Abstract
Huntington's disease (HD) is caused by the expansion of a polyglutamine tract in the N-terminal region of huntingtin (htt) and is characterized by selective neurodegeneration. In addition to forming nuclear aggregates, mutant htt accumulates in neuronal processes as well as synapses and affects synaptic function. However, the mechanism for the synaptic toxicity of mutant htt remains to be investigated. We targeted fluorescent reporters for the ubiquitin–proteasome system (UPS) to presynaptic or postsynaptic terminals of neurons. Using these reporters and biochemical assays of isolated synaptosomes, we found that mutant htt decreases synaptic UPS activity in cultured neurons and in HD mouse brains that express N-terminal or full-length mutant htt. Given that the UPS is a key regulator of synaptic plasticity and function, our findings offer insight into the selective neuronal dysfunction seen in HD and also establish a method to measure synaptic UPS activity in other neurological disease models.
Introduction
Protein misfolding and aggregation are the common pathological changes in age-dependent neurodegenerative disorders (Kopito, 2000; Goldberg, 2003). Of these disorders, Huntington's disease (HD) is caused by the expansion of a polyglutamine (polyQ) tract (>37 glutamines) in the N-terminal region of huntingtin (htt), a 350-kD protein that is predominantly distributed in the cytoplasm (Gusella and Macdonald, 2006). PolyQ expansion, which results in protein misfolding, aggregation, and cytotoxicity, also leads to selective neurodegeneration in distinct brain regions in eight other polyQ diseases (Orr and Zoghbi, 2007). Unlike most polyQ diseases, in which mutant proteins are mainly located in the nucleus, HD features the accumulation of mutant htt in neuronal processes and synapses in addition to nuclear inclusions. Although mounting evidence indicates that nuclear mutant htt induces neuropathology by altering gene expression, the effect of mutant htt in synapses remains unclear, despite its association with synaptic dysfunction (Smith et al., 2005; Li and Li, 2006).
The ubiquitin–proteasome system (UPS) plays an essential role in degrading damaged or misfolded proteins (Hershko and Ciechanover, 1998). Misfolded proteins and protein fragments generated by proteolysis are polyubiquitinated by ubiquitin ligases, a process that targets the substrates to the proteasome for degradation (Demartino and Gillette, 2007). The 26S proteasome, which consists of a 20S catalytic core particle and a 19S regulatory particle, selectively degrades ubiquitinated proteins (Pickart and Cohen, 2004). Thus, a major aspect of UPS function is the dynamic control of protein stability, which is important for a variety of cellular processes, including cell cycle control, transcription, chromatin remodeling, and protein trafficking. Recently, the UPS has emerged as a key regulator of synaptic plasticity and function (Korhonen and Lindholm, 2004; Yi and Ehlers, 2005; Patrick, 2006). In the presynaptic terminal, the UPS regulates presynaptic function through multi-ubiquitination and protein turnover, thereby altering protein activity and vesicle dynamics (Wilson et al., 2002; Chen et al., 2003; Speese et al., 2003). The UPS also critically controls postsynaptic remodeling and plasticity. For example, proteasomal inhibition prevents neuronal activity-regulated composition of the postsynaptic density (PSD) proteins (Ehlers, 2003), and the activity-dependent endocytosis of AMPA receptors is sensitive to proteasome inhibition (Colledge et al., 2003).
Cells can regulate proteasome function in response to changing physiological demands by altering the total number of proteasomes (Lecker et al., 2006) and/or by altering proteasomal subunit composition (Glickman and Raveh, 2005). The complex components of the proteasome, however, make it difficult to investigate potential changes in a large number of proteasomal subunits. Also, the proteasome exists as a heterogeneous group of structures in different subcellular regions (Brooks et al., 2000; Wójcik and DeMartino, 2003). Thus, functional assays of the proteasome with specific inhibitors or substrates have been widely used to assess a variety of pathological conditions. Although polyQ-expanded proteins were found to impair UPS function in various cell models or in vitro systems (Bence et al., 2001; Verhoef et al., 2002; Venkatraman et al., 2004; Bennett et al., 2005), in vivo studies of UPS function in polyQ disease mouse models have not yielded consistent results (Bowman et al., 2005; Bett et al., 2006; Bennett et al., 2007). These previous in vivo studies, however, did not examine proteasomal activity in different subcellular regions of neurons. Given that mutant htt accumulates in neuronal processes and synapses (DiFiglia et al., 1997; Gutekunst et al., 1999; Li et al., 2000) and affects synaptic function (Usdin et al., 1999; Cepeda et al., 2001; Zeron et al., 2002; Smith et al., 2005; Cummings et al., 2006; Fan et al., 2007), it is important to investigate the specific effect of mutant htt on the UPS in synaptic terminals.
In the present study, we targeted UPS reporters to presynaptic or postsynaptic terminals. In combination with synaptosomal fractionation and biochemical analysis, our studies demonstrate that mutant htt can inhibit the function of synaptic proteasomes. This finding offers important insight into the mechanism by which mutant htt affects synaptic function and neurotransmitter release. The assays used in our studies can also be applied to other neurological disorder models to examine possible changes in synaptic UPS activities.
Results
Generation of synaptic reporters for UPS activity
The fluorescent UPS reporter GFPu, which is a green fluorescent fusion protein tagged with a CL-1 degron sequence (16 amino acids) specific for ubiquitination and degradation by the proteasome, has been widely used to examine the activity of the proteasome in cells (Bence et al., 2001; Dong et al., 2004; Avraham et al., 2005; Bennett et al., 2005). To specifically measure synaptic UPS activity using this reporter, we fused the postsynaptic protein PSD95 or the presynaptic protein SNAP25 to GFPu in order to target this reporter to postsynaptic or presynaptic terminals. To ensure neuronal expression, the fusion proteins are expressed via an adenoviral vector under the control of the neuronal promoter for human synapsin 1, a synaptic vesicle-associated protein (Fig. 1 A). As controls, we also generated cDNAs encoding for DsRed fusion proteins (RFP), which provide red fluorescent signals but are not degraded rapidly by the UPS. Thus, the ratio of GFPu to RFP (GFPu/RFP) reflects the relative activity of the UPS when GFPu and RFP reporters are coexpressed in the same cells.
Figure 1.
Expression of synaptic UPS reporters in neurons. (A) Schematic map of DNA constructs for expressing fluorescent UPS reporters. RFP or GFPu is added to the C terminus of PSD95 or SNAP25 in an adenoviral vector to express fusion proteins under the control of the human synapsin 1 promoter. (B) Western blots showing the expression of these fusion proteins in HEK293 cells after adenoviral infection. (C) Expression of fluorescent UPS reporters in primary cultured cortical neurons after infection with adenoviral SNAP25-RFP vector. SNAP25-RFP was expressed in cultured primary neurons (NeuN-positive) but not in cultured glial cells (GFAP-positive). (D) In the mouse brain striatum injected with adenoviral SNAP25-RFP reporter, SNAP25-RFP was also expressed in neurons, but not glial cells. Immunostaining for GFAP or NeuN (green), nuclear staining (blue) by Hoechst, and SNAP25-RFP (red) were revealed by fluorescent microscopy. Bars, 5 μm (C) and 10 μm (D).
Using Western blotting, we first verified that these reporters were expressed at the correct molecular weights in HEK293 cells (Fig. 1 B). To confirm that the adenoviral vector we generated drives transgene expression exclusively in neurons, we infected primary cultures from embryonic rat brain with adenoviral SNAP25-RFP and verified that this reporter was expressed in neurons (NeuN-positive) but not in glial cells (GFAP-positive) (Fig. 1 C). Moreover, after injecting this viral reporter into the striatum of mouse brain, we also detected red fluorescence in NeuN-positive neurons but not GFAP-positive glial cells (Fig. 1 D). Thus, both in vitro and in vivo experiments prove the neuronal-specific expression of our adenoviral vector.
The ratio of GFPu to RFP is increased by inhibiting UPS activity
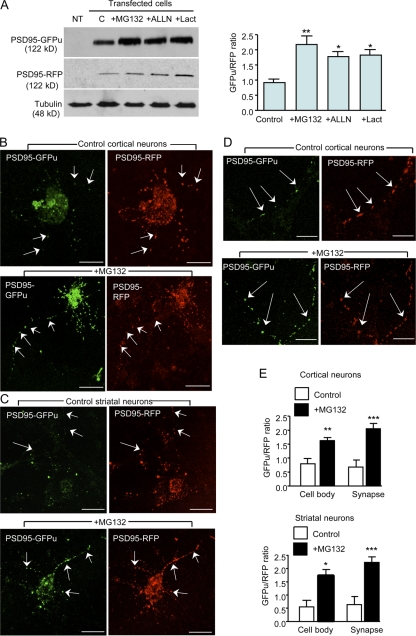
Next, we wanted to verify that the reporters can be used to measure UPS activity in synapses. We first expressed these reporters in HEK293 cells and then treated the cells with various proteasomal inhibitors including MG132, ALLN, and lactacystin (Lact) for 8 h. As expected, these inhibitors significantly increased the levels of GFPu compared with the RFP control reporter or tubulin (Fig. 2 A, left). Quantification of the ratio of GFPu to RFP confirmed this increase (Fig. 2 A, right). We then infected cultured primary cortical and striatal neurons with these adenoviral vectors and observed the localization of these reporters in synaptic structures, which are characterized by small puncta in neurites (Fig. 2, B–D). The ratio of GFPu to RFP in synaptic structures is simple to quantify because of the defined and selective fluorescence signals in synapses. Because transgenic reporters are overexpressed in the cell body, however, the diffuse fluorescent signals there are less readily quantified. Despite this we were able to note an increased GFPu/RFP ratio in the cell body after MG132 treatment (Fig. 2 E), which was more obvious in those images with a shorter exposure time (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200709080/DC1). Under conditions that allowed clear visualization of synaptic signals, we observed that the ratio of GFPu to RFP in synaptic puncta showed a greater increase than in the cell body after inhibiting proteasomal activity by MG132 (Fig. 2 E). Using the same approach, we confirmed that postsynaptic reporters (PSD95-GFPu/RFP) could also reflect changes in synaptic UPS activity (Fig. 3).
Figure 2.
Presynaptic UPS reporters (SNAP25-GFPu/-RFP) reflect UPS activity in cultured cells. (A) Western blot showing a significant increase in SNAP25-GFPu, but not SNAP25-RFP, in HEK293 cells after inhibition of proteasomal activity by MG132 (10 μM), ALLN (10 μg/ml), or lactacystin (lact,10 μM) for 8 h. The ratio (mean + SEM, n = 3) of GFPu to RFP was quantified (right). (B and C) In cortical or striatal neurons coinfected with adenoviral SNAP25-RFP and SNAP25-GFPu, the expression ratio of GFPu to RFP in synapses was increased after addition of MG132. Arrows indicate synapses. Bars, 5 μm. (D) Higher magnification showing the expression of SNAP25-GFPu in the synapses (arrows) of cultured cortical neurons. Bars, 2.5 μm. (E) The ratio (mean + SEM n = 45) of GFPu/RFP in the cell body or the synapses was analyzed. The ratio of GFPu to RFP signal was significantly increased after inhibiting the proteasome with 10 μM MG132. *, P < 0.05; **, P < 0.01. ***, P < 0.001 as compared with control.
Figure 3.
Postsynaptic UPS reporters (PSD95-GFPu and PSD95-RFP) reflect UPS activity in cultured cells. (A) Western blot showing a significant increase in PSD95-GFPu, but not PSD95-RFP, in HEK293 cells after inhibition of proteasomal activity by MG132 (10 μM), ALLN (10 μg/ml), or lactacystin (lact, 10 μM). The ratio (mean + SEM, n = 3) of GFPu to RFP is quantified (right). (B and C) In cortical or striatal neurons coinfected by adenoviruses of PSD95-RFP and PSD95-GFPu, the expression ratio of GFPu to RFP in synapses was increased after addition of MG132. Arrows indicate synapses. Bars, 5 μm. (D) Higher magnification showing the expression of PSD95-GFPu in the synapses (arrows) of cortical neurons. Bars, 2.5 μm. (E) The ratio (mean + SEM, n = 40) of GFPu to RFP signal was significantly increased after inhibiting the proteasome by 10 μM MG132. *, P < 0.05; **, P < 0.01. ***, P < 0.001 as compared with control.
Decreased UPS activity in synapses of HD neurons that express exon1 mutant htt
Having established the specificity of these fluorescent reporters to reflect synaptic UPS activity, we examined synaptic UPS activity in cultured neurons from wild-type and R6/2 mice that express exon1 mutant htt with 115-150Q (Davies et al., 1997). After culturing these neurons for 10 d, we infected them with adenoviral UPS reporters. 4–5 d later, we found that presynaptic SNAP25-GFPu was expressed at a lower level in synapses of wild-type neurons than in HD neurons (Fig. 4 A, left), suggesting that the UPS activity is higher in the former. Quantitative assessment of the ratio of GFPu/RFP demonstrated that the presynaptic ratio is greater in HD neurons than in wild-type neurons (Fig. 4 A, right). Using postsynaptic UPS reporters (PSD95-GFPu/RFP), we also observed an increased ratio of GFPu/RFP in the synapses of HD neurons compared with wild-type neurons (Fig. 4 B). Thus, these findings indicate that synaptic UPS activity is decreased in cultured neurons from HD mouse brains.
Figure 4.
Expression of synaptic UPS reporters in cultured primary neurons from R6/2 and wild-type mice. (A and B) Expression of SNAP25-GFPu/-RFP (A) and PSD95-GFP/-RFP (B) in cultured primary striatal neurons from embryonic wild-type (WT, top panels) and R6/2 (bottom panels) mice. The levels of GFPu in neuronal processes and synapses are higher in R6/2 neurons than in wild-type neurons. Right panels show the ratio (mean + SEM, n = 52) of GFPu/RFP. ***, P < 0.01. Bars, 2.5 μm.
To examine whether these reporters can also reflect changes in synaptic UPS activity in the brain, we performed stereotaxic injection of adenoviral reporters into the striatum of wild-type or R6/2 mice aged 7–8 wk and examined their expression in brain sections 7–8 d after injection. SNAP25-GFPu was colocalized with a number of neuropil htt aggregates in HD mouse brains (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200709080/DC1), suggesting that mutant htt in neuronal processes is localized to synapses. Similar to what we observed in cultured neurons (Fig. 4), presynaptic (Fig. 5 A) and postsynaptic (Fig. 5 B) GFPu/RFP ratios were increased in the synapses of HD mouse brains compared with those of wild-type mouse striatum. Although the ratio of GFPu/RFP in the cell body is not statistically different between HD and wild-type mice, this ratio in synapses of HD neurons was significantly higher than that in wild-type mouse brains. In situ hybridization did not reveal any significant difference in the expression of SNAP25-GFPu transcripts between wild-type and R6/2 mouse brains (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200709080/DC1). Thus, the increased GFPu/RFP ratio in HD mouse brains reflects decreased GFPu degradation in synapses.
Figure 5.
Expression of synaptic UPS reporters in the brain striatum. (A) Expression of presynaptic reporters (SNAP25-GFPu/-RFP) in the striatum of R6/2 mice at 9 wk of age 8 d after adenoviral injection. Right panels show high magnification graphs and quantitative data of the ratio (mean +SEM, n = 48) of GFPu/RFP in synapses (arrows). (B) Expression of PSD95-GFPu/-RFP in the striatum of R6/2 mice. The high magnification graphs and the ratio (mean + SEM, n = 43) of GFPu/RFP are also displayed in right panels. Bars, 5 μm. *, P < 0.05.
Because mutant htt may affect synaptic UPS activity, we examined whether there is a correlation in the accumulation of htt in neuronal processes (or neuropil aggregates) with decreased synaptic UPS activity. Similar to what we observed previously (Li et al., 1999), EM48 immunostaining of the striatum and cortex of R6/2 mice at 4 and 10 wk of age revealed a dramatic increase in neuropil aggregates over time (Fig. 6 A).Although the fluorescent UPS reporters allowed us to examine UPS activity in synapses, these reporters indirectly measure UPS activity, and their sensitivity to detect UPS activity in the cell body is limited by diffuse and intense fluorescent signals. To compare proteasomal activity in different subcellular regions, we isolated synaptosomal fractions and measured their chymotrypsin-like activity using the small fluorogenic peptide Suc-LLVY-AMC as a substrate. Synaptosomal fractions isolated from mouse brains were enriched in synaptic proteins such as synaptophysin but not the cytoplasmic protein GAPDH or the nuclear protein TFIIB (Fig. 6 B, top). Proteasomal core subunits β1i (LMP2), β5i (LMP7), and other core subunits (a5, a7, b1, b5i, and b7) of 20S were also present in synaptosomes, substantiating the presence of the proteasome in synapses. Comparison of chymotrypsin activity in different fractions from wild-type mouse brains revealed that nuclear proteasomal activity is lower than cytosolic activity and that the synaptosomes have the lowest activity (Fig. 6 C). We then examined synaptosomes isolated from the cortex and striatum of R6/2 mice at 4 and 10 wk of age. EM48 immunostaining revealed oligomeric htt in the stacking gel on Western blots (Fig. 6 D). More oligomeric htt was found in synaptosomes isolated from older HD mice than younger HD mice. These findings are consistent with the age-dependent increase of neuropil aggregates in R6/2 mouse brains (Fig. 6 A).
Figure 6.
Accumulation of aggregated htt is correlated with the decrease in synaptic proteasome activity in R6/2 mouse brain. (A) EM48 immunostaining showed more neuropil aggregates in R6/2 mice at 10 wk than at 4 wk. Bars, 10 μm. (B) Mouse brain extracts were fractionated to isolate cytosolic, nuclear, and synaptosomal fractions. Synaptosomal fraction is enriched for the synaptic protein synaptophysin but not the cytoplasmic protein GAPDH or the nuclear transcription factor TFIIB. Antibodies to proteasomal subunits LMP2, LMP7, and core subunits (a5, a7, b1, b5i, and b7) of 20S also detected the presence of these subunits in synaptosomes. (C) The chymotrypsin-like activity (mean + SEM, n = 4–5) of different fractions. (D) EM48 Western blotting showed more aggregated htt in synaptosomes (S) from younger mice at 4 wk of age. T: total lysates, C: cytosolic fraction, N: nuclear fraction. (E) The chymotrypsin-like activity (mean +SEM, n = 5–7) of total lysates (left) and synaptosomes (right) isolated from the striatum of R6/2 mice at 4 and 10 wk of age. Synaptosomal chymotrypsin-like activity in the striatum was lower than from the cortex, and synaptosomes isolated from HD brains also showed lower proteasomal activity than those from wild-type mouse brains. *, P < 0.05 as compared with wild type.
Consistent with a previous study that did not find any decrease in proteasomal activity in whole brain lysates of R6/2 mice (Bett et al., 2006), examination of the chymotrypsin-like activity of total cell lysates from the cortex and striatum did not reveal a significant difference between wild-type and R6/2 mouse brain samples (Fig. 6 E, left). We therefore focused on the proteasomal activity in isolated synaptosomes by measuring their chymotrypsin-like activity. These assays revealed that proteasomal activity was decreased in synaptosomes isolated from the cortex or striatum of HD mice compared with samples from wild-type mice (Fig. 6 E, right). Striatal synaptosomes showed lower proteasomal activity than cortical synaptosomes in both wild-type and HD mice. Thus, there is an intrinsic difference in synaptic proteasome activity between the striatum and cortex. Also, an age-dependent decrease in synaptic proteasomal activity occurs in the brain.
Decreased UPS activity in the synapses of HD knock-in mice
The above findings suggest that N-terminal mutant htt may impair UPS activity in the synapses of HD mice. It is important to investigate whether this defect occurs in HD knock-in mice in which full-length mutant htt is expressed under the control of the endogenous mouse htt promoter and is cleaved to small N-terminal htt fragments. In these mice, N-terminal mutant htt fragments preferentially form aggregates or inclusions in the striatum (Lin et al., 2001). Because of the weak staining of mouse mutant htt by EM48, we used 1C2 immunostaining to reveal neuropil aggregates in HD knock-in mice (Fig. 7 A). Neuropil aggregates are less abundant than nuclear aggregates in HD knock-in mice, suggesting that the accumulation of cleaved N-terminal htt fragments in distal neuronal processes requires more time, perhaps because transport of mutant htt to nerve terminals is necessary for this accumulation. However, there is a significant increase of neuropil aggregates from 4 to 12 mo (Fig. 7 A), allowing us to examine whether there is a correlated change in synaptic UPS activity. After injection of adenoviral UPS reporters into the striatum of HD knock-in mice aged 4, 8, and 12 mo, we observed a relative increase in presynaptic and postsynaptic GFPu signals in older mice (Fig. 7, B and C). Quantification of the ratio of GFPu/RFP in the synapses of HD mice at different ages also showed a time-dependent increase of this ratio, which likely reflects a decrease in synaptic UPS activity (Fig. 7 D). To confirm this increase, we used a biochemical assay to examine the chymotrypsin activity of isolated synaptosomes in HD and age-matched wild-type mice. Examining proteasomal activity in whole cell extracts did not reveal significant differences between wild-type and HD mice, except for decreased activity in the striatum of HD mice at 12 mo of age (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200709080/DC1). Although the biochemical assay for proteasomal activity is less sensitive than the fluorescent reporter at detecting changes in the proteasomal activity in subcellular regions, this assay indicated that cortical synaptosomes from HD mice at 12 mo of age had decreased proteasomal activity compared with samples from control littermates. However, the difference in cortical proteasomal activity is not statistically significant. Importantly, a statistically significant decrease in proteasomal activity was seen in synaptosomes from striatal neurons of HD knock-in mice and wild-type littermates at 12 mo of age (Fig. 7 E). A decrease in postglutamyl and trypsin-like activity was also detected in synaptosomes from the striatum of HD knock-in mice at 12 mo of age (Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200709080/DC1). Thus, synaptic proteasomal activity is inversely correlated with the age-dependent accumulation of N-terminal mutant htt fragments in striatal neurons (Lin et al., 2001).
Figure 7.
Neuropil aggregates and synaptic UPS activity in HD knock-in mouse brain. (A) Immunostaining of heterozygous HD knock-in mice with the 1C2 antibody showing an increase in neuropil aggregates in the striatum with age. Bars, 10 μm. (B and C) Expression of presynaptic (SNAP25-GFPu/-RFP) (B) and postsynaptic (PSD95-GFP/-RFP) (C) reporters in the striatum of HD knock-in mice at 12 mo of age 7 d after injection of adenoviral UPS reporters. Note the increase in GFPu signals over time. Bars, 10 μm. (D) The ratios of GFPu to RFP in synapses (30–48 each group) of HD knock-in mice at 4, 8, and 12 mo of age were quantified. **, P < 0.01. (E) Chymotrypsin-like proteasomal activity of synaptosomes isolated from the cortex and striatum of wild-type or heterozygous HD knock-in mice at 4, 8, and 12 mo of age. The data were obtained from five experiments and are represented as mean + SEM (n = 48). *, P < 0.05 as compared with wild-type samples. The chymotrypsin-like activity of brain homogenates and post-glutamyl and trypsin-like activity of synaptosomes are presented in Fig. S1 and Fig. 2.
Decreased synaptic ATP in HD knock-in mice
The activity of the UPS relies on adenosine triphosphate (ATP) generated by mitochondria. As the transport of mitochondria in neuronal processes is impaired by mutant htt (Trushina et al., 2004; Chang et al., 2006), we explored the possibility that the defect in UPS activity seen in HD synapses is associated with a lower level of ATP in synapses. Electron microscopic examination revealed the presence of organelles with disrupted structures (Fig. 8 A, single arrows), which are likely to be degenerated mitochondria, in some axonal terminals containing htt aggregates (Fig. 8 A, double arrows). This finding suggests that functional mitochondria could be reduced in the synapses of HD knock-in mice. To better quantify the amount of mitochondria in the synapses of HD mice, we isolated synaptosomes and measured the level of the mitochondrial protein succinate dehydrogenase (SDH) by Western blotting. Compared with synaptosomes from wild-type mice at the same age (14 mo), there was a decrease in the SDH level in HD knock-in synaptosomes (Fig. 8 B); however, the SDH levels in total cell lysates from wild-type and HD samples were no different. The decreased SDH level in HD synaptosomes was also verified by the ratio of SDH to the synaptic protein syntaxin (Fig. 8 C). Furthermore, by measuring the ATP content of striatal synaptosomes from wild-type and HD knock-in mice at the age of 14 mo, we also observed a decrease in ATP levels in the HD samples (Fig. 8 D). ATP levels in total cell lysates were ∼7-8–fold higher than in synaptosomes and were not significantly different between wild-type and HD samples, suggesting that mutant htt is more likely to affect ATP levels in synapses than in the cell body. Thus, a decrease in the amount of mitochondria and ATP in the synapses of HD mice may contribute to the reduced activity of synaptic UPS in HD mice.
Figure 8.
Decreased synaptic ATP level in HD knock-in mouse brain. (A) Electron micrograph of EM48 immunogold labeling of the striatum of a 12-mo-old HD 150Q knock-in mouse brain. Arrows indicate degenerating mitochondria-like organelles. Double arrows indicate htt aggregates that were labeled by EM48 immunogold particles. Bar, 250 nm. (B) Western blotting of total cell lysates (T), cytoplasmic (C), synaptosomal (S), and nuclear (N) fractions isolated from the brain of wild-type (WT) and HD 150Q knock-in (KI) mice at 14 mo of age. The blots were probed with 1C2 for htt and antibodies to the mitochondrial protein SDH, the cytoplasmic protein GAPDH, the synaptic protein syntaxin, and the nuclear protein TBP. Arrow indicates full-length mutant htt. (C) Densitometry analysis of the ratio (mean + SEM, n = 3) of SDH to GAPDH or syntaxin. (D) ATP content (raw luminescence [A.U.]) in total cell lysates and synaptosomes isolated from the striatum of wild-type and HD KI mice at 12–14 mo of age. The data were presented as mean + SEM (n = 5–6). *, P < 0.05 as compared with wild type.
Discussion
We generated adenoviral fluorescent reporters to measure synaptic UPS activity in vitro and in vivo and observed a significant decrease in synaptic UPS activity in HD mice. These findings offer important insight into the synaptic dysfunction and selective neuropathology seen in HD. Furthermore, these new reporters can serve as a tool for measuring the function of the synaptic UPS and its regulation under normal physiological or pathological conditions.
Cell models and in vitro studies have shown that polyQ-containing proteins can impair the activity of the UPS (Bence et al., 2001; Verhoef et al., 2002; Venkatraman et al., 2004; Bennett et al., 2005). These studies have provided valuable information regarding the mechanisms by which polyQ proteins affect UPS function. Early studies suggested that protein aggregation directly impairs the function of the UPS (Bence et al., 2001). It was also shown that the proteasome cannot digest and release polyQ-expanded peptides, which can impair the function of the proteasome (Venkatraman et al., 2004). Recent studies demonstrate that misfolded polyQ proteins or filamentous mutant htt can directly affect the function of proteasomes before the formation of inclusions (Bennett et al., 2005; Díaz-Hernández et al., 2006). Although these studies demonstrate that mutant htt can directly impair proteasomal function, they were performed under in vitro conditions in which the levels of mutant proteins might exceed the endogenous level of mutant protein in the HD brain. Thus, an important issue is whether UPS impairment also occurs in vivo.
Seo et al. (2004) examined postmortem brain tissues of HD patients and observed a decrease in proteasomal activity in the striatum and cortex. However, this decrease could be due to atrophy or degeneration already present in the HD patient brains. A transgenic approach was used to express a fluorescent UPS reporter (UbG76V-GFP) in mice (Lindsten et al., 2003). With this UPS reporter, mouse model was shown that prion pathology causes impairment of the UPS in the brain (Kristiansen et al., 2007). This transgenic mouse model led to the discovery that mutant polyQ proteins do not impair UPS activity in the retina of transgenic SCA7 mice (Bowman et al., 2005). In addition, biochemical assays on brain homogenates from HD mice that express exon1 mutant htt did not reveal a reduction in proteasomal activity or the levels of proteasomal subunits LMP2 and LMP7 in HD mice (Díaz-Hernández et al., 2003; Bett et al., 2006). Instead, increased chymotrypsin-like activity was seen in whole brain homogenates of 13-wk-old R6/2 mice (Bett et al., 2006) and in conditional HD (HD94) mice (Díaz-Hernández et al., 2003), which could be the indirect consequence of htt toxicity or cell stress. Nevertheless, measurement of polyubiquitin chains revealed global changes in polyubiquitination in HD mouse brains, suggesting the presence of impaired UPS activity in the HD brain (Bennett et al., 2007). It is possible that the effect of polyQ proteins on UPS activity is dependent on its accumulation and subcellular localization, which cannot be detected by examining whole cell homogenates. Because UPS activity varies in different types of cells and in subcellular regions, it is important to evaluate UPS activity in different subcellular compartments of neurons, especially in synapses in which mutant htt accumulates and affects neurotransmitter release or receptor function in the HD brain (Usdin et al., 1999; Cepeda et al., 2001; Zeron et al., 2002; Smith et al., 2005; Cummings et al., 2006; Fan et al., 2007).
Using adenoviral vectors that express synaptic fusion proteins containing fluorescent UPS reporters under the control of a neuronal promoter, we were able to target these reporters to presynaptic or postsynaptic terminals. Targeting of these reporters to defined synaptic structures allows for sensitive detection of changes in UPS activity at synapses. It is possible that GFPu can aggregate and that this aggregation renders resistance to proteasomal degradation of GFPu. In cultured primary neurons, however, proteasomal inhibitors caused a more dramatic increase in the ratio of GFPu to RFP in synapses than in the cell body (Figs. 2 and Figs.3). The selective localization of these reporters in small synaptic structures allows for more reliable quantification of fluorescent signals than quantification of diffuse signals in the cell body. Accordingly, these reporters allowed us to detect for the first time synaptic UPS activity in the brain and also revealed decreased UPS activity in the synapses of HD mouse brains. This decrease was confirmed by biochemical assays of proteasomal activity in isolated synaptosomes. Thus, the reporters we generated are effective in detecting synaptic UPS activity. Because these reporters can differentiate presynaptic and postsynaptic UPS activity, they will be valuable for the investigation of how presynaptic or postsynaptic UPS function is regulated under physiological or pathological conditions.
Expression of the fluorescence UPS reporters in the defined synaptic structure allowed us to uncover defective UPS activity in HD synapses. In addition to the direct effect of mutant htt on the UPS (Venkatraman et al., 2004; Bennett et al., 2005), additional mechanisms may account for the effect of mutant htt on synaptic UPS function. The activity of the UPS is critically dependent on mitochondrial ATP, and synaptic ATP levels are determined by the transport of mitochondria from the cell body to nerve terminals. Recent studies have demonstrated that mutant htt can impair the trafficking of mitochondria in neuronal processes (Trushina et al., 2004; Chang et al., 2006). Consistent with our earlier findings that degenerated mitochondria were present in axonal terminals of R6/2 (Yu et al., 2003) and HD 82Q KI (Li et al., 2001) mice, degenerating mitochondria-like organelles were also seen in the synapses of HD 150Q KI mice. Furthermore, we detected decreases in the levels of the mitochondrial protein SDH and ATP in synaptosomes from HD 150Q KI mouse brain. Thus, the reduced ATP levels in synapses can lead to a defect in synaptic UPS function. In addition, the accumulation of mutant htt in synapses can promote its effect on synaptic mitochondria via other mechanisms. For example, mutant htt was found to reduce ATP levels by activating caspases (Sánchez et al., 2003), enhancing Ca2+ influx through NMDA receptors (Seong et al., 2005), and reducing mitochondrial respiratory function (Milakovic and Johnson, 2005). All these effects could contribute to the preferential impairment of synaptic UPS function in HD mouse brains.
Because UPS activity is important in regulating the normal function and plasticity of synapses (Yi and Ehlers, 2005; Patrick 2006), decreased UPS activity may contribute to the synaptic dysfunction seen in HD mice. Previous studies have revealed the abnormal release of various neurotransmitters from synapses (Usdin et al., 1999; Cepeda et al., 2001; Zeron et al., 2002; Cummings et al., 2006) and other defects, such as trafficking of synaptic membrane receptors (Fan et al., 2007), in HD mice, suggesting that mutant htt may have broad adverse effects on various aspects of synaptic function. Further investigation of the relationship between postsynaptic or presynaptic UPS activity and synaptic htt should provide deeper insight into the mechanism for synaptic dysfunction in HD.
Another finding that supports the synaptic toxicity of mutant htt is the correlation between decreased synaptic UPS activity and the accumulation of mutant htt in synapses. Although this correlation suggests that mutant htt can impact UPS activity, additional factors might also influence UPS activity. In wild-type mice, synaptic UPS function decreases with age, suggesting that this age-dependent decrease in UPS activity can at least contribute to the age-dependent increase in synaptic htt aggregates. Interestingly, we have shown that proteasomal chymotrypsin-like activity in synapses is lower than in the cell bodies of mouse brains. Our previous studies also show that proteasome activity is lower in the nucleus than in the cytosolic fraction (Zhou et al., 2003). Thus, the intrinsically lower activity of the proteasome in both the nucleus and synapses favors the preferential accumulation of mutant htt in these two subcellular regions. The preferential accumulation of mutant htt in synapses is probably also determined by other cellular factors, such as transport of htt to nerve terminals. As mutant htt also accumulates in the nucleus and affects gene expression, effects from both nuclear and cytoplasmic mutant htt can cause HD pathology. Because the synapses are a unique structure of neurons, the accumulation of mutant htt in this important site may be particularly relevant to the selective neuropathology seen in HD.
Our findings also have implications for other neurological disorders. In Alzheimer's and Parkinson's diseases, synaptic dysfunction has been well documented (Klyubin et al., 2005). Parkinson's disease is also characterized by the presence of Lewy bodies that are formed by α-synuclein, a protein that is abundant in synapses. UPS impairment is thought to be involved in the pathogenesis of these diseases (Moore et al., 2003; Bossy-Wetzel et al., 2004; Ross and Pickart, 2004), yet whether misfolded proteins can affect synaptic UPS function in these diseases remains to be investigated. Development of the synaptic UPS reporters in our study will help the investigation of synaptic function in these diseases, as well as other neurological disorders that may also involve synaptic dysfunction or UPS impairment.
Materials and methods
Antibodies and reagents
The rabbit polyclonal (EM48) and mouse monoclonal (mEM48) antibodies to htt were generated by using the first 256 amino acids of human htt as described previously (Gutekunst et al., 1999; Zhou et al., 2003). 1C2, a mouse antibody to expanded polyQ tract, was obtained from Chemicon International. Other antibodies used for the following proteins were purchased from commercial sources: GFP (Av monoclonal antibody 632380), DsRed (RFP) (Clontech Laboratories, Inc.), 20S proteasome subunit β1i (LMP2) and β5i (LMP7) (BIOMOL International, L.P.); γ-tubulin and synaptophysin (Sigma-Aldrich); GAPDH, neuronal-specific nuclear protein (NeuN), and GFAP (Chemicon International); TFIIB (Santa Cruz Biotechnology, Inc.); SNAP25 (Transduction Laboratories), PSD95 (Millipore), SDH (Invitrogen), Syntaxin (Sigma-Aldrich), TBP (Santa Cruz Biotechnology, Inc.), and rabbit anti-20S Proteasome Core Subunits (a5, a7, b1, b5i, and b7) (EMD). Hoechst 33258 was obtained from Invitrogen.
HD mice
R6/2 mice [B6CBA-TgN (HDexon1) 62], which express the first exon of human Htt (67 amino acids) with an additional 150-glutamine repeat (Davies et al., 1997), were obtained from The Jackson Laboratory. HD 150CAG repeat knock-in mice (HdhCAG150) expressing full-length mouse htt with an expanded polyglutamine repeat (150Q) (Lin et al., 2001) were bred and maintained in the animal facility at Emory University as described previously (Yu et al., 2003). Genotyping of transgenic mice was performed using methods described previously (Yu et al., 2003). Heterozygous HD 150CAG knock-in mice were used in the study.
Primary neuronal culture
Striatal and cortical neurons were isolated from E18 rat brains. Dissected tissues were treated with 0.0625 mg/ml trypsin in 1xHBSS buffer without calcium or magnesium for 10 min at 37°C followed by triturating with a 1 ml pipette tip 20 times. Cells were then washed once with the tissue culture medium and spun down at 500 g for 3 min. Cells were plated on the glass coverslips that had been precoated with 0.1 mg/ml poly-d-lysine and 1 μg/ml laminin, and grown initially in neurobasal/B27 medium following the method used in our previous study (Li et al., 2000). To reduce glial proliferation, 5 μM cytosine arabinoside was added 12–16 h after plating.
Adenoviral vector construction and preparation
A CL-1 degron sequence (Bence et al., 2001) was added to the C terminus of GFP in the PRK vector to generate PRK-GFPu construct. cDNA sequences for DsRed (BD Biosciences) were used to replace GFPu to generate PRK-RFP construct. RT-PCR using RNAs from mouse brains was performed to isolate full-length cDNAs for mouse SNAP25 and PSD95. SNAP25 cDNA was isolated with a sense primer 5′-acatcgatATGGCCGAAGACGCAGACATG-3′ and an antisense primer 5′-acacgcgtACCACTTCCCAGCATCTTTG-3′. Full length of PSD95 cDNA was isolated using the sense primer 5′-acatcgatCCAACATGGACTGTCTCTGTATAG-3′ and the antisense primer 5′-atacgcgtGAGTCTCTCTCGGGCTGGGAC-3′. The PCR products were subcloned into the PRK-GFPu and PRK-RFP vectors to express fusion proteins that contain GFPu or RFP at the C terminus of SNAP25 or PSD95. The DNA fragments encoding these fusion proteins were inserted into the shuttle vector of the AdEasy vector system (Qbiogene). The CMV promoter was replaced by human synapsin-1 promoter provided by Dr. S. Kugler (University of Goettingen, Goettingen, Germany). Adenovirus amplification and purification were performed according to the method used in our previous study (Shin et al., 2005). Viral titer was determined by measuring the number of infected HEK293 cells expressing GFPu or RFP. All viral stocks were adjusted to 109 VP/ml.
Stereotaxic injection
HD mice described above and their wild-type littermates were used for this study. All experiments were performed in accordance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals and the Emory University Institutional Animal Care and Use Committee. In brief, mice were anesthetized with intraperitoneal injections of a combination of ketamine (90 mg/kg) and xylazine (13 mg/kg) and positioned in a stereotaxic apparatus. A small incision was made in the scalp, and the striatum was marked using the following stereotaxic coordinates, relative to Bregma: anterior–posterior +0.8 mm, medial–lateral −1.6 mm, and dorsal–ventral −3.3 mm. A small hole was drilled into the skull, and a 26-gauge needle attached to a 5-μl Hamilton syringe was lowered into the striatum according to the dorsal–ventral coordinate. A nanoinjector pump (World Precision Instrument) controlled the infusion of 2.0 μl of adenovirus at a rate of 0.4 μl/min, after which the needle was left in place for 5 min to ensure complete diffusion of the viruses. The mice were examined by immunocytochemistry 7–8 d after injection.
In situ hybridization
The EGFP antisense oligonucleotide probe (5′-GGTGTCGCCCTCGAACTTCACCTCGGCGCGGGTCTTGTAGTTGCC-3′) and the control scrambled EGFP probe (5′-CGATGTTCTTGTGGCGGCTCGGCTTCCACCTCAAGCTGCCGTGG-3′) were 3′ end-labeled with digoxigenin-dUTP and dATP using the DIG-Tailing kit (Roche). The tailed probes were purified by ethanol precipitation and detected by dot blot using a DIG Nucleic Acid Detection kit (Roche). Frozen brain tissues were cut and mounted onto microscope slides and fixed with cold (4°C) 4% paraformaldehyde in 0.1 M PB solution. The sections were washed in 1× PBS three times for 15 min, dipped quickly in water, dehydrated, and then air dried at least 2 h. Hybridization buffer (50 ml of 4× SSC, 20% dextran sulfate, 0.5× Denhardts, 50% deionized formamide, 0.1 M DTT, 0.25 mg/ml poly A, 0.25 mg/ml tRNA, and 0.25mg/ml ssDNA) containing 500 ng/ml DIG-tailed oligo probe was applied to each section for incubation overnight at 39°C. The sections were sequentially washed in 1 and 0.5× SSC/10 mM DTT at 55°C, buffer (0.1 M maleic acid, 0.15 M NaCl, pH 7.5). Anti-DIG-AP (1:300 dilution; Roche) and NBT/BCIP substrate were used to detect hybridization signals. Negative controls included the scrambled EGFP probe, hybridization buffer only, and RNase A treatment of the sections before hybridization.
Fractionation
Subcellular fractions of mouse brain tissues were prepared using previously described methods (Phillips et al., 2001). Cortical or striatal tissues from R6/2 or KI mice and control littermates were homogenized in 500 μl homogenization solution (0.32 M sucrose, 15 mM Tris-HCl pH 8.0, 60 mM KCl, 15 mM NaCl, 5 mM EDTA, 1 mM EGTA, and 2 mM ATP) at 4°C in an Eppendorf tube. The homogenate was centrifuged at 500 g for 2 min to remove unbroken tissue clumps and cells. The supernatant (S1) was centrifuged at 1,300 g for 10 min to precipitate nuclei fraction (P1). The (S1) supernatant was transferred to another microfuge tube and centrifuged at 10,000 g for 10 min to obtain a mitochondria- and synaptosome-enriched pellet (P2) and the supernatant (S2). The (P2) pellet was resuspended in 500 μl of 0.32 M sucrose and layered onto 750 μl of 0.8 M sucrose in a microfuge tube. The samples were centrifuged at 9,100 g for 15 min using a swinging bucket rotor. The 0.8-M sucrose layer and most of the loose pellet containing the synaptosomes were collected and separated from the mitochondrial pellet. The (S2) supernatant was centrifuged at 13,000 g for 25 min to yield the supernatant (S3) or cytosolic fraction.
Proteasome activity assay
For determining proteasome activity, clear whole-cell extracts or cell fractions were adjusted to 0.5 mg/ml total protein by dilution with homogenization buffer. All assays were done in triplicate. Chymotrypsin-like activity of 20S β5 was determined using the substrate Suc-LLVY-aminomethylcoumarin (AMC) (40 μM; Bilmol), trypsin-like activity of 20S β2 was determined using the substrate Boc-LRR-AMC (Bilmol; 100 μM), and postglutamyl activity of 20S β1 was determined using the substrate Z-LLE-AMC (400 μM; Bilmol). Equal amounts (10 μg) of the extracts were incubated with corresponding substrates in 100 μl proteasome activity assay buffer (0.05 M Tris-HCl, pH 8.0, 0.5 mM EDTA, 1 mM ATP, and 1 mM DTT) for 30–60 min at 37°C. The reactions were stopped by adding 0.8 ml of cold water and placing the reaction mixtures on ice for at least 10 min. The free AMC fluorescence was quantified by using the CytoFluor multi-well plate reader (FLUOstar; BMG Labtech) with excitation and emission wavelengths at 380 and 460 nm, respectively. All readings were standardized using the fluorescence intensity of an equal volume of free 7-amino-4-methyl-coumarin (AMC) solution (40 mM), normalized by the protein concentrations and expressed as nmol/min/mg protein.
ATP content assays
ATP levels were measured using a method described previously with minor modifications (Milakovic and Johnson, 2005) and a FLUOstar Galaxy microplate reader (BMG Labtech). Total cell lysates and synaptosomal fractions, which were isolated form striata of wild-type and HD mouse brains and stored at −80°C until analysis for ATP content. Total (25 μg) and synaptosomal (50 μg) samples were diluted to 50 μl with isolation buffer, added to 50 μl of Luciferase reaction buffer, and luminescence measured immediately. Each sample was analyzed in triplicate. Data are presented as raw luminescence (A.U.) + SEM.
Microscopy
All imaging was done at room temperature, ∼26°C. EM48 immunocytochemical analysis of cultured cells and mouse brain tissues was performed as described previously (Li et al., 2000, Shin et al., 2005). Light micrographs were taken using a microscope (Axiovert 200 MOT; Carl Zeiss, Inc.) equipped with a digital camera (Orca-100; Hamamatsu) and the image acquisition software OpenLAB (Improvision). A 20x (LD-Achroplan 0.4 NA) or 63x lens (0.75 NA) was used for light microscopy. Confocal imaging was performed using the 63x/1.4 NA oil immersion objective lens (Plan-apochromat) and an LSM 510 confocal microscope system (Carl Zeiss, Inc). For immunofluorescent staining, species-specific fluorescein- or Texas red–conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) were applied for 1 h at room temperature followed by counterstaining with Hoechst dye. Enhanced GFP was imaged using 488-nm excitation and a 500–530-nm band-pass filter, and RFP was imaged using 543-nm excitation and a 565–615-nm band-pass filter. The figures were created using Photoshop 7.0 software (Adobe) and, in some cases, where the brightness and contrast of the whole image needed adjustment, we used the brightness/contrast adjustment function.
Electron microscopic analysis of HD 150Q knock-in mouse brain tissue was performed with EM48 according to previously published procedures from our laboratory (Li et al., 2000). In brief, brain sections were incubated with EM48 in PBS containing 4% NGS for 24 h at 4°C and then with Fab fragments of goat anti–rabbit secondary antibodies (1:50) conjugated to 1.4-μm gold particles (Nanoprobes, Inc.) in PBS with 4% NGS overnight at 4°C. After rinsing in PBS, sections were fixed again in 2% glutaraldehyde in PB for 1 h, silver intensified using the IntenSEM kit (GE Healthcare; osmicated in 1% OsO4 in PB, and stained overnight in 2% aqueous uranyl acetate). Sections used for electron microscopy were dehydrated in ascending concentrations of ethanol and propylene oxide/Eponate 12 (1:1) and embedded in Eponate12 (Ted Pella). Ultrathin sections (60 nm) were cut using an ultramicrotome (Ultracut; Leica). Thin sections were counterstained with 5% aqueous uranyl acetate for 5 min followed by Reynolds lead citrate for 5 min and examined using an electron microscope (H-7500; Hitachi).
Quantitative fluorescence imaging analysis
Captured image files were loaded into the Image-Pro Plus (Media Cybernetics, Inc.) environment with the “count/size” function. Precise segmentation of bright spots was achieved by setting a lower threshold just above background emission, typically ∼23–25 grayscale units (gsu), and an upper threshold at the upper limit of the 8-bit grayscale (i.e., 255 gsu). Automated image analysis was performed with a custom-built algorithm. The algorithm performs a series of analyses on each image in both the red channel (for RFP) and the green channel (for GFPu) and then calculates quantitative output for fluorescence intensity. The algorithm first corrects background if necessary and attempts to offset background anomalies. Cell bodies were detected as objects around the nuclei, separated from the neurites by a watershed filter. The next step in the algorithm locates synapses. It starts by running a “Hi-Pass” filter across the image to enhance edges (replaces each pixel with a value that increases contrast with neighboring pixels) (Mitchell et al., 2007). Synapses (40–75/each group) and cell bodies (7–9/each group) are selected as any objects with a size and signal intensity greater than a user-defined minimum that is above a defined background. Image-Pro Plus generated a high-content array of measurements that included event number, area, sum of pixel intensities, and maximum and minimum pixel intensities. The sum of pixel intensities is selected as the parameter for statistical analysis.
Statistical analysis
Statistical significance (P < 0.05) was assessed using the t test whenever two groups were compared. When analyzing multiple groups, we used ANOVA with Scheffé's post hoc test to determine statistical significance. Data are presented as mean + SE. Calculations were performed with SigmaPlot 4.11 and Prism (version 4) software.
Online supplemental material
Fig. S1 shows that Inhibition of proteasomal activity increases the GFPu level in neuronal cell bodies. Fig. S2 shows colocalization of SNAP25-GFPu with neuropil htt aggregates in R6/2 mouse brain. Fig. S3 shows In situ hybridization analysis of the transcripts of SNAP25-GFPu in wild-type and R6/2 mouse brains. Fig. S4 shows chymotrypsin-like activities of the proteasome in whole cell extracts from the cortex or striatum of wild-type and HD knock-in mice. Fig. S5 shows post-glutamyl and trypsin-like activities of the synaptosomes isolated from the brain cortex or striatum of wild-type and HD knock-in mice. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200709080/DC1.
Acknowledgments
We thank G.Q. Sheng for helping with adenoviral purification, Z.H. Fang for immunocytochemistry and electron microscopy, S. Kugler for providing the human synapsin-1 promoter plasmid, and M. Friedman and C. Strauss for critical reading of the manuscript.
This work was supported by NIH grants AG019206 and NS036232 (to X.-J. Li), and grant NS045016 (to S.-H. Li).
Abbreviations used in this paper: HD, Huntington's disease; htt, huntingtin; polyQ, polyglutamine; PSD, postsynaptic density; RFP, DsRed fusion protein; SDH, succinate dehydrogenase; UPS, ubiquitin–proteasome system.
References
- Avraham, E., R. Szargel, A. Eyal, R. Rott, and S. Engelender. 2005. Glycogen synthase kinase 3beta modulates synphilin-1 ubiquitylation and cellular inclusion formation by SIAH: implications for proteasomal function and Lewy body formation. J. Biol. Chem. 280:42877–42886. [DOI] [PubMed] [Google Scholar]
- Bence, N.F., R.M. Sampat, and R.R. Kopito. 2001. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 292:1552–1555. [DOI] [PubMed] [Google Scholar]
- Bennett, E.J., N.F. Bence, R. Jayakumar, and R.R. Kopito. 2005. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol. Cell. 17:351–365. [DOI] [PubMed] [Google Scholar]
- Bennett, E.J., T.A. Shaler, B. Woodman, K.Y. Ryu, T.S. Zaitseva, C.H. Becker, G.P. Bates, H. Schulman, and R.R. Kopito. 2007. Global changes to the ubiquitin system in Huntington's disease. Nature. 448:704–708. [DOI] [PubMed] [Google Scholar]
- Bett, J.S., G.M. Goellner, B. Woodman, G. Pratt, M. Rechsteiner, and G.P. Bates. 2006. Proteasome impairment does not contribute to pathogenesis in R6/2 Huntington's disease mice: exclusion of proteasome activator REGgamma as a therapeutic target. Hum. Mol. Genet. 15:33–44. [DOI] [PubMed] [Google Scholar]
- Bossy-Wetzel, E., R. Schwarzenbacher, and S.A. Lipton. 2004. Molecular pathways to neurodegeneration. Nat. Med. 10(Suppl):S2–S9. [DOI] [PubMed] [Google Scholar]
- Bowman, A.B., S.Y. Yoo, N.P. Dantuma, and H.Y. Zoghbi. 2005. Neuronal dysfunction in a polyglutamine disease model occurs in the absence of ubiquitin-proteasome system impairment and inversely correlates with the degree of nuclear inclusion formation. Hum. Mol. Genet. 14:679–691. [DOI] [PubMed] [Google Scholar]
- Brooks, P., G. Fuertes, R.Z. Murray, S. Bose, E. Knecht, M.C. Rechsteiner, K.B. Hendil, K. Tanaka, J. Dyson, and J. Rivett. 2000. Subcellular localization of proteasomes and their regulatory complexes in mammalian cells. Biochem. J. 346:155–161. [PMC free article] [PubMed] [Google Scholar]
- Cepeda, C., M.A. Ariano, C.R. Calvert, J. Flores-Hernandez, S.H. Chandler, B.R. Leavitt, M.R. Hayden, and M.S. Levine. 2001. NMDA receptor function in mouse models of Huntington disease. J. Neurosci. Res. 66:525–539. [DOI] [PubMed] [Google Scholar]
- Chang, D.T., G.L. Rintoul, S. Pandipati, and I.J. Reynolds. 2006. Mutant huntingtin aggregates impair mitochondrial movement and trafficking in cortical neurons. Neurobiol. Dis. 22:388–400. [DOI] [PubMed] [Google Scholar]
- Chen, H., S. Polo, P.P. Di Fiore, and P.V. De Camilli. 2003. Rapid Ca2+-dependent decrease of protein ubiquitination at synapses. Proc. Natl. Acad. Sci. USA. 100:14908–14913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colledge, M., E.M. Snyder, R.A. Crozier, J.A. Soderling, Y. Jin, L.K. Langeberg, H. Lu, M.F. Bear, and J.D. Scott. 2003. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron. 40:595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings, D.M., A.J. Milnerwood, G.M. Dallerac, V. Waights, J.Y. Brown, S.C. Vatsavayai, M.C. Hirst, and K.P. Murphy. 2006. Aberrant cortical synaptic plasticity and dopaminergic dysfunction in a mouse model of Huntington's disease. Hum. Mol. Genet. 15:2856–2868. [DOI] [PubMed] [Google Scholar]
- Davies, S.W., M. Turmaine, B.A. Cozens, M. DiFiglia, A.H. Sharp, C.A. Ross, E. Scherzinger, E.E. Wanker, L. Mangiarini, and G.P. Bates. 1997. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 90:537–548. [DOI] [PubMed] [Google Scholar]
- Demartino, G.N., and T.G. Gillette. 2007. Proteasomes: machines for all reasons. Cell. 129:659–662. [DOI] [PubMed] [Google Scholar]
- Díaz-Hernández, M., F. Hernández, E. Martín-Aparicio, P. Gómez-Ramos, M.A. Morán, J.G. Castaño, I. Ferrer, J. Avila, and J.J. Lucas. 2003. Neuronal induction of the immunoproteasome in Huntington's disease. J. Neurosci. 23:11653–11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Hernández, M., A.G. Valera, M.A. Morán, P. Gómez-Ramos, B. Alvarez-Castelao, J.G. Castaño, F. Hernández, and J.J. Lucas. 2006. Inhibition of 26S proteasome activity by huntingtin filaments but not inclusion bodies isolated from mouse and human brain. J. Neurochem. 98:1585–1596. [DOI] [PubMed] [Google Scholar]
- DiFiglia, M., E. Sapp, K.O. Chase, S.W. Davies, G.P. Bates, J.P. Vonsattel, and N. Aronin. 1997. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 277:1990–1993. [DOI] [PubMed] [Google Scholar]
- Dong, X., J. Liu, H. Zheng, J.W. Glasford, W. Huang, Q.H. Chen, N.R. Harden, F. Li, A.M. Gerdes, and X. Wang. 2004. In situ dynamically monitoring the proteolytic function of the ubiquitin-proteasome system in cultured cardiac myocytes. Am. J. Physiol. Heart Circ. Physiol. 287:H1417–H1425. [DOI] [PubMed] [Google Scholar]
- Ehlers, M.D. 2003. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat. Neurosci. 6:231–242. [DOI] [PubMed] [Google Scholar]
- Fan, M.M., H.B. Fernandes, L.Y. Zhang, M.R. Hayden, and L.A. Raymond. 2007. Altered NMDA receptor trafficking in a yeast artificial chromosome transgenic mouse model of Huntington's disease. J. Neurosci. 27:3768–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman, M.H., and D. Raveh. 2005. Proteasome plasticity. FEBS Lett. 579:3214–3223. [DOI] [PubMed] [Google Scholar]
- Goldberg, A.L. 2003. Protein degradation and protection against misfolded or damaged proteins. Nature. 426:895–899. [DOI] [PubMed] [Google Scholar]
- Gusella, J.F., and M.E. Macdonald. 2006. Huntington's disease: seeing the pathogenic process through a genetic lens. Trends Biochem. Sci. 31:533–540. [DOI] [PubMed] [Google Scholar]
- Gutekunst, C.A., S.H. Li, H. Yi, J.S. Mulroy, S. Kuemmerle, R. Jones, D. Rye, R.J. Ferrante, S.M. Hersch, and X.J. Li. 1999. Nuclear and neuropil aggregates in Huntington's disease: relationship to neuropathology. J. Neurosci. 19:2522–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425–479. [DOI] [PubMed] [Google Scholar]
- Klyubin, I., D.M. Walsh, C.A. Lemere, W.K. Cullen, G.M. Shankar, V. Betts, E.T. Spooner, L. Jiang, R. Anwyl, D.J. Selkoe, and M.J. Rowan. 2005. Amyloid beta protein immunotherapy neutralizes Abeta oligomers that disrupt synaptic plasticity in vivo. Nat. Med. 11:556–561. [DOI] [PubMed] [Google Scholar]
- Kristiansen, M., P. Deriziotis, D.E. Dimcheff, G.S. Jackson, H. Ovaa, H. Naumann, A.R. Clarke, F.W. van Leeuwen, V. Menendez-Benito, N.P. Dantuma, et al. 2007. Disease-associated prion protein oligomers inhibit the 26S proteasome. Mol. Cell. 26:175–188. [DOI] [PubMed] [Google Scholar]
- Kopito, R.R. 2000. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10:524–530. [DOI] [PubMed] [Google Scholar]
- Korhonen, L., and D. Lindholm. 2004. The ubiquitin proteasome system in synaptic and axonal degeneration: a new twist to an old cycle. J. Cell Biol. 165:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecker, S.H., A.L. Goldberg, and W.E. Mitch. 2006. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J. Am. Soc. Nephrol. 17:1807–1819. [DOI] [PubMed] [Google Scholar]
- Li, H., S.H. Li, A.L. Cheng, L. Mangiarini, G.P. Bates, and X.J. Li. 1999. Ultrastructural localization and progressive formation of neuropil aggregates in Huntington's disease transgenic mice. Hum. Mol. Genet. 8:1227–1236. [DOI] [PubMed] [Google Scholar]
- Li, H., S.H. Li, H. Johnston, P.F. Shelbourne, and X.J. Li. 2000. Amino-terminal fragments of mutant huntingtin show selective accumulation in striatal neurons and synaptic toxicity. Nat. Genet. 25:385–389. [DOI] [PubMed] [Google Scholar]
- Li, H., S.H. Li, Z.X. Yu, P. Shelbourne, and X.J. Li. 2001. Huntingtin aggregate-associated axonal degeneration is an early pathological event in Huntington's disease mice. J. Neurosci. 21:8473–8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., and X.J. Li. 2006. Multiple pathways contribute to the pathogenesis of Huntington disease. Mol. Neurodegener. 1:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C.H., S. Tallaksen-Greene, W.M. Chien, J.A. Cearley, W.S. Jackson, A.B. Crouse, S. Ren, X.J. Li, R.L. Albin, and P.J. Detloff. 2001. Neurological abnormalities in a knock-in mouse model of Huntington's disease. Hum. Mol. Genet. 10:137–144. [DOI] [PubMed] [Google Scholar]
- Lindsten, K., V. Menendez-Benito, M.G. Masucci, and N.P. Dantuma. 2003. A transgenic mouse model of the ubiquitin/proteasome system. Nat. Biotechnol. 21:897–902. [DOI] [PubMed] [Google Scholar]
- Milakovic, T., and G.V. Johnson. 2005. Mitochondrial respiration and ATP production are significantly impaired instriatal cells expressing mutant huntingtin. J. Biol. Chem. 280:30773–30782. [DOI] [PubMed] [Google Scholar]
- Mitchell, P.J., J.C. Hanson, A.T. Quets-Nguyen, M. Bergeron, and R.C. Smith. 2007. A quantitative method for analysis of in vitro neurite outgrowth. J. Neurosci. Methods. 164:350–362. [DOI] [PubMed] [Google Scholar]
- Moore, D.J., V.L. Dawson, and T.M. Dawson. 2003. Role for the ubiquitin-proteasome system in Parkinson's disease and other neurodegenerative brain amyloidoses. Neuromolecular Med. 4:95–108. [DOI] [PubMed] [Google Scholar]
- Orr, H.T., and H.Y. Zoghbi. 2007. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 30:575–621. [DOI] [PubMed] [Google Scholar]
- Patrick, G.N. 2006. Synapse formation and plasticity: recent insights from the perspective of the ubiquitin proteasome system. Curr. Opin. Neurobiol. 16:90–94. [DOI] [PubMed] [Google Scholar]
- Phillips, G.R., J.K. Huang, Y. Wang, H. Tanaka, L. Shapiro, W. Zhang, W.S. Shan, K. Arndt, M. Frank, R.E. Gordon, et al. 2001. The presynaptic particle web: ultrastructure, composition, dissolution, and reconstitution. Neuron. 32:63–77. [DOI] [PubMed] [Google Scholar]
- Pickart, C.M., and R.E. Cohen. 2004. Proteasomes and their kin: proteases in the machine age. Nat. Rev. Mol. Cell Biol. 5:177–187. [DOI] [PubMed] [Google Scholar]
- Ross, C.A., and C.M. Pickart. 2004. The ubiquitin-proteasome pathway in Parkinson's disease and other neurodegenerative diseases. Trends Cell Biol. 14:703–711. [DOI] [PubMed] [Google Scholar]
- Sánchez, I., C. Mahlke, and J. Yuan. 2003. Pivotal role of oligomerization in expanded polyglutamine neurodegenerative disorders. Nature. 421:373–379. [DOI] [PubMed] [Google Scholar]
- Seo, H., K.C. Sonntag, and O. Isacson. 2004. Generalized brain and skin proteasome inhibition in Huntington's disease. Ann. Neurol. 56:319–328. [DOI] [PubMed] [Google Scholar]
- Seong, I.S., E. Ivanova, J.M. Lee, Y.S. Choo, E. Fossale, M. Anderson, J.F. Gusella, J.M. Laramie, R.H. Myers, M. Lesort, and M.E. MacDonald. 2005. HD CAG repeat implicates a dominant property of huntingtin in mitochondrial energy metabolism. Hum. Mol. Genet. 14:2871–2880. [DOI] [PubMed] [Google Scholar]
- Shin, J.Y., Z.H. Fang, Z.X. Yu, C.E. Wang, S.H. Li, and X.J. Li. 2005. Expression of mutant huntingtin in glial cells contributes to neuronal excitotoxicity. J. Cell Biol. 171:1001–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, R., P. Brundin, and J.Y. Li. 2005. Synaptic dysfunction in Huntington's disease: a new perspective. Cell. Mol. Life Sci. 62:1901–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speese, S.D., N. Trotta, C.K. Rodesch, B. Aravamudan, and K. Broadie. 2003. The ubiquitin proteasome system acutely regulates presynaptic protein turnover and synaptic efficacy. Curr. Biol. 13:899–910. [DOI] [PubMed] [Google Scholar]
- Trushina, E., R.B. Dyer, J.D. Badger II, D. Ure, L. Eide, D.D. Tran, B.T. Vrieze, V. Legendre-Guillemin, P.S. McPherson, B.S. Mandavilli, et al. 2004. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol. Cell. Biol. 24:8195–8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usdin, M.T., P.F. Shelbourne, R.M. Myers, and D.V. Madison. 1999. Impaired synaptic plasticity in mice carrying the Huntington's disease mutation. Hum. Mol. Genet. 8:839–846. [DOI] [PubMed] [Google Scholar]
- Venkatraman, P., R. Wetzel, M. Tanaka, N. Nukina, and A.L. Goldberg. 2004. Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol. Cell. 14:95–104. [DOI] [PubMed] [Google Scholar]
- Verhoef, L.G., K. Lindsten, M.G. Masucci, and N.P. Dantuma. 2002. Aggregate formation inhibits proteasomal degradation of polyglutamine proteins. Hum. Mol. Genet. 11:2689–2700. [DOI] [PubMed] [Google Scholar]
- Wilson, S.M., B. Bhattacharyya, R.A. Rachel, V. Coppola, L. Tessarollo, D.B. Householder, C.F. Fletcher, R.J. Miller, N.G. Copeland, and N.A. Jenkins. 2002. Synaptic defects in ataxia mice result from a mutation in Usp14, encoding a ubiquitin-specific protease. Nat. Genet. 32:420–425. [DOI] [PubMed] [Google Scholar]
- Wójcik, C., and G.N. DeMartino. 2003. Intracellular localization of proteasomes. Int. J. Biochem. Cell Biol. 35:579–589. [DOI] [PubMed] [Google Scholar]
- Yi, J.J., and M.D. Ehlers. 2005. Ubiquitin and protein turnover in synapse function. Neuron. 47:629–632. [DOI] [PubMed] [Google Scholar]
- Yu, Z.X., S.H. Li, J. Evans, A. Pillarisetti, H. Li, and X.J. Li. 2003. Mutant huntingtin causes context-dependent neurodegeneration in mice with Huntington's disease. J. Neurosci. 23:2193–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeron, M.M., O. Hansson, N. Chen, C.L. Wellington, B.R. Leavitt, P. Brundin, M.R. Hayden, and L.A. Raymond. 2002. Increased sensitivity to N-methyl-D-aspartate receptor-mediated excitotoxicity in a mouse model of Huntington's disease. Neuron. 33:849–860. [DOI] [PubMed] [Google Scholar]
- Zhou, H., F. Cao, Z. Wang, Z.X. Yu, H.P. Nguyen, J. Evans, S.H. Li, and X.J. Li. 2003. Huntingtin forms toxic NH2-terminal fragment complexes that are promoted by the age-dependent decrease in proteasome activity. J. Cell Biol. 163:109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]