Abstract
DNA sequencing has become central to the study of evolution. Comparing the sequences of individual genes from a variety of organisms has revolutionized our understanding of how single genes evolve, but the challenge of analyzing polygenic phenotypes has complicated efforts to study how genes evolve when they are part of a group that functions collectively. We suggest that biosynthetic gene clusters from microbes are ideal candidates for the evolutionary study of gene collectives; these selfish genetic elements evolve rapidly, they usually comprise a complete pathway, and they have a phenotype—a small molecule—that is easy to identify and assay. Because these elements are transferred horizontally as well as vertically, they also provide an opportunity to study the effects of horizontal transmission on gene evolution. We discuss known examples to begin addressing two fundamental questions about the evolution of biosynthetic gene clusters: How do they propagate by horizontal transfer? How do they change to create new molecules?
Darwin, in On the Origin of Species, said, “To suppose that the eye … could have been formed by natural selection, seems, I freely confess, absurd in the highest possible degree. Yet reason tells me, that if numerous gradations from a perfect and complex eye to one very imperfect and simple, each grade being useful to its possessor, can be shown to exist … then the difficulty of believing that a perfect and complex eye could be formed by natural selection … can hardly be considered real” (1). Complex small molecules like yersiniabactin (Fig. 1a) astonish chemists for the same reason that organs such as the eye intrigued Darwin (1): their incredible complexity makes us wonder how they came to be. Because the genes that are necessary and sufficient for producing yersiniabactin (2) (and many other small molecules) have been identified, we now have the tools to ascertain how the gene collectives that produce these complex phenotypes came to exist. Before discussing the evolution of small-molecule-producing gene collectives, we will briefly review what is known about their functional roles and genetic organization.
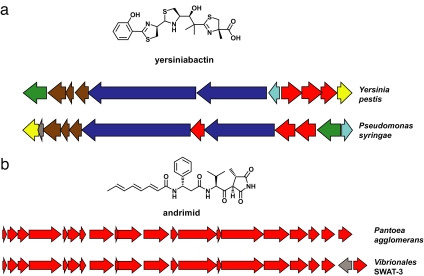
Fig. 1.
Propagation of gene clusters. (a) Horizontal transfer of the yersiniabactin gene cluster. The yersiniabactin gene clusters from Y. pestis KIM and P. syringae phaseolicola 1448A are shown, with related genes depicted in the same color to highlight intracluster gene rearrangements. (b) Horizontal transfer of the andrimid gene cluster. The andrimid gene clusters from Pantoea agglomerans CU2194 and Vibrionales bacterium SWAT-3 are shown; the only syntenic difference is the insertion of a single gene at the 3′ end of the latter cluster.
Bacteria and fungi produce a multitude of small molecules that are not used for primary (“housekeeping”) metabolism (3). These “secondary metabolites” play important and diverse roles in the ecology and physiology of microorganisms, particularly in mediating interactions both among microbial species (4, 5) and between microbes and multicellular organisms (6–10).
Given the basic metabolic capabilities of cellular life, many of these secondary metabolites are astoundingly complex in form (11). The biosynthetic pathways of secondary metabolites are similarly complex, sometimes composed of >40 genes (12) and 100 kb of DNA sequence (13). The set of proteins that comprise a complete biosynthetic pathway can be twice the size of the ribosome (13), even though the ribosome translates thousands of different proteins, whereas the biosynthetic pathway produces a small molecule. The metabolic cost of maintaining such a massive biosynthetic system is high, and the selective pressure fueling its maintenance must be correspondingly strong.
The genes that encode the biosynthetic pathway for a small molecule are almost always clustered in the genome of their microbial producer (14, 15), which undoubtedly reflects their evolutionary history through horizontal transmission (16). Because identifying one gene means the others are close by, cloning gene clusters for complete biosynthetic pathways is now straightforward and commonplace (17, 18). The considerable progress made over the last two decades in understanding the genetics and biochemistry of small-molecule synthesis is due in large part to the phenomenon of clustered genes.
Why Should We Study the Evolution of Biosynthetic Gene Clusters?
There are two reasons why biosynthetic gene clusters are an ideal class of genetic elements to study through an evolutionary lens. First, even from the limited set of gene cluster sequences in the database, it is apparent that biosynthetic gene clusters are among life's most diverse and rapidly evolving genetic elements. The speed with which they evolve is due in part to the relatively short replication times of their microbial hosts (19) and in part to their propensity for horizontal transfer among microbes (20–22). As such, they provide an opportunity to study genetic elements that evolve over shorter time scales than genes from higher organisms.
Second, the phenotypes of biosynthetic gene clusters are concrete and measurable. The small molecule(s) a gene cluster produces can be isolated, structurally characterized, and assayed for biological activity. Their biosynthetic pathways are understood in sufficient detail that the role of each gene in forming the small molecule can generally be pinpointed. Like the quantitative trait loci (23) that have advanced the study of evolution in plants and higher organisms, gene clusters provide a clear connection between genotype and phenotype.
Furthermore, because gene clusters are responsible for producing myriad human medicines (24) including antibiotics (25), antifungal agents (26), antitumor agents (27), immunosuppressants (28), and cholesterol-lowering agents (29), they represent a rich and promising source of new drugs. As our ability to genetically engineer microorganisms improves, the prospect of producing new molecules by modifying existing pathways (30)—or even building new pathways from scratch—may become a reality. To perform this task efficiently, we will have to know the rules that govern gene cluster evolution in the real world. By further developing our knowledge of the rules Nature uses to diversify its small molecules, we can facilitate the efforts of chemists to synthesize libraries of small molecules that are “natural product-like” and therefore, more likely to have useful biological activities (31). In what follows, we look forward by considering two fundamental questions: How do gene clusters propagate by horizontal transfer? How do they change to create new molecules? The examples below are not exhaustive, but rather a sampling of common themes in the evolution of gene collectives.
How Do Gene Clusters Propagate?
Biosynthetic gene clusters spread laterally because the small molecules they produce confer a selective advantage on their host. Like genes that confer antibiotic resistance (32), biosynthetic gene clusters are transmitted by selfish genetic elements like pathogenicity islands (33) and plasmids (34). Some restrictions on horizontal gene cluster transfer are imposed by the limited host ranges of genetic elements like conjugal plasmids (35), and other restrictions arise from differences in the metabolic infrastructure of the donor and recipient such as the availability of the three-carbon building block propionate (36) for biosynthesis of polyketides like erythromycin.
Nevertheless, certain gene clusters are widely distributed among phylogenetically distant species. The scarcity of iron often limits microbial growth, and the gene cluster responsible for producing the iron-scavenging agent yersiniabactin (Fig. 1a) has been found not just in the plague bacterium Yersinia pestis (2) from which it gets its name, but also in a menagerie of other bacteria (37) including the nematode symbiont Photorhabdus luminescens (38), the plant pathogen Pseudomonas syringae (39), pathogenic strains of Escherichia coli (40), and even the Gram-positive marine bacterium Salinispora tropica (41). In Y. pestis and E. coli, the yersiniabactin gene cluster resides on a ≈40-kb “high-pathogenicity island” (42) encoding a set of virulence-associated genes, and its propagation is facilitated by the en masse horizontal transfer of this entire element. The similarity between modes of transferring antibiotic resistance genes and small-molecule biosynthetic genes is exemplified by the 120-kb plasmid pRSB107 (34). Isolated from a sewage treatment plant, it harbors nine different antibiotic resistance genes in addition to the five-gene cluster responsible for producing the iron scavenger aerobactin (43).
Despite the limitations noted above, biosynthetic gene clusters appear to have crossed boundaries imposed by geography and ecology. The potent antibiotic andrimid (44) (Fig. 1b) has been isolated from a diverse assortment of Gram-negative bacteria, including a free-living marine strain of Vibrio cholera (45), a sponge-associated marine strain of Pseudomonas fluorescens (46), a terrestrial strain of Enterobacter (47) that is an endosymbiont of the brown planthopper, and a free-living terrestrial strain of Pantoea agglomerans (48). Andrimid's 20-kb gene cluster from P. agglomerans is flanked by a pseudogene that resembles a transposase, betraying its nomadic origin.
The gene clusters responsible for producing the β-lactam antibiotics (49) (e.g., penicillins and cephalosporins) are thought to have made an even larger horizontal jump between bacteria and fungi. Examples of prokaryote-to-eukaryote gene transfer (or vice versa) are still few in number (21, 50, 51), but biosynthetic gene clusters are promising candidates for future analyses.
How Do Gene Clusters Change?
Although limited in number, the biosynthetic gene clusters in the database reveal modes of diversification that have multiplied the members of several natural-product families. These examples cast mutation and natural selection as potent forces driving chemical innovation by creating new molecules with different biological activities from their ancestors. This diversification presents formidable challenges, because a nonfunctional biosynthetic gene cluster would have a limited evolutionary lifetime (52, 53). Likewise, all intermediate gene clusters must produce a molecule that justifies the cost of their existence before their evolutionary grace period expires. Here, we divide the modes of change into two categories: changes to individual genes and changes to the number and identity of genes that comprise the cluster.
Changes to Individual Genes.
The most straightforward manner in which biosynthetic genes change is through mutation. Examples of mutations in biosynthetic genes that lead to changes in product structure abound among terpene cyclases (54–56), enzymes that fold linear polymers of the five-carbon building block isopentenyl pyrophosphate into multiring hydrocarbon skeletons. High potential for mutation-induced diversity also arises when the precursor to a small molecule is a peptide translated by the ribosome. A seven-gene cluster found in isolates of the cyanobacterium Prochloron produces cyclic peptides like patellamide (57) that derive from short stretches of a single gene in the cluster; after translation, the peptide is modified and cleaved to release the products. In a family of these gene clusters (58), mutations in the small-molecule-encoding gene have produced a diverse family of cyclic peptides, whereas the other six genes in the cluster have remained nearly identical. The direct connection between gene sequence and small-molecule structure makes this biosynthetic family quite versatile, so it would not be surprising to find that patellamide-like gene clusters are widely distributed and that their products have diverse biological activities.
Mutation-induced diversity is also found in polyketide synthases (PKSs), a class of large, modular enzymes (14) that resemble an assembly line insofar as each module is responsible for incorporating one building block into the growing chain. Each module contains anywhere from zero to three “processing” domains—ketoreductase (KR), dehydratase (DH), and enoylreductase (ER)—that modify the building block incorporated by the acyltransferase (AT) domain. The processing domains in each module, and the order of the modules in the assembly line, determine the identity and order of the building blocks that comprise the small-molecule product. Diversity among PK structures is therefore derived from differences in the complement of these processing domains in each module. These differences commonly arise through point mutations in catalytic residues that render a processing domain inactive. For example, Fig. 2a shows two modules from the FK520 PKS (59) that have the same complement of modifying domains (DH + KR); whereas the DH domain in the red module is active, a mutation has rendered the DH domain in the blue module inactive, leading to the differing chemical structures of the three-carbon units incorporated by each module. This example demonstrates how loss- or gain-of-function mutations can be just as important for evolutionary diversification (60) as mutations that alter selectivity or change function.
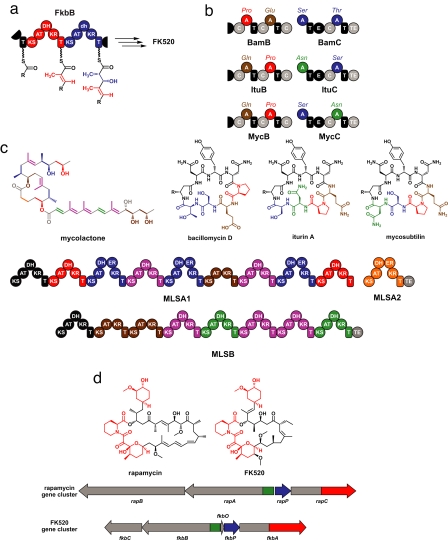
Fig. 2.
How individual genes in a cluster change. (a) A loss-of-function mutation leads to building-block diversity in FK520. Two modules from the FK520 PKS component FkbB are shown; the first module (red) has an active DH domain, whereas the second module (blue) has a mutationally inactivated DH domain (denoted by a lowercase “dh”). As a result, the first module modifies its three-carbon building block (red) so that it has a double bond, whereas the second module performs one fewer modification step to its building block (blue), leaving a hydroxyl group instead of a double bond. (b) Mutation and rearrangement of adenylation (A) domains creates diversity among the iturin family NRPs. The portions of the bacillomycin (Bam), iturin (Itu), and mycosubtilin (Myc) synthetases that insert the last 4 aa are shown at the top. A domains that are homologous to each other are depicted in the same color, and the building block they insert is listed above the domain. The chemical structures of bacillomycin D, iturin A, and mycosubtilin are shown at the bottom, and the residues are colored to correspond with the A domains. Mutation and divergent evolution of an ancestral A domain likely gave rise to the Glu/Gln- and Ser/Thr-inserting A domain families, whereas intragenic A domain rearrangement probably led to the differences among the three synthetases. (c) Module duplications within the mycolactone gene cluster. Modules that share >98% amino acid sequence identity are shown in the same color, and the chemical structure of mycolactone is shown with the building blocks resulting from the action of each module colored correspondingly. (d) Intergenic rearrangements lead to the chimerism between rapamycin and FK520. The chemical structures of rapamycin and FK520 are shown, with the identical “left halves” colored red and the distinct “right halves” colored black. Portions of the rapamycin and FK520 gene clusters are shown below, with homologous sections shown in the same color and divergent sections colored gray.
Other examples of mutation-induced diversity come from a class of small molecules known as nonribosomal peptides (NRPs). As their name suggests, these peptide-derived molecules are not produced by the ribosome but are synthesized by assembly-line enzymes (nonribosomal peptide synthetases, or NRPSs) that function similarly to the PKSs (14). Like PKSs, NRPSs are composed of modules. Unlike PKSs, however, processing domains in NRPSs are not the major source of building-block diversity. The diversity of NRPSs primarily derives from the building-block-inserting adenylation (A) domain in each module. For example, the Bacillus NRPSs that produce the related molecules bacillomycin, iturin, and mycosubtilin have the same A domain organization in their first halves, leading to incorporation of the same building blocks in the “top halves” of these molecules (shown in black); but differences in A domain building-block selectivity in their second halves lead to the incorporation of different building blocks in the “bottom halves” of the molecules (shown in color) (Fig. 2b). The serine (Ser)- and threonine (Thr)-incorporating A domains in this family are closely related, as are the A domains that incorporate asparagine (Asn) and aspartate (Asp) (61). This suggests that mutations in a common ancestor of these domains led to their divergent building-block selectivity, and ultimately to changes in the structure of their products. This theme is echoed in another family of NRPSs that produce the myxochromides (62). There are even A domains that activate multiple amino acid substrates (63); these promiscuous A domains may be an evolutionary intermediate between A domains with more rigid selectivity.
Core biosynthetic genes can also be altered through intragenic rearrangement. The positions of the A domains within the bacillomycin, iturin, and mycosubtilin NRPSs (Fig. 2b) appear to have been rearranged (61), leading to the difference in building-block order in the bottom halves of these molecules. Such rearrangements, and similar examples in PKSs (64), suggest that the modularity of NRPS and PKS genes renders them particularly amenable to evolution by recombination.
A more drastic mode of intragenic change is module duplication. The Mycobacterium ulcerans PKS responsible for producing the toxin mycolactone (65) comprises three proteins, MLSA1, MLSA2, and MLSB. Modules from these proteins cluster into clades with >98% sequence identity, which suggests that intragenic module duplication transformed a more diminutive ancestral gene into this gargantuan enzyme (Fig. 2c). Intragenic module duplication is also found among NRPSs. P. luminescens (38) harbors an as-yet-uncharacterized 49-kb gene that encodes an NRPS (Plu2670) from which module-encoding portions cluster into five clades featuring strong amino acid sequence identity (>85%) within each clade. Perhaps it is not coincidental that MLSA1 (16,990 aa) and Plu2670 (16,367 aa) are, to our knowledge, the largest known proteins from any cellular life form. Pairs of proteins that differ by the presence or absence of a module have also been observed, which could have resulted from either an intragenic deletion or insertion; these changes predictably create a pair of molecules like the PKs spinosyn and butenylspinosyn (66) and the NRPs microcystin and nodularin (67, 68), which differ according to the presence or absence of the building block encoded by the additional module.
The “left halves” of the immunosuppressants rapamycin (28) and FK520 (59) are nearly identical, whereas the “right halves” are different (Fig. 2d). Unlike the iturin-family NRPSs, this distinction did not arise from an intragenic rearrangement. Instead, it appears that an intergenic rearrangement caused portions of their core biosynthetic genes (part of two genes and all of a third) to reside in two different contexts. Significantly, this rearrangement has led to a clear and well characterized difference between the activities of rapamycin and FK520 (69), both of which exert their immunosuppressive effect by inducing the dimerization of two proteins that do not normally form a complex. The nearly identical left halves of these molecules bind the FK506-binding protein (FKBP12). In contrast, the right halves have different binding partners: rapamycin binds mTOR (70), and FK520 binds calcineurin (71). Nature's cutting and pasting was quite precise; those portions of genes required to produce an FKBP12-binding fragment were retained, and the rest have diverged elegantly to bind different partner proteins with high affinity. Phylogenetic analyses have implicated additional forms of intergenic rearrangement as primary modes of PKS evolution (64, 72, 73).
The examples in this section demonstrate that the modularity of PKSs and NRPSs genes has made them unusually evolvable. As we will discuss in the next section, the evolution of gene clusters is likewise connected to the modularity of their constituent subclusters. The nexus between modularity and evolvability is a theme found in other realms of evolutionary biology, including the rich diversity of animals with modular (segmented) body plans in the phyla Annelida and Arthropoda (74).
Changes to the Gene Roster.
The teicoplanin family antibiotics (75) (and their relatives in the vancomycin family) are NRPs that undergo modification both during and after their synthesis on the assembly-line enzyme. Three or four cross-linking events common to all family members form the cup structure that confers on these molecules the ability to bind the d-alanine-d-alanine termini of peptidoglycan monomers (76), preventing peptidoglycan cross-linking and thereby inhibiting bacterial growth. However, the teicoplanin family molecules differ in the complement of “tailoring” modifications they undergo after being released from the assembly-line enzyme. A47934 (77) has a sulfate group added, whereas teicoplanin (25) has three sugars attached (Fig. 3a), one of which is subsequently linked to a long-chain fatty acid. The discrepant tailoring of A47934 and teicoplanin results from the acquisition and loss of genes from their gene clusters (78) that encode tailoring enzymes.
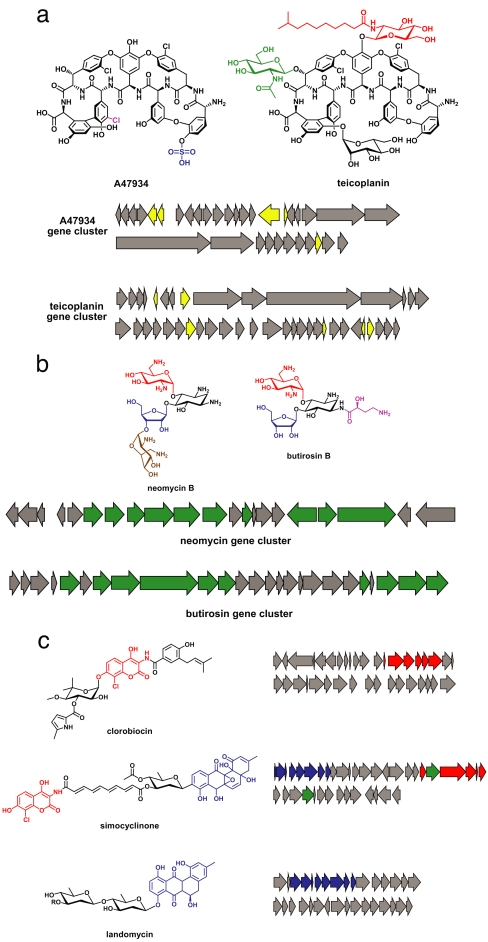
Fig. 3.
Changes to the number and identity of genes comprising a cluster. (a) Differences in the complement of peripheral genes encoding “tailoring” enzymes are a source of diversity in the glycopeptide antibiotics. The chemical structures of A47934 and teicoplanin are shown, with chemical groups unique to each molecule colored magenta, blue, green, or red. The A47934 and teicoplanin gene clusters are shown below, with the genes unique to each cluster colored yellow. (b) Differential tailoring of a common core in the aminoglycoside antibiotics. The chemical structures of neomycin B and butirosin B are shown with the 2-deoxystreptamine colored black, two peripheral components shared between these molecules colored red and blue, and peripheral components unique to each molecule colored brown and magenta. The neomycin and butirosin gene clusters are shown below, with genes shared between the two clusters colored green. (c) Subcluster joining gives rise to a new gene cluster. The chemical structures of clorobiocin, simocyclinone, and landomycin are shown with the aminocoumarin groups colored red and the anthracycline groups colored blue. The gene clusters for each molecule are shown at the right, with the aminocoumarin-producing subclusters colored red and the anthracycline-producing subclusters colored blue. Genes encoding putative conjugating enzymes are colored green.
Differential tailoring of a common core is also a hallmark of the aminoglycoside antibiotics (79), which perturb protein synthesis by binding to the 30S subunit of the bacterial ribosome. These molecules are based on the unusual sugar 2-deoxystreptamine (80), the scaffold that forms their core structure. However, the aminoglycosides are distinguishable by the identity of the sugars that are attached to the 2-deoxystreptamine and the position on the 2-deoxystreptamine to which they are attached (Fig. 3b). This core similarity and peripheral divergence is reflected in the gene clusters for aminoglycosides (15), which all share the enzymes responsible for production of 2-deoxystreptamine from the primary metabolite glucose-6-phosphate. These gene clusters differ, however, in the additional genes they encode, which are responsible for the synthesis and attachment of alternative peripheral sugars. Indeed, these alternative genes often reside in subclusters themselves, and it is likely that aminoglycoside gene clusters form primarily by joining semiindependently functioning subclusters. This mixing and matching of subclusters is another form of modularity that likely enhances the capacity of aminoglycoside gene clusters to evolve. The key enzymes that facilitate subcluster fusion are those that conjugate the products of the subclusters. For aminoglycosides, these enzymes are glycosyltransferases, and their evolutionary origins are particularly significant. Glycosyltransferase substitutions are responsible for the diversity in at least one other polysaccharide family, the O antigens of E. coli (81).
The process of joining subclusters often produces entirely new gene clusters. The antibiotic simocyclinone (Fig. 3c) consists of three chemical groups—a two-ring aminocoumarin, a four-ring anthracycline, and a linear polyene. Some small molecules, like the aminocoumarin clorobiocin and the anthracycline landomycin, contain only one of these chemical groups. The simocyclinone gene cluster (12, 82) contains three corresponding subclusters: one that is similar to the genes in the clorobiocin cluster that assemble the aminocoumarin, one that is related to the portion of the landomycin gene cluster that produces the anthracycline, and one that is likely to produce the polyolefinic linker. In all likelihood, the linkage of these three subclusters within a single genome triggered the “invention” of the hybrid small molecule simocyclinone. An important avenue of inquiry for simocyclinone (as for glycosyltransferases from aminoglycoside gene clusters) implicates the enzymes that link the three chemical groups together (83, 84). Because these conjugating enzymes are necessary for the functioning of a new “supercluster,” but would not have been required for the original gene clusters, their evolutionary origin is unclear. Some relatives of the glycosyltransferase- and acyltransferase-conjugating enzymes are promiscuous (85, 86), and this plasticity may have contributed to the evolution of conjugating enzymes.
Subcluster joining facilitates a particularly interesting form of chemical innovation in which distinct metabolic pathways merge, enabling new forms of structural diversity among small-molecule products by joining types of small-molecule fragments that normally are not linked. One compelling example of this process is leupyrrin (87), an unusual small molecule that comprises the products of four different metabolic pathways (88). Although its biosynthetic gene cluster has not yet been identified, it furnishes an intriguing system for studying the origins of subclusters and the conjugating enzymes that allow them to be joined functionally.
Divergent Biosynthetic Evolution: Never Far from a Functional Molecule.
The twin processes of gene duplication and functional divergence are central to the evolution of complexity because they permit one of the genes to veer off on a new course. Not surprisingly, they also play a prominent role in the evolution of new biosynthetic function, with two added twists: duplication can be intergenic (horizontal gene transfer) as well as intragenic, and entire gene clusters can diverge as a cohesive unit. We begin by discussing duplication and divergence of subgene fragments and progress to individual genes and gene clusters.
The duplication and divergence of subgene fragments was discussed above in the context of intragenic module duplications in PKSs and NRPSs. A phylogenetic analysis of PKSs from the bacterial genus Streptomyces demonstrates convincingly that similar intragenic module duplications, combined with recombination events, have given rise to much of the diversity of these PKSs (64). Moving up to the level of an individual gene, a separate phylogenetic analysis of fungal PKSs—which are often composed of a single multimodular gene—demonstrates that duplication and divergent evolution have led to large and widely distributed families of these genes (89). These studies illustrate how biosynthetic genes with common ancestry can diverge into broad families that produce richly diverse small molecules with a wide variety of biological activities.
Duplication and divergence also provide clues to a second mystery: Where did the biosynthetic genes for secondary metabolites originate? Perhaps not surprisingly, biosynthetic gene clusters often harbor enzymes with strong homology to a primary metabolic enzyme. For example, the enzyme RifG (90) cyclizes a linear intermediate as part of the pathway that forms 3-amino-5-hydroxybenzoic acid (AHBA), a precursor to the antibiotic rifamycin (91) and the antitumor agent geldanamycin (92) (Fig. 4a). RifG shares considerable homology with AroB (93), a primary metabolic enzyme that catalyzes a nearly identical reaction along the pathway to shikimic acid, the precursor of the aromatic amino acids. RifG almost certainly arose from duplication of the gene that encodes AroB and subsequent divergent evolution to alter its substrate selectivity. A more impressive change in gene function through divergent evolution is found in the enzyme β-lactam synthetase (94), which constructs the strained four-membered ring found in the β-lactamase inhibitor clavulanic acid. A combination of bioinformatics and structural biology (95) was used to demonstrate that β-lactam synthetase is closely related to the primary metabolic enzyme asparagine synthetase, indicating that functional divergence is not limited to minor changes in activity.
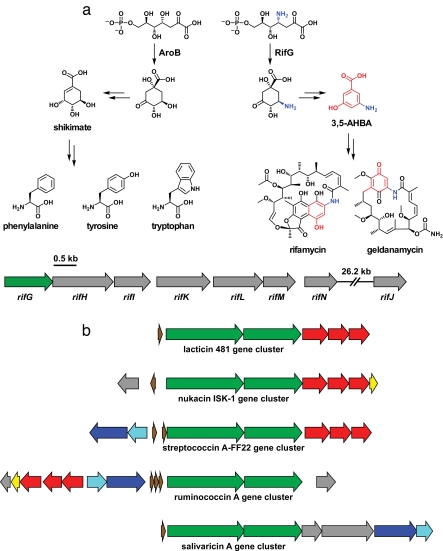
Fig. 4.
Divergent biosynthetic evolution. (a) Genes with origins in primary metabolism. The 3,5-AHBA biosynthetic enzyme RifG and its homolog in the shikimate biosynthetic pathway, AroB, catalyze similar reactions. Shikimate is the precursor to the aromatic amino acids, whereas 3,5-AHBA is a precursor to the polyketides rifamycin and geldanamycin. The 3,5-AHBA-producing subcluster from the rifamycin gene cluster is shown below, with rifG colored green. (b) Duplication and divergence of gene clusters. Five lantibiotic gene clusters from the lacticin 481 family are shown; related genes are depicted in the same color to highlight the common features of the clusters. Adapted from ref. 97.
Finally, entire gene clusters commonly undergo functional divergence after duplication, giving rise to families of related gene clusters that produced related small-molecule products. The gene clusters that produce lantibiotics (96, 97) (Fig. 4b), a large class of ribosomal peptide antibiotics that require posttranslational intrapeptide cross-linking to reach their active form (98), almost certainly had a common ancestor. So too do numerous other gene clusters, including those that produce nonribosomal peptide antibiotics of the glycopeptides (78) and lipopeptide (99) families. As more gene clusters are sequenced, gene clusters that are part of families that arose from duplication and divergence may become the norm.
Convergent Biosynthetic Evolution: More than One Molecule for the Job, and More than One Way to Get the Molecule.
Thus far we have limited our focus to how biosynthetic genes change, move about, and recombine to generate small-molecule diversity. Yet some of the most intriguing results of these analyses are the ways in which these processes can converge on different small molecules with the same function or different pathways to the small molecule.
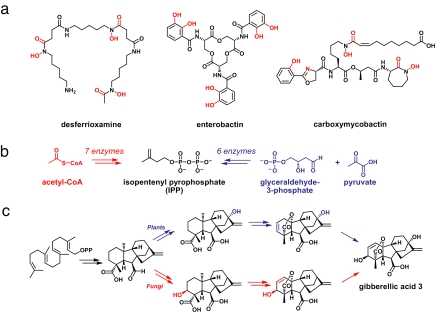
Examples of functional convergence—two or more unrelated gene clusters that produce a molecule with the same function—abound. As noted above, many bacteria and fungi produce iron-binding small molecules to scavenge iron from their environment, and three examples are depicted in Fig. 5a. Despite featuring structures and biosynthetic pathways that are unrelated, desferrioxamine, enterobactin, and carboxymycobactin all bind iron tightly and are used by their hosts for the same purpose.
Fig. 5.
Convergent biosynthetic evolution. (a) Desferrioxamine, enterobactin, and carboxymycobactin are functionally convergent in that these unrelated molecules all bind iron. Their chemical structures are with chemical groups that are ligands to the iron, which is colored red. (b) Two different pathways for the formation of the terpene building-block isopentenyl pyrophosphate (IPP). The mevalonate pathway (red) converts acetyl-CoA to IPP using seven enzymes, whereas the nonmevalonate pathway (blue) uses six unrelated enzymes to convert glyceraldehyde-3-phosphate and pyruvate to IPP. (c) Convergent biosynthesis of the gibberellins. The plant (blue) and fungal (red) biosynthetic pathways for giberellic acid 3 involve two different sequences of chemical tailoring steps that yield identical small-molecule products.
Just as unrelated protein folds can evolve convergently to catalyze the same reaction (100, 101), unrelated gene clusters can evolve to produce similar (in some cases, identical) molecules. These examples are more compelling than two unrelated protein folds converging on the same activity; often, in order for unrelated gene clusters to produce the same molecule, more than five proteins (or linked but independently folded protein domains) must converge coordinately because they function cooperatively to synthesize a small molecule. One example of convergent biosynthetic evolution emerges from the primary metabolic pathways (102) that produce the Δ2 and Δ3 isomers of isopentenyl pyrophosphate (IPP), the five-carbon building block from which cholesterol, steroid hormones, and terpenoid natural products are constructed (Fig. 5b). Eukaryotes and archaea use the “mevalonate pathway” (103), which converts acetyl-CoA into IPP through the action of seven enzymes. In contrast, most prokaryotes use the “nonmevalonate pathway” (104), which uses six enzymes unrelated to the mevalonate pathway enzymes to convert pyruvate and glyceraldehyde-3-phosphate into IPP. Every feature of these pathways—other than their endpoints—is completely dissimilar. Curiously, some prokaryotes use the mevalonate pathway (105), possibly suggesting that both pathways originated in the prokaryotic world. The evolutionary history of these pathways has begun to be elucidated (102, 106).
The gibberellins are a particularly striking example of convergent evolution (107, 108) (Fig. 5c). These terpenoid natural products are produced by plants as growth hormones; identical molecules are produced by fungi and bacteria to regulate (or dysregulate) plant growth processes. The fungal and plant pathways differ in the manner in which the precursor's carbon skeleton is oxidatively modified to form the final products (107, 108). The bacterial pathway is still unknown. It might either be similar to the fungal pathway (like the bacterial and fungal β-lactam biosynthetic pathways) or it might be a third unrelated pathway that converged on the same product. In the former case, the gibberellins would be a family of natural products in which convergent and divergent evolution have both occurred. In the latter case, they would be a singularly fascinating example of three biosynthetic systems converging on the same small-molecule product. In either case, it is remarkable that identical molecules are produced two different ways by unrelated gene clusters. This convergence is a testament to the evolvability of biosynthetic gene clusters, and it suggests that there may be additional examples of convergent biosynthetic evolution waiting to be discovered.
Conclusion: How Do New Gene Clusters Form?
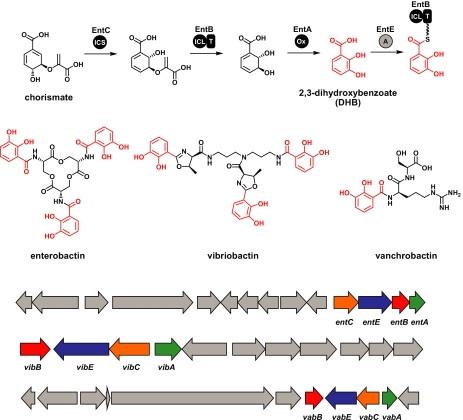
Ironically, the best way to begin studying how gene clusters evolve might be to ask how they came to exist in the first place. It is tempting to speculate that many of the enormous and complex gene clusters we observe in contemporary organisms arose largely from successive subcluster-joining events. “Vestiges” of these fragments still exist in the form of small clusters of genes that divert a primary metabolite to a widely used secondary metabolite. For example, a four-gene cluster (Fig. 6) that converts the primary metabolite chorismate into an activated form of the secondary metabolite 2,3-dihydroxybenzoate (DHB) is found in several genetic contexts (109–111). Not only is the DHB subcluster found in a variety of gene clusters that produce a DHB-containing siderophore, but DHB itself can act as a siderophore (112) (albeit not as effectively). Intriguingly, this suggests that DHB-incorporating siderophore gene clusters may have evolved by adding genes that would synthesize a small-molecule scaffold to link more than one DHB. Importantly, no point along this evolutionary path would require a transition through an intermediate gene cluster that did not produce an iron-chelating molecule.
Fig. 6.
Ancestral subclusters. A four-gene subcluster found in many different genetic contexts diverts the primary metabolite chorismate to an activated form of the secondary metabolite 2,3-dihydroxybenzoate (DHB). The chemical structures of the DHB-containing molecules enterobactin (ent), vibriobactin (vib), and vanchrobactin (vab) are shown with the DHB groups colored red. Their gene clusters are shown below, with the DHB subcluster genes shown in color. A portion of the vibriobactin gene cluster has been omitted for clarity.
Constructing sequence-based gene-cluster phylogenies presents formidable challenges. If we deconstruct gene clusters into fragments for analysis, should those fragments be subclusters, individual genes, or subportions of genes? How will phylogenies of these fragments be combined coherently to accurately reflect the evolutionary history of a gene cluster? Undoubtedly, these higher-order phylogenies will be richly interwoven and difficult to analyze. But if we can begin to construct the evolutionary histories of biosynthetic gene clusters, we may ultimately piece together what Darwin entrusted us to confirm: the story of how complexity evolved through natural selection.
Acknowledgments.
We thank Dan Hartl and Craig Townsend for their helpful comments on the manuscript. Work in the authors' laboratories is supported by National Institutes of Health Grants CA24487 and CA59021 (to J.C.) and GM20011 and GM49338 (to C.T.W). M.A.F. was supported by a fellowship from the Hertz Foundation.
Footnotes
The authors declare no conflict of interest.
References
- 1.Darwin CR. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. London: John Murray; 1859. [PMC free article] [PubMed] [Google Scholar]
- 2.Pelludat C, Rakin A, Jacobi CA, Schubert S, Heesemann J. J Bacteriol. 1998;180:538–546. doi: 10.1128/jb.180.3.538-546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanson JR. Natural Products: The Secondary Metabolites. Cambridge, UK: R Soc of Chem; 2003. [Google Scholar]
- 4.Straight PD, Fischbach MA, Walsh CT, Rudner DZ, Kolter R. Proc Natl Acad Sci USA. 2007;104:305–310. doi: 10.1073/pnas.0609073103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Straight PD, Willey JM, Kolter R. J Bacteriol. 2006;188:4918–4925. doi: 10.1128/JB.00162-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strobel G, Daisy B. Microbiol Mol Biol Rev. 2003;67:491–502. doi: 10.1128/MMBR.67.4.491-502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gil-Turnes MS, Hay ME, Fenical W. Science. 1989;246:116–118. doi: 10.1126/science.2781297. [DOI] [PubMed] [Google Scholar]
- 8.Long SR. Plant Cell. 1996;8:1885–1898. doi: 10.1105/tpc.8.10.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engel S, Jensen PR, Fenical W. J Chem Ecol. 2002;28:1971–1985. doi: 10.1023/a:1020793726898. [DOI] [PubMed] [Google Scholar]
- 10.Currie CR, Scott JA, Summerbell RC, Malloch D. Nature. 1999;398:701–704. [Google Scholar]
- 11.Clardy J, Walsh C. Nature. 2004;432:829–837. doi: 10.1038/nature03194. [DOI] [PubMed] [Google Scholar]
- 12.Trefzer A, Pelzer S, Schimana J, Stockert S, Bihlmaier C, Fiedler HP, Welzel K, Vente A, Bechthold A. Antimicrob Agents Chemother. 2002;46:1174–1182. doi: 10.1128/AAC.46.5.1174-1182.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McAlpine JB, Bachmann BO, Piraee M, Tremblay S, Alarco AM, Zazopoulos E, Farnet CM. J Nat Prod. 2005;68:493–496. doi: 10.1021/np0401664. [DOI] [PubMed] [Google Scholar]
- 14.Fischbach MA, Walsh CT. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 15.Flatt PM, Mahmud T. Nat Prod Rep. 2007;24:358–392. doi: 10.1039/b603816f. [DOI] [PubMed] [Google Scholar]
- 16.Lawrence J. Curr Opin Genet Dev. 1999;9:642–648. doi: 10.1016/s0959-437x(99)00025-8. [DOI] [PubMed] [Google Scholar]
- 17.Rachid S, Krug D, Kunze B, Kochems I, Scharfe M, Zabriskie TM, Blocker H, Muller R. Chem Biol. 2006;13:667–681. doi: 10.1016/j.chembiol.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Winter JM, Moffitt MC, Zazopoulos E, McAlpine JB, Dorrestein PC, Moore BS. J Biol Chem. 2007;282:16362–16368. doi: 10.1074/jbc.M611046200. [DOI] [PubMed] [Google Scholar]
- 19.Lenski RE, Travisano M. Proc Natl Acad Sci USA. 1994;91:6808–6814. doi: 10.1073/pnas.91.15.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawrence JG, Roth JR. Genetics. 1996;143:1843–1860. doi: 10.1093/genetics/143.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koonin EV, Makarova KS, Aravind L. Annu Rev Microbiol. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ochman H, Lawrence JG, Groisman EA. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 23.Frary A, Nesbitt TC, Grandillo S, Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB, Tanksley SD. Science. 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- 24.Newman DJ, Cragg GM, Snader KM. J Nat Prod. 2003;66:1022–1037. doi: 10.1021/np030096l. [DOI] [PubMed] [Google Scholar]
- 25.Sosio M, Kloosterman H, Bianchi A, de Vreugd P, Dijkhuizen L, Donadio S. Microbiology. 2004;150:95–102. doi: 10.1099/mic.0.26507-0. [DOI] [PubMed] [Google Scholar]
- 26.Brautaset T, Sekurova ON, Sletta H, Ellingsen TE, StrLm AR, Valla S, Zotchev SB. Chem Biol. 2000;7:395–403. doi: 10.1016/s1074-5521(00)00120-4. [DOI] [PubMed] [Google Scholar]
- 27.Tang L, Shah S, Chung L, Carney J, Katz L, Khosla C, Julien B. Science. 2000;287:640–642. doi: 10.1126/science.287.5453.640. [DOI] [PubMed] [Google Scholar]
- 28.Schwecke T, Aparicio JF, Molnar I, Konig A, Khaw LE, Haydock SF, Oliynyk M, Caffrey P, Cortes J, Lester JB, et al. Proc Natl Acad Sci USA. 1995;92:7839–7843. doi: 10.1073/pnas.92.17.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hendrickson L, Davis CR, Roach C, Nguyen DK, Aldrich T, McAda PC, Reeves CD. Chem Biol. 1999;6:429–439. doi: 10.1016/s1074-5521(99)80061-1. [DOI] [PubMed] [Google Scholar]
- 30.Walsh CT. Chembiochem. 2002;3:125–134. doi: 10.1002/1439-7633(20020301)3:2/3<124::AID-CBIC124>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 31.Arya P, Joseph R, Chou DT. Chem Biol. 2002;9:145–156. doi: 10.1016/s1074-5521(02)00105-9. [DOI] [PubMed] [Google Scholar]
- 32.Holden MT, Feil EJ, Lindsay JA, Peacock SJ, Day NP, Enright MC, Foster TJ, Moore CE, Hurst L, Atkin R, et al. Proc Natl Acad Sci USA. 2004;101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 34.Szczepanowski R, Braun S, Riedel V, Schneiker S, Krahn I, Puhler A, Schluter A. Microbiology. 2005;151:1095–1111. doi: 10.1099/mic.0.27773-0. [DOI] [PubMed] [Google Scholar]
- 35.Courvalin P. Antimicrob Agents Chemother. 1994;38:1447–1451. doi: 10.1128/aac.38.7.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dayem LC, Carney JR, Santi DV, Pfeifer BA, Khosla C, Kealey JT. Biochemistry. 2002;41:5193–5201. doi: 10.1021/bi015593k. [DOI] [PubMed] [Google Scholar]
- 37.Bultreys A, Gheysen I, de Hoffmann E. Appl Environ Microbiol. 2006;72:3814–3825. doi: 10.1128/AEM.00119-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duchaud E, Rusniok C, Frangeul L, Buchrieser C, Givaudan A, Taourit S, Bocs S, Boursaux-Eude C, Chandler M, Charles JF, et al. Nat Biotechnol. 2003;21:1307–1313. doi: 10.1038/nbt886. [DOI] [PubMed] [Google Scholar]
- 39.Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, Dodson RJ, Deboy RT, Durkin AS, Kolonay JF, et al. Proc Natl Acad Sci USA. 2003;100:10181–10186. doi: 10.1073/pnas.1731982100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koczura R, Kaznowski A. J Med Microbiol. 2003;52:637–642. doi: 10.1099/jmm.0.05219-0. [DOI] [PubMed] [Google Scholar]
- 41.Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W, Jensen PR, Moore BS. Proc Natl Acad Sci USA. 2007;104:10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carniel E. Int Microbiol. 1999;2:161–167. [PubMed] [Google Scholar]
- 43.Vokes SA, Reeves SA, Torres AG, Payne SM. Mol Microbiol. 1999;33:63–73. doi: 10.1046/j.1365-2958.1999.01448.x. [DOI] [PubMed] [Google Scholar]
- 44.Freiberg C, Brunner NA, Schiffer G, Lampe T, Pohlmann J, Brands M, Raabe M, Habich D, Ziegelbauer K. J Biol Chem. 2004;279:26066–26073. doi: 10.1074/jbc.M402989200. [DOI] [PubMed] [Google Scholar]
- 45.Long RA, Rowley DC, Zamora E, Liu J, Bartlett DH, Azam F. Appl Environ Microbiol. 2005;71:8531–8536. doi: 10.1128/AEM.71.12.8531-8536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Needham J, Kelly MT, Ishige M, Andersen RJ. J Org Chem. 1994;59:2058–2063. [Google Scholar]
- 47.Fredenhagen A, Tamura SY, Kenny PTM, Komura H, Naya Y, Nakanishi K, Nishiyama K, Sugiura M, Kita H. J Am Chem Soc. 1987;109:4409–4411. [Google Scholar]
- 48.Jin M, Fischbach MA, Clardy J. J Am Chem Soc. 2006;128:10660–10661. doi: 10.1021/ja063194c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liras P, Rodriguez-Garcia A, Martin JF. Int Microbiol. 1998;1:271–278. [PubMed] [Google Scholar]
- 50.Doolittle RF, Feng DF, Anderson KL, Alberro MR. J Mol Evol. 1990;31:383–388. doi: 10.1007/BF02106053. [DOI] [PubMed] [Google Scholar]
- 51.Salzberg SL, White O, Peterson J, Eisen JA. Science. 2001;292:1903–1906. doi: 10.1126/science.1061036. [DOI] [PubMed] [Google Scholar]
- 52.Lawrence JG, Hendrix RW, Casjens S. Trends Microbiol. 2001;9:535–540. doi: 10.1016/s0966-842x(01)02198-9. [DOI] [PubMed] [Google Scholar]
- 53.Ochman H, Moran NA. Science. 2001;292:1096–1099. doi: 10.1126/science.1058543. [DOI] [PubMed] [Google Scholar]
- 54.Phillips DR, Rasbery JM, Bartel B, Matsuda SP. Curr Opin Plant Biol. 2006;9:305–314. doi: 10.1016/j.pbi.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 55.Sawai S, Akashi T, Sakurai N, Suzuki H, Shibata D, Ayabe S, Aoki T. Plant Cell Physiol. 2006;47:673–677. doi: 10.1093/pcp/pcj032. [DOI] [PubMed] [Google Scholar]
- 56.Yoshikuni Y, Ferrin TE, Keasling JD. Nature. 2006;440:1078–1082. doi: 10.1038/nature04607. [DOI] [PubMed] [Google Scholar]
- 57.Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Proc Natl Acad Sci USA. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Donia MS, Hathaway BJ, Sudek S, Haygood MG, Rosovitz MJ, Ravel J, Schmidt EW. Nat Chem Biol. 2006;2:729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- 59.Wu K, Chung L, Revill WP, Katz L, Reeves CD. Gene. 2000;251:81–90. doi: 10.1016/s0378-1119(00)00171-2. [DOI] [PubMed] [Google Scholar]
- 60.Olson MV. Am J Hum Genet. 1999;64:18–23. doi: 10.1086/302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moyne AL, Cleveland TE, Tuzun S. FEMS Microbiol Lett. 2004;234:43–49. doi: 10.1016/j.femsle.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 62.Wenzel SC, Meiser P, Binz TM, Mahmud T, Muller R. Angew Chem. 2006;45:2296–2301. doi: 10.1002/anie.200503737. [DOI] [PubMed] [Google Scholar]
- 63.Konz D, Doekel S, Marahiel MA. J Bacteriol. 1999;181:133–140. doi: 10.1128/jb.181.1.133-140.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jenke-Kodama H, Borner T, Dittmann E. PLoS Comput Biol. 2006;2:e132. doi: 10.1371/journal.pcbi.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stinear TP, Mve-Obiang A, Small PL, Frigui W, Pryor MJ, Brosch R, Jenkin GA, Johnson PD, Davies JK, Lee RE, et al. Proc Natl Acad Sci USA. 2004;101:1345–1349. doi: 10.1073/pnas.0305877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hahn DR, Gustafson G, Waldron C, Bullard B, Jackson JD, Mitchell J. J Indus Microbiol Biotechnol. 2006;33:94–104. doi: 10.1007/s10295-005-0016-9. [DOI] [PubMed] [Google Scholar]
- 67.Moffitt MC, Neilan BA. Appl Environ Microbiol. 2004;70:6353–6362. doi: 10.1128/AEM.70.11.6353-6362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rantala A, Fewer DP, Hisbergues M, Rouhiainen L, Vaitomaa J, Borner T, Sivonen K. Proc Natl Acad Sci USA. 2004;101:568–573. doi: 10.1073/pnas.0304489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clardy J. Proc Natl Acad Sci USA. 1995;92:56–61. doi: 10.1073/pnas.92.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Choi J, Chen J, Schreiber SL, Clardy J. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 71.Kissinger CR, Parge HE, Knighton DR, Lewis CT, Pelletier LA, Tempczyk A, Kalish VJ, Tucker KD, Showalter RE, Moomaw EW, et al. Nature. 1995;378:641–644. doi: 10.1038/378641a0. [DOI] [PubMed] [Google Scholar]
- 72.Ginolhac A, Jarrin C, Robe P, Perriere G, Vogel TM, Simonet P, Nalin R. J Mol Evol. 2005;60:716–725. doi: 10.1007/s00239-004-0161-1. [DOI] [PubMed] [Google Scholar]
- 73.Jenke-Kodama H, Sandmann A, Muller R, Dittmann E. Mol Biol Evol. 2005;22:2027–2039. doi: 10.1093/molbev/msi193. [DOI] [PubMed] [Google Scholar]
- 74.Dawkins R. Climbing Mount Improbable. New York: Norton; 1996. [Google Scholar]
- 75.Kahne D, Leimkuhler C, Lu W, Walsh C. Chem Rev. 2005;105:425–448. doi: 10.1021/cr030103a. [DOI] [PubMed] [Google Scholar]
- 76.Hubbard BK, Walsh CT. Angew Chem. 2003;42:730–765. doi: 10.1002/anie.200390202. [DOI] [PubMed] [Google Scholar]
- 77.Pootoolal J, Thomas MG, Marshall CG, Neu JM, Hubbard BK, Walsh CT, Wright GD. Proc Natl Acad Sci USA. 2002;99:8962–8967. doi: 10.1073/pnas.102285099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Donadio S, Sosio M, Stegmann E, Weber T, Wohlleben W. Mol Genet Genom. 2005;274:40–50. doi: 10.1007/s00438-005-1156-3. [DOI] [PubMed] [Google Scholar]
- 79.Llewellyn NM, Spencer JB. Nat Prod Rep. 2006;23:864–874. doi: 10.1039/b604709m. [DOI] [PubMed] [Google Scholar]
- 80.Busscher GF, Rutjes FP, van Delft FL. Chem Rev. 2005;105:775–791. doi: 10.1021/cr0404085. [DOI] [PubMed] [Google Scholar]
- 81.Cheng J, Wang Q, Wang W, Wang Y, Wang L, Feng L. Curr Microbiol. 2006;53:470–476. doi: 10.1007/s00284-006-0032-7. [DOI] [PubMed] [Google Scholar]
- 82.Galm U, Schimana J, Fiedler HP, Schmidt J, Li SM, Heide L. Arch Microbiol. 2002;178:102–114. doi: 10.1007/s00203-002-0429-z. [DOI] [PubMed] [Google Scholar]
- 83.Luft T, Li SM, Scheible H, Kammerer B, Heide L. Arch Microbiol. 2005;183:277–285. doi: 10.1007/s00203-005-0770-0. [DOI] [PubMed] [Google Scholar]
- 84.Pacholec M, Freel Meyers CL, Oberthur M, Kahne D, Walsh CT. Biochemistry. 2005;44:4949–4956. doi: 10.1021/bi047303g. [DOI] [PubMed] [Google Scholar]
- 85.O'Brien PJ, Herschlag D. Chem Biol. 1999;6:R91–R105. doi: 10.1016/S1074-5521(99)80033-7. [DOI] [PubMed] [Google Scholar]
- 86.Langenhan JM, Griffith BR, Thorson JS. J Nat Prod. 2005;68:1696–1711. doi: 10.1021/np0502084. [DOI] [PubMed] [Google Scholar]
- 87.Bode HB, Irschik H, Wenzel SC, Reichenbach H, Muller R, Hofle G. J Nat Prod. 2003;66:1203–1206. doi: 10.1021/np030109v. [DOI] [PubMed] [Google Scholar]
- 88.Bode HB, Wenzel SC, Irschik H, Hofle G, Muller R. Angew Chem. 2004;43:4163–4167. doi: 10.1002/anie.200454240. [DOI] [PubMed] [Google Scholar]
- 89.Kroken S, Glass NL, Taylor JW, Yoder OC, Turgeon BG. Proc Natl Acad Sci USA. 2003;100:15670–15675. doi: 10.1073/pnas.2532165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yu TW, Muller R, Muller M, Zhang X, Draeger G, Kim CG, Leistner E, Floss HG. J Biol Chem. 2001;276:12546–12555. doi: 10.1074/jbc.M009667200. [DOI] [PubMed] [Google Scholar]
- 91.August PR, Tang L, Yoon YJ, Ning S, Muller R, Yu TW, Taylor M, Hoffmann D, Kim CG, Zhang X, et al. Chem Biol. 1998;5:69–79. doi: 10.1016/s1074-5521(98)90141-7. [DOI] [PubMed] [Google Scholar]
- 92.Rascher A, Hu Z, Viswanathan N, Schirmer A, Reid R, Nierman WC, Lewis M, Hutchinson CR. FEMS Microb Lett. 2003;218:223–230. doi: 10.1016/S0378-1097(02)01148-5. [DOI] [PubMed] [Google Scholar]
- 93.Carpenter EP, Hawkins AR, Frost JW, Brown KA. Nature. 1998;394:299–302. doi: 10.1038/28431. [DOI] [PubMed] [Google Scholar]
- 94.Bachmann BO, Li R, Townsend CA. Proc Natl Acad Sci USA. 1998;95:9082–9086. doi: 10.1073/pnas.95.16.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Miller MT, Bachmann BO, Townsend CA, Rosenzweig AC. Nat Struct Biol. 2001;8:684–689. doi: 10.1038/90394. [DOI] [PubMed] [Google Scholar]
- 96.Stein T, Borchert S, Conrad B, Feesche J, Hofemeister B, Hofemeister J, Entian KD. J Bacteriol. 2002;184:1703–1711. doi: 10.1128/JB.184.6.1703-1711.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dufour A, Hindre T, Haras D, Le Pennec JP. FEMS Microbiol Rev. 2007;31:134–167. doi: 10.1111/j.1574-6976.2006.00045.x. [DOI] [PubMed] [Google Scholar]
- 98.Patton GC, van der Donk WA. Curr Opin Microbiol. 2005;8:543–551. doi: 10.1016/j.mib.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 99.Baltz RH, Miao V, Wrigley SK. Nat Prod Rep. 2005;22:717–741. doi: 10.1039/b416648p. [DOI] [PubMed] [Google Scholar]
- 100.Bork P, Sander C, Valencia A. Protein Sci. 1993;2:31–40. doi: 10.1002/pro.5560020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stebbins CE, Galan JE. Mol Cell. 2000;6:1449–1460. doi: 10.1016/s1097-2765(00)00141-6. [DOI] [PubMed] [Google Scholar]
- 102.Boucher Y, Doolittle WF. Mol Microbiol. 2000;37:703–716. doi: 10.1046/j.1365-2958.2000.02004.x. [DOI] [PubMed] [Google Scholar]
- 103.Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Nat Biotechnol. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 104.Rohdich F, Kis K, Bacher A, Eisenreich W. Curr Opin Chem Biol. 2001;5:535–540. doi: 10.1016/s1367-5931(00)00240-4. [DOI] [PubMed] [Google Scholar]
- 105.Wilding EI, Brown JR, Bryant AP, Chalker AF, Holmes DJ, Ingraham KA, Iordanescu S, So CY, Rosenberg M, Gwynn MN. J Bacteriol. 2000;182:4319–4327. doi: 10.1128/jb.182.15.4319-4327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lange BM, Rujan T, Martin W, Croteau R. Proc Natl Acad Sci USA. 2000;97:13172–13177. doi: 10.1073/pnas.240454797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hedden P, Phillips AL, Rojas MC, Carrera E, Tudzynski B. J Plant Growth Regul. 2001;20:319–331. doi: 10.1007/s003440010037. [DOI] [PubMed] [Google Scholar]
- 108.Tudzynski B. Appl Microbiol Biotechnol. 2005;66:597–611. doi: 10.1007/s00253-004-1805-1. [DOI] [PubMed] [Google Scholar]
- 109.May JJ, Wendrich TM, Marahiel MA. J Biol Chem. 2001;276:7209–7217. doi: 10.1074/jbc.M009140200. [DOI] [PubMed] [Google Scholar]
- 110.Wyckoff EE, Stoebner JA, Reed KE, Payne SM. J Bacteriol. 1997;179:7055–7062. doi: 10.1128/jb.179.22.7055-7062.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Crosa JH, Mey AR, Payne SM. Iron Transport in Bacteria. Washington, DC: Am Soc Microbiol; 2004. [Google Scholar]
- 112.Lopez-Goni I, Moriyon I, Neilands JB. Infect Immun. 1992;60:4496–4503. doi: 10.1128/iai.60.11.4496-4503.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]