Abstract
DNA-methylation changes in human cancer are complex and vary between the different types of cancer. Capturing this epigenetic variability in an atlas of DNA-methylation changes will be beneficial for basic research as well as translational medicine. Hypothesis-free approaches that interrogate methylation patterns genome-wide have already generated promising results. However, these methods are still limited by their quantitative accuracy and the number of CpG sites that can be assessed individually. Here, we use a unique approach to measure quantitative methylation patterns in a set of >400 candidate genes. In this high-resolution study, we employed a cell-line model consisting of 59 cancer cell lines provided by the National Cancer Institute and six healthy control tissues for discovery of methylation differences in cancer-related genes. To assess the effect of cell culturing, we validated the results from colon cancer cell lines by using clinical colon cancer specimens. Our results show that a large proportion of genes (78 of 400 genes) are epigenetically altered in cancer. Although most genes show methylation changes in only one tumor type (35 genes), we also found a set of genes that changed in many different forms of cancer (seven genes). This dataset can easily be expanded to develop a more comprehensive and ultimately complete map of quantitative methylation changes. Our methylation data also provide an ideal starting point for further translational research where the results can be combined with existing large-scale datasets to develop an approach that integrates epigenetic, transcriptional, and mutational findings.
Keywords: colon cancer, DNA methylation, NCI-60, MALDI-TOF
DNA methylation has become an increasingly important field in cancer research. The availability of technologies providing more comprehensive overviews now allows us to acquire data about DNA methylation changes more rapidly. It is evident that the DNA methylation changes in cancer are complex and multifaceted in nature. The two best known changes are perhaps promoter-specific hypermethylation of individual genes with consequent decreases in expression (1) and global hypomethylation of repetitive elements within the genome (2).
We compiled a set of >400 cancer-relevant genes and used these for a high-resolution scan of DNA methylation. The genes were selected to include a majority of cancer consensus genes as described by Futreal et al. (3) and a subset of known imprinted genes (www.geneimprint.com/). All genes were analyzed in 59 cell lines derived from nine different tumor types and control DNA from six normal tissues. The cancer cell lines are compiled by the National Cancer Institute (NCI) as the NCI-60 panel, which has been widely used for in vitro anticancer drug testing. Over the years, the NCI-60 set has become one of the best characterized cell line sets available. These also have been analyzed by using a variety of methods including transcriptional profiling (refs. 4 and 5, and see http://dtp.nci.nih.gov/index.html), spectral karyotyping (6), and proteomic profiling (7). In addition, cytotoxicity profiling has been documented for >100,000 chemical compounds by the NCI's Developmental Therapeutics Program (DTP) (http://dtp.nci.nih.gov/index.html and ref. 8).
Here, we report the results of a large-scale DNA methylation profiling study, which includes the quantitative analysis of >500 genomic target regions representing >400 genes in 59 cell lines with confirmation of a subset of targets in 48 colorectal cancer/normal tissue pairs. The resulting data provide a comprehensive panel of cancer-related DNA methylation changes and can be integrated with previous datasets on mutational, transcriptional, and proteomic profiles to obtain a more comprehensive understanding of neoplastic transformations.
Results
We studied DNA methylation by gene-specific amplification of bisulfite-treated DNA followed by in vitro transcription and matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) analysis (9). The amplification regions were primarily designed to cover CpG islands (CGIs) overlapping with the 5′ UTR of the target genes. When no CGI was annotated by the UCSC genome browser in close proximity, we used the sequence directly surrounding the 5′ UTR for primer design, and if no CpG dinucleotides were found in this region, we used the next upstream flanking CGI in a 10-kb window.
The initial methylation data were filtered to exclude poor quality measurements. Poor quality was defined as amplicons with data available for <75% of all samples. These regions were excluded from further analysis. The filtered dataset contained 531 amplification regions, representing 430 genes. For excluded amplicons, PCR was identified as the leading cause of reaction failure.
All autosomal chromosomes and the X chromosome are represented in the current gene set. The median amplicon length was 413 bp (range = 171–683 bp) and the median CpG content per amplicon was 33 CpG/amplicon (range = 6–81 CpG per amplicon). For each sample, a total of 11,723 CpG sites were analyzed. The analytical method used herein uses a biochemistry that does not always allow quantitative read out of methylation values for every single CpG in an amplification region. Some values represent the methylation state of a short stretch of subsequent CpG sites, which we refer to as CpG units. In this study, the 11,723 CpG sites were represented by 7,216 CpG units. To reduce the complexity of the dataset, we build amplicon-specific mean methylation values for each sample, which were used for the later analysis. Sequence-specific details are given in supporting information (SI) Table 2. We analyzed DNA methylation in the NCI-60 panel composed of 59 cell lines and used six commercially available DNAs from adult tissues to represent “healthy” control samples.
Stability.
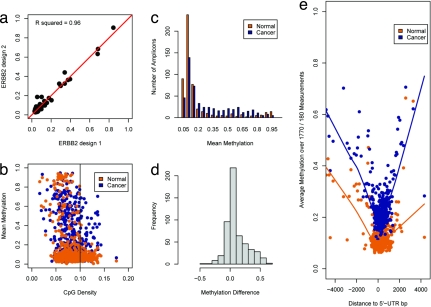
All bisulfite-based methylation analysis methods suffer from a considerable amount of measurement variability introduced by the chemical treatment of genomic DNA. To assess the degree of this inherent variability, we previously dissected the method into its four components and measured the variability for each step in the process (10). The results demonstrated that the greatest source of process-dependent variability is the bisulfite conversion reaction (SD = 10–15%). To determine whether the previous results are applicable to the model system used in this study, we performed duplicate measurements of four control DNAs in 96 amplicons and observed sufficient data stability (R2 = 0.98, SI Fig. 5). We also were interested in evaluating the effect of primer design on the quantitative measurements. We designed two different but overlapping amplification regions for the ERBB2 gene. The quantitative values from both reactions were almost identical and showed a high correlation (R2 = 0.96; see also Fig. 1a).
Fig. 1.
Descriptive analysis of methylation data for normal and tumor cell-line samples. (a) A scatterplot depicts the results from a replicate analysis of ERBB2 by using two different primer designs. The quantitative measurements are highly concordant. (b) Relationship between CpG density and mean methylation levels in cancer and normal samples. CpG density of amplicons was calculated as the fraction of CpG nucleotides within the total amplicon sequence. The mean methylation value for each amplicon was generated by using all individual CpG sites' methylation values. Amplicons with >10% CpG content are likely to have lower methylation values in normal tissues. In cancer cell lines, DNA methylation is observed more frequently in these amplicons. (c) Amplicons were binned based on their average methylation values. Each bin contained amplicons within a 5% range of methylation values. Bins from 15% to 85% average methylation contain more amplicons in the set of cancer cell lines. (d) Histogram of methylation differences. For each amplicon, we calculated the difference in mean methylation between the group of normal samples and the group of tumor cell lines. Positive values translate to hypermethylation in cancer cell lines, whereas negative values indicate that the mean methylation was higher in the group of normal samples. The distribution of methylation differences is skewed toward hypermethylation. (e) DNA methylation in relation to the closest 5′ UTR. The distance from the 5′ UTR was calculated for every individual CpG site. Each data point contains 1,770 individual methylation values for the cancer cell lines and 180 values for the normal samples. It is necessary to adjust the number of data points in each group because of the difference in sample numbers in the two sets. A window of 1 kbp around the 5′ UTR shows low methylation values in the normal and the cancer cell-line samples. Methylation values in the cancer cell-line samples are generally elevated.
Methylation Data Distribution for Cell Lines.
The analyzed target regions were primarily designed within annotated CGIs, which are frequently unmethylated in normal tissue (2). In this part of the analysis, the methylation values, which were measured for each sample individually, were averaged for the group of normal samples and the group of cancer cell-line samples. Our data are in agreement with previous findings for normal tissue samples and confirm that CGIs with a CpG density >10% are generally unmethylated (<15% methylation) (11) (Fig. 1b). In the normal tissue samples, 76% of all amplicons showed mean methylation values <15% methylation; only 5% of amplicons showed methylation levels >85% methylation, and the remaining 19% of amplicons showed methylation levels between 15% and 85%. In the group of cancer cell lines, we observed a shift toward medium (between 20% and 80%) but not high (>80%) methylation levels. Here, only 49% of all amplicons show methylation levels <15% methylation, 2% show methylation levels >85%, but 49% showed methylation between 15% and 85%. Differences in the distribution of CGI methylation between normal samples and cancer cell lines were further assessed by grouping the analyzed genomic regions into 10 bins based on methylation value deciles. In cancer samples, all bins containing genes with methylation values between 20% and 80% showed a 2- to 4-fold increase in the number of amplicons allocated to this bin. The groups <20% and >80% contained fewer amplicons in cancer samples compared with normal samples (Fig. 1c). This increased heterogeneity of methylation values in cancer cell lines might reflect the differences introduced by tumor-specific methylation. We then built the pair-wise differences for each amplicon between normal and cancer samples and found that hypermethylation of promoter CGIs in the cancer samples is the most frequently observed event (Fig. 1d).
Recent studies have shown a decrease of epigenetic marking in a 1-kb window around the transcription start site. In active Drosophila melanogaster promoters, histone occupancy is decreased, and in normal human tissue samples, DNA methylation is reduced within this core region (11, 12). To further investigate this relationship, we mapped the distance from the 5′ UTR for each measured CpG in the dataset (>700,000 data points). CpG methylation in normal samples showed the expected core window of unmethylated CpG sites within 1 kb around the 5′ UTR (Fig. 1e). In cancer cell lines, methylation averages are generally elevated, but the same symmetrical methylation decrease is observed. Thus, these results confirm previous findings and expand their applicability to cancer cell lines (see also SI Text and SI Fig. 6).
Methylation-Based Cell-Line Clustering.
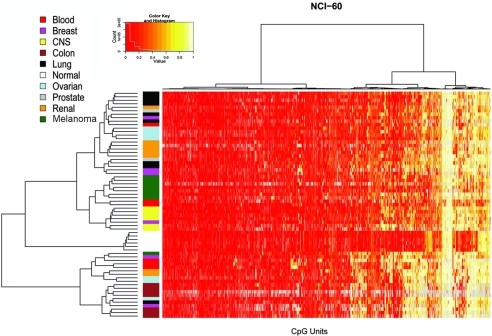
To examine relationships among cell lines and CpG sites, we performed an unsupervised two-dimensional hierarchical clustering analysis, which provides an unbiased view on these relationships (Fig. 2).
Fig. 2.
Two-way hierarchical cluster analysis of 59 tumor cell-line samples and 6 samples from normal tissues (rows) and DNA-methylation of CpG Units in 531 promoter regions (columns). DNA-methylation values are depicted in this false-color image on a continuous scale from red (nonmethylated) to yellow (100% methylated). Poor quality data are in gray. Samples are color-coded according to their cell-line tissue origin (legend, upper left) to simplify identification of potential sample clusters. Strong sample-cluster formation is observed for the group of normal samples, the group of colon cancer samples (brown), melanoma samples (green), and CNS tumors (yellow). Less-dominant clustering is observed in lung cancers (black), renal carcinoma (orange), and ovarian cancer (blue). The cell-line samples derived from breast cancer (pink), leukemia (red), and prostate cancer (gray) do not form obvious clusters. The normal samples are characterized by consistent low methylation levels. The cancer cell-line samples show more-variable methylation patterns. Sample annotations for the final branches of the tree are shown in SI Fig. 10.
The most notable feature of the resulting sample clusters is a separation into different terminal branches of the tree of all normal tissue samples from all cancer cell-line samples. The normal samples are likely to comprise DNA from non-tissue-specific sources, like connective tissue, which might account for some of the difference between the normal samples and the cancer cell-line samples. The differences between the various normal samples are minimal, and, despite their different tissues origins, they cluster tightly together. The cancer cell-line samples are found in two major groups. A group of 19 samples splits as the first branch in the tree. This cluster is characterized by higher methylation levels in a number of genes and includes all seven colon cancer samples, three ovarian and leukemia samples, two breast and kidney cancer samples, one melanoma, and one lung cancer sample. When analyzing the relationship within the cell lines, it becomes apparent that cluster formation is associated with the cancer tissue origin. Strongest cluster formation is observed for colon cancer, CNS, and melanoma. Some group formation is found for non-small-cell lung cancer, renal carcinoma, and ovarian cancer, although more samples are spread across the dendrogram. Both acute lymphoblastic leukemia (ALL) samples and one sample derived from large-cell immunoblastic lymphoma cluster together. Methylation patterns of breast cancer samples do not show strong similarities to each other. These results are comparable to findings from early gene-expression studies or clustering based on copy-number variations (8, 13). It was reported that gene-expression patterns of one breast cancer cell line (MDA-MB-435) showed more similarities with melanoma samples than breast cancer samples (14, 15). Our methylation patterns do not show this behavior. MDA-MB-435 is located outside of any noticeable group formation. Recently, the use of SNP arrays suggested that two cell-line pairs are derived from a single individual each (the glioblastoma cell lines SNB-19 and U251 are derived from one and the NCI/ADR-RES line and the ovarian line OVCAR-8 are derived another individual) (13). Our results support those findings. Both cell-line pairs have remarkably similar methylation patterns and are located next to each other in end-terminal branches of the dendrogram. Also, in concordance with expression data from another study, our findings for SN12C (renal CA), PC3 (prostate CA), and DU-145 (prostate CA) did not show tissue-specific patterns (5).
Confirmation of Cell-Line Results in Colon Cancer Samples.
It remains unclear whether the observed methylation differences are a consequence of the manipulation of cell lines during in vitro growth or whether they represent cancer-specific characteristics. Accordingly, our model system bears the risk of overinterpreting the detected methylation differences. To explore the validity of our findings, we chose the colon cancer cell-line models for confirmation in clinical samples. A set of 50 genes was selected that showed significant differential methylation (ΔM >20%, P < 0.001, two-sided t test) in the colon cancer cell lines. To assess the specificity of our finding, we also selected 14 genes that did not show any cell line methylation differences. We investigated the methylation status of these genes in 48 matched sample pairs of colon cancer tissue and adjacent normal colon tissue. The majority of patients were male [male (M) = 30, female (F) = 18]; the median age at diagnosis was 65 years (range 46–83). Fourteen patients had experienced local or distant cancer recurrence, and all stages (I–IV) were evenly represented. The analysis of methylation differences between the normal and cancer tissue samples confirmed the previous cell-line findings for the majority of genes. In the set of differentially methylated genes, we found 42 of 50 (84%) genes to be significantly differentially methylated in the clinical tissue samples. Additionally, all 14 genes that did not show a methylation difference were still not differentially methylated in the clinical samples (SI Table 2).
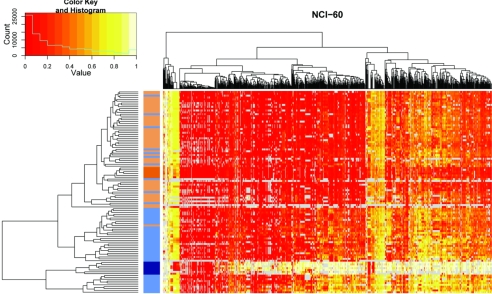
We next used the methylation patterns to characterize relationships among the colon cancer samples and to explore potential associations to their clinical features. None of the clinical features showed a strong correlation to the resulting colon cancer methylation groups (SI Fig. 7a). We explored the degree of similarity between methylation patterns derived from cell-line samples and their tissue counterparts by using hierarchical clustering (Fig. 3). As expected, the normal tissues grouped with the normal colon tissue samples, and the colon cancer cell lines grouped with the colon cancer tissue samples. However, the segregation of normal and colon cancer tissue samples was not perfect. A subset (n = 10) of colon cancer samples is found in the group of normal tissue samples (Fig. 3).
Fig. 3.
Two-way hierarchical cluster analysis of 48 colon cancer samples, 48 adjacent normal colon tissue samples, 7 colon cancer cell lines, and 6 normal DNA samples (rows) and DNA-methylation of CpG Units in 64 promoter regions (columns). DNA-methylation values are depicted in this false-color image on a continuous scale from red (nonmethylated) to yellow (100% methylated). Poor quality data are annotated in gray. Samples are color-coded according to their origin (dark blue, colon cancer cell line; dark orange, normal control DNA; light blue, colon cancer tissue; light orange, normal colon tissue) to simplify identification of potential sample clusters. The hierarchical cluster algorithm separates colon cancer samples from normal tissue samples. Although they are clustered tightly together in a terminal branch of the dendrogram, all colon cancer cell lines are found among colon cancer tissue samples. The same applies to normal tissue samples within the group of normal colon tissue samples. Some (n = 10) colon cancer tissues are located in the group of normal tissue samples. Note that methylation differences for colon cancer cell lines tend to be higher than for colon cancer tissues.
Although the colon cancer samples cluster together with the colon cancer cell-line samples, there are obvious differences in their methylation patterns. The cell-line samples tend to show extreme methylation values. The majority of CpG sites are either not methylated or they are fully methylated. The colon cancer tissue samples show much more heterogeneity and a much broader spectrum of methylation values. In general, methylation differences between normal and cancer were larger in the cell-line models compared with the clinical tissue samples. We find that although we can confirm methylation differences, which have been identified between normal samples and colon cancer cell lines, the magnitude of the effect is much smaller in clinical tissue specimens. This effect might be caused by heterogeneity of the clinical samples, which contained other cell types, like connective tissue or inflammatory cells.
Finally, we compared our findings to results from a recent methylation study of colon cancer tumors that analyzed DNA methylation with a different technology (16). A total of 38 genes were shared by both datasets. The results of both studies are in good agreement (92% concordance). Nine genes were found to be hypermethylated in colon cancer in both datasets, 26 genes showed no colon cancer-specific methylation in both datasets. Two genes were identified as hypermethylated only by the previous study, and one gene was found to be hypermethylated only in our study.
Differentially Methylated Genes.
Tissue-specific DNA methylation has been observed in normal tissues (17), and several cancer-specific methylation markers have been described. The specificity of such cancer markers to a single cancer type remains unclear. Several markers have been found to be differentially methylated in multiple cancer types. These markers might be more universally involved in the progression of cancer. Here, we attempted to identify groups of genes that are differentially methylated between each type of cancer cell line and normal tissues. We then examined the individual groups and determined which genes overlap in multiple cancer types and which are found in specific tumor types only.
Because several genetic loci were tested in many separate runs (one for each cell line), the results will contain false positives that arise from multiple testing in high-dimensional datasets. Although this does not completely erase the issue, we included a minimum difference of 20% as an additional selection criterion to filter out false positives. We classified a gene to be differentially methylated when the difference in methylation values between the normal samples and the subset of cancer cell lines was >20% and the P value for a two-sided t test was <0.001.
The results for the group of leukemia samples and the group of prostate cancer cell lines should be viewed with some caution, because the group of leukemia samples represent a biologically heterogeneous group. Prostate cancer is the smallest subgroup containing only two cell-line samples, which, in addition, do not show prostate cancer-specific gene-expression signatures (5). Hence, their results are less likely to be representative.
A total of 71 genes were statistically significantly hypermethylated in at least one tumor type. A large fraction of these genes (n = 30, 42%) were found only in one tumor type, and nearly 10% were found in more than five tumor types (TSPYL, PAX8, LEP, PHOX2B, and TMPRSS2 were found in five tumor types; MYOD1 was found in six tumor types; PAX5 was found in eight tumor types).
Seven genes were hypomethylated (TCL1A, SLC22A2, TRPM5, IGF2, PEG3 were found in one tumor type; 2 KCNQ1, DLK1 were found in two tumor types). As suggested from our previous analysis depicted in SI Fig. 8, CNS neoplasms (n = 4) and melanomas (n = 3) had the highest number of hypomethylated genes. Interestingly almost all of the hypomethylated genes are known to be imprinted, which might point to a loss of imprinting in these cases (SI Table 3).
PRC2 Target Identification for Colon Cancer and All Others.
A retrospective analysis of DNA methylation by Widschwendter et al. provided evidence that genes targeted by the Polycomb repressive complex 2 are silenced in human colon cancer (18).
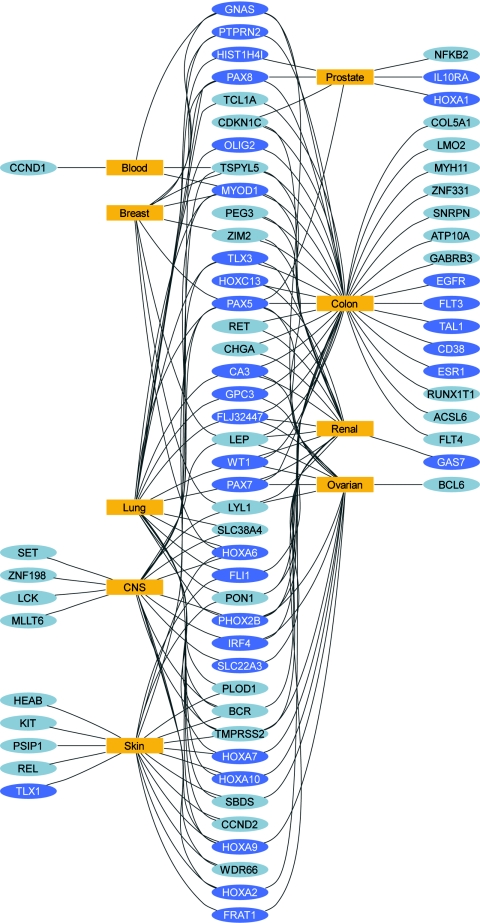
We were able to retrieve information about PRC2 binding sites for 440 amplicons, including 79 amplicons with more than one PRC2 -binding site. We calculated the fraction of amplicons that contain one or more PRC2 binding sites for both: The set of genes that did not show significant methylation differences and the set of genes that did show significant methylation differences in cancer cell lines versus normal tissue. Our findings show a significant (P < 0.001, Fisher's exact test) enrichment for PRC2 targets in the set of significantly hypermethylated genes in six of the nine tumor types. In the group of tumor cell lines with sufficient numbers of samples (excluding leukemia and prostate), we find that only the melanoma-specific gene set is not enriched for PRC2 targets. All other tumor types are 2- to 6-fold enriched for PRC2 targets (Table 1). A graphical representation of gene–tumor associations reveals that highly connected genes also tend to be PRC2 targets (Fig. 4 and SI Fig. 9).
Table 1.
Fraction of PRC2 target and test for statistical significance
| Tissue | Fraction PRC2 target in nonsignificant genes | Fraction PRC2 target in significant genes | P value, Fisher's exact test |
|---|---|---|---|
| Blood | 0.15 | 0.33 | 0.237 |
| Breast | 0.12 | 0.67 | 0.000 |
| CNS | 0.12 | 0.52 | 0.000 |
| Colon | 0.12 | 0.56 | 0.000 |
| Lung | 0.13 | 0.73 | 0.000 |
| Ovarian | 0.15 | 0.70 | 0.000 |
| Prostate | 0.18 | 0.71 | 0.003 |
| Renal | 0.12 | 0.50 | 0.000 |
| Skin | 0.17 | 0.44 | 0.003 |
| Normal | 0.19 | NA | NA |
NA, not available.
Fig. 4.
This network diagram illustrates the relationship between significantly differentially methylated genes and the cell-line tumor types. Genes are shown as colored ellipses (yellow, PRC2 targets; gray, no PRC2 binding site) and cell-line types are shown as blue rectangles. A connection is shown between a cell-line tumor type and a gene when a statistically significant methylation difference was identified between the tumor type and the normal samples. Genes located on the outside are connected to a single tumor type. Genes located between tumor types are connected to at least two different tumor types. Most highly connected genes tend to be PRC2 targets, whereas genes connected to a single tumor type are less likely to be a target of PRC2. Fifty-eight percent of all genes connected to more than one tumor type are PCR2 targets, whereas only 30% of genes connected to a single tumor type are PRC2 targets.
Discussion
Our dataset provides a comprehensive assessment of the methylation status for a group of cancer-related genes in the NCI 60 cell-line set. Although it does not provide a genome-wide representation it will be helpful in understanding the interaction among epigenetic, transcriptional, and mutational status. Multiple datasets are available for the NCI 60 cell lines, including microarray expression, spectral karyotyping, SNP array, and comparative genome hybridization data (refs. 5, 6, 13, 19; also see http://dtp.nci.nih.gov/index.html). We are currently exploring the integration of information obtained from those datasets. Preliminary results indicate that copy-number variants might be associated with methylation levels in the Wilm's tumor gene (WT1). We have previously shown that combining microarray expression data with DNA methylation data remains challenging (20). However, integrating these two datasets might become one of the most interesting opportunities resulting from this analysis.
We are aware that the use of normal tissue DNA samples for comparison to cancer cell-line samples represents a great limitation of our study. It remains unclear what fraction of observed methylation changes has to be attributed to cell-line transformation in vitro. In cultured embryonic stem cells, epigenetic instability has been reported (21), but high-resolution scans have not been performed in cancer cell lines. It has been shown that expression profiles of glioblastoma cell lines (U251 and U87) can diverge remarkably when cultured in vitro versus growth in vivo as s.c. or intracerebral xenografts (22). To assess the impact of these limitations and to evaluate the practical applicability of our findings, we chose to verify the results by using the colon cancer model. Using 96 clinical samples from 48 patients, we were able to confirm ≈85% of the differentially methylated genes detected in the cell-line model. Eight genes no longer showed a significant methylation difference, which might be attributed to the fact that the observed methylation differences between colon cancer tissue samples and normal colon cancer samples were generally smaller than those observed in cell lines. To exclude overinterpretation of the results based on nonspecific promoter hypermethylation, we also analyzed a set of genes that did not show methylation differences in the cell-line model and one gene that showed hypomethylation. We confirmed indifferent methylation in 12 genes with low methylation levels (<20%) and in two genes with higher methylation levels (RUNX3 = 40% and TRPM5 = 80%). KCNQ1, a gene known to be imprinted, was hypomethylated in colon cancer cell lines and colon cancer tissues.
We further cross-compared our results with findings from another comprehensive methylation study performed on colon cancer tissue samples (16). The results from both studies are highly concordant. The few observed disparities might simply be caused by using different interrogation sites. Unfortunately, we were unable to investigate this issue, because the publication did not reveal the exact genomic location of the analyzed amplicons.
One of the most interesting results of this study was that we were able to integrate our results into a recently developed biological model of the polycomb repressive complex 2. Most functional studies have so far been performed on stem cells and association studies mainly focused on colon cancer samples or colon cancer cell lines (18, 23). Our study provides quantitative methylation data for at least seven different tumor types (excluding leukemias and prostate cancer). For every tumor type, a different but not exclusive set of genes was identified that showed higher methylation levels compared with normal samples. Interestingly, six of these seven sets of hypermethylated genes were enriched for PRC2 target genes. These findings suggest that PRC2 target methylation is a common event in cancer and is not limited to individual tumor types.
Our study also confirms the feasibility of the current method for large-scale quantitative high-resolution mapping of DNA methylation. This approach provides an ideal method for large-scale validation of genome-wide methylation studies or to expand the current dataset by analyzing additional regions.
We hope that the availability of this large-scale methylation dataset will initiate interdisciplinary research that integrates multiple available datasets. This information might help to design and execute experiments that will elucidate some of questions that remained unsolved.
Methods
Cells Lines and Tissue Samples.
The NCI-60 panel of 59 cell lines was provided by NCI Division of Cancer Treatment and Diagnosis Tumor Repository (NCI-DCTD). For individual cancer types, normal cell lines or tissue-derived genomic DNA was commercially purchased. Tissue samples from brain (catalogue nos. D1234035 and D123062), breast (D1234086), colon (D1234090), kidney (D1234142), leukocyte (D1234148), and lung (D1234152) were obtained from BioChain.
Clinical Colon Cancer Tissue.
Tissue colorectal cancer and normal colon tissue samples were obtained from the Royal Melbourne Hospital (RMH) Tissue Bank as part of the Ludwig Colon Cancer Initiative biomarker project. Samples were obtained with informed consent under the approved protocol from patients having a resection for histologically confirmed colorectal cancer, and the normal matched tissue was obtained from the same resection specimen at a site adjacent to the tumor.
Tissue samples were snap-frozen in liquid nitrogen within 30 min of collection and stored in a −80°C freezer. Matched tissue sample pairs were cut into 2-mm (see www.geneimprint.com/) cube sections weighing ≈10–15 mg. After manual dissection, DNA was extracted from the tissue sections by using a Qiagen DNeasy blood and tissue kit. Briefly, samples were first lysed by using a Proteinase K digestion for 3 h at 56°C followed by selective binding of DNA to a membrane; final steps involving a spin-column procedure allowed for the washing and subsequent elution of DNA with precipitated DNA resuspended in a buffer solution. DNA was quantified by using a biophotometer, with A260/A280 ratio in the range of 1.7- 2.0. DNA samples were normalized to a concentration of 50 μg/ml.
Clinical data regarding patients and histopathological data for tumor samples were derived from the Australian Comprehensive Clinical Outcomes and Research Database (ACCORD) of Bio21 Molecular Medicine Informatics Model (Bio21:MMIM) (http://mmim.ssg.org.au). This is a unique resource of datasets, physically located at various organizations that are able to be integrated, searched, and queried seamlessly via a federated data integrator. All patients' consent to their data being captured (data are deidentified where appropriate) and all data collection and linkage are approved by the relevant human research ethics committees.
DNA Methylation Analysis.
Bisulfite treatment.
Genomic DNA sodium bisulfite conversion was performed by using EZ-96 DNA methylation kit (Zymo Research). The manufacture's protocol was followed by using 1 μg of genomic DNA and the alternative conversion protocol (a two-temperature DNA denaturation).
Methylation analysis.
Sequenom's MassARRAY platform was used to perform quantitative methylation analysis. This system utilizes MALDI-TOF mass spectrometry in combination with RNA base specific cleavage (MassCLEAVE). A detectable pattern is then analyzed for methylation status. PCR primers were designed by using Methprimer (www.urogene.org/methprimer/). When it was feasible, amplicons were designed to cover CGIs in the same region as the 5′ UTR. For each reverse primer, an additional T7 promoter tag for in vivo transcription was added, as well as a 10-mer tag on the forward primer to adjust for melting-temperature differences. The MassCLEAVE biochemistry was performed as previously described (for details also see SI Text) (9). Mass spectra were acquired by using a MassARRAY Compact MALDI-TOF (Sequenom) and spectra's methylation ratios were generated by the Epityper software v1.0 (Sequenom).
Statistical Methods.
All statistical analyses were performed by using the R statistical environment (www.r-project.org). Distances from gene start sites have been calculated by using the RMySQL package and the SQL database version of the UCSC genome browser (http://genome.ucsc.edu/cgi-bin/hgGateway). Two-dimensional clustering has been performed by using the heatmap.2 function in the gregmisc package. Classical multidimensional scaling has been performed by using the cmdscale function, and visualization was done through the scatterplot3d function in the same-named package. Tests for statistical significance (t test, Wilcox test, and Fisher's exact test) have been used with standard function in R build into the stats package.
For sequence-motif detection, we used a permutation-based method. We randomly sampled n sequences from the pool of all analyzed sequences (n is equal to the number of sequences in the low- or high-methylation group). We then counted how often every possible 6-mer (n = 4,096) is present in the sampled subset. One thousand permutations were performed for each analysis. A sequence motif was identified as being overrepresented if it occurred more often in the analyzed group of sequences than in any of the 1,000 random draws. Graphical representation of the gene-tissue relationships was performed by using the dot algorithm implemented in Graphviz.
Supplementary Material
Footnotes
Conflict of interest statement: M.E., J.T., C.C., and D.v.d.B. are shareholders and full-time employees of Sequenom, Inc.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712251105/DC1.
References
- 1.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 2.Rollins RA, et al. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16:157–163. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Futreal PA, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Staunton JE, et al. Chemosensitivity prediction by transcriptional profiling. Proc Natl Acad Sci USA. 2001;98:10787–10792. doi: 10.1073/pnas.191368598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H, et al. Comparative analysis and integrative classification of NCI60 cell lines and primary tumors using gene expression profiling data. BMC Genomics. 2006;7:166. doi: 10.1186/1471-2164-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roschke AV, et al. Karyotypic complexity of the NCI-60 drug-screening panel. Cancer Res. 2003;63:8634–8647. [PubMed] [Google Scholar]
- 7.Nishizuka S, et al. Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc Natl Acad Sci USA. 2003;100:14229–14234. doi: 10.1073/pnas.2331323100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scherf U, et al. A gene expression database for the molecular pharmacology of cancer. Nat Genet. 2000;24:236–244. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- 9.Ehrich M, et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci USA. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehrich M, Zoll S, Sur S, van den Boom D. A new method for accurate assessment of DNA quality after bisulfite treatment. Nucleic Acids Res. 2007;35:e29. doi: 10.1093/nar/gkl1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckhardt F, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 13.Garraway LA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 14.Ross DT, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 15.Ellison G, et al. Further evidence to support the melanocytic origin of MDA-MB-435. Mol Pathol. 2002;55:294–299. doi: 10.1136/mp.55.5.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weisenberger DJ, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 17.Shiota K. DNA methylation profiles of CpG islands for cellular differentiation and development in mammals. Cytogenet Genome Res. 2004;105:325–334. doi: 10.1159/000078205. [DOI] [PubMed] [Google Scholar]
- 18.Widschwendter M, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 19.Bussey KJ, et al. Integrating data on DNA copy number with gene expression levels and drug sensitivities in the NCI-60 cell line panel. Mol Cancer Ther. 2006;5:853–867. doi: 10.1158/1535-7163.MCT-05-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ehrich M, et al. Cytosine methylation profiles as a molecular marker in non-small cell lung cancer. Cancer Res. 2006;66:10911–10918. doi: 10.1158/0008-5472.CAN-06-0400. [DOI] [PubMed] [Google Scholar]
- 21.Humpherys D, et al. Abnormal gene expression in cloned mice derived from embryonic stem cell and cumulus cell nuclei. Proc Natl Acad Sci USA. 2002;99:12889–12894. doi: 10.1073/pnas.192433399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Camphausen K, et al. Influence of in vivo growth on human glioma cell line gene expression: convergent profiles under orthotopic conditions. Proc Natl Acad Sci USA. 2005;102:8287–8292. doi: 10.1073/pnas.0502887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlesinger Y, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.