Abstract
The S-phase checkpoint activated at replication forks coordinates DNA replication when forks stall because of DNA damage or low deoxyribonucleotide triphosphate pools. We explore the involvement of replication forks in coordinating the S-phase checkpoint using dun1Δ cells that have a defect in the number of stalled forks formed from early origins and are dependent on the DNA damage Chk1p pathway for survival when replication is stalled. We show that providing additional origins activated in early S phase and establishing a paused fork at a replication fork pause site restores S-phase checkpoint signaling to chk1Δ dun1Δ cells and relieves the reliance on the DNA damage checkpoint pathway. Origin licensing and activation are controlled by the cyclin–Cdk complexes. Thus, oncogene-mediated deregulation of cyclins in the early stages of cancer development could contribute to genomic instability through a deficiency in the forks required to establish the S-phase checkpoint.
Introduction
Cell cycle checkpoints coordinate the maintenance of genomic integrity with cell division. In Saccharomyces cerevisiae, the coordination between S phase and chromosome segregation is ensured by the S-phase checkpoint. This signal transduction pathway monitors the presence of replication forks that are stalled by DNA damage lesions, low deoxyribonucleotide triphosphate (dNTP) pools, or mutations in the components of the DNA replication machinery (Nyberg et al., 2002), and its activation is dependent on the protein kinase Mec1p and the downstream kinase Rad53p (Atr and Chk2/Cds1 in humans; Branzei and Foiani, 2005). The S-phase checkpoint effector functions can be subdivided according to their execution point. Intra–S-phase checkpoint effector functions are performed within S phase, whereas the S/M checkpoint restrains mitosis. Rad53p activation in response to replication blocks results in slowing of S-phase progression, delayed activation of late replication origins, stabilization of stalled replication forks, induction of a transcriptional response through the kinase Dun1p, and restraint of mitosis (Allen et al., 1994; Santocanale and Diffley, 1998; Shirahige et al., 1998; Lopes et al., 2001; Alvino et al., 2007).
Stalled or paused replication forks occur at every S phase (Deshpande and Newlon, 1996; Wang et al., 2001) and checkpoint mutant cells have increased gross chromosomal rearrangements (GCRs), suggesting an intrinsic role for the S-phase checkpoint in every cell cycle to resolve these forks (Myung et al., 2001). In addition, Mec1p plays a role in averting mutagenic double-strand breaks by promoting fork progression through slow replicating regions when dNTP pools are reduced (Branzei and Foiani, 2005).
The formation of replication forks, which must be present to activate the S-phase checkpoint (Michael et al., 2000; Tercero et al., 2003), is dependent on origin licensing. This process occurs when prereplication complexes (preRCs) form during the period of low cyclin–Cdk activity at the end of mitosis and in G1. Origin activation is executed by cyclin–Cdk and Cdc7p/Dbf4p kinases and the recruitment of Cdc45p, which initiates DNA unwinding and recruitment of RPA and the DNA polymerase α/primase (Zou and Stillman, 2000), resulting in the formation of two replication forks.
An important factor influencing the number of replication forks present at a given time in S phase is the timing of origin activation. In S. cerevisiae, a subset of the ∼400 sequences defined as autonomously replicating sequences (ARSs) function as replication origins. Origin activation occurs in a continuum with some origins activated early, the bulk activated in mid–S phase, and some activated late (Deshpande and Newlon, 1992; Raghuraman et al., 2001). Therefore, the generation of signal to elicit the S-phase checkpoint is dependent on both preRC formation and origin activation. Mutations or deficiencies in genes encoding preRC components or deregulation of cyclins confer genomic instability or checkpoint defects (Kelly et al., 1993; Saka and Yanagida, 1993;Labib et al., 2000; Shimada et al., 2002; Huang and Koshland, 2003).
The DNA damage checkpoint, which is activated when damage is sensed in late S or G2 phases outside the context of a replication fork, is not required to maintain viability during replication blocks. In this case, some of the signaling components from the S-phase checkpoint, in addition to the kinase Chk1p, inhibit spindle elongation and chromosome segregation in response to DNA damage (Weinert et al., 1994; Gardner et al., 1999; Sanchez et al., 1999; Searle et al., 2004).
In this paper, we report that dun1Δ cells have a deficiency in the number of stalled forks that are formed from early origins. Previously, we showed that replication blocks activate the DNA damage checkpoint in these cells, which are dependent on the checkpoint kinase Chk1p for survival on the ribonucleotide reductase inhibitor hydroxyurea (HU; Schollaert et al., 2004). We show that the lethality of chk1Δ dun1Δ cells on HU is suppressed by the introduction of episomal replication origins that are activated in early S phase. In addition, we demonstrate that the timing of episomal origin activation is critical to their ability to restore checkpoint signaling and viability to chk1Δ dun1Δ cells. Advancing the timing of a late episomal replication origin activation by targeting an acetylase to the origin resulted in suppression of chk1Δ dun1Δ cells to a similar extent as the early episomal origin. Similarly, advancing the timing of late chromosomal replication origin activation globally by RPD3 deletion suppresses the HU sensitivity of chk1Δ dun1Δ cells to a similar extent as the episomal origin. In addition, we find that the forks from the early episomal origins must encounter a replication pause site to generate the S-phase checkpoint signal.
Results
A deficiency of stalled forks in early S phase causes dependence on the DNA damage checkpoint to survive replication blocks
Although chk1Δ mutants exhibit no sensitivity to high HU concentrations (Sanchez et al., 1999) and dun1Δ mutants exhibit sensitivity only to chronic exposure to high levels of HU (Zhou and Elledge, 1993), Chk1p is essential for survival of dun1Δ cells under such conditions. This dependence on Chk1p is because of a defect other than the role of Dun1p in the regulation of dNTP pools. chk1Δ dun1Δ cells have a defect in recovering from replication blocks. In fact, the sensitivity of chk1Δ dun1Δ cells to HU is similar to that of rad53 cells (Schollaert et al., 2004).
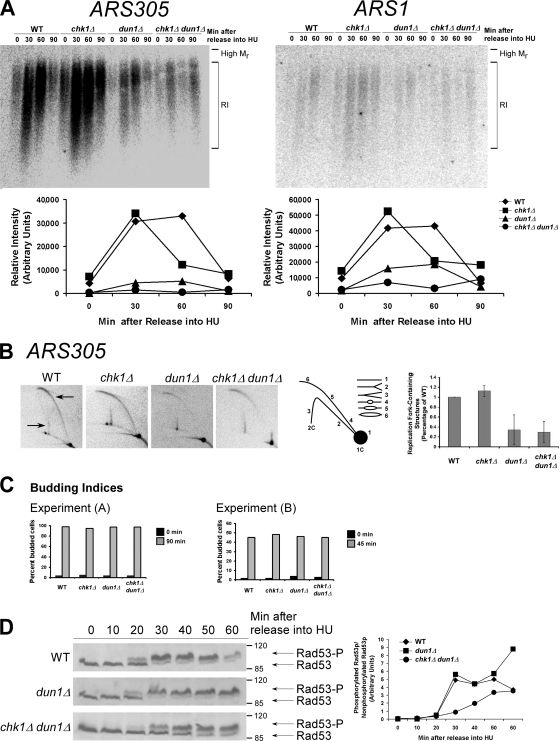
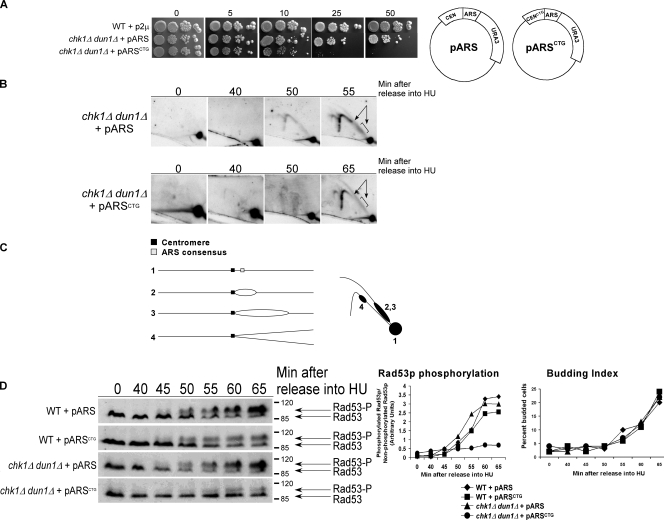
The forks formed from early replication origins, which are not under checkpoint control, establish the S-phase checkpoint signal. Cells deficient in early origin activation fail to activate the S-phase checkpoint and become dependent on the DNA damage checkpoint for survival (Branzei and Foiani, 2005). We hypothesized that the defect in dun1Δ cells that makes CHK1 essential after replication blocks was a deficiency in stalled forks from early replication origins. To determine the relative number of cells within a population that formed stalled forks from early chromosomal replication origins, we examined the accumulation of replication intermediates at ARS305 and ARS1 in cells released from G1 into medium containing HU (Santocanale and Diffley, 1998). Both ARS305 and ARS1 are activated in early S phase (Trep = 12.6 min for ARS305 and 23.3 min for ARS1, where Trep is the time after release from G1 at which half of the cells within the population have replicated that particular sequence; Raghuraman et al., 2001). Fewer dun1Δ and chk1Δ dun1Δ cells showed replication intermediates from both origins compared with wild-type cells (Fig. 1 A). For both ARS305 and ARS1, the relative signal intensity for wild-type cells was approximately seven times greater than that for dun1Δ and chk1Δ dun1Δ cells at the time of maximum intensity for all strains. The budding index indicated that dun1Δ cells entered the cell cycle with the same kinetics as wild-type cells (Fig. 1 C, left). The few dun1Δ and chk1Δ dun1Δ cells that activated the early origins did so with the same timing as wild-type cells.
Figure 1.
The number of stalled chromosomal replication forks early in S phase is reduced in dun1Δ and chk1Δ dun1Δ cells in response to HU and correlates with a defect in S-phase checkpoint activation. (A, top) Cells were released from G1 into medium containing 200 mM HU. Chromosomal DNA was subjected to alkaline gel electrophoresis. RIs were monitored by Southern blot analysis using a probe for the indicated replication origin. (A, bottom) Relative intensity of RI signal normalized to the amount of DNA loaded. Seven independent experiments showed similar trends and one typical experiment is shown for each ARS. (B) Cells were treated as in A. (B, left) Southern blot analysis of chromosomal DNA prepared 45 min after release into HU and subjected to 2D gel electrophoresis was performed using a probe for ARS305. Arrows designate signal that represents replication fork–containing structures. (B, middle) Illustration of restriction fragment structures and the corresponding Southern blot patterns. (B, right) Mean ratio of signal for replication fork–containing structures to linear DNA as a percentage of the wild-type (WT) ratio for three experiments. Error bars represent one standard deviation from the mean. (C) Budding index. 100 cells from A and B were scored for buds. Additional data that support this trend were observed in other replicates (Tables S2 and S3, available at http://www.jcb.org/cgi/content/full/jcb.200706009/DC1). (D, left) Cells were released from G1 into medium containing 200 mM HU. Rad53p phosphorylation was monitored by Western analysis. The Molecular masses (in kilodaltons) are indicated to the right of each panel. (D, right) The ratio of phosphorylated to unphosphorylated Rad53p signal.
To confirm the results obtained from the alkaline gel analysis, 2D gel analysis was performed to monitor replication structures from the early origin ARS305. Wild-type and chk1Δ cells activated ARS305 45 min after release from G1 into HU, as indicated by prominent bubble arc and late Y arc signals (Fig. 1 B, arrows;Santocanale and Diffley, 1998). However, despite similar kinetics of cell cycle entry (Fig. 1 C, right), dun1Δ and chk1Δ dun1Δ cells had reduced signal representing replication fork–containing structures (bubble and late Y arcs) at the same time point in three independent experiments (Fig. 1 B, right). This was not because of premature fork formation in dun1Δ or chk1Δ dun1Δ cells at earlier time points (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200706009/DC1).
Because dun1Δ cells had fewer stalled forks in early S phase, we next examined the timing of S-phase checkpoint activation in these cells by monitoring Rad53p phosphorylation (Sanchez et al., 1996). Rad53p was partially modified by 20 min and fully modified by 30 min in wild-type cells released from G1 into HU. Under these conditions, Rad53p modification in dun1Δ cells had similar kinetics as in wild-type cells, whereas in chk1Δ dun1Δ cells, Rad53p was not modified until 30 min after release, and modification increased by 40 min but never reached full activation, as indicated by the unphosphorylated species present at the later time points (Fig. 1 D). This delay was confirmed by calculating the signal ratio for phosphorylated/unphosphorylated Rad53p (Fig. 1 D, right) and was not caused by differences in cell cycle entry, as confirmed by DNA content and budding index (Fig. S1, B and C). We have shown that Chk1 is required to promote recovery from replication blocks in dun1Δ cells (Schollaert et al., 2004). One possible explanation for our findings that Rad53p activation was normal in dun1Δ cells despite defects in formation of replication intermediates from early origins is that Chk1p could be promoting a process at the replication fork that would keep the few forks in a configuration that would be competent for Rad53p signaling. This process could be physical, involving prevention or promotion of the processing of the structures that arise at stalled forks.
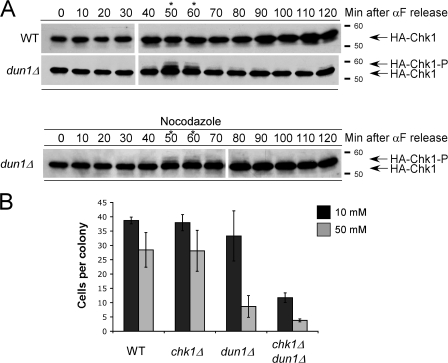
dun1Δ cells have increased numbers of GCRs, which suggests that conditions exist at every S phase that could result in DNA damage checkpoint activation (Myung et al., 2001). We monitored Chk1p modification as an indicator of DNA damage checkpoint activation in wild-type and dun1Δ cells in the absence of exogenous sources of replication stress. Chk1p was activated in dun1Δ cells but not wild-type cells in late S phase/G2 (Fig. 2 A and Fig S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200706009/DC1). Dephosphorylation of Chk1p occurred as dun1Δ cells progressed to mitosis, as indicated by DNA content (Fig. 2 A and Fig. S2 B), and before cells reached a nocodazole-induced preanaphase block (Fig. 2 A, bottom). The phosphorylation of Chk1p in dun1Δ cells did not require Mre11p, which has a role in the checkpoint response to double-strand breaks and replication stress (Fig. S2 A; D'Amours and Jackson, 2001; Grenon et al., 2001; Usui et al., 2001). Phosphorylation of Chk1p in dun1Δ cells required the signal amplifier Rad9p but not Mrc1p (Fig. S2 A; Schollaert et al., 2004), supporting the argument that dun1Δ deficiency leads to the activation of the Rad9p-mediated DNA damage checkpoint.
Figure 2.
Chk1p is activated in dun1Δ cells in late S phase and G2 during an unperturbed cell cycle. (A, top) Cells were released from G1. Western blot analysis was performed to monitor HA-Chk1p protein migration. (A, bottom) dun1Δ cells were treated as in the top panel, except cells were released into medium containing nocodazole. The asterisks emphasize the time points at which HA-Chk1p phosphorylation was observed. The Molecular masses (in kilodaltons) are indicated to the right of each panel. (B) Cells were plated on YPD medium containing either 10 or 50 mM HU and incubated at 30°C for 24 h, and the number of cells per colony was counted for 50 colonies. Bars represent the mean number of cells per colony. Error bars represent one standard deviation from the mean.
Because Chk1p phosphorylation occurred in late S/G2 in dun1Δ cells, we next used a microcolony assay to determine whether the lethality in chk1Δ dun1Δ cells occurred in the same cell cycle or required passage through mitosis. chk1Δ dun1Δ cells formed microcolonies containing, on average, 15 or 4 cells 24 h after being plated on 10 or 50 mM HU, respectively, indicating that the cells undergo at least one division before death (Fig. 2 B). These results are consistent with our finding that chk1Δ dun1Δ cells transiently treated with HU (6 h) and allowed to proceed through mitosis exhibited loss of viability that was suppressed by concomitant treatment with nocodazole to block mitotic progression (Fig. S2 C). This suggests that failure to activate the S-phase checkpoint fully in dun1Δ cells results in lesions that require the DNA damage checkpoint for their resolution before mitosis and that these lesions may not be lethal until chk1Δ dun1Δ cells go through mitosis.
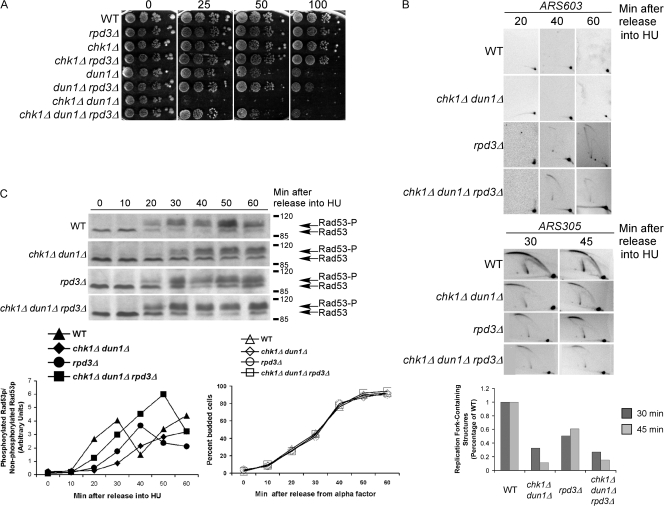
Advancing global timing of origin activation by RPD3 deletion suppressed the HU sensitivity of dun1 Δ and chk1Δ dun1Δ cells
Reaching the threshold of stalled forks in early S phase may depend on the efficiency of formation of replication forks from the limited number of chromosomal origins that are activated at that time (Branzei and Foiani, 2005). We predicted that increasing the number of origins activated early in S phase in dun1Δ cells would restore S-phase checkpoint signaling and relieve the requirement of the DNA damage checkpoint for survival on HU. HU treatment delays late origin activation in wild-type cells and this is dependent on the deacetylase Rpd3p, which controls the timing of activation of late origins (Vogelauer et al., 2002; Aparicio et al., 2004). RPD3 deletion was shown to advance the activation of late chromosomal origins, even in HU, and to restore checkpoint signaling to mec1, rad9, and rad53 cells in response to UV and, to a lesser extent, suppressed the HU-sensitivity of mec1 cells (Scott and Plon, 2003). Deletion of RPD3 suppressed the HU sensitivity of both dun1Δ and chk1Δ dun1Δ cells nearly to the same extent as the early activating episomal origin (Fig. 3 A) presumably by increasing the number of forks in early S phase.
Figure 3.
Deletion of RPD3, which encodes a histone deacetylase, suppresses the HU sensitivity of chk1Δ dun1Δ cells. (A) Cells were spotted onto YPD medium containing the indicated concentration of HU (mM). (B) Analyses of replication fork–containing structures were performed as in Fig. 1 B, using a probe for the late chromosomal origin ARS603 (top) or the early chromosomal origin ARS305 (bottom). (C, top)Rad53p phosphorylation in response to HU treatment is restored to wild-type levels and kinetics in chk1D dun1D cells by additional replication origins that are activated in early S phase. Rad53p phosphorylation was monitored as in Fig. 1 D, except that the cells were released at 18°C. (C, bottom left) Ratio of phosphorylated to nonphosphorylated Rad53p. (C, bottom right) Budding index.
We next confirmed that deletion of RPD3 allowed activation of the late chromosomal origin ARS603 in chk1Δ dun1Δ cells. Like wild-type cells, chk1Δ dun1Δ cells showed no bubble arc signal for the late origin ARS603 up to 60 min after release from G1 into HU, indicating that these cells are competent for the suppression of late replication origins; however, bubble arc structures were apparent in rpd3Δ cells 40 and 60 min after release from G1 (Fig. 3 B; Aparicio et al., 2004). chk1Δ dun1Δ rpd3Δ cells activated ARS603 in HU similar to rpd3Δ cells (Fig. 3 B), confirming that late origins are not suppressed in chk1Δ dun1Δ rpd3Δ cells. Deletion of RPD3 in the chk1Δ dun1Δ background did not advance the timing or increase the efficiency of early chromosomal origin activation (Fig. 3 B).
Altering the activity of the cyclin–CDK complexes that control preRC formation and replication timing causes a checkpoint-mediated delay before anaphase (Lengronne and Schwob, 2002; Gibson et al., 2004). Cells lacking either S-phase cyclin Clb5p or 6p are proficient for early origin activation, whereas cells lacking Clb5p have a defect in late origin activation (Donaldson et al., 1998). Deletion of CLB5 makes cells dependent on Rad53p for viability, even in the absence of replication blocks, and is synthetic sick with a CHK1 deletion (Gibson et al., 2004). We found that deletion of CLB5 was synthetic with deletion of DUN1 for HU sensitivity (Fig. S3 G, available at http://www.jcb.org/cgi/content/full/jcb.200706009/DC1). In addition, overexpression of CLB6, which regulates early origin activation, partially suppressed the HU sensitivity of chk1Δ dun1Δ cells (Fig. S3 H). Our data suggest that the combination of a defect in late origin activation caused by lack of Clb5p exacerbates a defect in the formation or maintenance of replication intermediates from early origins because of lack of Dun1p.
We next examined the kinetics of Rad53p activation in response to HU in chk1Δ dun1Δ cells with increased numbers of forks in early S phase by RPD3 deletion. Rad53p activation in chk1Δ dun1Δ rpd3Δ cells occurred with similar kinetics to wild-type cells (Fig. 3 C and Fig. S3 B). The budding index from these experiments showed that the difference in Rad53p phosphorylation kinetics was not caused by altered timing of cell cycle entry (Fig. 3 C, bottom). These experiments were performed at 18°C to slow down cell cycle progression; however, at a higher temperature, the same trend of Rad53p activation was apparent (Fig. S3 B). These results are consistent with a model in which increasing the number of replication forks in early S phase allows the checkpoint to be established at a relatively early time during S phase.
An episomal early replication origin restores the ability of checkpoint-defective mutants to survive replication blocks
We next examined whether our system could be used to reconstitute S-phase checkpoint signaling from episomal origins of replication that differed in their timing of activation. This would allow us to dissect the contribution of the timing of activation and/or numbers of forks formed in early S phase to checkpoint signaling and survival after replication stress.
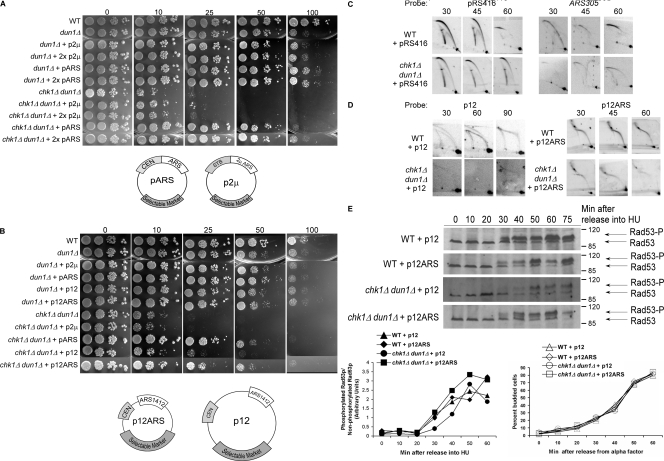
We observed that the HU sensitivity of both dun1Δ and chk1Δ dun1Δ cells was suppressed by the introduction of an episomal shuttle vector that contains the histone H4 replication origin (ARSH4) and a centromeric sequence (CEN6; Fig. 4 A; Sikorski and Hieter, 1989). This type of shuttle vector, pARS (yeast centromeric plasmid), is maintained at a copy number of 1–2 per cell (Hsiao and Carbon, 1981) and is replicated and segregated like other yeast chromosomes. chk1Δ dun1Δ cells grew equally well on high concentrations of HU whether transformed with one or two pARS; however, another episomal vector, p2μ (yeast episomal plasmid), which contains a canonical ARS but lacks a CEN (Christianson et al., 1992), or one lacking both CEN and ARS sequences (yeast integrative plasmid; Sikorski and Hieter, 1989) did not confer suppression (Fig. 4, A and B; and not depicted). These data suggest that the replication origin and/or the centromeric sequences are the likely elements restoring growth on HU. Because cells containing the p2μ had similar phenotype to cells lacking any vector, we have used cells containing the p2μ as negative controls in our studies.
Figure 4.
Suppression of the HU sensitivity of chk1Δ dun1Δ cells by an episomal replication origin activated early in S phase. (A and B) Cells containing the indicated number of copies of pARS (pRS416, pRS415), p2μ (pRS426, pRS425), p12, or p12ARS were spotted onto YPD medium containing the indicated concentration of HU (in millimoles). (C and D) Analyses of replication fork–containing structures were performed as in Fig. 1 B, using probes for the indicated plasmid or ARS305. All cells analyzed in D contained pRS416. (E)Rad53p phosphorylation in response to HU treatment is restored to wild-type levels and kinetics in chk1D dun1D cells by additional replication origins that are activated in early S phase. Rad53p phosphorylation was monitored as in Fig. 3 C. The Molecular masses (in kilodaltons) are indicated to the right of each panel. (E, bottom left) Ratio of phosphorylated to nonphosphorylated Rad53p. (E, bottom right) Budding index.
To test whether the timing of episomal replication fork formation was critical to restoring checkpoint signaling, we used two pARS that contain the same origin sequence, ARS1412, with different timing of activation (Friedman et al., 1996). ARS1412 with chromosomal flanking sequences maintains its late activation time on an episome (p12; Friedman et al., 1996). Deletion of sequence flanking ARS1412 in p12 resulted in p12ARS with the origin now activated at the same time as ARS305, which is one of the earliest activated origins (Reynolds et al., 1989; Friedman et al., 1996; Raghuraman et al., 2001). dun1Δ and chk1Δ dun1Δ cells carrying p12ARS (early) grew on HU-containing medium to the same extent as cells carrying pARS. However, chk1Δ dun1Δ cells carrying p12 (late) did not grow on HU (Fig. 4 B). Our data suggest that the additional signal from stalled episomal forks formed in early S phase compensates for a lower number of chromosomal stalled forks. In addition, chk1Δ dun1Δ rpd3Δ cells containing the episomal origin showed no difference in growth on higher concentrations of HU (Fig. S3 F). These results suggest that the episomal origin and rpd3Δ mutation restore viability to dun1Δ and chk1Δ dun1Δ cells on HU by a similar mechanism.
Similar to chk1Δ dun1Δ cells, dun1Δ cells lacking the signal amplifier Rad9p exhibit sensitivity to chronic HU exposure (Fig. S2 D; Schollaert et al., 2004). The viability of dun1Δ rad9Δ and chk1Δ dun1Δ rad9Δ cells was restored by the episomal replication origin to nearly the same extent as observed for chk1Δ dun1Δ cells, suggesting that Rad9p is not required for growth on HU under these circumstances (Fig. S2 D).
Sensing of UV DNA damage lesions in S phase requires components of the replication machinery (Navas et al., 1996; Wang and Elledge, 1999). chk1Δ dun1Δ cells also exhibit additive sensitivity to UV irradiation (Sanchez et al., 1999) that could be caused by a defect in establishing an S-phase checkpoint signal. The UV sensitivity of dun1Δ and chk1Δ dun1Δ cells was suppressed by the presence of either episome with early activating origins (pARS and p12ARS) but not by the p2μ vector or p12 (Fig. S2 E). These findings suggest that the additional replication forks provided by the episomal early origins contribute to sensing UV-induced DNA damage to activate the S-phase checkpoint. Therefore, the episomal origin of replication relieved the dependence of dun1Δ cells on DNA damage checkpoint proteins for survival of forks stalled by low dNTPs or DNA damage.
We then confirmed the replication timing of these episomes and pARS in checkpoint mutant cells released from G1 into HU. A bubble arc was detected for pARS by 30 min after release, which is consistent with the early activation of ARSH4 (Fig. 4 C). Furthermore, introduction of the episomal forks did not restore the strength of activation to the chromosomal ARS305 in the chk1Δ dun1Δ cells (Fig. 4 C). Maximum replication intermediates signal for p12 was observed 60 min after release in wild-type cells, and a faint signal was present at 60 and 90 min in the chk1Δ dun1Δ cells, which is consistent with late activation timing (Fig. 4 D). In contrast we detected bubble arcs for p12ARS as early as 30 min, which is consistent with early activation timing (Fig. 4 D).
Our data suggested that the activation of the episomal and chromosomal origins were under different control. To test this, the origin ARS305 and minimal flanking sequences were cloned into a vector backbone containing a CEN. Despite the fact that fewer stalled forks form from the chromosomal ARS305 in HU-treated dun1Δ and chk1Δ dun1Δ cells, the HU sensitivity of these cells was suppressed by the episomal ARS305 to a greater extent than pARS (Fig. S1 D), suggesting that the episomal ARS305 was activated with the correct kinetics.
We next examined the kinetics of Rad53p activation in response to HU in chk1Δ dun1Δ cells with increased numbers of forks in early S phase by p12ARS. chk1Δ dun1Δ cells with p12ARS, but not p12, showed wild-type kinetics of Rad53p activation in response to HU (Fig. 4 E and Fig. S3 A). The budding index from these experiments showed that the difference in Rad53p phosphorylation kinetics was not because of altered timing of cell cycle entry (Fig. 4 E, bottom). These experiments were performed at 18°C to slow down cell cycle progression; however, at a higher temperature, the same trend of Rad53p activation was apparent (Fig. S3 A). These results are consistent with a model in which increasing the number of replication forks in early S phase allows the checkpoint to be established at a relatively early time during S phase.
The timing of replication fork formation determines whether the S-phase checkpoint is established
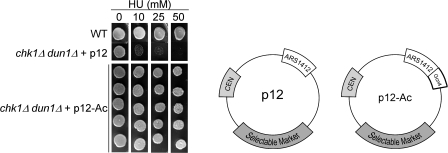
Because the amount of flanking sequence around ARS1412 is significantly different in p12 and p12ARS, we used p12 (late) to examine whether changing the timing of activation of ARS1412 via a different mechanism was sufficient to restore checkpoint signaling in chk1Δ dun1Δ cells. Recruitment of the acetylase Gcn5p by Gcn4p to the chromosomal ARS1412 was shown to advance its activation timing (Vogelauer et al., 2002). Thus, we inserted Gcn4p binding sites (total of 500 bp) adjacent to ARS1412 in p12 resulting in p12-Ac. Because expression of Gcn4p is induced by amino acid starvation, we introduced a constitutive GCN4 expression construct into the same cells to keep the Gcn4 levels constant in all backgrounds. We observed that the episome with the late origin flanked by Gcn4p–Gcn5p binding sites restored viability to the chk1Δ dun1Δ cells 100% of the time (Fig. 5). These findings suggest that the critical characteristics of the episome that restored viability on HU were the timing of replication fork formation.
Figure 5.
The timing of origin activation and of the episomal replication fork pause is critical to restore viability to chk1Δ dun1Δ cells. Targeting the acetylase Gcn5p–Gcn4p complex to the late activating origin ARS1412 restored the S-phase checkpoint in chk1D dun1D cells with constitutive expression of Gcn4p. Cells containing a high copy vector expressing Gcn4p and the indicated episomal origin were grown in SC-ura-leu media and spotted onto YPD medium containing the indicated concentration of HU (in millimoles). (bottom) Five representative chk1Δ dun1Δ transformants containing a high copy vector expressing Gcn4p and p12-Ac show results typical for 43/43 chk1Δ dun1Δ transformants containing the same constructs. p12, episome with late activating origin that failed to suppress chk1Δ dun1Δ; p12-Ac, same episome containing binding sites for the Gcn4p–Gcn5p acetylase complex.
A replication fork pause at the centromere plays a role in restoring checkpoint signaling independent of spindle dynamics
In addition to the case of dNTP depletion by HU, replication forks pause when they encounter nonhistone multiprotein complexes bound to the DNA template. Replication forks pause at centromeres (Greenfeder and Newlon, 1992) as well as at tRNA genes actively transcribed in a direction opposite of fork movement (Deshpande and Newlon, 1996).
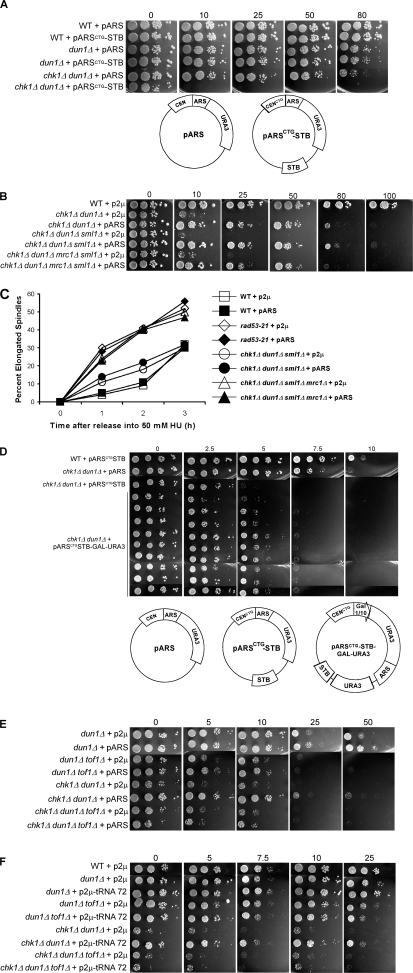
We first investigated whether eliminating a functional centromere on the pARS episome abolished its ability to restore viability to chk1Δ dun1Δ cells by introducing a point mutation into the CDEIII element of the pARS CEN (CCG→CTG) to obtain the construct pARSCTG. The CDEIII element, which is required for proper centromere chromatin structure and chromosome segregation, is rendered nonfunctional by this mutation (McGrew et al., 1986). pARSCTG failed to restore viability to chk1Δ dun1Δ cells on HU (see Fig. 7 A). Addition of the STB sequence from the p2μ episome to provide a segregation mechanism into pARSCTG failed to restore viability to the cells (Fig. 6 A) despite improving the segregation efficiency of the episome (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200706009/DC1).
Figure 7.
The timing of the episomal replication fork pause is critical to restore viability to chk1Δ dun1Δ cells. (A) Cells containing the indicated episome were spotted onto YPD medium containing the indicated concentration of HU (in millimoles). (B) chk1Δ dun1Δ cells containing the indicated episome were treated as in Fig. 1 B, except that they were released at 18°C. Southern blot analysis of chromosomal DNA prepared at the indicated times after release into HU and subjected to 2D gel electrophoresis was performed using a probe for pARS (pRS416). (C) Restriction fragment shapes that correspond to the observed Southern blot pattern. (D, left) In a separate experiment, cells were treated as in Fig. 3 C and processed for Western analysis to monitor Rad53p phosphorylation. The Molecular masses (in kilodaltons) are indicated to the right of each panel. (D, right) The ratio of phosphorylated to unphosphorylated Rad53p signal and budding index.
Figure 6.
Replication fork pause plays a role in suppression of HU sensitivity of chk1Δ dun1Δ cells. (A) Cells containing the indicated episome were spotted onto YPD medium containing the indicated concentration of HU (in millimoles). (B) Suppression of checkpoint mutant HU sensitivity by an episomal replication origin is independent of spindle dynamics regulation.Cells containing either p2μ (pRS426) or pARS (pRS416) were spotted onto YPD medium containing the indicated concentration of HU (in millimoles). (C) Cells were released from G1 into medium containing 50 mM HU, fixed, and stained with DAPI and anti-tubulin to visualize nuclei and spindles. For each strain, 100 cells for each time point were scored as having either short (<3 μm) or elongated (≥3 μm) spindles. Data points indicate the percentage of cells with elongated spindles. (D–F) The episomal replication fork pause is critical to restore viability to chk1Δ dun1Δ cells.Cells containing the indicated episomes were spotted onto YPD medium containing the indicated concentration of HU (in millimoles). (A and D, bottom) Schematics of episomes used in these studies.
Rad53p plays a critical role in the execution of the checkpoints operating during S phase, including blocking premature spindle elongation when replication is slowed down by HU. Bachant, et al. (2005) showed that several copies of pARS restored spindle regulation to rad53-21 cells in HU, likely by establishing tension from the kinetochores formed at the replicated CEN, which is juxtaposed to the early activating origin in pARS and is replicated early. We also found that the CEN was required for suppression in our system; however, our data suggested that restoration of spindle elongation delay was not the sole mechanism responsible for restoring growth on HU. We found that additional copies of the pARS (two to four copies) that had been shown by Bachant, et al. (2005) to prevent premature spindle elongation in rad53-21 cells in a high concentration of HU (200 mM) did not suppress their growth defect on HU (Fig. S3, C and D). This was not surprising, considering the essential role of Rad53p in the S-phase checkpoint response in preventing irreversible fork collapse. We also observed that at a lower concentration of HU (50 mM) that slows down but does not block DNA replication, introduction of four copies of pARS only slightly altered the spindle elongation kinetics of rad53-21 cells (Fig. S3 E). In addition, a single copy of pARS was sufficient to restore checkpoint signaling to chk1Δ dun1Δ cells without altering their spindle elongation kinetics (Fig. 6 C and not depicted).
We next addressed the correlation of spindle elongation with viability of cells. To do this, we examined the spindle elongation kinetics of chk1Δ dun1Δ mrc1Δ cells. mrc1Δ cells elongate their spindles in high concentrations of HU, despite their ability to form colonies when chronically exposed to HU (Alcasabas et al., 2001; Tanaka and Russell, 2001; Osborn and Elledge, 2003). Deletion of MRC1 increased the sensitivity of dun1Δ cells to HU, although not to the extent of a RAD9 deletion (unpublished data). chk1Δ dun1Δ mrc1Δ cells are inviable (unpublished data); however, they are viable when the gene encoding the ribonucleotide reductase inhibitor Sml1p is deleted. sml1Δ deletion had been shown to suppress the lethality of mec1 and rad53 deletions and the lethality of simultaneous loss of Mrc1p and Rad9p (Zhao et al., 1998; Gibson et al., 2004). Episomal early replication origins suppressed the HU sensitivity of chk1Δ dun1Δ cells lacking Mrc1p (Fig. 6 B), yet the pARS did not alter the premature spindle elongation of chk1Δ dun1Δ mrc1Δ sml1Δ cells in HU. chk1Δ dun1Δ mrc1Δ sml1Δ cells containing either p2μ or pARS elongated their spindles with kinetics similar to rad53-21 cells containing either pARS or p2μ, whereas chk1Δ dun1Δ sml1Δ cells containing either vector elongated their spindles with delayed kinetics compared with rad53-21 and chk1Δ dun1Δ mrc1Δ sml1Δ cells (Fig. 6 C). chk1Δ dun1Δ mrc1Δ sml1Δ cells containing pARS illustrate an example in which viability of cells exposed to HU is restored while having little effect on spindle elongation kinetics.
Our results also indicated that the suppression by pARS did not require Mrc1p. Recent studies using a frog extract system suggested that in order for a stalled fork to signal, unwinding of DNA by the minichromosome maintenance proteins must occur ahead of the stalled polymerases. The single-stranded DNA generated leads to recruitment of RPA and the sensor kinase Mec1p. In those studies, Claspin, an Mrc1p-like protein in vertebrates, was required for the generation of the checkpoint signal from unwound DNA generated at a stalled fork (Byun et al., 2005). Our data suggest that the checkpoint signal is being restored without the Rad9p or Mrc1p mediators/signal amplifiers or that Rad9p and Mrc1p can each compensate for the absence of the other. Alternatively, a mediator that has been shown to travel with the replication fork, such as Tof1p (see the next section), could also be involved in signaling from the episomal forks (Katou et al., 2003).
To further investigate whether a fork pause could restore checkpoint signaling, we engineered a highly active GAL1-GAL10 promoter driving expression of the URA3 gene into the pARSCTG construct and introduced this construct into chk1Δ dun1Δ cells. Growth on galactose drives transcription from this promoter in the opposite orientation to one of the replication forks. The movement of the transcription machinery opposes the direction of the movement of the replication fork originating from the ARS, which would lead to a replication fork pause. The pARSCTG construct containing the GAL-URA3 insert restored viability of chk1Δ dun1Δ cells exposed to HU in the presence of galactose, although to a lesser extent than the episome containing the ARS (Fig. 6 D). It should be noted that the mutant cells exhibit greater sensitivity to HU when grown on galactose, even when containing the pARS.
Tof1p is required for replication forks to pause at the centromere and the replication fork barrier in the ribosomal DNA locus, an actively transcribed tRNA (Calzada et al., 2005; Mohanty et al., 2006; Hodgson et al., 2007). The pARS failed to restore the viability of chk1Δ dun1Δ cells lacking Tof1p (Fig. 6 E), suggesting that the fork pause site at the centromere plays a role in suppression by the pARS. The model that the replication fork pause may contribute to the suppression by the episomal early origin was also supported by the discovery that a genomic clone that contained a tRNA gene (tE(UUC)C) acted as a high copy suppressor of chk1Δ dun1Δ HU sensitivity (Fig. 6 F). Additionally, this high copy suppressor failed to restore viability when TOF1 was deleted in the chk1Δ dun1Δ background (Fig. 6 F).
Activation of Rad53 after fork pause at CEN correlates with viability of chk1Δ dun1Δ cells
Elimination of a functional centromere on the pARS episome by introducing a point mutation into the CDEIII element of the pARS CEN (CCG→CTG) to obtain the construct pARSCTG abolished its ability to restore viability to chk1Δ dun1Δ cells (Fig. 7 A). This mutation of the CDEIII element had been shown to abolish the replication fork pause at the centromere (Greenfeder and Newlon, 1992). We examined whether the timing of replication fork pause at the episomal centromere correlated with the restored activation timing of Rad53p in chk1Δ dun1Δ cells containing either pARS or pARSCTG. Replication of pARS had initiated by 50 min after release from G1 into HU as indicated by the presence of Y-shaped replication intermediates (Fig. 7, B and C). Western analysis had shown that under these conditions Rad53p activation was evident 50 min after release into HU, at which time the replication fork pause caused by the centromere was apparent by 2D gel analyses (Fig. 7 D). The signal representing the replication fork pause at the centromere was absent in the replication intermediates of pARSCTG. The lack of replication fork pause correlates with a later time of Rad53p activation cells carrying pARSCTG, despite the fact that replication of the episome had occurred by that time. Collectively, our findings suggest that the critical characteristics of the episome that restored viability on HU were the timing of replication fork formation or the time during S phase that the pause is encountered.
Discussion
In this paper, we demonstrate that episomal origins activated in early S phase restore checkpoint signaling and viability to checkpoint mutants that have a deficiency in the number of stable replication forks present during early S phase. We show that the timing of replication origin activation is critical for restoration of the S-phase checkpoint response. Furthermore, we provide evidence that reconstitution of a few forks present in the first few minutes of S phase is sufficient to establish a checkpoint signal, which eliminated the requirement for the DNA damage checkpoint to survive replication blocks.
The S-phase checkpoint signals from replication forks at pause sites and centromeres to coordinate S phase, spindle elongation, and recovery from replication blocks
The functions of the S-phase checkpoint that allow recovery from blocked or paused replication forks include the role of Mec1p and Rad53p in stabilizing the association of polymerases α and ε with the replication fork, promoting fork progression in replication slow zones under conditions of low dNTPs, and preventing formation of pathological structures at forks arising from fork reversal (Branzei and Foiani, 2005). Several studies suggested that the inability to maintain replication complex and fork integrity is the lethal event in mec1 and rad53 mutants (Desany et al., 1998; Tercero et al., 2003).
In wild-type cells, replication blocks induced by HU prevent spindle elongation, whereas spindles proceed to elongate in rad53 cells (Allen et al., 1994; Krishnan et al., 2004; Bachant et al., 2005). Bachant, et al. (2005) showed that this defect in rad53 cells was suppressed by several copies of the pARS, presumably by providing tension from kinetochores formed at the replicated episomal CEN. In contrast, in our system, suppression of the HU sensitivity of chk1Δ dun1Δ cells by pARS involved activation of Rad53p and was independent of spindle elongation. These two studies (Bachant et al., 2005; this paper) suggest two signals that coordinate the response to replication blocks. One provides S-phase checkpoint signal from forks that allows recovery from replication blocks by maintaining fork integrity, stabilizing replication complexes, and possibly promoting fork progression through replication slow zones. The other prevents premature spindle elongation by providing replicated centromeric sequences that attach to the spindle and generate tension.
Our data suggest that stalled forks at replication fork pause sites are sufficient to provide checkpoint signal in chk1Δ dun1Δ cells. Simply changing the timing of activation of the episomal origin, and presumably the time that the fork encounters the pause site, determined whether the S-phase checkpoint was established. Previous studies have characterized the proteins involved in the stability of paused forks, including Tof1p, and have proposed a model for recovery of these forks that is independent of Rad53p (Calzada et al., 2005). We observed that the kinetics of Rad53p activation were restored by the episomal origin. It is also possible that the fork pause site at the centromere may simply lengthen the time it takes to replicate the episome allowing for the forks to stall and activate Rad53p. The model that a stalled fork at a pause site can signal through Rad53p remains to be tested because of the many roles that Rad53p plays in the S-phase checkpoint.
Replication of the 2-μm episome initiates from an ARS consensus sequence (Brewer and Fangman, 1987) only once per S-phase during the first third of S phase (Zakian et al., 1979). The 2-μm episome uses a segregation system that is distinct from the centromere (Kikuchi, 1983). We speculate that the reason that the 2-μm episome does not suppress the HU sensitivity of the chk1Δ dun1Δ cells, despite the fact that the episome provides additional replication forks, is because of the absence of a pause site (centromere) on the episome.
Coordination of the S-phase and DNA damage checkpoints for genomic stability
Deficiencies in the number of forks present during early S phase results in the activation of the DNA damage checkpoint by replication blocks and dependence on the DNA damage checkpoint for viability. Defects in dun1Δ cells lead to the activation of the Rad9p-mediated DNA damage checkpoint that is not normally required to survive replication blocks. In fact, Rad9p localizes to replication forks that are presumably collapsed, but only in cells lacking S-phase checkpoint proteins (Katou et al., 2003).
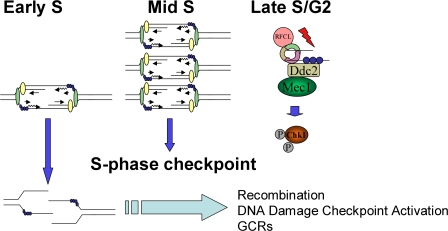
The GCRs in dun1Δ cells in the absence of exogenous stress could be formed in G2 by recombination-mediated repair of lesions incurred in S phase and could require the DNA damage checkpoint to provide time for their resolution before mitosis (Fig. 8). Because recombination and checkpoint signaling have been shown to occur simultaneously (Aylon et al., 2004; Ira et al., 2004), this could explain the activation of Chk1p in late S/G2 in dun1Δ cells (Fig. 8, model).
Figure 8.
Model. A deficiency of replication forks from early origins results in activation of the DNA damage checkpoint. RFCL is the checkpoint clamp loader. Ddc2, also known as Lcd1 and in mammals as Atrip, localizes Mec1 to DNA damage lesions (Nyberg et al., 2002; see text for details).
Inappropriate cyclin regulation or cyclin–Cdk complex activation by oncogenes in the early stages of cancer development deregulates the length of M/G1 and transition into S phase. Failure to coordinate the relative timing of these phases interferes with preRC formation and activation, decreasing the potential number of forks and ultimately leading to genomic instability (Spruck et al., 1999; Sidorova and Breeden, 2003; Ekholm-Reed et al., 2004). In fact, cyclin deregulation results in DNA damage checkpoint activation in mammals, which is similar to what we have observed in yeast (Bartkova et al., 2005). Therefore, the lessons learned from the yeast model on how early origin activation orchestrates the S-phase and DNA damage checkpoints are valuable to our understanding of the factors that affect genomic stability in human cancer.
Materials and methods
Yeast strains
Yeast strains generated using standard genetic techniques and constructs used in this study are listed in Table S4 (available at http://www.jcb.org/cgi/content/full/jcb.200706009/DC1). Selected strains, as noted, were provided by D. Koshland (Carnegie Institute of Washington, Baltimore, MD) and S. Elledge (Harvard University Medical School, Boston, MA).
Plasmid construction
p2μ-Clb6.
Chromosome VII coordinates 705169–706886 were amplified by the PCR using primers engineered to introduce a SacI restriction site at the 5′ end of the product and a BamHI restriction site at the 3′ end of the product. The PCR product was subjected to restriction digest and ligated into the corresponding sites in the polylinker of pRS426.
pARSCTG.
This construct was generated by a PCR mutagenesis strategy resulting in the introduction of a point mutation (CCG→CTG or C4432T, according to GenBank annotation for pRS416) in the essential triplet found within CDEIII of the centromeric sequence of the vector pRS416.
pARSCTG-STB.
Coordinates 4315–4612 of pRS426 (according to GenBank annotation for pRS426) were amplified by the PCR using primers engineered to introduce a BamHI restriction site at the 5′ end of the product and a SalI restriction site at the 3′ end of the product. This sequence was ligated into the corresponding sites in the polylinker of pARSCTG.
p12-Ac.
This construct was generated using a PCR strategy to insert chromosome XV sequence coordinates 721577–721772, which includes the HIS3 promoter sequence containing five Gcn4p binding sites, immediately adjacent to ARS1412 in the p12 construct (Friedman et al., 1996; provided by J. Diller, Pennsylvania State University, University Park, PA; and W. Fangman, University of Washington, Seattle, WA). The specified chromosome XV coordinates were inserted between the sequence in p12 that corresponds to chromosome XIV coordinates 196102 and 196103.
p2μ-Gcn4.
Chromosome V sequence 138761–140226, which includes the GCN4 sequence, was inserted into the HindIII and SacI sites of pRS425.
p2μ-tRNA.
This clone was isolated from the S. cerevisiae AB320 genomic library (American Type Culture Collection), which was constructed in yeast episomal plasmid 13. The clone contains ∼8 kb of chromosome III sequence centromere proximal to coordinate 80321. This sequence includes tE((UUC)C), which encodes tRNA-Glu.
pARS305.
Chromosome III 38498–341933 was amplified by PCR, cut with BamHI and HindIII, and ligated into pBSKS, resulting in pJP25. The BamHI, HindIII fragment was then cut out of pJP25 and ligated into BamHI–HindIII-digested p13ARS (Friedman et al., 1996; provided by J. Diller and W. Fangman).
pARSCTG-STB-GAL-URA3.
GAL1-GAL10 promoter sequence (chromosome II, 278357–279016 [Saccharomyces Genome Database annotation]) was PCR amplified using primers engineered to introduce SmaI sites at both the 3′ and 5′ ends of the product. The primers also introduced HindIII and BglII sites at the 3′ end of the product. This sequence was ligated into the unique PmlI site in pARSCTG-STB to produce pARSCTG-STB-GAL. URA3 sequence (chromosome V, 116167–117048 [Saccharomyces Genome Database annotation]) was then PCR amplified using primers engineered to introduce a HindIII restriction site at the 5′ end and a BglII restriction site at the 3′ end of the product. This sequence was then ligated into the corresponding sites in pARSCTG-STB-GAL.
Growth conditions
Cells were grown on normal or modified YPD, SC-Leu, SC-Ura, SC-Trp, SC-Ura-Leu, SC-Ura-Leu-Trp, or SC-Ura-Leu-Trp-His medium. Cell viability was measured by spotting cells in serial dilutions of 5,000, 500, 50, and 5 cells onto the indicated medium and growing at 30°C. For G1 synchrony, cells were arrested in YPD or SC-Ura medium, pH 3.9, at 24°C for replication intermediates (RI) and cdc15-2 or at 30°C for all other experiments with 10 μg/ml α-factor (Protein Chemistry Core Laboratory, Baylor College of Medicine, Houston, TX). Cells were released from G1 by washing twice with YPD or SC-Ura medium and resuspending in medium containing the indicated HU concentration at 24°C for RI, 37°C for cdc15-2, or 30°C for all other experiments. 10 μg/ml nocodazole was added where indicated.
RI analysis
RI experiments were performed as previously described (Santocanale and Diffley, 1998; http://science.cancerresearchuk.org/sci/chromrep/pdf/alkalineriassay.pdf?version=1) with the following modifications: cells were treated with 50 μg/ml oxalyticase (Enzogenetics) for 1 h at 30°C before DNA isolation. DNA was transferred to Hybond-N+ membrane (GE Healthcare) by the neutral transfer method. Membranes were hybridized with an α-[32P]dCTP–labeled probe prepared with the Random Primers kit (Invitrogen) and 25 ng of template DNA amplified from the following chromosomal coordinates: ARS305 ChIII 38995–40048; and ARS1 ChIV 461969–463011. RI signal from each lane was quantified using a phosphor imager (Molecular Diagnostics) and ImageQuant software (GE Healthcare). As loading control, an equal aliquot of DNA was digested with EcoRI and subjected to Southern blot analysis using a probe generated, as described in this section, from 25 ng PCR-generated template (ChIII 256040–257211). The loading control signal was used to normalize the signal for the RI to the relative amount of DNA loaded for each sample.
2D gel analysis
Cells were prepared as previously described (Deshpande and Newlon, 1992) using ice-cold azide stop buffer (0.5 M NaOH, 0.4 M Na2EDTA, and 0.2% sodium azide). DNA isolation was performed as previously described (Huberman et al., 1987). Neutral–neutral 2D gel electrophoresis was performed as previously described (Brewer and Fangman, 1987; Deshpande and Newlon, 1992). Approximately 3 μg DNA was digested for each sample with NcoI (ARS305), BamHI and NcoI (ARS603), EcoRI (plasmid RI), or PvuII and EcoRV (pARS plasmid RI for RFP). DNA was transferred to Hybond-XL membrane as previously described (Schollaert et al., 2004). Probes were generated by random priming or by the PCR, as described in Materials and methods, from the following templates: ARS305: NcoI–BamHI restriction fragment of plasmid A6C110 (Newlon et al., 1991); ARS603 (PCR product of ChVI 68233–69364); p12 and p12ARS (EcoRI–MscI restriction fragment of p12ARS [a gift from J. Diller]); pRS416 (EcoRI-linearized pBluescript II [KS]); and pRS416, for RFP at CEN (PvuII–SspI [1873 bp] restriction fragment of pBluescript II [KS]). Probes were hybridized as previously described (Deshpande and Newlon, 1992). Membranes were exposed to phosphor screens, and RI signal was quantified as described in RI analysis.
Western analyses
Cell protein extracts, prepared as previously described (Schollaert et al., 2004), were separated on 10% acrylamide and 0.067% bis-acrylamide gels, transferred to nitrocellulose membrane, and probed with anti-HA (16B12; Covance) or anti-Rad53 (yC-19; Santa Cruz Biotechnology, Inc.) antibodies.
Flow cytometry and immunofluorescence
Cell cycle analyses by flow cytometry, staining nuclei, and visualizing spindles were performed as previously described (Sanchez et al., 1999).
Online supplemental material
Fig. S1 shows activation of chromosomal and episomal origin ARS305.Fig. S2 shows that the episomal origin relieves the dependence of dun1Δ cells on DNA damage checkpoint proteins for survival of forks stalled by low dNTPs or DNA damage. Fig. S3 shows that suppression of HU sensitivity of checkpoint mutants by an episomal replication origin requires Rad53p and is independent of the regulation of spindle dynamics. Table S1 shows that addition of the STB sequence to the pARSCTG episome improves its mitotic stability.Table S2 shows budding indices for alkaline gel experiments (replicates of the experiment in Fig. 1 A). Table S3 shows budding indices for 2D gel experiments (replicates of the experiment in Fig. 1 B). Table S4 lists the strains used in these studies. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200706009/DC1.
Acknowledgments
We are grateful to Ted Weinert, David Cortez, Craig Tomlinson, Elizabeth Pereira, and members of the Sanchez laboratory for helpful comments and discussions. We thank Douglas Koshland and Stephen Elledge for strains, Walton Fangman and John Diller for p12 and p12ARS constructs, Sandy Schwemberger for expert flow cytometric analyses, and Reta Gibbons and Alice L. Givan for help with data analyses.
J.M. Caldwell is a Ryan Fellow. This work was funded in part by National Institutes of Health grant RO1 GM35679 to C.S. Newlon, grant 8470 from Shriners of North America to G.F. Babcock, National Institutes of Health/National Cancer Institute grant RO1 CA84463, Department of Defense grant DAMD 17-01-1-020, The Ohio Cancer Research Associates and the Pew Scholars Program in the Biomedical Sciences grant to Y. Sanchez, and grants P30 ES06096 and U01 ES11038 from the National Institute of Environmental Health Sciences.
The authors declare no competing financial interests.
J.M. Caldwell's present address is Division of Allergy and Immunology, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, 45220
Abbreviations used in this paper: ARS, autonomously replicating sequence; dNTP, deoxyribonucleotide triphosphate; GCR, gross chromosomal rearrangement; HU, hydroxyurea; preRC, prereplication complex; RI, replication intermediate.
References
- Alcasabas, A.A., A.J. Osborn, J. Bachant, F. Hu, P.J. Werler, K. Bousset, K. Furuya, J.F. Diffley, A.M. Carr, and S.J. Elledge. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3:958–965. [DOI] [PubMed] [Google Scholar]
- Allen, J.B., Z. Zhou, W. Siede, E.C. Friedberg, and S.J. Elledge. 1994. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 8:2401–2415. [DOI] [PubMed] [Google Scholar]
- Alvino, G.M., D. Collingwood, J.M. Murphy, J. Delrow, B.J. Brewer, and M.K. Raghuraman. 2007. Replication in hydroxyurea: it's a matter of time. Mol. Cell. Biol. 27:6396–6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio, J.G., C.J. Viggiani, D.G. Gibson, and O.M. Aparicio. 2004. The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:4769–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon, Y., B. Liefshitz, and M. Kupiec. 2004. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 23:4868–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachant, J., S.R. Jessen, S.E. Kavanaugh, and C.S. Fielding. 2005. The yeast S phase checkpoint enables replicating chromosomes to bi-orient and restrain spindle extension during S phase distress. J. Cell Biol. 168:999–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova, J., Z. Horejsi, K. Koed, A. Kramer, F. Tort, K. Zieger, P. Guldberg, M. Sehested, J.M. Nesland, C. Lukas, et al. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 434:864–870. [DOI] [PubMed] [Google Scholar]
- Branzei, D., and M. Foiani. 2005. The DNA damage response during DNA replication. Curr. Opin. Cell Biol. 17:568–575. [DOI] [PubMed] [Google Scholar]
- Brewer, B.J., and W.L. Fangman. 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 51:463–471. [DOI] [PubMed] [Google Scholar]
- Byun, T.S., M. Pacek, M.C. Yee, J.C. Walter, and K.A. Cimprich. 2005. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19:1040–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzada, A., B. Hodgson, M. Kanemaki, A. Bueno, and K. Labib. 2005. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 19:1905–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson, T.W., R.S. Sikorski, M. Dante, J.H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene. 110:119–122. [DOI] [PubMed] [Google Scholar]
- D'Amours, D., and S.P. Jackson. 2001. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 15:2238–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desany, B.A., A.A. Alcasabas, J.B. Bachant, and S.J. Elledge. 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 12:2956–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande, A.M., and C.S. Newlon. 1992. The ARS consensus sequence is required for chromosomal origin function in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:4305–4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande, A.M., and C.S. Newlon. 1996. DNA replication fork pause sites dependent on transcription. Science. 272:1030–1033. [DOI] [PubMed] [Google Scholar]
- Donaldson, A.D., M.K. Raghuraman, K.L. Friedman, F.R. Cross, B.J. Brewer, and W.L. Fangman. 1998. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol. Cell. 2:173–182. [DOI] [PubMed] [Google Scholar]
- Ekholm-Reed, S., J. Mendez, D. Tedesco, A. Zetterberg, B. Stillman, and S.I. Reed. 2004. Deregulation of cyclin E in human cells interferes with prereplication complex assembly. J. Cell Biol. 165:789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, K.L., J.D. Diller, B.M. Ferguson, S.V. Nyland, B.J. Brewer, and W.L. Fangman. 1996. Multiple determinants controlling activation of yeast replication origins late in S phase. Genes Dev. 10:1595–1607. [DOI] [PubMed] [Google Scholar]
- Gardner, R., C.W. Putnam, and T. Weinert. 1999. RAD53, DUN1 and PDS1 define two parallel G2/M checkpoint pathways in budding yeast. EMBO J. 18:3173–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, D.G., J.G. Aparicio, F. Hu, and O.M. Aparicio. 2004. Diminished S-phase cyclin-dependent kinase function elicits vital Rad53-dependent checkpoint responses in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:10208–10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfeder, S.A., and C.S. Newlon. 1992. Replication forks pause at yeast centromeres. Mol. Cell. Biol. 12:4056–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenon, M., C. Gilbert, and N.F. Lowndes. 2001. Checkpoint activation in response to double-strand breaks requires the Mre11/Rad50/Xrs2 complex. Nat. Cell Biol. 3:844–847. [DOI] [PubMed] [Google Scholar]
- Hodgson, B., A. Calzada, and K. Labib. 2007. Mrc1 and Tof1 regulate DNA replication forks in different ways during normal S phase. Mol. Biol. Cell. 18:3894–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao, C.L., and J. Carbon. 1981. Characterization of a yeast replication origin (ars2) and construction of stable minichromosomes containing cloned yeast centromere DNA (CEN3). Gene. 15:157–166. [DOI] [PubMed] [Google Scholar]
- Huang, D., and D. Koshland. 2003. Chromosome integrity in Saccharomyces cerevisiae: the interplay of DNA replication initiation factors, elongation factors, and origins. Genes Dev. 17:1741–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman, J.A., L.D. Spotila, K.A. Nawotka, S.M. el-Assouli, and L.R. Davis. 1987. The in vivo replication origin of the yeast 2 microns plasmid. Cell. 51:473–481. [DOI] [PubMed] [Google Scholar]
- Ira, G., A. Pellicioli, A. Balijja, X. Wang, S. Fiorani, W. Carotenuto, G. Liberi, D. Bressan, L. Wan, N.M. Hollingsworth, et al. 2004. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 431:1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou, Y., Y. Kanoh, M. Bando, H. Noguchi, H. Tanaka, T. Ashikari, K. Sugimoto, and K. Shirahige. 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 424:1078–1083. [DOI] [PubMed] [Google Scholar]
- Kelly, T.J., G.S. Martin, S.L. Forsburg, R.J. Stephen, A. Russo, and P. Nurse. 1993. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 74:371–382. [DOI] [PubMed] [Google Scholar]
- Kikuchi, Y. 1983. Yeast plasmid requires a cis-acting locus and two plasmid proteins for its stable maintenance. Cell. 35:487–493. [DOI] [PubMed] [Google Scholar]
- Krishnan, V., S. Nirantar, K. Crasta, A.Y. Cheng, and U. Surana. 2004. DNA replication checkpoint prevents precocious chromosome segregation by regulating spindle behavior. Mol. Cell. 16:687–700. [DOI] [PubMed] [Google Scholar]
- Labib, K., J.A. Tercero, and J.F. Diffley. 2000. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science. 288:1643–1647. [DOI] [PubMed] [Google Scholar]
- Lengronne, A., and E. Schwob. 2002. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G(1). Mol. Cell. 9:1067–1078. [DOI] [PubMed] [Google Scholar]
- Lopes, M., C. Cotta-Ramusino, A. Pellicioli, G. Liberi, P. Plevani, M. Muzi-Falconi, C.S. Newlon, and M. Foiani. 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 412:557–561. [DOI] [PubMed] [Google Scholar]
- McGrew, J., B. Diehl, and M. Fitzgerald-Hayes. 1986. Single base-pair mutations in centromere element III cause aberrant chromosome segregation in Saccharomyces cerevisiae. Mol. Cell. Biol. 6:530–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael, W.M., R. Ott, E. Fanning, and J. Newport. 2000. Activation of the DNA replication checkpoint through RNA synthesis by primase. Science. 289:2133–2137. [DOI] [PubMed] [Google Scholar]
- Mohanty, B.K., N.K. Bairwa, and D. Bastia. 2006. The Tof1p-Csm3p protein complex counteracts the Rrm3p helicase to control replication termination of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 103:897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung, K., A. Datta, and R.D. Kolodner. 2001. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell. 104:397–408. [DOI] [PubMed] [Google Scholar]
- Navas, T.A., Y. Sanchez, and S.J. Elledge. 1996. RAD9 and DNA polymerase epsilon form parallel sensory branches for transducing the DNA damage checkpoint signal in Saccharomyces cerevisiae. Genes Dev. 10:2632–2643. [DOI] [PubMed] [Google Scholar]
- Newlon, C.S., L.R. Lipchitz, I. Collins, A. Deshpande, R.J. Devenish, R.P. Green, H.L. Klein, T.G. Palzkill, R.B. Ren, S. Synn, et al. 1991. Analysis of a circular derivative of Saccharomyces cerevisiae chromosome III: a physical map and identification and location of ARS elements. Genetics. 129:343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyberg, K.A., R.J. Michelson, C.W. Putnam, and T.A. Weinert. 2002. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36:617–656. [DOI] [PubMed] [Google Scholar]
- Osborn, A.J., and S.J. Elledge. 2003. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 17:1755–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman, M.K., E.A. Winzeler, D. Collingwood, S. Hunt, L. Wodicka, A. Conway, D.J. Lockhart, R.W. Davis, B.J. Brewer, and W.L. Fangman. 2001. Replication dynamics of the yeast genome. Science. 294:115–121. [DOI] [PubMed] [Google Scholar]
- Reynolds, A.E., R.M. McCarroll, C.S. Newlon, and W.L. Fangman. 1989. Time of replication of ARS elements along yeast chromosome III. Mol. Cell. Biol. 9:4488–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka, Y., and M. Yanagida. 1993. Fission yeast cut5+, required for S phase onset and M phase restraint, is identical to the radiation-damage repair gene rad4+. Cell. 74:383–393. [DOI] [PubMed] [Google Scholar]
- Sanchez, Y., B.A. Desany, W.J. Jones, Q. Liu, B. Wang, and S.J. Elledge. 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 271:357–360. [DOI] [PubMed] [Google Scholar]
- Sanchez, Y., J. Bachant, H. Wang, F. Hu, D. Liu, M. Tetzlaff, and S.J. Elledge. 1999. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science. 286:1166–1171. [DOI] [PubMed] [Google Scholar]
- Santocanale, C., and J.F. Diffley. 1998. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 395:615–618. [DOI] [PubMed] [Google Scholar]
- Schollaert, K., J.M. Poisson, J.S. Searle, J.A. Schwanekamp, C.R. Tomlinson, and Y. Sanchez. 2004. A novel role for Saccharomyces cerevisiae Chk1p in the response to replication blocks. Mol. Biol. Cell. 15:4051–4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, K.L., and S.E. Plon. 2003. Loss of Sin3/Rpd3 histone deacetylase restores the DNA damage response in checkpoint-deficient strains of Saccharomyces cerevisiae. Mol. Cell. Biol. 23:4522–4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle, J.S., K.L. Schollaert, B.J. Wilkins, and Y. Sanchez. 2004. The DNA damage checkpoint and PKA pathways converge on APC substrates and Cdc20 to regulate mitotic progression. Nat. Cell Biol. 6:138–145. [DOI] [PubMed] [Google Scholar]
- Shimada, K., P. Pasero, and S.M. Gasser. 2002. ORC and the intra-S-phase checkpoint: a threshold regulates Rad53p activation in S phase. Genes Dev. 16:3236–3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahige, K., Y. Hori, K. Shiraishi, M. Yamashita, K. Takahashi, C. Obuse, T. Tsurimoto, and H. Yoshikawa. 1998. Regulation of DNA-replication origins during cell-cycle progression. Nature. 395:618–621. [DOI] [PubMed] [Google Scholar]
- Sidorova, J.M., and L.L. Breeden. 2003. Precocious G1/S transitions and genomic instability: the origin connection. Mutat. Res. 532:5–19. [DOI] [PubMed] [Google Scholar]
- Sikorski, R.S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruck, C.H., K.A. Won, and S.I. Reed. 1999. Deregulated cyclin E induces chromosome instability. Nature. 401:297–300. [DOI] [PubMed] [Google Scholar]
- Tanaka, K., and P. Russell. 2001. Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nat. Cell Biol. 3:966–972. [DOI] [PubMed] [Google Scholar]
- Tercero, J.A., M.P. Longhese, and J.F. Diffley. 2003. A central role for DNA replication forks in checkpoint activation and response. Mol. Cell. 11:1323–1336. [DOI] [PubMed] [Google Scholar]
- Usui, T., H. Ogawa, and J.H. Petrini. 2001. A DNA damage response pathway controlled by tel1 and the mre11 complex. Mol. Cell. 7:1255–1266. [DOI] [PubMed] [Google Scholar]
- Vogelauer, M., L. Rubbi, I. Lucas, B.J. Brewer, and M. Grunstein. 2002. Histone acetylation regulates the time of replication origin firing. Mol. Cell. 10:1223–1233. [DOI] [PubMed] [Google Scholar]
- Wang, H., and S.J. Elledge. 1999. DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 96:3824–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., M. Vujcic, and D. Kowalski. 2001. DNA replication forks pause at silent origins near the HML locus in budding yeast. Mol. Cell. Biol. 21:4938–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert, T.A., G.L. Kiser, and L.H. Hartwell. 1994. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8:652–665. [DOI] [PubMed] [Google Scholar]
- Zakian, V.A., B.J. Brewer, and W.L. Fangman. 1979. Replication of each copy of the yeast 2 micron DNA plasmid occurs during the S phase. Cell. 17:923–934. [DOI] [PubMed] [Google Scholar]
- Zhao, X., E.G. Muller, and R. Rothstein. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell. 2:329–340. [DOI] [PubMed] [Google Scholar]
- Zhou, Z., and S.J. Elledge. 1993. DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell. 75:1119–1127. [DOI] [PubMed] [Google Scholar]
- Zou, L., and B. Stillman. 2000. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol. Cell. Biol. 20:3086–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]