sforza LL, Feldman MW. An evaluation of genetic distances for use with microsatellite loci. Genetics. 1995;139:463–471. doi: 10.1093/genetics/139.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kremer A, Zanetto A, Ducousso A. Multilocus and multitrait measures of differentiation for gene markers and phenotypic traits. Genetics. 1997;145:1229–1241. doi: 10.1093/genetics/145.4.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falconer DS. Introduction to Quantitative Genetics. Essex, UK: Longman Group Limited; 1981. [Google Scholar]
- 29.Lynch M. The rate of polygenic mutation. Genet Res. 1988;51:137–148. doi: 10.1017/s0016672300024150. [DOI] [PubMed] [Google Scholar]
- 30.Whitlock MC, Fowler K. The changes in genetic and environmental variance with inbreeding in Drosophila melanogaster. Genetics. 1999;152:345–353. doi: 10.1093/genetics/152.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Howells WW. Cranial Variation in Man: A Study by Multivariate Analysis of Patterns of Difference Among Recent Human Populations. Cambridge, MA: Harvard University; 1973. [Google Scholar]
- 32.Howells WW. Skull Shapes and the Map: Craniometric Analyses in the Dispersion of Modern Homo. Cambridge, MA: Harvard University; 1989. [Google Scholar]
- 33.Howells WW. Who's Who in Skulls: Ethnic Identification of Crania from Measurements. Cambridge, MA: Harvard University; 1995. [Google Scholar]
- 34.Efron B, Gong G. A leisurely look at the bootstrap, the jacknife, and cross-validation. Am Stat. 1983;37:36–48. [Google Scholar]
- 35.Lande R. Quantitative genetic analysis of multivariate evolution, applied to brain:body size allometry. Evolution (Lawrence, Kans) 1979;33:402–416. doi: 10.1111/j.1558-5646.1979.tb04694.x. [DOI] [PubMed] [Google Scholar]
- 36.Ackermann RR, Cheverud JM. Discerning evolutionary processes in patterns of Tamarin (Genus Saguinus) craniofacial variation. Am J Phys Anthropol. 2002;117:260–271. doi: 10.1002/ajpa.10038. [DOI] [PubMed] [Google Scholar]
- 37.Ackermann RR, Cheverud JM. Detecting genetic drift versus selection in human evolution. Proc Natl Acad Sci USA. 2004;101:17946–17951. doi: 10.1073/pnas.0405919102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roseman CC, Weaver TD. Multivariate apportionment of global human craniometric diversity. Am J Phys Anthropol. 2004;125:257–263. doi: 10.1002/ajpa.10424. [DOI] [PubMed] [Google Scholar]
- 39.Schillaci MA, Froehlich JW. Nonhuman primate hybridization and the taxonomic status of Neanderthals. Am J Phys Anthropol. 2001;115:157–166. doi: 10.1002/ajpa.1065. [DOI] [PubMed] [Google Scholar]
- 40.Carson EA. Maximum likelihood estimation of human craniometric heritabilities. Am J Phys Anthropol. 2006;131:169–180. doi: 10.1002/ajpa.20424. [DOI] [PubMed] [Google Scholar]
- 41.Houle D, Morikawa B, Lynch M. Comparing mutational variabilities. Genetics. 1996;143:1467–1483. doi: 10.1093/genetics/143.3.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ho SYW, Phillips MJ, Cooper A, Drummond AJ. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol Biol Evol. 2005;22:1561–1568. doi: 10.1093/molbev/msi145. [DOI] [PubMed] [Google Scholar]
- 43.Zhivotovsky LA, et al. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet. 2004;74:50–61. doi: 10.1086/380911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hublin J-J. In: Neandertals and Modern Humans in Western Asia. Akazawa T, Aoki K, Bar-Yosef O, editors. New York: Plenum; 1998. pp. 295–310. [Google Scholar]
- 45.Santa Luca AP. A re-examination of presumed Neandertal-like fossils. J Hum Evol. 1978;7:619–636. [Google Scholar]
- 46.Arsuaga JL, Martínez I, Gracia A, Lorenzo C. The Sima de los Huesos crania (Sierra de Atapuerca, Spain). A comparative study. J Hum Evol. 1997;33:219–281. doi: 10.1006/jhev.1997.0133. [DOI] [PubMed] [Google Scholar]
- 47.Stringer CB, Hublin J-J. New age estimates for the Swanscombe hominid, and their significance for human evolution. J Hum Evol. 1999;37:873–877. doi: 10.1006/jhev.1999.0367. [DOI] [PubMed] [Google Scholar]
- 48.Bischoff JL, et al. High-resolution U-series dates from the Sima de los Huesos hominids yields 600 kyrs: implications for the evolution of the early Neanderthal lineage. J Archaeol Sci. 2007;34:763–770. [Google Scholar]
- 49.Bermúdez de Castro J-M, et al. A hominid from the Lower Pleistocene of Atapuerca, Spain: Possible ancestor to Neandertals and modern humans. Science. 1997;276:1392–1395. doi: 10.1126/science.276.5317.1392. [DOI] [PubMed] [Google Scholar]
- 50.Rosenberg NA, Feldman MW. In: Modern Developments in Theoretical Population Genetics. Slatkin M, Veuille M, editors. Oxford, UK: Oxford Univ Press; 2002. pp. 130–164. [Google Scholar]
- 51.Krings M, et al. Neandertal DNA sequences and the origin of modern humans. Cell. 1997;90:19–30. doi: 10.1016/s0092-8674(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 52.Green RE, et al. Analysis of one million base pairs of Neanderthal DNA. Nature. 2006;444:330–336. doi: 10.1038/nature05336. [DOI] [PubMed] [Google Scholar]
- 53.Noonan JP, et al. Sequencing and analysis of Neanderthal genomic DNA. Science. 2006;314:1113–1118. doi: 10.1126/science.1131412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ovchinnikov IV, et al. Molecular analysis of Neanderthal DNA from the northern Caucasus. Nature. 2000;404:490–493. doi: 10.1038/35006625. [DOI] [PubMed] [Google Scholar]
- 55.Beerli P, Edwards SV. When did Neanderthals and modern humans diverge? Evol Anthropol Suppl. 2002;1:60–63. [Google Scholar]
- 56.Arsuaga J-L, et al. A complete human pelvis from the Middle Pleistocene of Spain. Nature. 1999;399:255–258. doi: 10.1038/20430. [DOI] [PubMed] [Google Scholar]
- 57.Ruff CB, Trinkaus E, Holliday TW. Body mass and encephalization in Pleistocene Homo. Nature. 1997;387:173–176. doi: 10.1038/387173a0. [DOI] [PubMed] [Google Scholar]
- 58.Bruner E, Manzi G, Arsuaga J-L. Ecephalization and allometric trajectories in the genus Homo: evidence from the Neandertal and modern lineages. Proc Natl Acad Sci USA. 2003;100:15335–15340. doi: 10.1073/pnas.2536671100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lynch M. The rate of morphological evolution in mammals from the standpoint of neutral expectation. Am Nat. 1990;136:727–741. [Google Scholar]
- 60.Gura T. Bones, molecules or both? Nature. 2000;406:230–233. doi: 10.1038/35018729. [DOI] [PubMed] [Google Scholar]