Abstract
X chromosome inactivation (XCI) is an essential mechanism for dosage compensation of X-linked genes in female cells. We report that subcultures from lines of female human embryonic stem cells (hESCs) exhibit variation (0–100%) for XCI markers, including XIST RNA expression and enrichment of histone H3 lysine 27 trimethylation (H3K27me3) on the inactive X chromosome (Xi). Surprisingly, regardless of the presence or absence of XCI markers in different cultures, all female hESCs we examined (H7, H9, and HSF6 cells) exhibit a monoallelic expression pattern for a majority of X-linked genes. Our results suggest that these established female hESCs have already completed XCI during the process of derivation and/or propagation, and the XCI pattern of lines we investigated is already not random. Moreover, XIST gene expression in subsets of cultured female hESCs is unstable and subject to stable epigenetic silencing by DNA methylation. In the absence of XIST expression, ≈12% of X-linked promoter CpG islands become hypomethylated and a portion of X-linked alleles on the Xi are reactivated. Because alterations in dosage compensation of X-linked genes could impair somatic cell function, we propose that XCI status should be routinely checked in subcultures of female hESCs, with cultures displaying XCI markers better suited for use in regenerative medicine.
Keywords: culture variation, DNA methylation, gene regulation
Human embryonic stem cells (hESCs) are regarded as one of the most promising stem cells for regenerative medicine because of their unusual capacity of self-renewal and pluripotency (1). However, given the variations in the derivation and propagation of hESCs in different laboratories, it is imperative to establish a common set of criteria for the quality control of hESCs. Efforts have been devoted to characterizing whether established lines of hESCs carry inherent differences in gene expression and epigenetic modifications such as DNA methylation (2). Although different lines of hESCs can exhibit a common set of stem cell markers, differences in gene expression are observed including allelic expression of several imprinted genes and XIST, a crucial gene for X-inactivation (2). Several studies also demonstrated that in vitro cultures or differentiation of hESCs can contribute to changes in CpG methylation patterns and genome stability in different lines of hESCs (2–4). Thus, routine and thorough characterization of genetic and epigenetic stability in hESCs is a necessary step to ensure the quality of hESCs for regenerative medicine.
X chromosome inactivation (XCI) is required for dosage compensation of X-linked genes in female cells (5). So far, only a few studies have examined XCI in female hESCs and conflicting data exist regarding the nature of XCI. It has been reported that ≈50% of all established female hESC lines exhibit XCI markers such as XIST expression and/or punctate histone H3 lysine 27 trimethylation (H3K27me3) staining on the inactive X chromosome (Xi), whereas other lines do not (2, 6–9). Moreover, discrepancies in detecting XIST expression exist in different laboratories even for subcultures of the same lines of hESCs such as H7, H9, and HES1 cells (2, 6–9).
The initiation and maintenance of XCI is extremely important for embryogenesis and adult cell physiology (10). Because many X-linked loci are associated with mental retardation disease, proper expression of X-linked genes at the right dosage is essential for brain function and social skill development (11). In addition, disruption of XCI is often found in pathological conditions such as female cancer cells (12).
Concerning the maintenance of XCI, once XCI is fully established, Xist/XIST RNA appears to be dispensable in dosage compensation in differentiated somatic cells (13, 14). However, recent studies also showed that conditional deletion of the Xist gene in mouse somatic cells can influence the frequency of reactivation of previously silenced X-linked alleles and the genome stability, suggesting that Xist/XIST expression in differentiated female cells still plays a role in the maintenance of XCI (13, 15, 16).
In this study, we focus on the characterization of XCI status and XIST expression in three well studied female hESC lines (H7, H9, and HSF6). We find that culture conditions can influence the expression of XCI markers, including coating of one X chromosome by XIST and H3K27me3 staining. By comparing subcultures of female hESCs with or without XCI markers, we found that loss of XCI markers is correlated with demethylation of promoter CpG islands and an increased level of mRNAs for a significant portion of X-linked genes, some of which are involved in gene regulation and developmental processes. Our results highlight the need to routinely monitor XCI markers as a quality control in the established lines of female hESCs.
Results
Differential Expression of XCI Markers in Subcultures of Female hESC Lines.
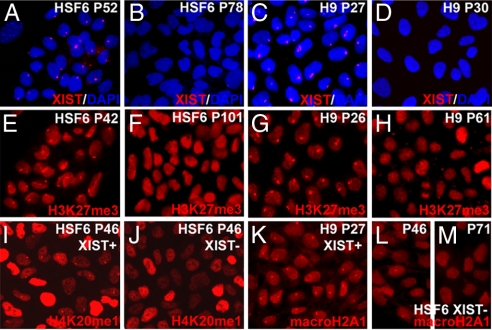
By detection of XIST RNA coating in cis by using FISH analysis, or by the punctate immunostaining for H3K27me3 (9, 17), subcultures of female H9 and HSF6 cells exhibit 0–100% of XCI markers at either early or late passages, depending on the source of cells and passaging history from different laboratories (Fig. 1). The absence of XIST expression is not due to the loss of an X chromosome in subcultures of H9 and HSF6, because the identification of numerous SNP polymorphisms across the entire X chromosome indicates the presence of two X chromosomes (data not shown). In addition, standard G-band karyotyping and DNA FISH analysis showed two intact X chromosomes in XIST-negative (XIST−) HSF6 cells [supporting information (SI) Fig. 5]. Taken together, our results confirmed that subcultures of the same line of hESCs can exhibit different XIST expression and H3K27me3 staining patterns. We also checked other XCI markers such as H4K20me1 (18) and macroH2A1 (19) in XIST-positive (XIST+) and XIST− hESCs. H4K20me1 staining is consistent with H3K27me3 staining (Fig. 1 I and J). However, we observed partial punctate staining of macroH2A (Fig. 1L) in early passaged cells (P46) but not in late passaged XIST− hESCs (P71) (Fig. 1M). H3K27me3 and H4K20me1 punctate staining patterns in hESCs are closely coupled with XIST expression, whereas punctate macroH2A1 staining could persist for a short period in XIST− hESCs.
Fig. 1.
Different subcultures of hESCs (HSF6 and H9) exhibit varied XCI status. (A–D) XIST RNA FISH signal (red) shows XIST RNA coating on the Xi. Immunostaining of hESCs with antibodies against H3K27me3 (red) (E–H), H4K20me1 (red) (I and J), and macroH2A1 (red) (K–M). Punctate XIST FISH signals and foci of H3K27me3, H4K20me1, and macroH2A1 stainings indicate the presence of an Xi. Please note that, for XIST− hESCs, the punctate staining pattern of H4K20me1 in some hESCs cannot be seen because of overexposure of the image to compensate for the weakly stained cells.
To ascertain whether certain culture parameters can influence the expression of XCI markers in female hESCs, we tested the impact of different enzymatic treatments (trypsin vs. collagenase IV and dispase) and freezing/thaw cycles on the stability of XCI markers. Under standard culture conditions, we detected XIST expression and H3K27me3 focus staining in HSF6 cells for >100 passages over a 2-year period. XCI markers are not affected by withdrawal of basic fibroblast growth factor (bFGF) treatment, or different enzymatic digestions, or repetitive freeze/thaw cycles of cells. However, when subcultures of hESCs exhibit excessive cell death during passaging and display abnormal nuclear morphology, they tend to lose XCI markers such as the H3K27me3 focus staining (SI Fig. 6) and XIST expression. In subsequent expansion of these subcultures, we observed colonies containing mixed cells with or without XCI markers, or homogenous and stable populations of HSF6 hESCs without XCI markers. Subcloning from the mixed parental population can also yield homogenous population of cells with or without XCI markers under standard passage conditions. Although we still do not know exactly how XCI markers are lost in subcultures of HSF6 and H9 hESCs, our observations favor the possibility that transient exposure to stress or suboptimal conditions may lead to epigenetic silencing of XIST expression in hESCs (see below). Finally, loss of these XCI markers in female hESCs appears irreversible, because HSF6 cells without XCI markers do not reexpress XIST even upon differentiation (data not shown).
Female hESC Lines Exhibit the Nonrandom XCI Pattern Regardless of the Presence or Absence of XIST Expression in Subcultures.
If random XCI occurred during hESC derivation without clonal expansion, one would expect the detection of both X-linked alleles in a population of hESCs. This can be verified by sequence analysis of multiple polymorphic cDNAs of X-linked genes. We identified all SNPs in the coding regions of the X-linked genes in H9 and HSF6 cells by using Affymetrix 500K genotyping array (see Materials and Methods and Table 1). Because ≈15% of X-linked genes are known to escape XCI in human female somatic cells (20), we first chose eight polymorphic X-linked genes for H9 and seven for HSF6 hESCs that are known to be subjected to X-inactivation. Surprisingly, cDNA sequencing analysis showed that each set of polymorphic X-linked genes is monoallelically expressed in the population of H9 and HSF6 hESCs that express XIST (Table 1), indicating that X-inactivation is not random in these cells. To further determine that polymorphic monoallelic expression of X-linked genes is from one X chromosome in cis, we genotyped eight polymorphic genes for a subline of H9 cells that carry only one X chromosome (XO karyotyping). Our genotyping result indicated that the polymorphic specificity of eight X-linked genes in this subline of XO H9 cells is exactly the same as we found in cDNAs of H9 cells with XIST expression (Table 1), confirming that monoallelic expression of X-linked genes is from one X chromosome in cis.
Table 1.
Genomic SNP genotyping and polymorphic cDNA analysis of a subset of X-linked genes in HSF6, H9, and H7 hESCs
| Gene name | SNP ID | Genotyping | Allelic expression in XIST+ hESCs | Allelic expression in XIST– hESCs | Genotyping (XO) |
|---|---|---|---|---|---|
| H9 | |||||
| DMD | rs228406 | C/T | T (10T) | C/T (6C/4T) | T |
| GK | rs6526997 | A/G | A (8A) | A/G (8A/3G) | A |
| HS6ST2 | rs5933220 | T/C | C | C | C |
| FGF13 | rs2267628 | T/C | T | T | T |
| AR | rs4827545 | A/C | A | A | A |
| UIP1 | rs933190 | A/G | A | A | A |
| FHL | rs7061270 | C/T | T | T | T |
| WDR44 | rs10521584 | C/T | C | C | C |
| HSF6 | |||||
| CXORF22 | rs6632450 | C/T | T (9T) | C/T (6T/4G) | |
| CXORF12 | rs7350355 | A/G | A (10A) | A/G (14A/6G) | |
| AFF2 | rs6641482 | A/G | A | A | |
| ATP7A | rs2227291 | C/G | C | C | |
| WDR44 | rs10521584 | C/T | C | C | |
| FHL1 | rs9018 | A/G | G | G | |
| UIP1 | rs933190 | A/G | A | A or A/G* | |
| H7 | |||||
| POLA1 | rs929313 | A/C | A/C (7C/3A) | ||
| AFF2 | rs6641482 | A/G | G | ||
| FHL | rs7061270 | C/T | T | ||
| FMR1 | rs29282 | C/T | T |
Genes in red show reactivation of the second allele in hESCs without XCI markers, and genes in black are monoallelic expressed in both XIST+ and XIST− hESCs.
*Note that UIP1 is observed either monoallelic or biallelic expressed in different batches of XIST– HSF6 subcultures.
We next determined whether these polymorphic X-linked genes are monoallelically or biallelically expressed in subsets of XIST− H9 and HSF6 cells. We found that two polymorphic genes exhibited biallelic expression patterns (Table 1). This result suggests that a portion of the previously silenced X-linked allele can be reactivated in the absence of XIST expression in female hESCs. However, a majority of polymorphic X-linked genes (71–75%) still maintained monoallelic expression in these cells.
Previous studies were unclear whether absence of XIST expression in H7 hESCs is before the occurrence of XCI or because of loss of XCI markers after the completion of XCI. To distinguish these two possibilities, we performed cDNA polymorphic sequencing analysis with H7 cells at passage 27 (P27) that already do not exhibit XCI markers. Three of four polymorphic genes maintain monoallelic gene expression (Table 1). This result is consistent with the notion that H7 cells have completed XCI, but loss of XIST expression can result in reactivation of a small subset of X-linked alleles as indicated by the biallelic expression of the X-linked genes (POLA1) (Table 1). Taken together, our data indicate that all three lines of female hESCs studied have undergone XCI during the derivation and/or expansion process.
Loss of XCI Markers Is Associated with Hypermethylation in the XIST Promoter.
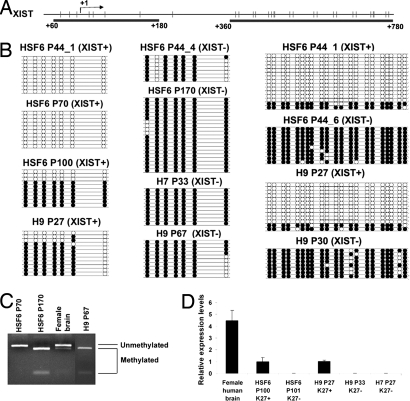
To determine whether the loss of XIST expression in female hESCs is associated with stable epigenetic change, we first examined DNA methylation on the XIST promoter. Bisulfite genomic sequencing and combined bisulfite restriction analysis (COBRA) assay (21) were performed to quantify the levels of DNA methylation in the promoter/first exon of the XIST gene. In female somatic cells, the XIST promoter is methylated and silenced on the active X chromosome (Xa), but remains unmethylated and actively transcribed on the Xi (22). We observed partial methylation in H9 female hESCs with XCI markers, consistent with methylation only on Xa (Fig. 2 B and C). Surprisingly, we found that the XIST promoter in female HSF6 hESCs with XCI markers is fully unmethylated in both alleles at moderate passages (P44–P70) (Fig. 2B). Only in late passage (P101) of HSF6 cells did we observe partial methylation in subcultures with XCI markers. These results are consistent with the idea that XIST promoter methylation is a late event of the epigenetic cascade during the completion of XCI. Thus, XCI in HSF6 cells could be in an intermediate state at P40–P70 without any XIST promoter methylation, but becomes more complete by acquiring monoallelic XIST promoter methylation at P101 or above. In contrast, in subcultures of HSF6, H7, and H9 cells that no longer express XIST, the XIST promoter is 100% methylated, indicating biallelic methylation (Fig. 2B). A similar methylation change is also observed in the first exon (Fig. 2B), consistent with the spreading of DNA methylation in the regions surrounding the transcription initiation site in the silenced XIST allele. Biallelic methylation of the XIST promoter can occur quite early (P33–P67) suggesting that DNA hypermethylation is coupled with the loss of XIST expression (Fig. 2 B and C). The levels of XIST expression in hESCs is also lower than that in differentiated somatic cells, consistent with the possibility that the regulation of XIST gene expression in hESCs is not as complete as in somatic cells (Fig. 2D).
Fig. 2.
DNA methyation analysis of the XIST promoter and real-time RT-PCR analysis of XIST RNA levels. (A) Schematic diagram of XIST promoter/first exon and a further downstream region analyzed. CpG sites are presented in vertical bars, and the arrow indicates the transcription initiation site of XIST. CpG sites analyzed are underlined by black bars. (B) Bisulfite sequencing analysis reveals methylation patterns of promoter/first exon and a further downstream region of XIST gene in XIST+ and XIST− H9 and HSF6 hESCs and XIST− H7 hESCs in various passages. Each filled black dot represents one methylated CpG site, and an open dot represents an unmethylated CpG. (C) COBRA assay showing the XIST promoter for XIST− H9 and HSF6 are fully methylated, whereas the XIST promoter of XIST+ HSF6 is hypomethylated compared with normal female brain DNA. (D) Real-time quantitative PCR showing relative XIST expression levels in various samples of HSF6, H9, and H7 hESCs.
Changes of Promoter CpG Island Methylation in X-Linked Genes in the Absence of XCI Markers.
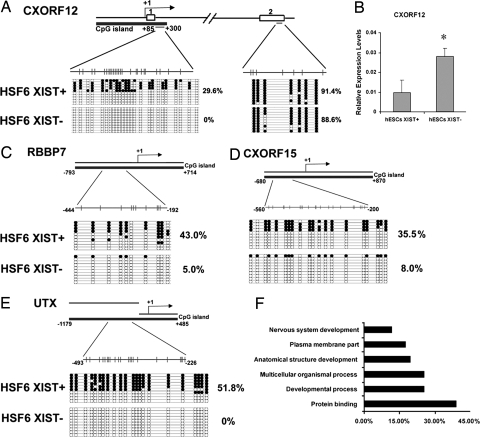
X-linked genes frequently contain a CpG island promoter that is unmethylated on the expressed alleles and methylated on the silenced alleles (23–25). We were interested in identifying whether the XCI-mediated silencing of one of the alleles is correlated with CpG island methylation in hESCs. Furthermore, we wanted to determine whether reactivation of a subset of X-linked genes is associated with demethylation in promoter CpG islands. We first performed bisulfite sequencing analysis of the promoter CpG island in the CXORF12 gene, which is reactivated in XIST− HSF6 cells (Table 1). Whereas methylation in the exon region was similar in both hESCs with or without XCI markers, the CXORF12 promoter was ≈50% methylated in XIST+ HSF6 cells, but became totally unmethylated in XIST− HSF6 cells (Fig. 3A). This result indicates that reactivation of the silenced allele in female hESCs is coupled with selective demethylation of the promoter CpG island. Moreover, in the absence of XIST expression, CXORF12 mRNA increased 2- to 3-fold compared with XIST+ HSF6 cells (Fig. 3B), indicating that demethylation of this CpG island promoter is associated with an increase in gene expression.
Fig. 3.
Analysis of methylation levels at promoter CpG islands in female hESCs in the presence or absence of XIST expression. (A) Bisulfite genomic sequencing analysis of the CpG island promoter and an exon region of CXORF12 gene in XIST+ and XIST− HSF6 cells. Note the promoter is 50% methylated in XIST+ HSF6 cells and becomes unmethylated in XIST− cells. In contrast, the exon region is 100% methylated in both XIST+ and XIST− cells. (B) Real-time quantitative PCR showing that the expression level of CXORF12 is significantly higher in XIST+ HSF6 cells compared with XIST− cells. *, P < 0.01. (C–E) Bisulfite methylation analysis in CpG islands of RBBP7, UTX, and CXORF15 genes. (F) Gene ontology analysis of 51 X-linked genes with decreased methylation levels in promoter CpG islands in XIST− hESCs (P < 0.05).
To systematically identify those X-linked genes that are either already reactivated or poised to be reactivated in XIST− hESCs, we examined whether CpG island promoter demethylation takes place on the entire X chromosome. We therefore carried out high-throughput CpG island methylation profiling by using methylated DNA immunoprecipitation in combination with microarray hybridization (mDIP-ChIP) with a pair of HSF6 hESCs with or without XIST expression at the same passage (P101). Statistical analysis of microarray data indicated that of the total 419 annotated X-linked promoters on the Agilent CpG island microarray, 51 (12.2% of CpG islands) genes showed a decrease in CpG island methylation in XIST− hESCs (SI Table 2). These 51 genes are distributed across the entire X chromosome, indicating that demethylation of promoter CpG islands is not limited to a particular segment of the X chromosome. Among these 51 genes, we randomly selected three genes (RBBP7, UTX, and CXORF15) for bisulfite genomic sequencing and confirmed that DNA demethylation in CpG island promoters does take place in XIST− hESCs (Fig. 3 C–E). Thus, our data suggest that up to 12.2% of X-linked genes could be reactivated in the absence of XCI marker in hESCs. Gene ontology analysis suggested these genes are enriched for regulatory proteins and developmental processes (Fig. 3F).
Loss of Dosage Compensation for a Subset of X-Linked Genes in Female hESCs in the Absence of XIST Expression.
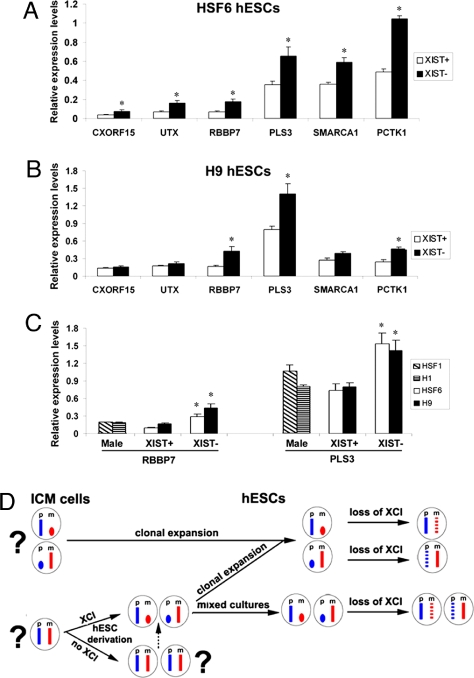
We next used gene expression profiling to identify the X-linked genes that exhibit mRNA level changes in XIST− hESCs. Microarray analysis of whole genome gene expression indicated that of the total 1,141 annotated X-linked genes, 44 (3.8%) exhibited at least a 1.5-fold increase in mRNA levels (P < 0.01) (SI Table 3). Real-time RT-PCR analysis confirmed that the expression levels of X-linked PLS3, RBBP7, UTX, CXORF15, SMARCA1, and PCTK1 are significantly increased by ≈2-fold in XIST− HSF6 cells compared with XIST+ HSF6 cells (Fig. 4A). Comparing the list of up-regulated X-linked genes (SI Table 3) with the list of demethylated genes (SI Table 2), we find 12 genes (12 of 44 or 27.3%) overlap, confirming that a subset of promoter-demethylated genes is up-regulated in hESCs. We suspect that this overlap could be even higher because a subset of demethylated genes could be expressed below detection sensitivity of the microarray. For example, we detected a significant increase in CXORF12 mRNA with real-time RT-PCR analysis (Fig. 4B), but not by microarray analysis.
Fig. 4.
Relative gene expression levels of a subset of X-linked genes using pairs of HSF6 and H9 cells with or without XCI markers. (A) Real-time RT-PCR demonstrates increased mRNAs in HSF6 cells without XCI markers. Six X-linked genes including CXORF15, UTX, RBBP7, PLS3, SMARCA1, and PCTK1 were analyzed. (B) Real-time RT-PCR results of the same six genes for a pair of XIST+ and XIST− H9 cells. *, P < 0.01. (C) Real-time RT-PCR results of the RBBP7 and PLS6 for male hESCs (H1 and HSF1) and XIST+ and XIST− female hESCs (H9 and HSF6) cells. *, P < 0.05. (D) Models of dynamic regulation of XCI in female hESCs. Xa is shown in full-length X chromosome. Xi is depicted in an oval shape. Dotted X chromosome indicates partial reactivation. Paternal (p) and maternal (m) X are shown in blue and red.
Among the six genes we analyzed by real-time PCRs, three of them showed ≈2-fold increase in expression in H9 hESCs, which is consistent with reactivation in HSF6 cells. However, the other three genes did not show any significant change expression level (Fig. 4B). This result implies that each individual female hESC may have a unique profile of gene reactivation for a subset of X-linked genes in XIST− cells because of the inherent genetic and epigenetic differences.
We further directly compared mRNA levels between two lines of male hESCs (H1 and HSF1) and female hESCs (H9 and HSF6) with or without XCI markers. Real-time RT-PCR assays showed that levels of mRNAs of both RBBP7 and PLS3 are similar between male hESCs and female XIST+ hESCs. However, XIST− female hESCs exhibited significantly higher levels of mRNAs than male hESCs, confirming the disruption of dosage compensation for these two X-linked genes in these cells (Fig. 4C).
Discussion
In our study, several classic X-inactivation markers are readily detected in human female hESCs. Under optimal culture conditions, XCI status can be stably maintained in female hESCs over 100 passages. However, we also observed XCI instability in subcultures of female hESCs, presumably because of suboptimal culture conditions. Importantly, a majority of X-linked genes are monoallelically expressed regardless of the presence or absence of XCI markers in all three established female hESC lines studied (H7, H9, and HSF6). This result suggests that established lines of female hESCs have already acquired XCI even at moderate passages (e.g., P25–P35). Furthermore, in female hESCs devoid of XCI markers, a subset of previously silenced X-linked genes (up to 10–15% of inactivated genes) become reactivated (or poised to be reactivated), leading to biallelic expression patterns.
Our findings reconcile controversial data in the literature concerning the varying XCI status within established lines of female hESCs (2, 7–9). According to our model (Fig. 4D), established lines of female hESCs have completed XCI by moderate passage numbers (P20–P30). The inconsistent observations of XIST expression for identical lines of female hESCs are most likely due to loss of XIST expression in subcultures, perhaps caused by culture variations among individual laboratories. Additionally, subcultures may exist as a mixed population of cells with or without XCI markers, leading to graded levels of XIST expression observed in RT-PCR assays (2, 7).
It is still unclear why the loss of XCI markers occurs in subcultures of these established lines of female hESCs. One possibility is that selection pressure favors the survival and cell proliferation of female hESCs in the absence of XCI markers. This scenario is consistent with a cell adaptation event such as the loss of XCI marker or aneuploidity that takes place in hESCs over long-term cultures, perhaps under suboptimal or stressed culture conditions (9, 26). Loss of XCI markers can occur in a relatively short time window within several cell passages from XIST+ cultures (Y.S. and G.F., unpublished work). Our data suggest that loss of XCI markers is not simply a passive event in which XIST− cells gradually takes over XIST+ cells from mixed cultures. In fact, when we compared cell proliferation rate between sister cultures of XIST+ and XIST− hESCs in consecutive passages, we did not find differences between these two populations in percentage of cells undergoing mitosis (SI Fig. 7).
One unresolved question in our study is when and how female hESCs acquire uniform XCI in a “nonrandom” pattern during the process of derivation and expansion (Fig. 4D). Enver et al. (9) reported that H7 cells express XIST RNA in very early passages, but lose it because of adaptive culture conditions, arguing that XCI occurs quite early. This raises the possibility that the uniform XCI pattern in a line of established hESCs could be simply the outcome of clonal expansion of a single ancestor cell that has undertaken random XCI during the derivation/expansion process (Fig. 4D). However, it is also possible that female hESCs may achieve “imprinted” and nonrandom X-inactivation through a mechanism of paternal XCI as seen in mouse trophectoderm (27) (Fig. 4D). Finally, it remains to be determined whether human female inner cell mass cells exhibit random XCI or maintain two active X chromosomes as is the case for mouse ICM cells or female embryonic stem cells (5) (Fig. 4D). Also, the possibility exists that the earliest passage of female hESCs may have two active X chromosomes and subsequently acquire XCI because of culture selection pressure (Fig. 4D).
The monoallelic expression pattern for a majority of X-linked genes (≈75%) in XIST− hESCs is consistent with the notion that once XCI is completed, it is rather stable for most genes even without XIST expression (13). However, loss of XIST expression and other XCI markers does significantly destabilize the inactive state and cause gene reactivation (28). We found that gene reactivation profiles of different lines of female hESCs appear to be different (Fig. 4). Such a difference could be due to the random nature of gene reactivation in the absence of XCI markers or because of inherent genetic/epigenetic differences. Thus, the exact list of reactivated genes in each female line may have to be determined individually.
We demonstrate that DNA hypermethylation of the XIST promoter is one of the epigenetic factors that correlate with the silencing of XIST expression. In XIST+ hESCs, the XIST promoter can be completely unmethylated in early passages. This differs from somatic cells in which human XIST or mouse Xist promoter is only methylated on the active X chromosome (22, 29, 30). The unmethylated status for both alleles of the XIST promoter implies that XCI in established hESCs may initially exist at an intermediate stage when XIST coating of Xi and dosage compensation for X-linked genes is completed, but before methylation of the XIST promoter on Xa. Nevertheless, allelic specific methylation of the XIST promoter is eventually achieved with subsequent passages, suggesting that the completion of a cascade of epigenetic modifications on Xa and Xi is a gradual process. Finally, when XCI markers disappear, we observe biallelic DNA hypermethylation of the XIST promoter, suggesting that it is prone to epigenetic alterations.
The potential impact of the altered expression of a portion of X-linked genes on survival and differentiation of hESCs remains to be examined. Gene ontology analysis of up-regulated X-linked genes in female hESCs without XCI markers reveals clusters of genes involved in developmental processes and gene regulation. For example, mRNA transcripts of SMARCA1 and UTX are up-regulated in XIST− HSF6 cells. SMARCA1 is shown to play a role in chromatin remodeling (31), which may impact the regulation of other genes. UTX encodes a histone H3K27 demethylase involved in autosomal HOX regulation during development (32). Indeed, our gene expression profiling experiments show a significant change for many autosomal genes in XIST− hESCs, such as HOXA4 (Y.S. and G.F., unpublished work). In view of the relaxation of dosage compensation for a subset of functionally important X-linked genes in XIST− hESCs, we propose that the status of XCI markers in female hESCs and their derivatives needs to be examined routinely. Furthermore, female hESCs displaying XCI markers would be the better choice for understanding basic mechanisms of development and for future applications in regenerative medicine.
Materials and Methods
Cultures of hESCs and Directed Neural Differentiation of hESCs in Vitro.
hESC culture and neural differentiation procedures were described previously with bFGF (10 ng/ml) supplement (21). This research project was approved by University of California at Los Angeles (UCLA) Embryonic Stem Cell Research Oversight and Institutional Review Board committees.
HSF6 hESCs used in this article are batch 1 HSF6 from University of California at San Francisco (UCSF) except otherwise mentioned. For Fig. 1 I, J, and L, P46, and Fig. 2B, P44, we used the batch 2 HSF6 from UCSF. XIST+ H9 and XIST− H7 hESCs were obtained from WiCell. All of the above hESCs were cultured in the Fan Laboratory at UCLA. The XIST− H9 hESCs were obtained from WiCell and cultured in the Xu Laboratory at University of Connecticut.
Immunohistochemistry and RNA-FISH.
Immunostaining procedures were described in ref. 21. Antibodies used were as follows: polyclonal H3K27me3 (1:1,000; a gift from Yi Zhang, University of North Carolina, Chapel Hill, NC), polyclonal H4K20me1 (1:1,000 from Upstate), polyclonal macroH2A1 (1:100; a gift from Kathrin Plath at UCLA), and monoclonal H3-phosphor-ser10 (1:5,000; Upstate). Coverslips were then incubated with fluorochrome-conjugated secondary antibodies for 1 h at room temperature. XIST RNA-FISH was performed as described in ref. 33 by using three 50-mer DNA probes designed from consensus sequences of map positions 6183–6232, 6234–6283, and 6368–6417 (accession no. L04961), which are in repeat D of XIST.
Bisulfite Genomic Sequencing Analysis and COBRA Assay.
Bisulfite sequencing and COBRA assay were performed as described in ref. 21. For COBRA assay, PCR products of bisulfite-treated DNA (XIST promoter, 300 bp) were digested with HpyCH4IV, which if its target sites are methylated yields 50- and 250-bp bands.
Identification of SNPs Through Affymetrix SNP Genotyping Microarray and the Analysis of Allelic Expression Pattern of X-Linked Genes.
Affymetix GeneChip Human Mapping 500K Array Set was used to map SNP sites in H7, H9, and HSF6 cells. Hybridization was carried out in the UCLA Microarray Core. For genotyping confirmation and analysis of allelic expression of X-linked genes, either genomic DNA or cDNA converted from DNase I-treated RNA samples was used for PCR amplification and direct sequencing. H9 XO genomic DNA was generously provided by Nissim Benvenisty (Jerusalem).
Agilent Human Whole Genome Gene Expression Array.
HSF6 hESCs P101 (XIST+ and XIST−) RNA were used for expression array. The detailed procedure was described in ref. 34. A list of significantly up-regulated genes (>1.5-fold) in XIST− hESCs was generated by using Focus (http://microarray.genetics.ucla.edu/focus/). In addition, a t test was performed across three arrays, and differentially expressed genes were generated with P value of <0.01 and >1.5-fold difference. By combining these two lists, a list of genes that are significantly up-regulated in XIST− hESCs is generated.
mDIP-ChIP and Data Analysis.
mDIP-ChIP procedure was done as described in ref. 34, by using Agilent human whole genome CpG island arrays. t tests between two sets of samples (XIST+ or XIST− hESCs) for each individual probe were performed. To evaluate whether the collection of t scores for a CpG island is significant, Z scores were computed by using the following formula: Z score = [mean(t score of CpG island probes) − mean(t score for all probes)] *square_root(number of CpG island probes)/standard_deviation(all probes). A positive Z score means a higher probability of higher methylation levels in XIST+ hESCs and vice versa.
Supplementary Material
Acknowledgments.
We thank L. Hutnick and Y. Marahrens for critically reading the article. This work is supported by National Institutes of Health (NIH) Grants R01 NS044405 and NS051411 and California Institute for Regenerative Medicine Comprehensive Research Grant RC01-00111-01 (to G.F.), and Connecticut Stem Cell Research Grant 06SCB14 (to R.X.). Y.S. is supported by a California Institute for Regenerative Medicine predoctoral fellowship. S.D.F. is supported by NIH Award 1F31MH070204.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE9637).
This article contains supporting information online at www.pnas.org/cgi/content/full/0712018105/DC1.
References
- 1.Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- 2.Adewumi O, et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- 3.Maitra A, et al. Genomic alterations in cultured human embryonic stem cells. Nat Genet. 2005;37:1099–1103. doi: 10.1038/ng1631. [DOI] [PubMed] [Google Scholar]
- 4.Allegrucci C, et al. Restriction landmark genome scanning identifies culture-induced DNA methylation instability in the human embryonic stem cell epigenome. Hum Mol Genet. 2007;16:1253–1268. doi: 10.1093/hmg/ddm074. [DOI] [PubMed] [Google Scholar]
- 5.Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- 6.Sperger JM, et al. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc Natl Acad Sci USA. 2003;100:13350–13355. doi: 10.1073/pnas.2235735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhara SK, Benvenisty N. Gene trap as a tool for genome annotation and analysis of X chromosome inactivation in human embryonic stem cells. Nucleic Acids Res. 2004;32:3995–4002. doi: 10.1093/nar/gkh746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffman LM, et al. X-inactivation status varies in human embryonic stem cell lines. Stem Cells. 2005;23:1468–1478. doi: 10.1634/stemcells.2004-0371. [DOI] [PubMed] [Google Scholar]
- 9.Enver T, et al. Cellular differentiation hierarchies in normal and culture-adapted human embryonic stem cells. Hum Mol Genet. 2005;14:3129–3140. doi: 10.1093/hmg/ddi345. [DOI] [PubMed] [Google Scholar]
- 10.Tomkins DJ, McDonald HL, Farrell SA, Brown CJ. Lack of expression of XIST from a small ring X chromosome containing the XIST locus in a girl with short stature, facial dysmorphism and developmental delay. Eur J Hum Genet. 2002;10:44–51. doi: 10.1038/sj.ejhg.5200757. [DOI] [PubMed] [Google Scholar]
- 11.Skuse DH. X-linked genes and mental functioning. Hum Mol Genet. 2005;14:R27–R32. doi: 10.1093/hmg/ddi112. (Spec. No. 1) [DOI] [PubMed] [Google Scholar]
- 12.Ganesan S, et al. Abnormalities of the inactive X chromosome are a common feature of BRCA1 mutant and sporadic basal-like breast cancer. Cold Spring Harb Symp Quant Biol. 2005;70:93–97. doi: 10.1101/sqb.2005.70.045. [DOI] [PubMed] [Google Scholar]
- 13.Csankovszki G, Panning B, Bates B, Pehrson JR, Jaenisch R. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat Genet. 1999;22:323–324. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- 14.Brown CJ, Willard HF. The human X-inactivation centre is not required for maintenance of X-chromosome inactivation. Nature. 1994;368:154–156. doi: 10.1038/368154a0. [DOI] [PubMed] [Google Scholar]
- 15.Zhang LF, Huynh KD, Lee JT. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell. 2007;129:693–706. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- 16.Diaz-Perez S, et al. The element(s) at the nontranscribed Xist locus of the active X chromosome controls chromosomal replication timing in the mouse. Genetics. 2005;171:663–672. doi: 10.1534/genetics.105.043026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plath K, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 18.Kohlmaier A, et al. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2004;2:E171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costanzi C, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- 20.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 21.Shen Y, Chow J, Wang Z, Fan G. Abnormal CpG island methylation occurs during in vitro differentiation of human embryonic stem cells. Hum Mol Genet. 2006;15:2623–2635. doi: 10.1093/hmg/ddl188. [DOI] [PubMed] [Google Scholar]
- 22.Norris DP, et al. Evidence that random and imprinted Xist expression is controlled by preemptive methylation. Cell. 1994;77:41–51. doi: 10.1016/0092-8674(94)90233-x. [DOI] [PubMed] [Google Scholar]
- 23.Heard E. Recent advances in X-chromosome inactivation. Curr Opin Cell Biol. 2004;16:247–255. doi: 10.1016/j.ceb.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 24.Lock LF, Takagi N, Martin GR. Methylation of the Hprt gene on the inactive X occurs after chromosome inactivation. Cell. 1987;48:39–46. doi: 10.1016/0092-8674(87)90353-9. [DOI] [PubMed] [Google Scholar]
- 25.Yen PH, Patel P, Chinault AC, Mohandas T, Shapiro LJ. Differential methylation of hypoxanthine phosphoribosyltransferase genes on active and inactive human X chromosomes. Proc Natl Acad Sci USA. 1984;81:1759–1763. doi: 10.1073/pnas.81.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baker DE, et al. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol. 2007;25:207–215. doi: 10.1038/nbt1285. [DOI] [PubMed] [Google Scholar]
- 27.Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- 28.Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol. 2001;153:773–784. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beard C, Li E, Jaenisch R. Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev. 1995;9:2325–2334. doi: 10.1101/gad.9.19.2325. [DOI] [PubMed] [Google Scholar]
- 30.Hendrich BD, Brown CJ, Willard HF. Evolutionary conservation of possible functional domains of the human and murine XIST genes. Hum Mol Genet. 1993;2:663–672. doi: 10.1093/hmg/2.6.663. [DOI] [PubMed] [Google Scholar]
- 31.Peterson CL. Multiple SWItches to turn on chromatin? Curr Opin Genet Dev. 1996;6:171–175. doi: 10.1016/s0959-437x(96)80047-5. [DOI] [PubMed] [Google Scholar]
- 32.Agger K, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 33.Michienzi A, Rossi JJ. Intracellular applications of ribozymes. Methods Enzymol. 2001;341:581–596. doi: 10.1016/s0076-6879(01)41178-5. [DOI] [PubMed] [Google Scholar]
- 34.Fouse S, et al. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.