Abstract
In all probability, natural selection began as ancient marine microorganisms were required to compete for limited resources. These pressures resulted in the evolution of diverse genetically encoded small molecules with a variety of ecological and metabolic roles. Remarkably, many of these same biologically active molecules have potential utility in modern medicine and biomedical research. The most promising of these natural products often derive from organisms richly populated by associated microorganisms (e.g., marine sponges and ascidians), and often there is great uncertainty about which organism in these assemblages is making these intriguing metabolites. To use the molecular machinery responsible for the biosynthesis of potential drug-lead natural products, new tools must be applied to delineate their genetic and enzymatic origins. The aim of this perspective is to highlight both traditional and emerging techniques for the localization of metabolic pathways within complex marine environments. Examples are given from the literature as well as recent proof-of-concept experiments from the authors' laboratories.
Extensive drug discovery and small molecule screening programs over the past two decades have shown marine organisms to be rich sources of structurally diverse and highly bioactive natural products (1). The molecular architectures of marine metabolites are distinct from those of their terrestrial relatives in that the physicochemical requirements of adaptation to an aqueous world, the biosynthetic pathways used, and even the elements employed in crafting their arsenal of defensive molecules are quite different (2). As a consequence of their structural diversity and uniqueness, marine natural products are providing a prominent share of the recent clinical and preclinical lead compounds for the treatment of various diseases, most prominently cancer (3).
At first analysis, the sources of these important leads are taxonomically diverse and include sponges, tunicates, corals, mollusks, fungi, and sediment-derived bacteria. However, there is growing recognition that the “collected source” for these molecules is not necessarily the “metabolic source.” That is, marine bacteria and cyanobacteria, either assimilated by an invertebrate grazer (e.g., sea hare grazing on cyanobacteria) or growing in association with an invertebrate host in a symbiotic or commensal relationship, are frequently the true origin of these molecules (4–6). The growing appreciation of microbial metabolism in the production of many of the marine pharmaceutical leads comes from evidence in several sectors. Ultimately, firm knowledge of the natural product-producing organism, the “holder of the genes,” is essential to numerous branches of science and technology, including ecology, biosynthesis, and natural product drug development.
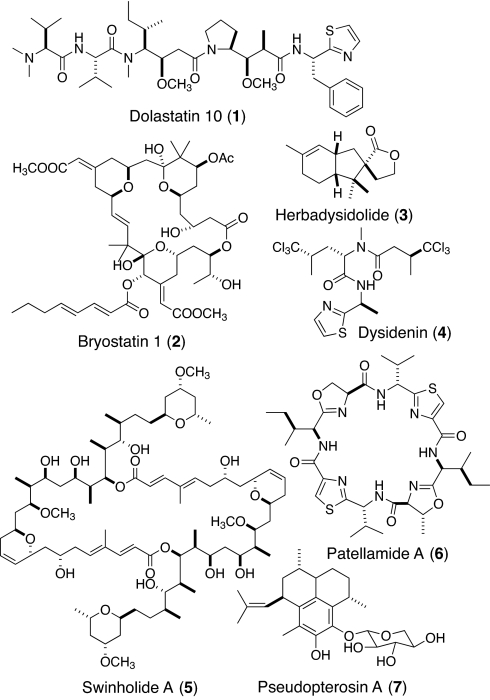
Of 20 compounds deriving from (or inspired by) marine natural products that are currently or were recently in clinical trials for the treatment of cancer (5), 15 were isolated from sponges, tunicates, and mollusks, with only 5 coming directly from a microorganism. However, based on biosynthetic parallels, distribution in taxonomically diverse organisms, and, in a few cases, subsequent direct isolation from a producing microorganism, 16 of these anticancer molecules actually derive from microbial sources and only 4 derive from macroorganisms. For example, dolastatin 10 (1; Fig. 1), originally isolated from the marine gastropod Dolabella auricularia, was later shown to originate from cyanobacteria of the genera Symploca and Lyngbya upon which they feed (7–10). The powerful PKC-activating cancer cell toxin bryostatin (2) was isolated from the Californian bryozoan Bugula neritina, but the biosynthetic capacity to make this unique macrolide has been localized to an associated bacterium Endobugula sertula (11). Although the ecological roles of most marine natural products are not well understood, most appear to be involved in defense against predation or competition for space and nutrients (12–14).
Fig. 1.
Structures 1–7.
It has been surprisingly difficult to demonstrate that an invertebrate-associated microorganism is unequivocally responsible for the production of a given secondary metabolite (6). However, recent advances on a number of fronts are providing new and powerful methods for investigating these relationships. For example, in the plant-endophyte arena, the anticancer compounds podophyllotoxin and camptothecin were recently discovered as products of endophytic microbes (15, 16). New advances in cell sorting, culturing methods, and methods of chemical detection are enabling the physical separation of associated species for subsequent chemical analysis. Additionally, mass spectral imaging is proving exceptionally powerful for identifying the location of natural products in mixed species assemblages. Finally, gene probing techniques, such as catalyzed reporter deposition (CARD)–FISH or in situ hybridization, can identify those organisms with the genetic capacity to produce specific molecules of interest. This article is a review of advances made in each of these three areas as reported in the literature and provides some new and original results from our laboratories on these subjects.
Results and Discussion
Microorganism Isolation Approaches.
Confronted with a mixture of species that produces important secondary metabolites, the simplest approach is to separate cell types and then analyze these for the compound(s) of interest. It is plausible, however, that the natural product under study is produced in one species, excreted, and then assimilated by a second species. For example, efforts to localize peptides in the tunicate Lissoclinum patella showed that they occur predominately in the ascidian tunic and not in the symbiotic cyanobacterium Prochloron sp. (17). However, subsequent heterologous expression and genome sequence analysis indicated that these peptides are indeed genetically encoded for and biosynthesized by the cyanobacterium (18, 19). In a few cases, and despite considerable technological difficulties, successful segregation of cell types and their cultivation has allowed isotope incorporation studies to demonstrate new biosynthesis and, hence, identification of the producing organism (4, 6, 20).
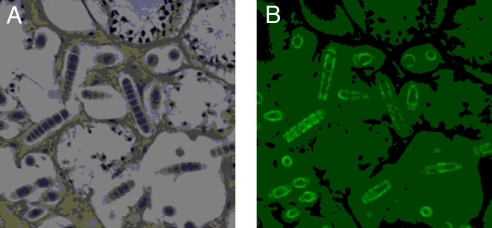
Sponge tissues are extremely rich in bacteria that can comprise up to 40% of their biomass (21). Early efforts to localize natural products to specific cell types used formalin or glutaraldehyde-fixed sponge-cyanobacterial tissues from the tropical sponge Dysidea herbacea (Fig. 2). The tissues were disrupted, and individual cells were separated by using a fluorescence-activated cell sorter (FACS). Chemical analysis of the sorted cell types showed sesquiterpenoid compounds (e.g., 3) to be physically associated with the sponge cells, whereas chlorinated peptides (e.g., 4) were localized to the associated cyanobacterium, Oscillatoria spongeliae (22). In studies of the cellular localization of several complex metabolites isolated from the sponge Theonella swinhoei, including swinholide A (5) and the peptide theopalauamide, a combination of dissection and differential centrifugation gave cell preparations that were chemically analyzed. Curiously, theopalauamide was found in a filamentous bacterial population morphologically related to Beggiatoa, whereas swinholide A (6) was localized to a mixed unicellular bacterial fraction (23, 24). This latter finding is at odds with our subsequent isolation of swinholide A from a free-living marine cyanobacterium, Geitlerinema sp. (25).
Fig. 2.
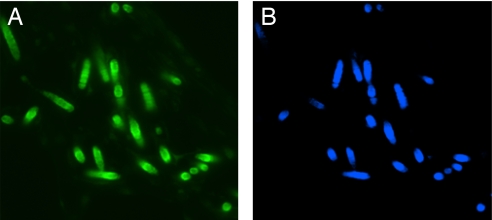
Photomicrographs of a thin section of sponge tissue showing embedded cyanobacterial cells. (A) Cross-section of the sponge Dysidea herbacea stained with O-toluidine showing composition of cells types. (B) Autofluorescence (522 nm) of cyanobacterial cells due to chlorophyll. [Reprinted with permission from ref. 92 (Copyright 2005, Springer).]
The microbial communities associated with various sponges have been investigated by using both culture-dependent and culture-independent methodologies (4). Recently, a culturable α-proteobacterial symbiont was obtained from several sponges, including Mycale laxissima (26). A fluorescently labeled 16S rRNA gene probe was prepared for FISH analysis to visualize gene distribution. Intriguingly, the bacteria were concentrated on the surfaces of the sponge larvae, suggesting vertical transmission of symbionts between sponge generations. However, no natural products have been reported from this bacterium to date (27).
In another recent example, the spatial distribution of bacteria within the sponge Tethya aurantium was mapped by using denaturing gradient gel electrophoresis (DGGE) and 16S rDNA clone library analysis. These studies showed that a new phylotype of Flexibacteria occurred in the sponge cortex whereas the endosome contained the cyanobacterium Synechococcus sp. Interestingly, 16S rDNA sequences for a β-proteobacteria were found throughout the endosome and cortex (27).
Various gene probing methods (e.g., FISH, PCR screening, Southern analysis, and genome sequencing) can demonstrate the occurrence of a biosynthetic gene in a particular isolated cell type. For example, the Haygood laboratory demonstrated a reduction in the abundance of a ketosynthase gene from the putative bryostatin gene cluster in B. neritina larvae that had been treated with antibiotics (28). In a second example, a genome sequence of the cyanobacterium Prochloron sp., a symbiont in the tunicate Lissoclinum patella, identified the cyanobacterium as the biosynthetic source of the ribosomally encoded peptides patellamides A (6) and C (18, 19).
Eukaryotic dinoflagellates are common coral symbionts, and soft corals in the Gorgonacea are particularly well studied for their bioactive diterpenoids. The pseudopterosins (e.g., 7) for example are potent antiinflammatory terpenes obtained from the soft coral Pseudopterogorgia elisabethae and are used in commercially available skin care products. Investigation of the site of biosynthesis of the pseudopterosins by using differential centrifugation produced a 99% pure preparation of the dinoflagellate Symbiodinium sp. This macroalgal preparation was found to contain compound 7, and provision of either [14C]NaHCO3 or 3H-geranylgeraniol pyrophosphate led to radioactive HPLC peaks corresponding to the pseudopterosins (29). From these and other data, it was concluded that the dinoflagellate was responsible for pesudopterosin biosynthesis. However, a subsequent patent has appeared that identifies associated bacteria as the biosynthetic source of the pseudopterosins, pointing out the danger of concluding too much from a cell separation followed by chemical analysis or undiluted radiolabeling experiments (30).
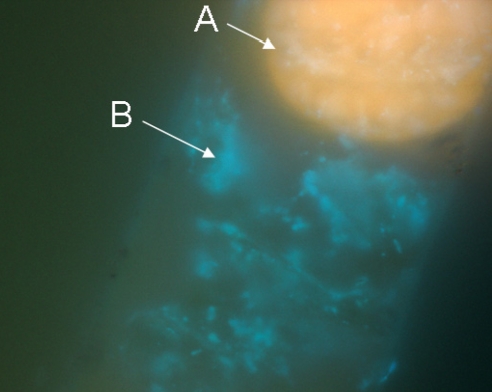
Although most marine microorganisms are not easily separated from complex assemblages by direct manipulation, filamentous cyanobacteria grow to dimensions more amenable to manual isolation (e.g., Lyngbya majuscula filaments are up to 100 μm wide and several centimeters long). However, the tough polysaccharide sheath surrounding cyanobacterial filaments can harbor a multitude of heterotrophic bacteria that are difficult to remove (31–33) (Fig. 3). We and others find that the dominating heterotrophic bacteria associated with these sheaths are gram-negatives from the phyla Proteobacteria and Bacteroidetes. These include the α-proteobacteria (Caulobacter, Thalassobius, and Rhodospirillaceae), β-proteobacteria (Achromobacter), γ-proteobacteria (Pseudomonas and Oceanospirillum), and Bacteroidetes (Flavobacterium and Flexibacter) (34, 35). Interestingly, many of these same bacterial groups are also commonly associated with sponges and tunicates (36, 37).
Fig. 3.
Trichome structure of the filamentous marine cyanobacterium Lyngbya majuscula-3L. Shown is DAPI staining and epifluorescent imaging at ×1,000 magnification using a Zeiss Axioskop (filter set #2, emission 420 nm+). A, cyanobacterial phycoerythrin in filament tip (orange); B, filament sheath-associated bacterial DNA (blue).
Creating axenic cultures of cyanobacteria is extraordinarily difficult, and relatively few are reported in the literature. Previous investigations proposed that axenic cultures were produced by using irradiation and antibiotics; however, proof of their monoculture state used lack of bacterial growth on multiple media types as evidence (31–33, 38). Because the media, nutrient, and environmental combinations were not exhaustive, doubt remains as to whether these cultures were truly free of bacteria. Many oligotrophic organisms grow very slowly and to low cell densities; therefore, their presence can be easily overlooked by classic culture-dependant tests for axenicity (39). Hence, it remains uncertain whether sheath-associated bacteria are involved in the production or modification of natural products reported from cyanobacteria.
Chemical Detection Methods.
Microanalysis by mass spectrometry (MS).
In conjunction with the physical and genetic approaches described above, chemical detection methods have improved significantly, in particular MS, and these are becoming highly useful in locating the biosynthetic origin of specific metabolites. Particularly noteworthy improvements in mass spectrometry include the ability to analyze small quantities of material while increasing both resolution and mass accuracy. These developments have been largely driven by the proteomics community, usually in the context of discovering disease biomarkers (40–43). These efforts have spurred considerable development of new pre-separation methods, new ionization approaches, improved mass filters, faster scan rates for improved time scale detection, and new detection methods, only some of which have found their way into the routine analysis of natural products. These developments in MS forecast that soon we may be able to perform rigorous structure elucidation on submicrogram quantities within certain metabolite classes. In the following sections, we present several examples from the authors' laboratories that demonstrate how some of these very sensitive MS technologies can be used in the discovery of new marine natural products, and how variations in these methods can help in identifying the true biosynthetic sources of these compounds.
Electrospray ionization (ESI) and MALDI MS are revolutionary soft ionization methods that enable MS measurements of chemically sensitive biomolecules (44). However, for either of these techniques to be effective, there is an absolute requirement for ionization. ESI has become the most common approach of ionization in the natural product chemist's arsenal because it can be interfaced with LC capabilities, thereby connecting molecular mass data with elution characteristics of a particular molecule. These data greatly expedite the assignment of a natural product as either novel or known (dereplication).
In addition to traditional HPLC-ESI-MS, capillary LC-ESI-MS has proven extraordinarily useful in proteomic research. Capillary LC requires extremely small sample sizes (<1.0 μg) with the ability to interface with many types of chromatographic supports such as reverse phase (C18, C4, etc.), hydrophilic interaction liquid chromatography (HILIC), cationic exchange, or even combinations of multiple resins [see supporting information (SI) Fig. 8] (45, 46). For example, we detected (and subsequently isolated and confirmed by NMR) the anticancer lead compound iejimalide A (8; Fig. 4) from a 0.1-μg crude cyanobacterial extract. This was significant because this macrolide had only previously been found from an Okinawan tunicate (47, 48). Detection was achieved by using a 100-μm fused silica capillary column loaded with 10 cm of C18 resin and a 5-μm tip. The estimated detection limit of iejimalide A by this technique is in the picomole-to-femtomole range. By contrast, traditional LC-MS with an LCQ instrument and 4.6-mm-diameter HPLC column required >100 μg of iejimalide-containing crude extract for detection (estimated detection limit in the micromole-to-nanomole range).
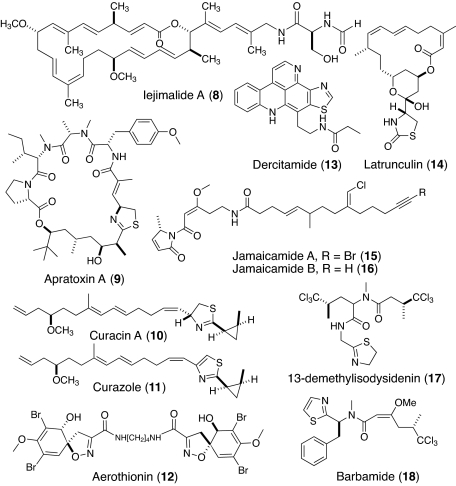
Fig. 4.
Structures 8–18.
Nanospray Fourier transform (FT)–ion cyclotron resonance MS (ICRMS).
High-resolution MS is another extremely useful technique for analyzing crude mixtures for the presence of a specific natural product (49). We used a complement of nanospray and FT-ICRMS to analyze the crude extract of the iejimalide A-producing cyanobacteria from Papua New Guinea. Despite the fact that the crude extract showed >40 ions in the region m/z 675–705, the sodiated iejimalide A peak at m/z 701.418 (calculated m/z 701.414) was well resolved and firmly identified the presence of iejimalide A (8; see SI Fig. 9). In total, this extract displayed several hundred unique molecular ions in a single spectrum (including several consistent with novel hydroxylated analogs of 8), and these were only resolvable because of the high-resolution attributes of FT-ICRMS. It should be noted that additional characterization of these metabolites can be achieved by combining the high resolution of FT-ICRMS with tandem MS (49). This “snapshot” of the secondary metabolome of a cyanobacterium will surely become an important discovery tool as natural product scientists learn to process and use such rich datasets.
MALDI-TOF-MS.
The species diversity associated with complex marine assemblages is well suited for analysis by another powerful MS technique, matrix-assisted laser desorption/ionization–time-of-flight MS (MALDI-TOF-MS). The utility of MALDI-MS for the analysis of biological samples (e.g., single cells and tissues) has been demonstrated for both biomedical and natural science applications. For example, MALDI-techniques were applied to investigate entire colonies of the cyanobacterium Microcystis to understand the diversity and distribution of hepatotoxic oligopeptides from these natural communities (50). Recently, “intact cell MALDI-TOF” (ICM) was used to assess the diversity of microbes from the tissues of marine invertebrates (e.g., sponges) by “proteometric clustering,” allowing both the dereplication of known secondary metabolites and providing a taxonomic characterization of the species present (51).
We have extended the utility of MALDI-TOF-MS to the analysis of single filaments of marine cyanobacteria for the identification of known and new secondary metabolites. The analysis of intact filament assemblages enables the localization of specific secondary metabolites within specific filament types. MALDI-TOF-MS analysis was performed on fresh single filaments of a strain of L. bouillonii known to produce apratoxin A (9), a potent mammalian cell cytotoxin (52, 53). In this example, a single filament was placed on a MALDI target plate and coated with an α-cyano-4-hydroxycinnamic acid matrix. Data analysis readily gave a strong pseudomolecular ion at m/z 840 (see SI Fig. 10), a value corresponding to the molecular formula of apratoxin A [C45H69N5O8S + H].
We also used MALDI-MS in combination with ESI-FTMS, LC-ESI-MS, and NMR to detect and characterize a novel analog of curacin A (10) (54–56). Aside from two double-bond isomers, curacins B and C (57), and the 8-desmethyl analog curacin D (58), no other naturally occurring analogs have yet been isolated. The biosynthesis of the distinctive cyclopropyl ring has been the focus of detailed molecular genetic and mechanistic biochemistry investigations because it appears to involve the intermediacy of a cryptic chlorination reaction (59–61). MALDI analysis of a single L. majuscula filament from Curaçao revealed a pseudomolecular ion at m/z 374 [M+H]+, consistent with the presence of curacin A. Another peak at m/z 372 [M+H]+ was also observed that did not correspond to any known cyanobacterial metabolite. FT-ICRMS of the extract from this strain allowed characterization of the 372 peak as exactly 2.015 Da less than curacin A, for a deduced molecular formula of C23H33NOS; thus, it represented a potentially intriguing structural homolog of curacin A. The pure compound was isolated and characterized by NMR and other spectroscopic techniques and shown to be an oxidized form of curacin A with a thiazole rather than a thiazoline ring. Previously, we had produced analog 11 via semisynthesis from natural curacin A and given it the trivial name “curazole” (62). Based on detection of the [M+H] ion from intact cyanobacterial filaments and through direct analysis of unfractionated crude extracts, it seems certain that curazole (11) is a genuine natural product of this L. majuscula strain. Contrary to literature precedent (60, 63), our sequence analysis revealed that the oxidase required for the thiazoline-to-thiazole transformation is not present in the curacin A gene cluster, nor is it found immediately up- or downstream of the cluster (60). This suggests that this transformation of the thiazoline to the thiazole is not encoded by a dedicated protein or protein domain.
There are pitfalls in the direct MALDI analysis of biological tissues, as illustrated by our attempt to detect iejimalide A (8) from cyanobacterial filaments. As noted, iejimalide A had been previously detected by traditional organic extraction of the collected biomass followed by NMR and FT-ICRMS approaches. MALDI-TOF-MS of single filaments from this sample failed to detect any 8. Consistent with this finding, pure iejimalide A, once it became available, also failed to provide a detectable ion signal by MALDI-TOF, likely because of its lack of basic nitrogen atoms. This example demonstrates that MALDI and ESI are complementary approaches and that both should be used in natural product dereplication and discovery programs.
Other important MS advances stem from the development of novel ionization methods and new mass analyzers. For example, desorption electrospray ionization (DESI), electrospray-assisted laser desorption ionization (ELDI), and direct analysis in real time (DART) are newly developed ionization methods that analyze samples directly and with minimal preparation (64–68). The new orbitrap Fourier transform (FT) mass analyzer can provide high resolution without the required expertise that an FT-ICR instrument requires (69, 70), a characteristic that has made this the instrument of choice in metabolomic studies (71). Although none of these more recent methods have yet been applied to the study of cyanobacterial natural products or complexes of coexisting species, it is likely that some will offer advantages over current methods.
Imaging Techniques.
Background.
In 1983, Faulkner and colleagues (72) used x-ray analysis coupled with transmission electron microscopy (TEM) to examine the location of two brominated metabolites, aerothionin (12) and homoaerothionin, within a sponge–microorganism complex. Although these experiments recorded increased bromine in a particular cell type (spherulous sponge cells), it was not certain that the detected bromine was actually part of the target compounds. Subsequently, the location of the polyaromatic alkaloid dercitamide (13) was directly imaged by using epifluorescence and laser-scanning confocal microscopy in conjunction with TEM. Interestingly, this study localized the natural product exclusively to the sponge cells (Oceanapia sagittaria) and not to the associated microbes as was expected (73). In another early study, the imaging of latrunculin B (14) in the sponge Negombata magnifica used immunogold staining with light microscopy and TEM. These studies also revealed that sponge cells were the site of storage and presumed biosynthesis (74). In light of subsequent natural product and biosynthetic investigations, these results with latrunculin B are particularly intriguing because related compounds have been shown to be produced by microorganisms using assembly-line type synthases typical of bacteria (2, 11).
MALDI imaging.
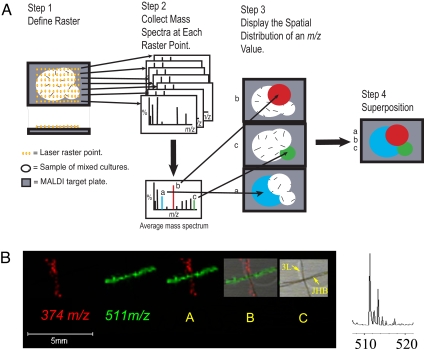
It is clear that single-filament MALDI-TOF analysis is a powerful method for directly observing both new and known secondary metabolites. Can MALDI-TOF also provide insight into the identity of metabolite-producing species found in complex environmental assemblages? As noted, this has been an extraordinarily difficult phenomenon to study because coexisting strains are difficult to grow as single pure strains, and many artifacts are introduced during sample preparation. An emerging strategy showing promise is MALDI-TOF imaging (75–78). MALDI imaging allows the spatial localization of precise molecular masses to particular cell types that are then observed through assignment of specific colors to a given mass (Fig. 5). Although this approach has not previously been applied to the analysis of natural products, it has been used to identify biomarkers of disease and the accumulation of therapeutics in whole-animal histological preparations (79, 80).
Fig. 5.
MALDI-TOF imaging to map the distribution of natural products in biological tissues. (A) Schematic for procedure of MALDI-TOF imaging. (B) From left to right, MALDI-TOF imaging of protonated curacin A (12) at m/z 374 (red), sodiated jamaicamide B (19) at m/z 511 (green), the simultaneous superposition of both m/z 374 and 511, superposition of both masses plus photograph of filaments, photograph of both filaments identified as strains “3L” (= curacin A producer) and JHB[ = jamaicamide A (18) and B (19) producer], and partial mass spectrum of JHB strain showing the molecular ion cluster of jamaicamide B with its distinctive chlorine isotope pattern.
Here, we briefly demonstrate the power and utility of this technology in two preliminary studies (E.E., R.C.C., T.L.S., D.G., W.H.G., and P.C.D., unpublished work). First, two L. majuscula strains (JHB from Jamaica and 3L from Curaçao) were mixed and analyzed by MALDI-TOF imaging. It is known that the JHB strain produces jamaicamide B (16) and the 3L strain produces curacin A (10) (54, 62, 81). Sodiated jamaicamide B has an m/z of 511 and is colored green on the image, whereas protonated curacin A has an m/z of 374 and is shown in red (Fig. 5). The MALDI-TOF image clearly distinguishes the two producing strains by the secondary metabolites they contain. Moreover, it is clear that jamaicamide B is chlorinated from the observed 35Cl/37Cl isotopic ratio.
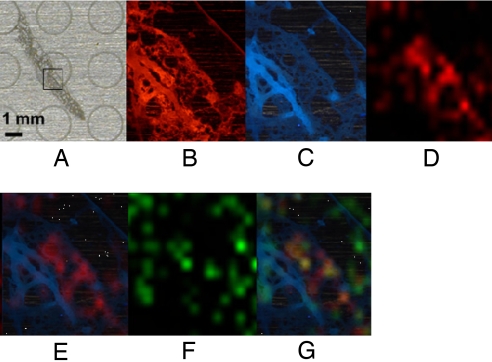
In a second example, we analyzed the marine sponge Dysidea herbaceae. Because of its relative availability, abundance of described natural products, and associated cyanobacterium (Oscillatoria spongeliae), D. herbaceae has developed as a useful model for secondary metabolite and biosynthetic gene cluster localization studies (see below, Gene-based imaging methods). Here, we prepared thin sections of sponge-cyanobacterium tissue using a cryogenic microtome for analysis by epifluorescence microscopy and MALDI imaging. Fig. 6 shows the portion of a thin section of sponge-cyanobacterial tissue used in the imaging analysis by autofluorescence at two wavelengths and then by MALDI-TOF. The mass at m/z 530 (Fig. 6 D, E, and G) and molecular ion isotope cluster (data not shown) are consistent with a hexachlorinated peptide such as 13-demethylisodysidenin (17). Another mass, m/z 1,028 (Fig. 6 F and G), is for an unknown metabolite with a very different distribution in the sponge matrix and clearly illustrates the power of MALDI-TOF imaging in differentially localizing secondary metabolites from complex environments.
Fig. 6.
MALDI-TOF imaging of the marine sponge Dysidea herbaceae. (A) MALDI target plate with thin section in place. (B) Autofluorescence of D. herbaceae thin section at 590 nm+. (C) Autofluorescence of D. herbaceae thin section at 420 nm+. (D) MALDI image of m/z 530. This mass and the complex molecular ion isotope cluster suggests its identity as the hexachlorinated peptide 13-demethylisodysidenin (20). (E) Autofluorescence (420 nm) image overlaid by MALDI image at m/z 530 indicating localization of compound with this mass. (F) MALDI image of unknown compound m/z 1,028. (G) Autofluorescence (420 nm) image overlaid with m/z 530 (red) and m/z 1,028 (green) showing the differential localization of these two molecules.
At present, MALDI imaging is limited by its spatial resolution, due in part to the requirement for a uniform crystalline matrix coating. The matrix coating allows for molecular diffusion of the analyte in the crystalline lattice, limiting the spatial resolution to the size of the crystals or the width of a typical N2 laser (≈100 μm). With improvements in both matrix deposition and laser technologies, it is anticipated that resolution in the range of 7–10 μm will become available (82). Finally, two recent advances that will allow structural characterization in conjunction with imaging are MALDI imaging-FT-ICR and the LTQ imaging system with tandem MS capabilities (77, 83).
Gene-based imaging methods.
The Haygood laboratory has examined the biosynthetic source of the anticancer bryo-statins (e.g., 2). Originally isolated from the bryozoan B. neritina, the bryostatin structures are consistent with a microbial polyketide synthase origin. To investigate the metabolic origin of 2, this group (28) used degenerate primers to clone a component of the polyketide synthase, the ketosynthase (KS) domain, from both larval and adult forms of the bryozoan. A total of 9 unique KS sequences were recovered, and one predominated in the larvae. This latter sequence was used to produce a biotin-labeled RNA gene probe that was subsequently visualized with an avidin-horseradish peroxidase detection system. Larvae treated with this probe were labeled only in a thin band, the pallial sinus, which was shown to contain a new γ-proteobacterial strain, Endobugula sertula. Although it was not conclusively demonstrated at this point whether the KS was specifically involved in bryostatin biosynthesis, subsequent gene cloning work indicated that it very likely was (11). Our analysis shows a 98% sequence identity at the DNA base level between the original KS and the BryB KS present in the subsequently cloned and sequenced bryostatin gene cluster.
In 1996, we reported the structure of the chlorinated peptide, barbamide (18), from a Curaçao collection of the marine cyanobacterium L. majuscula (84). Barbamide is strongly molluscicidal to the intermediate snail host involved in schistosomiasis, Biomphaleria glabrata. The halogen atoms comprising the unique trichloromethyl group are located at a position without an adjacent activating functionality, and this suggested a novel route for their biochemical incorporation (85, 86). These barbamide halogenases and related enzymes have since been characterized as a new class of “radical halogenases” capable of direct halogenation of unreactive carbon atoms in natural products, such as methyl groups (87–89). Early on, we recognized the striking structural similarity of barbamide to a number of Dysidea“sponge” chlorinated peptides. We and others proposed that the cyanobacterial symbiont (Oscillatoria spongeliae) found growing among the sponge cells was likely responsible for chlorinated peptide production (90, 91). Cell sorting experiments, as noted earlier, supported this hypothesis as the peptides were recovered from the purified cyanobacterial fraction (90, 91).
The DNA sequences of the barbamide halogenases BarB1 and BarB2 allowed a gene-based approach to investigate and confirm the identity of the producing organism in this symbiosis (92). First, homology-based cloning from the mixed sponge/cyanobacterial tissue (Fig. 2) yielded a barB1 homolog, dysB1, encoding a protein sequence with 93% amino acid identity to BarB1. The dysB1 sequence in combination with cyanobacterial-specific 16S rDNA sequences was used in FISH experiments to probe thin sections of the sponge containing the associated cyanobacterial symbiont. The cyanobacteria were identified in the thin sections by their chlorophyll fluorescence and by localization of the fluorescently labeled cyanobacterial-specific 16S rDNA gene probe. The crucial experiment showed colocalization of the fluorescently labeled dysB1 probe and 16S rDNA probe in cyanobacterial cells but not in sponge cells (Fig. 7). Cyanobacterial cells were thereby unequivocally shown to possess the halogenase mRNA necessary for chlorinated peptide biosynthesis.
Fig. 7.
CARD-FISH analysis of Dysidea herbacea showing autofluorescence (522 nm) of cyanobacterial cells due to chlorophyll (A) and probe-specific hybridization with the complimentary oligonucleotide to dysB1, a homolog of the barbamide barB1 biosynthetic gene (B). [Reprinted with permission from ref. 92 (Copyright 2005, Springer).]
Conclusions
The presence of a natural product within a particular cell type can provide only circumstantial evidence as to its biosynthetic origin. Above, various older techniques for showing the presence of a natural product have been reviewed and newer experimental approaches discussed. Some of these are appropriate for analysis of isolated cells, including HPLC, NMR, and, most powerfully, MS. Other techniques are well suited for sample imaging, such as x-ray or fluorescence microscopy, and MALDI approaches. Nevertheless, all of these approaches fall short of providing unequivocal proof for the site of biosynthesis of a given compound.
Conceptually, demonstrating the biosynthesis of a natural product in a given cell type can be achieved on chemical, protein, or genetic levels. At the chemical level, demonstration of new compound synthesis in an isolated cell type is compelling and may involve any of a number of analytical chemical approaches, possibly involving isotope-labeled precursors. At the genetic level, DNA or RNA sequences encoding the biosynthetic enzymes can be localized, either in isolated cells or through imaging techniques. The CARD-FISH analysis of the sponge/cyanobacterium complex Dysidea herbacea/Oscillatoria spongeliae provides an example of RNA detection (92); uncovering the gene for patellamide biosynthesis in isolated cells of Prochloron sp. from the tunicate Lissoclinum patella is an example of detection at the DNA level (18, 19). A considerable hurdle in applying most of these gene-based technologies is that the biosynthesis needs to be sufficiently understood such that the genes can be recognized with certainty. The standard methods for demonstrating that a given gene is involved in the biosynthesis of a compound involve either gene knockout experiments or heterologous expression of the biosynthetic pathway. Both of these methods require genetic manipulations that can be extremely challenging in a little studied marine organism. To date, the function of most marine natural product biosynthetic genes have been deduced through bioinformatic analysis.
At the protein level, little has been achieved because there is so little knowledge of biosynthetic proteins in general and even less so from members of complex marine environments. However, knowledge and techniques are emerging that will allow powerful interrogation of the site of biosynthesis using biosynthetic proteins. An increasing number of marine biosynthetic pathways are being cloned and characterized. Complementing this is a dramatic increase in the number and diversity of marine genome and metagenome sequences (93–95). In principle, this genetic sequence information can be used to identify diagnostic peptide fragments from putative biosynthetic enzymes that can be liberated by protease digestion and localized by subsequent MALDI imaging.
Exciting hybrid technologies are becoming available that will increase the rigor and ease of demonstrating the site of biosynthesis in complex assemblages. Cell isolations and genome determinations are now possible on a single cell [e.g., using multiple displacement polymerase chain reaction technology (96)], from which biosynthetic clusters can be deduced and candidate structures generated by using established biosynthetic logic. Structures of sufficient structural or anticipated biological interest can be located in cultured or collected biomass with increasing facility (97). Alternatively, synthetic genes of substantial size (e.g., 50,000 bp) can be produced with optimized codon usage and promoter sequences for effective heterologous expression (98–100). Indeed, the coming years will witness amazing technological advances and allow us to ask and answer long standing, profound, and currently unimagined questions in chemical biology.
Details of the experiments described in this article can be found in SI Methods and Data.
Supplementary Material
Acknowledgments.
We thank P. Crews and K. Tenney (University of California, Santa Cruz) for samples of the sponge Dysidea herbaceae, B. Palenik and B. Brahamsha for use of their microscopes, L. Gerwick for bioinformatics analysis of the curacin A gene cluster, R. Grindberg for analysis of BryB in the bryostatin gene cluster, T. Matainaho and the captains/crews of the Golden Dawn and Teleta Dive Boats for their assistance with cyanobacterial collections, and the governments of Papua New Guinea and Curaçao for permission to make these collections. This work was supported by National Institutes of Health Grants CA52955 and CA100851.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709851105/DC1.
References
- 1.Newman DJ, Cragg GM. J Nat Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 2.Ramaswamy AV, Flatt PM, Edwards DJ, Simmons TL, Han B, Gerwick WH. In: Frontiers in Marine Biotechnology. Proksch P, Müller WEG, editors. Norfolk, England: Horizon Bioscience; 2006. pp. 175–224. [Google Scholar]
- 3.Simmons TL, Andrianasolo E, McPhail K, Flatt P, Gerwick WH. Mol Cancer Ther. 2005;4:333–342. [PubMed] [Google Scholar]
- 4.Piel J. Curr Med Chem. 2006;13:39–50. [PubMed] [Google Scholar]
- 5.Simmons TL, Gerwick WH. In: Oceans and Human Health. Walsh P, Solo-Gabriele H, Fleming LE, Smith SL, Gerwick WH, editors. New York: Elsevier; 2008. in press. [Google Scholar]
- 6.Hildebrand M, Waggoner LE, Liu H, Sharp KH, Ridley CP, Haygood MG. Nat Prod Rep. 2004;21:122–142. doi: 10.1039/b302336m. [DOI] [PubMed] [Google Scholar]
- 7.Pettit GR, Kamano Y, Herald CL, Tuinman AA, Boettner FE, Kizu H, Schmidt JM, Baczynski L, Tomer K, Bontems RJ. J Am Chem Soc. 1987;109:6883–6885. [Google Scholar]
- 8.Pettit GR, Kamano Y, Dufresne C, Cerny RL, Herald CL, Schmidt JM. J Org Chem. 1989;54:6005–6006. [Google Scholar]
- 9.Leusch H, Moore RE, Paul VJ, Mooberry SL, Corbett TH. J Nat Prod. 2001;64:907–910. doi: 10.1021/np010049y. [DOI] [PubMed] [Google Scholar]
- 10.Williamson RT, Chapin EL, Carr AW, Gilbert JR, Graupner PR, Lewer P, McKamey P, Carney JR, Gerwick WH. Org Lett. 2000;2:289–292. doi: 10.1021/ol991239r. [DOI] [PubMed] [Google Scholar]
- 11.Sudek S, Lopanik NB, Waggoner LE, Hildebrand M, Anderson C, Liu H, Patel A, Sherman DH, Haygood MG. J Nat Prod. 2007;70:67–74. doi: 10.1021/np060361d. [DOI] [PubMed] [Google Scholar]
- 12.Paul VJ, Puglisi MP, Ritson-Williams R. Nat Prod Rep. 2006;23:153–180. doi: 10.1039/b404735b. [DOI] [PubMed] [Google Scholar]
- 13.Pawlik JR. Chem Rev. 1993;93:1911–1922. [Google Scholar]
- 14.Faulkner DJ, Ghiselin MT. Mar Ecol Prog Ser. 1983;13:295–301. [Google Scholar]
- 15.Eyberger AL, Dondapati R, Porter JR. J Nat Prod. 2006;69:1121–1124. doi: 10.1021/np060174f. [DOI] [PubMed] [Google Scholar]
- 16.Puri SC, Verma V, Amna T, Qazi GN, Spiteller M. J Nat Prod. 2005;68:1717–1719. doi: 10.1021/np0502802. [DOI] [PubMed] [Google Scholar]
- 17.Salomon CE, Faulkner DJ. J Nat Prod. 2002;65:689–692. doi: 10.1021/np010556f. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Proc Natl Acad Sci USA. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milne BF, Long PF, Starcevic A, Hranueli D, Jaspers M. Org Biomol Chem. 2006;4:631–638. doi: 10.1039/b515938e. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi J, Ishibashi M. Chem Rev. 1993;93:1753–1769. [Google Scholar]
- 21.Friedrich AB, Merkert H, Fendert T, Hacker J. Mar Biol. 1999;134:461–470. [Google Scholar]
- 22.Ridley CP, Bergquist PR, Harper MK, Faulkner DJ, Hooper JN, Haygood MG. Chem Biol. 2005;12:397–406. doi: 10.1016/j.chembiol.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Bewley CA, Holland ND, Faulkner DJ. Experientia. 1996;52:716–722. doi: 10.1007/BF01925581. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt EW, Bewley CA, Faulkner DJ, Haygood MG. J Org Chem. 1998;63:1254–1258. [Google Scholar]
- 25.Andrainansolo EH, Gross H, Goeger D, Musafija-Girt M, McPhail K, Leal RM, Mooberry SL, Gerwick WH. Org Lett. 2005;7:1375–1378. doi: 10.1021/ol050188x. [DOI] [PubMed] [Google Scholar]
- 26.Enticknap JJ, Kelly M, Peraud O, Hill RT. Appl Environ Microbiol. 2006;72:3724–3732. doi: 10.1128/AEM.72.5.3724-3732.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thiel V, Neulinger SC, Staufenberger T, Schmaljohann R, Imhoff JF. FEMS Microbiol Ecol. 2007;59:47–63. doi: 10.1111/j.1574-6941.2006.00217.x. [DOI] [PubMed] [Google Scholar]
- 28.Davidson SK, Allen SW, Lim GE, Anderson CM, Haygood MG. Appl Environ Microbiol. 2001;67:4531–4537. doi: 10.1128/AEM.67.10.4531-4537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coleman AC, Kerr RG. Tetrahedron. 2000;56:9569–9574. [Google Scholar]
- 30.Kerr RG, Brueck T. PCT Int Appl WO. 2007 20077037791. [Google Scholar]
- 31.Adams DG. Microbiol Today. 2001;28:131–133. [Google Scholar]
- 32.Ferris MJ, Hirsch CF. Appl Environ Microbiol. 1991;57:1448–1452. doi: 10.1128/aem.57.5.1448-1452.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerwick WH, Roberts MA, Proteau PJ, Chen J. J Appl Phycol. 1994;6:143–149. [Google Scholar]
- 34.Paerl HW. In: The Biology of Cyanobacteria. Carr NG, Whitton BA, editors. Berkeley: Univ of California Press; 1982. pp. 441–461. [Google Scholar]
- 35.Zehr JP, Mellon M, Braun S, Litaker W, Steppe T, Paerl HW. Appl Environ Microbiol. 1995;61:2527–2532. doi: 10.1128/aem.61.7.2527-2532.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bewley CA, Faulkner DJ. Angew Chem Int Ed. 1998;37:2162–2178. doi: 10.1002/(SICI)1521-3773(19980904)37:16<2162::AID-ANIE2162>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 37.Perez-Matos AE, Rosado W, Govind NS. Antonie van Leeuwenhoek. 2007;92:155–164. doi: 10.1007/s10482-007-9143-9. [DOI] [PubMed] [Google Scholar]
- 38.Kraus MP. Nature. 1966;211:310–311. [Google Scholar]
- 39.Giovannoni SJ, Stingl U. Nature. 2005;437:343–348. doi: 10.1038/nature04158. [DOI] [PubMed] [Google Scholar]
- 40.Tyers M, Mann M. Nature. 2003;422:193–197. doi: 10.1038/nature01510. [DOI] [PubMed] [Google Scholar]
- 41.Marshall AG, Hendrickson CL, Emmett MR, Rodgers RP, Blakney GT, Nilsson CL. Eur J Mass Spectrom. 2007;13:57–59. doi: 10.1255/ejms.846. [DOI] [PubMed] [Google Scholar]
- 42.Liu E, Gerszten RE. Cardiovasc Biomarkers. 2006:575–586. [Google Scholar]
- 43.Coon JJ, Syka JEP, Shabanowitz J, Hunt DF. BioTechniques. 2005;38:519–523. doi: 10.2144/05384TE01. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J. Proteomics Clin Appl. 2007;1:805–819. doi: 10.1002/prca.200700081. [DOI] [PubMed] [Google Scholar]
- 45.Boersema PJ, Divecha N, Heck AJR, Mohammed S. J Proteome Res. 2007;6:937–946. doi: 10.1021/pr060589m. [DOI] [PubMed] [Google Scholar]
- 46.Motoyama A, Venable JD, Ruse CI, Yates JR., III Anal Chem. 2006;78:5109–5118. doi: 10.1021/ac060354u. [DOI] [PubMed] [Google Scholar]
- 47.Nozawa K, Tsuda M, Ishiyama H, Sasaki T, Tsuruo T, Kobayashi J. Bioorg Med Chem. 2006;14:1063–1067. doi: 10.1016/j.bmc.2005.09.033. [DOI] [PubMed] [Google Scholar]
- 48.Kobayashi J, Cheng J, Ohta T, Nakamura H, Nozoe S, Hirata Y, Ohizumi Y, Sasaki T. J Org Chem. 1988;53:6147–6150. [Google Scholar]
- 49.Feng X, Siegel MM. Anal Bioanal Chem. 2007;389:1341–1363. doi: 10.1007/s00216-007-1468-8. [DOI] [PubMed] [Google Scholar]
- 50.Welker M, Brunke M, Preussel K, Lippert I, von Döhren H. Microbiology. 2004;150:1785–1796. doi: 10.1099/mic.0.26947-0. [DOI] [PubMed] [Google Scholar]
- 51.Dieckmann R, Graeber I, Kaesler I, Szewzyk U, von Döhren H. Appl Microbiol Biotechnol. 2005;67:539–548. doi: 10.1007/s00253-004-1812-2. [DOI] [PubMed] [Google Scholar]
- 52.Luesch H, Yoshida WY, Moore RE, Paul VJ, Corbett TH. J Am Chem Soc. 2001;123:5418–5423. doi: 10.1021/ja010453j. [DOI] [PubMed] [Google Scholar]
- 53.Luesch H, Chandra SK, Raya RM, DeJesus PD, Orth AP, Walker JR, Belmonte JCI, Schultz PG. Nat Chem Biol. 2006;2:158–167. doi: 10.1038/nchembio769. [DOI] [PubMed] [Google Scholar]
- 54.Gerwick WH, Proteau PJ, Nagle DG, Hamel E, Blohkin A, Slate DL. J Org Chem. 1994;59:1243–1245. [Google Scholar]
- 55.Verdier-Pinard P, Sitachitta N, Rossi JV, Sackett DL, Gerwick WH, Hamel E. Arch Biochem Biophys. 1999;370:51–58. doi: 10.1006/abbi.1999.1363. [DOI] [PubMed] [Google Scholar]
- 56.Wipf P, Reeves JT, Day BW. Curr Pharm Des. 2004;10:1417–1437. doi: 10.2174/1381612043384853. [DOI] [PubMed] [Google Scholar]
- 57.Yoo H-D, Gerwick WH. J Nat Prod. 1995;58:1961–1965. [Google Scholar]
- 58.Marquez B, Verdier-Pinard P, Hamel E, Gerwick WH. Phytochemistry. 1998;49:2387–2389. doi: 10.1016/s0031-9422(98)00365-3. [DOI] [PubMed] [Google Scholar]
- 59.Vaillancourt FH, Yeh E, Vosburg DA, O'Connor SE, Walsh CT. Nature. 2005;436:1191–1194. doi: 10.1038/nature03797. [DOI] [PubMed] [Google Scholar]
- 60.Chang Z, Sitachitta N, Rossi JV, Roberts MA, Flatt PM, Jia J, Sherman DH, Gerwick WH. J Nat Prod. 2004;67:1356–1367. doi: 10.1021/np0499261. [DOI] [PubMed] [Google Scholar]
- 61.Gu L, Jia J, Liu H, Håkansson K, Gerwick WH, Sherman DH. J Am Chem Soc. 2006;128:9014–9015. doi: 10.1021/ja0626382. [DOI] [PubMed] [Google Scholar]
- 62.Verdier-Pinard P, Lai J-Y, Yoo H-D, Yu J, Marquez B, Nagle DG, Nambu M, White JD, Falck JR, Gerwick WH, et al. Mol Pharmacol. 1998;53:62–76. doi: 10.1124/mol.53.1.62. [DOI] [PubMed] [Google Scholar]
- 63.Ming L-J, Epperson JD. J Inorg Biochem. 2002;91:46–58. doi: 10.1016/s0162-0134(02)00464-6. [DOI] [PubMed] [Google Scholar]
- 64.Keil A, Talaty N, Janfelt C, Noll RJ, Gao L, Ouyang Z, Cooks RG. Anal Chem. 2007;79:7734–7739. doi: 10.1021/ac071114x. [DOI] [PubMed] [Google Scholar]
- 65.Cooks RG, Ouyang Z, Takats Z, Wiseman JM. Science. 2006;311:1566–1570. doi: 10.1126/science.1119426. [DOI] [PubMed] [Google Scholar]
- 66.Shiea J, Huang M-Z, HSu H-J, Lee C-Y, Yuan C-H, Beech I, Sunner J. Rapid Commun Mass Spectrom. 2005;19:3701–3704. doi: 10.1002/rcm.2243. [DOI] [PubMed] [Google Scholar]
- 67.Haefliger OP, Jeckelmann N. Rapid Commun Mass Spectrom. 2007;21:1361–1366. doi: 10.1002/rcm.2969. [DOI] [PubMed] [Google Scholar]
- 68.Sampson JS, Hawkridge AM, Muddiman DC. J Am Soc Mass Spectrom. 2006;17:1712–1716. doi: 10.1016/j.jasms.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 69.Lim H-K, Chen J, Sensenhauser C, Cook K, Subrahmanyam V. Rapid Commun Mass Spectrom. 2007;21:1821–1832. doi: 10.1002/rcm.3024. [DOI] [PubMed] [Google Scholar]
- 70.Hu Q, Noll RJ, Li H, Makarov A, Hardman M, Cooks RG. J Mass Spectrom. 2005;40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- 71.Breitling R, Pitt AR, Barrett MP. Trends Biotechnol. 2006;24:543–548. doi: 10.1016/j.tibtech.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 72.Thompson JE, Barrow KD, Faulkner DJ. Acta Zool. 1983;64:199–210. [Google Scholar]
- 73.Salomon CE, Deerinck T, Ellisman MH, Faulkner DJ. Mar Biol. 2001;139:313–319. [Google Scholar]
- 74.Gillor O, Carmeli S, Rahamim Y, Fishelson Z, Ilan M. Mar Biotechnol. 2000;2:213–223. doi: 10.1007/s101260000026. [DOI] [PubMed] [Google Scholar]
- 75.McDonnell LA, Heeren RMA. Mass Spectrom Rev. 2007;26:606–643. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- 76.Signor L, Varesio E, Staack RF, Starke V, Richter WF, Hopfgartner G. J Mass Spectrom. 2007;42:900–909. doi: 10.1002/jms.1225. [DOI] [PubMed] [Google Scholar]
- 77.Taban IM, Altelaar AFM, van der Burgt YEM, McDonnell LA, Heeren RMA, Fuchser J, Baykut G. J Am Soc Mass Spectrom. 2007;18:145–151. doi: 10.1016/j.jasms.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 78.McLean JA, Ridenour WB, Caprioli RM. J Mass Spectrom. 2007;42:1099–1105. doi: 10.1002/jms.1254. [DOI] [PubMed] [Google Scholar]
- 79.Cornett DS, Reyzer ML, Chaurand P, Caprioli RM. Nat Methods. 2007;4:828–833. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- 80.Hsieh Y, Chen J, Korfmacher WA. J Pharmacol Toxicol Methods. 2007;55:193–200. doi: 10.1016/j.vascn.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 81.Edwards DJ, Marquez BL, Nogle LM, McPhail K, Goeger DE, Roberts MA, Gerwick WH. Chem Biol. 2004;11:817–833. doi: 10.1016/j.chembiol.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 82.Chaurand P, Schriver KE, Caprioli RM. J Mass Spectrom. 2007;42:476–489. doi: 10.1002/jms.1180. [DOI] [PubMed] [Google Scholar]
- 83.Verhaert PD, Conaway MCP, Pekar TM, Miller K. Int J Mass Spectrom. 2007;260:177–184. [Google Scholar]
- 84.Orjala J, Gerwick WH. J Nat Prod. 1996;59:427–430. doi: 10.1021/np960085a. [DOI] [PubMed] [Google Scholar]
- 85.Sitachitta N, Rossi J, Roberts MA, Gerwick WH, Fletcher M, Willis CL. J Am Chem Soc. 1998;120:7131–7132. [Google Scholar]
- 86.Sitachitta N, Marquez BM, Williamson RT, Rossi J, Roberts MA, Gerwick WH, Nguyen V-A. Tetrahedron. 2000;56:9103–9113. [Google Scholar]
- 87.Chang Z, Flatt P, Gerwick WH, Nguyen V-A, Willis CL, Sherman DL. Gene. 2002;296:235–247. doi: 10.1016/s0378-1119(02)00860-0. [DOI] [PubMed] [Google Scholar]
- 88.Flatt PM, O'Connell SJ, McPhail KL, Zeller G, Willis CL, Sherman DL, Gerwick WH. J Nat Prod. 2006;69:938–944. doi: 10.1021/np050523q. [DOI] [PubMed] [Google Scholar]
- 89.Galonic DP, Vaillancourt FH, Walsh CT. J Am Chem Soc. 2006;128:3900–3901. doi: 10.1021/ja060151n. [DOI] [PubMed] [Google Scholar]
- 90.Faulkner DJ, Unson MD, Bewley CA. Pure Appl Chem. 1994;66:1983–1990. [Google Scholar]
- 91.Flowers AE, Garson MJ, Webb RI, Dumdei EJ, Charan RD. Cell Tissue Res. 1998;292:597–607. doi: 10.1007/s004410051089. [DOI] [PubMed] [Google Scholar]
- 92.Flatt PM, Gautschi JT, Thacker RW, Musafija-Girt M, Crews P, Gerwick WH. Mar Biol. 2005;147:761–774. [Google Scholar]
- 93.Cheng Q, Xiang L, Izumikawa M, Meluzzi D, Moore BS. Nat Chem Biol. 2007;3:557–558. doi: 10.1038/nchembio.2007.22. [DOI] [PubMed] [Google Scholar]
- 94.Dunlap WC, Battershill CN, Liptrot CH, Cobb RE, Bourne DG, Jaspers M, Long PF, Newman DJ. Methods. 2007;42:358–376. doi: 10.1016/j.ymeth.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 95.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, et al. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lasken RS. Curr Opin Microbiol. 2007;10:1–7. doi: 10.1016/j.mib.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 97.McAlpine JB, Bachmann BO, Piraee M, Tremblay S, Alarco A-M, Zazopoulos E, Farnet CM. J Nat Prod. 2005;68:493–496. doi: 10.1021/np0401664. [DOI] [PubMed] [Google Scholar]
- 98.Menzella HG, Reisinger SJ, Welch M, Kealey JT, Kennedy J, Reid R, Tran CQ, Santi DV. J Ind Microbiol Biotechnol. 2006;33:22–28. doi: 10.1007/s10295-005-0038-3. [DOI] [PubMed] [Google Scholar]
- 99.Kodumal SJ, Patel KG, Reid R, Menzella HG, Welch M, Santi DV. Proc Natl Acad Sci USA. 2004;101:15573–15578. doi: 10.1073/pnas.0406911101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Smith HO, Hutchison CA, III, Pfannkoch C, Venter JC. Proc Natl Acad Sci USA. 2003;100:15440–15445. doi: 10.1073/pnas.2237126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.