Abstract
The formation of clathrin-coated endocytic vesicles is driven by a complex and highly dynamic molecular machinery. In this issue, Idrissi et al. (Idrissi, F.-Z., H. Grötsch, I.M. Fernández-Golbano, C. Presciatto-Baschong, H. Riezman, and M.-I. Geli. 2008. J. Cell Biol. 180:1219–1232) reveal some of the secrets of this machinery by analyzing the localizations of nine endocytic proteins during vesicle budding in yeast using quantitative immunoelectron microscopy.
More than 50 different proteins are thought to have roles in the formation of clathrin-coated endocytic vesicles. These proteins assemble together at the plasma membrane, forming the molecular machinery that drives budding of endocytic vesicles. Although clathrin-mediated endocytosis has been studied already for more than four decades, an understanding of the molecular mechanisms of the process is still quite limited. The difficulty of unraveling the molecular mechanisms is not only a result of the large number of involved proteins but is also a result of the dynamic nature of the endocytic machinery. Endocytic proteins are recruited to the site of vesicle formation in a sequential manner, each protein having its specific times of arrival and departure. The composition of the endocytic machinery can change in a matter of seconds. Many of the recent insights into the process of clathrin-mediated endocytosis have come from imaging of fluorescently labeled proteins in living cells using fluorescence microscopy. Light microscopy provides a good temporal resolution of dynamic events, but its spatial resolution is quite limiting when studying endocytic vesicle budding. On the other hand, electron microscopy offers much better spatial resolution but only provides still images.
In this issue, one study (see Idrissi et al. on p. 1219) uses immunoelectron microscopy to study the localizations of nine different proteins at sites of endocytosis in yeast cells. Most of the proteins involved in clathrin-mediated endocytosis in yeast are conserved throughout eukaryotes, including mammals, making yeast a good model system for studying the basic mechanisms of endocytosis. However, only a few studies have addressed the organization of the endocytic machinery at the ultrastructural level in yeast (Mulholland et al., 1994; Young et al., 2004; Rodal et al., 2005).
Idrissi et al. (2008) start by looking at clathrin, Pan1 (Eps15 homologue), and Sla1 (intersectin-like protein), which, when visualized in living cells by fluorescence microscopy, show similar behaviors. These proteins accumulate at the plasma membrane, forming small fluorescent spots that are initially nonmotile but then move ∼200 nm from the surface toward the interior of the cell at a constant speed for ∼10 s, after which the spots are rapidly disassembled (Kaksonen et al., 2005; Newpher et al., 2005). Idrissi et al. (2008) show by immunoelectron microscopy that clathrin, Pan1, and Sla1 each localize to tips of plasma membrane invaginations, which are ∼50 nm in diameter and have variable lengths up to 180 nm (Fig. 1). This confirms the earlier hypothesis that the movement of these proteins seen in living cells corresponds to the invagination of a clathrin-coated pit, not to the movement of an already budded vesicle. Importantly, these observations show that the length of the endocytic invagination can be used as an indicator for its age.
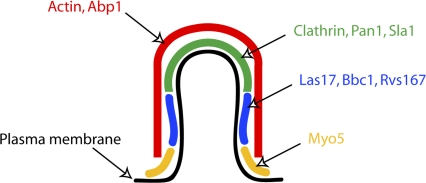
Figure 1.
Schematic model showing the localization of nine proteins on an endocytic invagination. An invagination of intermediate length (∼100 m) is depicted. The coat proteins, including clathrin, coat the tip of the invagination. Rvs167, Las17, and Bbc1 occupy the neck region below the tip. Myo5 concentrates to the base of the invagination. Actin and actin-binding protein Abp1 form a shell covering the whole invagination.
The quantitative immunoelectron microscopy analysis is then applied to six other proteins involved in endocytosis: Rvs167, Las17, Bbc1, Myo5, actin, and Abp1 (Fig. 1). Rvs167 is a homologue of mammalian amphiphysin, a protein involved in pinching vesicles off from the tips of clathrin-coated pits (Takei et al., 1999). The other studied proteins are regulators or components of the actin cytoskeleton, which, in yeast, is essential for endocytosis, specifically for the movement of clathrin and other coat-associated proteins away from the cell surface (i.e., for the membrane invagination; Kubler and Riezman, 1993; Kaksonen et al., 2003). This analysis reveals many exciting details about the dynamic organization of the endocytic machinery. The yeast amphiphysin homologue Rvs167 is shown to localize to the tubular area of the membrane invagination, just below its clathrin-coated tip. Las17 (yeast Wiskott-Aldrich syndrome protein), a strong activator of the actin filament nucleator Arp2/3 (Winter et al., 1999), and Bbc1, an inhibitor of Las17 (Rodal et al., 2003), both localize to the same area as Rvs167. Myo5, a type I myosin, which is both an actin-dependent molecular motor and an activator of the Arp2/3 complex (Sun et al., 2006), localizes mostly to the base of the invagination, where the membrane has a negative curvature. Genetic experiments together with live cell imaging have suggested that Las17 and Myo5 are both needed sequentially for actin-driven invagination of the membrane (Sirotkin et al., 2005; Sun et al., 2006; Galletta et al., 2008). Las17 has a role in initiating the actin polymerization at endocytic sites, whereas Myo5 is needed for the subsequent internalization process. Interestingly, these two major activators of the Arp2/3 complex localize slightly differently: Myo5 closer to the base of the invagination and Las17 in the middle. This suggests that actin polymerization may be spatially restricted to different areas during different stages of endocytosis. Analysis of actin and actin filament–binding protein Abp1 reveals that they are localized throughout the invagination. However, when compared with the other proteins, immunogold labeling for actin and Abp1 is significantly further away from the lipid bilayer, suggesting that the actin cytoskeleton forms an outer shell covering the rest of the endocytic protein machinery.
Using the invagination length as an indicator for the age of the endocytic site, Idrissi et al. (2008) are able to add the time dimension to their data, revealing some interesting temporal dynamics of protein localizations. The temporal order of protein recruitment derived from the electron microscopy data matches observations made using live cell imaging, but the localizations can now be seen at much higher resolution and in relation to the shape of the membrane. Bbc1 and Rvs167 colocalize with Las17, but they appear only on longer, older invaginations. Similarly, Las17 localization precedes Myo5 accumulation, which is consistent with their postulated order of function. The shortest invaginations (<50 nm) show very little labeling for actin. This may mean that the initial membrane bending is independent of actin and could be caused by clathrin or other coat proteins. In older invaginations, actin shows an intriguing distribution. The initial continuous labeling is split into two. Part of the staining localizes to the base of the invagination, and another part localizes to the tip. Similar behavior is also described for Myo5, which initially is concentrated at the base of the invagination but later also appears at the tip. It is not clear whether this staining pattern reflects two separate structures or whether one structure breaks into two. However, this finding shows that the organization of the actin cytoskeleton associated with the endocytic sites may be more complex than previously thought.
One of the key events on the endocytic pathway, vesicle scission, still escapes analysis. Scission and the following disassembly are probably too transient to be caught in fixed cells frequently enough to yield sufficient data for analysis. Other very transient events may also go undetected because they could get smeared as a result of the averaging of data from tens of different invaginations. For these very transient events, live cell imaging is likely to remain the method of choice (Merrifield et al., 2005). However, the superior resolution offered by electron microscopy will clearly continue to provide critical insights. Idrissi et al. (2008) analyzed localizations of nine different proteins. At least 40 yeast proteins involved in endocytic internalization remain to be studied. The rich collections of endocytic mutants will also provide many interesting samples for analysis. What happens to the organization of the endocytic machinery when one of the Arp2/3 activators is mutated? Does the shape or size of the invagination change if one of the coat components is deleted? What would be the effect of inhibiting the motor activity of Myo5? These are just a few examples of exciting questions that can now be addressed.
References
- Galletta, B.J., D.Y. Chuang, and J.A. Cooper. 2008. Distinct roles for Arp2/3 regulators in actin assembly and endocytosis. PLoS Biol. 6:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idrissi, F.-Z., H. Grötsch, I.M. Fernández-Golbano, C. Presciatto-Baschong, H. Riezman, and M.-I. Geli. 2008. Distinct acto/myosin-I structures associate with endocytic profiles at the plasma membrane. J. Cell Biol. 180:1219–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen, M., Y. Sun, and D.G. Drubin. 2003. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 115:475–487. [DOI] [PubMed] [Google Scholar]
- Kaksonen, M., C.P. Toret, and D.G. Drubin. 2005. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 123:305–320. [DOI] [PubMed] [Google Scholar]
- Kubler, E., and H. Riezman. 1993. Actin and fimbrin are required for the internalization step of endocytosis in yeast. EMBO J. 12:2855–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield, C.J., D. Perrais, and D. Zenisek. 2005. Coupling between clathrin-coated-pit invagination, cortactin recruitment, and membrane scission observed in live cells. Cell. 121:593–606. [DOI] [PubMed] [Google Scholar]
- Mulholland, J., D. Preuss, A. Moon, A. Wong, D. Drubin, and D. Botstein. 1994. Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J. Cell Biol. 125:381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newpher, T.M., R.P. Smith, V. Lemmon, and S.K. Lemmon. 2005. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev. Cell. 9:87–98. [DOI] [PubMed] [Google Scholar]
- Rodal, A.A., A.L. Manning, B.L. Goode, and D.G. Drubin. 2003. Negative regulation of yeast WASp by two SH3 domain-containing proteins. Curr. Biol. 13:1000–1008. [DOI] [PubMed] [Google Scholar]
- Rodal, A.A., L. Kozubowski, B.L. Goode, D.G. Drubin, and J.H. Hartwig. 2005. Actin and septin ultrastructures at the budding yeast cell cortex. Mol. Biol. Cell. 16:372–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirotkin, V., C.C. Beltzner, J.B. Marchand, and T.D. Pollard. 2005. Interactions of WASp, myosin-I, and verprolin with Arp2/3 complex during actin patch assembly in fission yeast. J. Cell Biol. 170:637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y., A.C. Martin, and D.G. Drubin. 2006. Endocytic internalization in budding yeast requires coordinated actin nucleation and myosin motor activity. Dev. Cell. 11:33–46. [DOI] [PubMed] [Google Scholar]
- Takei, K., V.I. Slepnev, V. Haucke, and P. De Camilli. 1999. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat. Cell Biol. 1:33–39. [DOI] [PubMed] [Google Scholar]
- Winter, D., T. Lechler, and R. Li. 1999. Activation of the yeast Arp2/3 complex by Bee1p, a WASP-family protein. Curr. Biol. 9:501–504. [DOI] [PubMed] [Google Scholar]
- Young, M.E., J.A. Cooper, and P.C. Bridgman. 2004. Yeast actin patches are networks of branched actin filaments. J. Cell Biol. 166:629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]