Abstract
We examined the role of the actin nucleation promoters neural Wiskott-Aldrich syndrome protein (N-WASP) and WAVE2 in cell protrusion in response to epidermal growth factor (EGF), a key regulator in carcinoma cell invasion. We found that WAVE2 knockdown (KD) suppresses lamellipod formation and increases filopod formation, whereas N-WASP KD has no effect. However, simultaneous KD of both proteins results in the formation of large jagged protrusions with lamellar properties and increased filopod formation. This suggests that another actin nucleation activity is at work in carcinoma cells in response to EGF. A mammalian Diaphanous–related formin, mDia1, localizes at the jagged protrusions in double KD cells. Constitutively active mDia1 recapitulated the phenotype, whereas inhibition of mDia1 blocked the formation of these protrusions. Increased RhoA activity, which stimulates mDia1 nucleation, was observed in the N-WASP/WAVE2 KD cells and was shown to be required for the N-WASP/WAVE2 KD phenotype. These data show that coordinate regulation between the WASP family and mDia proteins controls the balance between lamellar and lamellipodial protrusion activity.
Introduction
The initial step of cancer cell migration is the formation of leading edge protrusions toward the source of tumor microenvironment-produced chemoattractants (Condeelis and Segall, 2003). These protrusions are driven by actin polymerization at the leading edge of the cell (Condeelis et al., 2005). Malignant tumor cells migrate toward EGF because of enhanced activation of signaling pathways that regulate actin polymerization resulting in chemotaxis (Wang et al., 2004). The dissection of the molecular and cellular underpinnings of these chemotactic responses is fundamental to our understanding of cancer biology and the process of metastasis.
The Wiskott-Aldrich syndrome protein (WASP) family plays essential roles in the regulation of actin polymerization (Takenawa, 2001). There are five WASP family members; WASP, neural WASP (N-WASP), WAVE1 (WASP family verprolin-homologous protein 1), WAVE2, and WAVE3. The WASP family proteins share a conserved C-terminal region called the verprolin homology, cofilin homology (or central), and acidic domain. The verprolin homology domain associates directly with actin, and the cofilin homology acidic region interacts with the actin-related protein (Arp) 2/3 complex, forming a nucleation site for actin polymerization (Miki et al., 1996, 1998b; Miki and Takenawa, 1998; Machesky et al., 1999; Rohatgi et al., 1999; Suetsugu et al., 1999). The WASP family proteins activate the Arp2/3 complex in response to signals that induce cell migration (Rohatgi et al., 1999; Miki et al., 2000; Fukuoka et al., 2001; Suetsugu et al., 2001). The Arp2/3 complex nucleates actin filaments and forms branched actin filament networks. This dendritic nucleation activity is caused by the actin filament side binding of the Arp2/3 complex itself (Mullins et al., 1997). The elongation of filaments in the dendritic network results in the pushing force necessary for some cell protrusions (Mogilner and Oster, 2003).
There are other actin nucleators which play an important role in cytoskeletal rearrangements. The mammalian Diaphanous-related formins (mDia) act as effectors for Rho family small GTPases (Wallar and Alberts, 2003; Higgs, 2005). Similar to N-WASP, mDia proteins are autoregulated actin filament assembly factors controlled by intramolecular interactions. mDia autoregulation is mediated through the binding of the Dia-inhibitory (Higgs, 2005) and Dia-autoregulatory (Alberts, 2001) domains that flank the formin homology 2 (FH2) domain conserved in all formins. Activated Rho proteins interact with a GTPase-binding domain located adjacent to the Dia-inhibitory domain and interfere with binding to the Dia-autoregulatory domain, effectively activating the FH2 domain, which is then free to nucleate, processively elongate, and (in some cases) bundle nonbranched actin filaments (Harris and Higgs, 2004; Higgs, 2005; Kovar, 2006).
In this paper, we investigated the role of N-WASP, WAVE1, WAVE2, and mDia formin proteins in the reorganization of the cytoskeleton in MTLn3 rat adenocarcinoma cells. We found that WAVE2 is the main regulator of lamellipod formation. Inhibiting WAVE2 decreases lamellipod formation and increases filopod formation, whereas N-WASP seems to have no function in these processes. Inhibition of both WAVE2 and N-WASP increased RhoA-GTP levels and resulted in the formation of mDia1-dependent jagged protrusions and filopods. Therefore, mDia1 is responsible for an underlying pathway, usually not seen directly during N-WASP and WAVE2 activity, which contributes to protrusion of the lamella and filopods during EGF stimulation.
Results
Expression of WASP family members in MTLn3 carcinoma cells
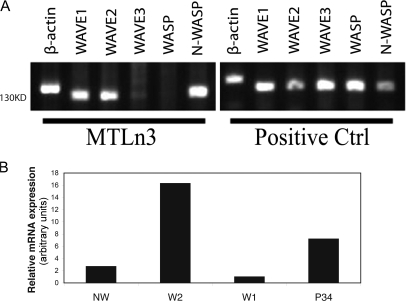
In MTLn3 cells, expression of the WASP family members was shown using three methods, RT-PCR, real-time PCR, and Western blotting. RT-PCR showed that N-WASP, WAVE1, and WAVE2 are the only three family members detected in these cells in culture (Fig. 1 A). Quantitative real-time PCR was used to observe the amounts of WASP family members' mRNA relative to each other in MTLn3 cells (Fig. 1 B). In comparison to WAVE1, WAVE2 mRNA is 16-fold and N-WASP is 3-fold higher. Through Western blot analysis we quantified the relative amounts of protein expression. Expression of WAVE1 was shown to be ∼40-fold lower than that of WAVE2 (Fig. S1 C, available at http://www.jcb.org/cgi/content/full/jcb.200708123/DC1). The low expression of WAVE1 protein and mRNA in comparison to WAVE2 and N-WASP suggests that WAVE1 may not be a major actin polymerization regulator in MTLn3 cells. Experiments knocking down WAVE1 using siRNA showed no change in lamellipod protrusion activity (Fig. S1 A). Therefore, WAVE1 has no detectable role in EGF-stimulated protrusive activity in MTLn3 cells in culture.
Figure 1.
Relative expression of WASP family members and p34 in carcinoma cells. (A) Expression of N-WASP, WAVE1, and WAVE2 was detected in MTLn3 cells by RT-PCR on mRNA isolated from MTLn3 cells and positive controls (mixed population of rat mRNA). (B) Relative abundance of WASP family member mRNA was measured by quantitative real-time PCR.
EGF stimulation of MTLn3 cells results in the recruitment of WAVE2
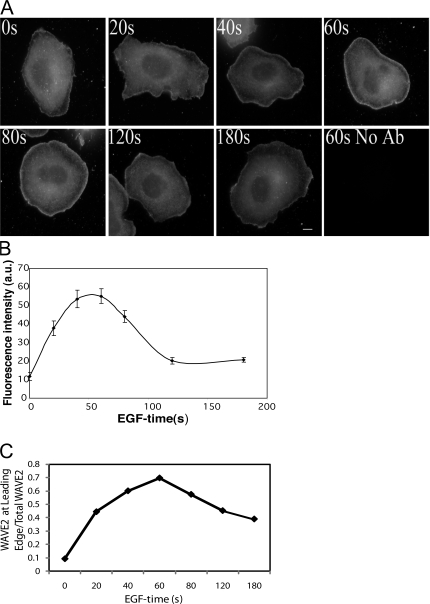
Changes in the localization of WAVE2 after EGF stimulation were determined using immunofluorescence and compared with that previously determined for N-WASP (Sukumvanich et al., 2004). In unstimulated cells the distribution of these proteins is diffuse throughout the cytoplasm. After EGF stimulation these proteins are both recruited to the leading edge (Fig. 2 A). N-WASP (Sukumvanich et al., 2004) and WAVE2 (Fig. 2, B and C) have similar recruitment kinetics during EGF-stimulated protrusion. Recruitment of both proteins to the leading edge shows a peak at 60 s after EGF stimulation, followed by a gradual decrease.
Figure 2.
WAVE2 is recruited to the leading edge of extending lamellipods after stimulation with EGF. (A) Cells were stimulated with EGF and stained with the C-14 anti-WAVE2 antibody. Bar, 10 μm. (B) Quantification of recruitment of WAVE2 in the leading edge. Mean fluorescent intensity of C-14 staining was measured at the leading edge and plotted versus time of EGF stimulation. (C) Ratio of WAVE2 fluorescent intensity at the leading edge to total cellular WAVE2 fluorescent intensity. Error bars are ±SEM of a total of 26 cells from three independent experiments.
WAVE2 is the major regulator of lamellipod formation
In carcinoma cells, such as MTLn3, growth factor–induced lamellipod formation is achieved by actin polymerization, some of which is contributed by the Arp2/3 complex–dependent pathway (Bailly et al., 2001; DesMarais et al., 2004). WAVE2 has been shown to be responsible for lamellipod formation in Cos7 cells (Suetsugu et al., 1999). To determine the role of WAVE2 in MTLn3 cells, we silenced WAVE2 using siRNA (WAVE2 knockdown [KD]). Western blot analysis shows decreased levels of WAVE2 expression at a mean of 70% in the WAVE2 KD at 48 h (Fig. 3 C and Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200708123/DC1). Lamellipod formation (area analysis) was visualized using fluorescence images of GFP-actin–expressing MTLn3 at 0 and 3 min after EGF stimulation, which are the minimal and maximal time points of cell protrusion (Fig. 3 A). Cells were starved for 3 h previous to EGF stimulation to obtain a time zero for the scoring of lamellipod and filopod formation. WAVE2 KD cells had a 40% mean decrease in lamellipod protrusion in comparison to control cells after EGF stimulation (Fig. 3 D). Rescue of the WAVE2 KD phenotype was achieved by stably expressing wild-type human WAVE2 lacking the target sequence in rat-specific WAVE2 siRNA-treated cells. The expression of human WAVE2 resulted in rescue of lamellipod protrusion (Fig. 3 D). Knocking down both WAVE2 and 1 gave a similar decrease in lamellipod formation as seen in the single WAVE2 KD (Fig. S1 B). These results indicate that WAVE2, and not WAVE1, is the main regulator of lamellipod formation in MTLn3 cells.
Figure 3.
WAVE2 siRNA inhibits lamellipod protrusion after EGF stimulation. GFP–β actin–expressing MTLn3 cells were treated with no oligofectamine (control), scrambled siRNA, rat-specific WAVE2 siRNA, or rat-specific WAVE2 siRNA + human WAVE2 plasmid (W2 rescue). (A) Representative time-lapse images of cells at 0 and 3 min after EGF stimulation. Bar, 10 μm. (B) Representative images of filopods being scored are indicated by arrows in scrambled control cells (left) and WAVE2 KD cells (right). (C) Western blot analysis of cells treated with WAVE2 rat-specific siRNA (W2si) or Scrambled siRNA (Scr) in cells with control plasmid (C) or WAVE2 human plasmid (W2 wt). Westerns were blotted with α-WAVE2 and α–β actin antibodies. (D) Quantification of cell area fold increase during lamellipod extension. Graph shows the fold increase in area of control, scrambled siRNA, WAVE2 siRNA, and human WAVE2-expressing cells treated with WAVE2 siRNA. Results are from a total of 67 cells from five independent experiments. (E) Quantification of filopod formation after EGF stimulation. Results are from a total of 43 cells from six independent experiments. (F) Knocking down Abi1 results in the destabilization of the WAVE2 complex and a WAVE2 KD phenotype. Representative Western blot of Abi1 KD at 48 h. Westerns were blotted with α-WAVE2, anti-Abi1 α-tubulin, and α–β actin antibodies. (G) Quantification of lamellipod extension at 0, 1, and 3 min. (H) Quantification of filopod formation. Error bars indicate ±SEM of 27 cells from three independent experiments.
Decreased WAVE2 expression increases filopod formation
The initial paradigm for actin-based protrusion was that Cdc42 activates N-WASP, which induces filopods, whereas Rac activates WAVE2, which induces lamellipods (Miki et al., 1998a; Miki and Takenawa, 1998; Takenawa, 2001). New evidence has challenged this simple model. Fibroblasts derived from N-WASP–deficient mice still retain ∼50% of their filopod formation (Snapper et al., 2001). This new data suggests that N-WASP is not the sole regulator of filopod formation. In Drosophila melanogaster cells, the KD of WAVE inhibited the formation of filopods (Biyasheva et al., 2004). This result led to the proposal of a new model in D. melanogaster, where Scar/WAVEs initiate lamellipods, which are subsequently transformed into filopods by actin filament-bundling proteins (Biyasheva et al., 2004). These competing models opened the question as to which WASP family member regulates filopod formation in MTLn3 cells. MTLn3 cells were treated with either WAVE2 or N-WASP siRNA. Quantification of filopod formation was achieved by observing time-lapse phase and fluorescent images (10-s intervals) of GFP-actin in MTLn3 cells. Filopods were scored after EGF stimulation, scoring only protruding actin-containing needle-shaped structures (i.e., retraction fibers were excluded from the scoring). MTLn3 control cells produce filopods on an average of two to five per cell over the time-lapse video duration (Fig. 3). Filopod quantification was normalized to one to compare different conditions. The images shown are representative of the filopods scored. The arrows point to the filopods observed as they formed in scrambled control cells (Fig. 3 B and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200708123/DC1). WAVE2 KD cells show a 2.4-fold increase in filopod formation in comparison to control cells (Fig. 3 E). WAVE2 KD cells show a decreased lamellipod formation and increased filopod formation. Therefore, filopod formation does not require WAVE2-induced lamellipods, which does not support the model of Biyasheva et al. (2004). In addition, the N-WASP KD had no effect on filopod formation (Fig. 4 C). Hence, our results suggest that another pathway may be responsible for filopod formation in tumor cells.
Figure 4.
N-WASP KD does not affect lamellipod or filopod formation. (A) Representative images of cells treated with scrambled or N-WASP siRNA. Cells were fixed and stained with Rhodamine phalloidin at 0 and 3 min after EGF stimulation. Bar, 10 μm. (B) Quantification of lamellipod extension (cell area). Cell areas were measured at 0 and 3 min after EGF stimulation. The graph shows the fold increase in area after EGF stimulation of scrambled siRNA and N-WASP siRNA. A total of 41 cells pooled from three independent experiments are shown. (C) Quantification of filopod formation. A total of 30 cells from three independent experiments are shown. (D) Western blot of cells treated with N-WASP–specific or scrambled (Scr) siRNA and blotted with α–N-WASP and α–β actin antibodies. Error bars indicate ±SEM.
WAVE2 has been shown to exist in a complex composed of Abi1, Nap1, PIR121 (Sra-1), and HSPC300. This WAVE2 complex stabilizes its components and protects them from degradation. Ablation of WAVE2 or any of the WAVE2 complex components has been shown to lead to the destabilization of the WAVE2 complex (Innocenti et al., 2004; Kheir et al., 2005). Abi1 has been implicated in the assembly of the WAVE2 complex and its localization at the leading edge in vivo (Innocenti et al., 2004). To determine if KD of Abi1 caused the same phenotype as WAVE2 KD, we suppressed the expression of Abi1. Knocking down Abi1 also knocked down WAVE2 expression (Fig. 3 F; and Fig. S2, E and F), as previously shown (Innocenti et al., 2004; Kheir et al., 2005). Lamellipod extension was inhibited in these cells (Fig. 3 G). Filopod formation was increased 2.5-fold, as seen in the WAVE2 KD cells (Fig. 3 H). These results suggest that knocking down Abi1 has a phenotype similar to the WAVE2 KD, which is consistent with a role for Abi1 in stabilizing the WAVE2 complex (Innocenti et al., 2004). In addition, the absence of Abi1 also resulted in the activation of blebbing, which was not seen in the WAVE2 KD, suggesting that Abi1 functions in more than one pathway. It has been reported previously that silencing other components of this complex, such as Sra-1 and Nap1, causes blebbing at the cell surface (Steffen et al., 2004), which is consistent with our findings.
Decreased N-WASP expression has no effect on lamellipod or filopod formation
N-WASP localizes in lamellipods and invadopods in EGF-stimulated carcinoma cells (Sukumvanich et al., 2004) and is required for invadopod formation in these cells (Yamaguchi et al., 2005). N-WASP has also been shown to use the Arp2/3 complex which polymerizes actin in vitro (Rohatgi et al., 1999). N-WASP's localization at the leading edge of lamellipods and its downstream activation of the Arp2/3 complex suggests that N-WASP might have a role in lamellipod protrusion. To investigate the role of N-WASP in lamellipod formation, using siRNA against N-WASP (N-WASP KD), N-WASP protein expression was decreased an average of 70% from normal levels (Fig. 4 D and Fig. S2 B). The decrease of expression had no significant affect on lamellipod formation compared with controls. However, N-WASP KD cells looked jagged when cells were quiescent but cells recovered to their normal lamellipod phenotype 3 min after EGF stimulation (Fig. 4, A and B). N-WASP KD cells also gave no change in filopod formation compared with control cells (Fig. 4 C). Therefore, N-WASP does not seem to have an effect on either lamellipod or filopod formation in response to EGF in MTLn3 cells.
siRNA knockout of both WAVE2 and N-WASP reveals an underlying pathway for polymerization and protrusion
Incomplete inhibition of lamellipods and increased filopod production in WAVE2 KD cells suggested that additional pathways were causing protrusions in the absence of WAVE2. To completely rule out N-WASP, both N-WASP and WAVE2 were knocked down simultaneously by siRNA (N-WASP/WAVE2 KD; Fig. 5 F; and Fig. S2, A and B). However, unlike the WAVE2 KD, which inhibited protrusion by 40%, decreasing the expression of both WAVE2 and N-WASP together changed cell shape and increased protrusion activity (Fig. 5 B and Fig. 3 D). In fact, the increase in protrusion area after EGF stimulation in N-WASP/WAVE2 KD cells was similar to that in control cells (Fig. 5 B), indicating that a gain of protrusion activity over that in WAVE2 KD cells that is independent of N-WASP and WAVE2. The N-WASP/WAVE2 KD cells had jagged (saw tooth) protrusions rather than a smooth leading edge (Fig. 5 A and Video 2, available at http://www.jcb.org/cgi/content/full/jcb.200708123/DC1). The jagged protrusions appeared to result from the merging and bundling of actin filaments present within the lamella (Fig. 5 A, compare side panels with electron microscopy image on the bottom right). Many jagged protrusions had filopods that elongated from the distal tips of the jagged protrusions (Fig. 5 A, right insets; and Video 2). Scoring of filopod formation after EGF stimulation showed a fourfold increase in filopods in the N-WASP/WAVE2 KD cells (Fig. 5 C) compared with a 2.5-fold increase in the absence of EGF (Fig. 5 D). There was also an increase in filopod length and persistence in N-WASP/WAVE2 KD cells compared to control cells (Tables I and II). Forming protrusions can be seen in Video 2.
Figure 5.
The simultaneous suppression of both WAVE2 and N-WASP results in a new type of protrusion during EGF stimulation. (A) Representative images of GFP–β actin–expressing MTLn3 cells treated with scrambled siRNA or double transfected with WAVE2 and N-WASP siRNA at 0 and 3 min after EGF addition. Bar, 10 μm. Side panels show a time-lapse sequence of filopod formation in an N-WASP/WAVE2 KD cell. An electron microscopy image, which is representative of the structure of actin in a filopod like that shown in the time lapse, is shown at the bottom right. The large image at the bottom is an enlargement of this. Bar, 1 μm. (B) Quantification of cell area fold increase during lamellipod extension. Cell area was measured at 0 and 3 min after EGF stimulation. Bar graph shows scrambled and N-WASP/WAVE2 siRNA. (C) Quantification of filopod formation after EGF. Error bars in B and C indicate ±SEM of a total of 30 cells from three independent experiments. (D) Filopods in the absence of EGF. P-values are in comparison to control. Error bars are ±SEM of a total of 36 cells from three independent experiments. (E) Stress fiber density (measured as total F-actin fluorescence intensity) was analyzed by measuring phalloidin staining in fixed cells. Error bars are ±SEM of a total of 58 cells from three independent experiments. (F) Western blot representative of scramble (Scr) and N-WASP/WAVE2 (NWW2) KD blotted with N-WASP, WAVE2, and actin antibodies. (G–J) Cells were treated with either p34-directed siRNA (Arp2/3si), scrambled siRNA, or p34 plus mDia1 siRNA. (G) Images representative of cells stimulated with EGF at 0 and 3 min. Bar, 10 μm. (H) Western blot of cell lysates treated with P34 (Arp2/3 si) or scrambled (Scr) siRNA and blotted with α-Arp3 and α–β actin antibodies. (I) Quantification of cell area fold increase during lamellipod extension. Cell area was measured at 0 and 3 min after EGF stimulation. Graph shows scrambled and Arp2/3-directed p34 siRNA. (J) Cells were treated with scrambled, p34, or p34 + mDia1 siRNA. Quantification of filopod formation. Error bars are ±SEM of a total of 32 cells from three independent experiments.
Table I. The length of filopods increases in N-WASP/WAVE2 double KD cell.
|
|
Length (μm)
|
|
|---|---|---|
| Scr | N-WASP/WAVE2 | |
| Mean | 1.77 | 2.48 |
| SEM | 0.18 | 0.17 |
Cells were treated with either scrambled (Scr) or N-WASP and WAVE2 siRNA. Filopod length was quantified from time-lapse videos after EGF stimulation. The maximum length of filopods was used in analysis. P = 8.31 × 10−6. P-values are in comparison to control.
Table II. The persistence of filopods increases in N-WASP/WAVE2 KD cells.
|
|
Persistence (s)
|
|
|---|---|---|
| Scr | N-WASP/WAVE2 | |
| Mean | 111.38 | 139.13 |
| SEM | 8.52 | 5.42 |
Cells were treated with either scrambled (Scr) or N-WASP and WAVE2 siRNA. Filopod length was quantified from time-lapse videos after EGF stimulation. Persistence was measured by lifetime of filopods throughout EGF stimulation. Lifetime is defined as the total duration of a filopod from first appearance to final disappearance in the time-lapse movie. P-values are in comparison to control. P = 1.73 × 10−5.
The N-WASP/WAVE2 KD cells also showed a 20% increase in stress fibers in unstimulated cells measured as total phalloidin staining (Fig. 5 E). This method was used because the majority of phalloidin staining is found in stress fibers in MTLn3 cells.
Both N-WASP and WAVE2 are upstream regulators of the Arp2/3 complex. Therefore, we investigated if the double KD phenotype was a result of inhibiting activation of the Arp2/3 complex pathway. We knocked down the Arp2/3 complex by using siRNA (Arp2/3 KD) designed against its P34 subunit, which has been shown to knock down the entire complex (Fig. 5 H and Fig. S2 G; Kempiak and Segall, 2004; Sidani et al., 2007). Unlike the increase in protrusions seen in the N-WASP/WAVE2 KD cells, Arp2/3 KD resulted in inhibition of protrusion formation (Fig. 5, G and I). In addition, Arp2/3 KD resulted in an increase in filopod formation (Fig. 5 J), but these were not generated from jagged protrusions as seen in the N-WASP/WAVE2 KD cells. Therefore, the phenotype seen in the N-WASP/WAVE2 KD cells is not a direct result of inhibition of the Arp2/3 complex but involves an additional pathway that is activated in the absence of N-WASP and WAVE2.
To determine the effects of the N-WASP/WAVE2 KD on actin polymerization, we used the barbed end (BE) assay, which records the number of growing actin filaments throughout the cell including leading edge and focal adhesions associated with the ends of stress fibers (Chan et al., 1998). BEs in the WAVE2 KD cells were inhibited 59% compared with control cells (Fig. 6, A and B). There was no significant difference in BEs in N-WASP KD cells (Fig. 6, A and B). In N-WASP/WAVE2 KD cells, there was only a small inhibition of BEs compared with control cells after EGF stimulation (Fig. 6 B), indicating that a BE-generating activity was present even in the absence of WAVE2. The Arp2/3 KD results in inhibition of BEs were similar to those of the WAVE2 KD (Fig. 6, A and D). These results demonstrate that by simultaneously inhibiting both the WAVE2 and N-WASP expression, another actin nucleation pathway, independent of the Arp2/3 complex, is up-regulated which results in the production of BEs, jagged protrusions, and filopods.
Figure 6.
Actin polymerization is inhibited by WAVE2 siRNA but restored in N-WASP/WAVE2 double KD cells. (A) Representative images of actin polymerization were measured using the BE assay of cells (left columns) and total F-actin (right columns) treated with scrambled (control), WAVE2, N-WASP, N-WASP/WAVE2, N-WASP/WAVE2 + {YFP-DNmDia1 plasmid }, and Arp2/3 siRNA at 0 and 1 min after EGF stimulation, the time of maximum early generation of BE formation. Bar, 10 μm. (B) The relative number of BEs (arbitrary units of fluorescence intensity) at the leading edge in control, WAVE2, N-WASP, and N-WASP/WAVE2 is shown. Error bars are ±SEM of 80 cells from three independent experiments. (C) The relative number of BEs at the leading edge in control, N-WASP/WAVE2 siRNA with YFP-vector, and N-WASP/WAVE2 siRNA with YFP-DNmDia1 plasmid is shown. P-values are in comparison to control. Error bars are ±SEM of a total of 50 cells from at least three independent experiments. (D) The relative number of BEs at the leading edge in scrambled or Arp2/3 siRNA is shown. P-values are shown in comparison to control. Error bars are ±SEM of a total of 37 cells from three independent experiments.
The Role of mDia in N-WASP and WAVE2 double KD cells
The mDia formins are proposed to be an important actin polymerization factor in the lamella of migrating cells (Gupton et al., 2005). Therefore, jagged protrusions formed in the N-WASP/WAVE2 KD cells were investigated for the presence of the lamellar marker tropomyosin (LC24 Ab; DesMarais et al., 2002). The jagged protrusions were found to be tropomyosin-rich structures (Fig. S3, A and B, available at http://www.jcb.org/cgi/content/full/jcb.200708123/DC1), which defines them as being part of the lamella (DesMarais et al., 2002; Gupton et al., 2005).
Next we wished to investigate if mDia formins are involved in the pathway responsible for the protrusions and BEs in the lamella of N-WASP/WAVE2 KD cells. Therefore, we inhibited mDia using a dominant-negative (DN) mDia1. We used DN-mdia1 (mDia1 F2ΔN1 YFP) because it functions as a pan-DN by inhibiting actin polymerization of both mDia1 and mDia2 (Copeland et al., 2004). DN-mDia1 was expressed in N-WASP/WAVE2 KD cells. Protrusion activity was compared with cells expressing an empty YFP vector either in control (scrambled siRNA) or N-WASP/WAVE2 KD cells. The expression of DN-mDia1 resulted in the inhibition of filopod production by the N-WASP/WAVE2 KD cells (Fig. 7 B).
Figure 7.
mDia1 is responsible for protrusions formed in N-WASP/WAVE2 double KD cells. (A) Relative expression of mDia family members in carcinoma cells. (B) Quantification of filopod formation. Cells were transfected with scrambled siRNA + YFP-vector, N-WASP/WAVE2 siRNA, and with either YFP-DN-mDia1 (NWW2 + DNmDia1) or YFP-vector (NWW2). Error bars indicate ±SEM of a total of 25 cells from three independent experiments. (C and D) Cells were treated with the following siRNAs: Scrambled (Scr); N-WASP/WAVE2 (NWW2); N-WASP/WAVE2 and mDia1 (NWW2 + mDia1); N-WASP/WAVE2 and mDia2 (NWW2 + mDia2). (C) Quantification of filopod formation. (D) Quantification of cell area fold increase during lamellipod extension. Error bars indicate ±SEM of a total of 30 cells from at least three independent experiments. (E) Western blot of cells treated with Scr, N-WASP/WAVE2, N-WASP/WAVE2+mDia1, or N-WASP/WAVE2+mDia2 siRNA and probed with anti-WAVE2, –N-WASP, -mDia1, or -mDia2 (right to left blot) and tubulin and β actin antibodies. P-values are in comparison to scrambled control.
A BE assay was used to determine if the actin polymerization seen in the N-WASP/WAVE2 KD cells was caused by mDia formins. The DN-mDia1 inhibited BE formation in the protrusions formed by the N-WASP/WAVE2 KD cells (Fig. 6 C). To determine the effect of expression of the DN-mDia1 on control cells, DN-mdia1 was expressed in otherwise untreated MTLn3 cells and was found to have no affect on BE formation (Fig. S4, A and B, available at http://www.jcb.org/cgi/content/full/jcb.200708123/DC1). These results suggest that both N-WASP and WAVE2 pathways must be inhibited to activate the mDia pathway to produce BEs, which result in production of jagged protrusions and filopod formation.
mDia1 is responsible for the formation of filopods and jagged protrusions in the absence of N-WASP and WAVE2
There are three known mammalian mDia proteins, mDia1, mDia2, and mDia3. We measured mRNA expression levels of all three in MTLn3 cells using quantitative real-time PCR and found that mDia1 and 2 are the only family members expressed in MTLn3 cells (Fig. 7 A). Therefore, we focused on mDia1 and 2 as potential regulators of actin polymerization in MTLn3 cells.
To determine which mDia was responsible for the protrusions and filopod formation in N-WASP/WAVE2 KD cells, we knocked down either mDia1 or 2 in these cells using siRNA (Fig. 7 E; and Fig. S2, C and D). Only mDia1, not mDia2, siRNA treatment inhibited the formation of filopods produced by the N-WASP/WAVE2 KD cells (Fig. 7 C). When general protrusion activity was measured, only mDia1 inhibited the protrusions formed in N-WASP/WAVE2 KD cells (Fig. 7 D). In addition, suppression of mDia1 expression with siRNA in Arp2/3 KD cells did not inhibit the formation of filopods seen in the Arp KD cells (Fig. 5 J), indicating that the filopods in the Arp2/3 KD cells do not result from mDia1 activity. Therefore, mDia1 activity is responsible for the formation of filopods, and their associated jagged protrusions, only when produced in the absence of N-WASP and WAVE2.
To determine the effect of suppression of expression of mDia1 and 2 in normal cells expressing N-WASP and WAVE2, mDia1 and 2 were knocked down individually by siRNA in otherwise untreated control cells (mDia1 KD and mDia2 KD, respectively). The expression of mDia1 was decreased ∼80% and that of mDia2 by ∼70% and was specific for each mDia (Fig. S2, C and D; and Fig. S4 D). In both cases, there was no significant affect on the protrusion of lamellipods (Fig. S4, C and E) and there was only a small increase in filopod number (Fig. S4 F), unlike the large effects seen in N-WASP/WAVE2 KD cells (Fig. 5). These results indicate that suppression of either mDia1 or 2 does not generate the same phenotype as seen in N-WASP/WAVE2 KD cells.
The jagged protrusions formed in the N-WASP/WAVE2 KD cells were found to be tropomyosin-rich structures (Fig. S3, A and B), which defined them as being part of the lamella (DesMarais et al., 2002; Gupton et al., 2005). To investigate the role of mDia1 in lamellar formation, tropomyosin was localized in mDia1 KD cells. The reduction in tropomyosin staining at the cell edge in mDia1 KD cells suggests that there is a decrease in lamellar formation in the absence of mDia1 (Fig. S3, C and D).
In carcinoma cells, mDia1 activates the formation of filopods and jagged protrusions
The localization of endogenous mDia1 was determined by staining with antibodies specific for mDia1. mDia1 was localized throughout the leading edge of control cells. In N-WASP/WAVE2 KD cells, mDia1 was localized at the leading edge of the jagged protrusions and in filopods (Fig. 8 A). mDia1's localization is consistent with its role in the formation of filopods and jagged protrusions in N-WASP/WAVE2 KD cells.
Figure 8.
mDia1 localization and activity. (A) Localization of mDia1 by antibody staining in N-WASP/WAVE2 double KD cells and control cells. Cells were treated with Scrambled or N-WASP/WAVE2 siRNA and stained with anti-mDia1 antibody (green) at 3 min after EGF stimulation. Rhodamine phalloidin was used to stain for actin (red). Insets show actin protrusion with filopods in N-WASP/WAVE2 KD cells. Bar, 10 μm. (B) Constitutively active mDia1 activation gives similar phenotype to N-WASP/WAVE2 double KD cells. Images of cells transfected with control GFP empty vector (top, red) or GFP-ΔGBD-mDia1 (bottom, red), a DA mDia1, for 6 h. Images were taken of cells fixed at 0, 1, and 3 min after EGF stimulation. Rhodamine phalloidin was used to stain for actin (green). Insets (indicated by dashed boxes) show active mDia1 localized at tips of filopods. Bar, 10 μm. (C) Quantification of filopod formation. P-values are compared with control GFP vector cells. (D) Total F-actin fluorescence intensity (percent) was measured using the mean value of phalloidin staining of unstimulated cells transfected with GFP or DA-mDia1 vector. P-values are in comparison to control. Error bars indicate ±SEM of a total of 19 cells from three independent experiments.
Constitutively activated (DA) mDia1 (ΔGBD-mDia1) was expressed in otherwise untreated MTLn3 cells to investigate the effect of mDia1 activity on cell morphology. Expression of the DA-mDia1 reproduced the N-WASP/WAVE2 KD phenotype. Cells formed jagged protrusions throughout the 3-min motility cycle of MTLn3 cells after EGF stimulation (Fig. 8 B). Expression of DA-mDia1 also increased the number of filopods fivefold compared with control cells (Fig. 8 C). DA-mDia1 was localized at the tips of the filopods (Fig. 8 B, inset), which is consistent with it being involved in the production of filopods in MTLn3 cells. There was also a 35% increase in stress fibers, as measured by total F-actin content per cell, in cells expressing DA-mDia1 cells compared with the GFP control cells (Fig. 8 D). The reproduction of the N-WASP/WAVE2 KD phenotype and the localization of the DA-mDia1 suggest that mDia1 is responsible for the N-WASP/WAVE2 KD phenotype and that mDia1 is being activated in the absence of N-WASP and WAVE2.
mDia1-dependent protrusions are correlated with, and require, RhoA activity
Several formins are effectors for Rho family GTPases. Both mDia1 and 2 bind to activated RhoA, whereas mDia2 also interacts with Cdc42 (Alberts et al., 1998; Watanabe et al., 1999; Alberts, 2001). One of the most studied interactions is between mDia1 and RhoA. RhoA binding relieves autoinhibition of mDia1 (Watanabe et al., 1999), thereby activating mDia1. Because the protrusions in N-WASP/WAVE2 KD cells are mDia1 dependent, we measured the activity of RhoA in our siRNA-treated cells. RhoA activity was quantified using a GST–Rho binding domain (RBD; binding domain from the RhoA effector Rhotekin) pulldown assay (Fig. 9 A). N-WASP/WAVE2 KD cells showed a significant increase in RhoA activation compared with control cells and cells singly knocked down for either WAVE2, N-WASP (Fig. 9 B), or Arp2/3 complex (Fig. 9, C and D). In addition, RhoA activity showed a small but statistically significant increase after EGF stimulation (Fig. 9 B). Given the short time of 3–4 h that the cells were starved in serum-free medium, a starvation time chosen to maintain viability, it is not surprising that the differences in activity before and after EGF would be small because residual activation of pathways leading from the EGF receptor usually occur.
Figure 9.
RhoA activity is increased in N-WASP/WAVE2 KD cells and is required for filopod production in N-WASP/WAVE2 double KD cells. (A–F) MTLn3 cells were transfected with scrambled control, WAVE2, N-WASP, N-WASP/WAVE2, or Arp2/3 siRNA as indicated in blots and graphs. (A and B) The cells were then starved for 3 h and stimulated with EGF for the indicated times. (A) Representative Western blots of GST-RBD pulldowns showing Rho activation in EGF-stimulated MTLn3 cells. Proteins bound to immobilized GST-RBD were immunoblotted with anti-RhoA,B,C antibody. The amount of total Rho in cell lysates is shown at the bottom. (B) Quantification of the Western blot data by densitometry (arbitrary units) for RhoA in A. P-values are in comparison to control at each time point or in comparison to the zero time point (brackets). Activation is calculated as percent of the maximum within each experiment. Error bars indicate ±SEM of three independent experiments (C) Western blot representative of RhoA pulldown (top) and total cell lysate (bottom) of cells treated with scrambled and Arp2/3 siRNA at steady state. (D) Quantification of the Western blot data by densitometry (arbitrary units) for RhoA in C. (E) Inhibition of RhoA results in a decrease in the filopods produced in N-WASP/WAVE2 KD cells. Quantification of filopod formation. Cells were transfected with either N-WASP/WAVE2 or scrambled siRNA for 48 h. N-WASP/WAVE2 siRNA cells were treated with (NWW2+C3) or without (NWW2) tat-C3 toxin. Error bars indicate ±SEM from a total of 24 cells from three independent experiments. (F) Dominant-active RhoA increases filopod formation in MTLn3 cells. Quantification of filopod formation. Cells were transfected with either N-WASP/WAVE2 or scrambled siRNA for 48 h. N-WASP/WAVE2 siRNA cells were also transfected with CFP or CFP-DA-RhoA (NWW2+DA-RhoA). Error bars indicate ±SEM, from a total of 21 cells, from three independent experiments.
We also tested activation of the related G proteins Rac and Cdc42 in these KD cells during stimulation. GTP-Cdc42 levels were unchanged in all KD cells except in the Arp2/3 KD, where Cdc42 was decreased (Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200708123/DC1). GTP-Rac levels were elevated in the WAVE2, N-WASP, and N-WASP/WAVE2 KDs but suppressed in the Arp2/3 KD cells (Fig. S5). Because Rac activity is elevated in N-WASP and WAVE2 single KD cells as well as the N-WASP/WAVE2 KD, it cannot account for the phenotype that is unique to the N-WASP/WAVE2 KD cells. It has also previously been shown that the KD of Rac in MTLn3 does not affect area protrusion in EGF-stimulated MTLn3 cells (Yip et al., 2007). These results suggest that the increased RhoA activity, which is unique to the N-WASP/WAVE2 KD cells, results in the activation of mDia1.
Increased filopod formation in N-WASP/WAVE2 KD cells is an mDia1-dependent phenotype. To investigate if filopod production in N-WASP/WAVE2 KD cells requires increased RhoA activity, we determined the effects of the inhibition and activation or RhoA on filopod formation in these cells. RhoA was inhibited in N-WASP/WAVE2 KD cells by incubating cells with Clostridium botulinum TAT-C3 exoenzyme for 12 h (Sahai and Olson, 2006). TAT-C3 exoenzyme resulted in a decrease in filopod formation compared with the N-WASP/WAVE2 KD controls (Fig. 9 E). In addition, we transfected N-WASP/WAVE2 KD cells with CFP-tagged DA RhoA (NWW2+DA-RhoA), which resulted in an increase in filopod formation compared with cells expressing a CFP-empty vector (Fig. 9 F). Therefore, the increase in filopod formation in the N-WASP/WAVE2 KD cells requires RhoA activity (Tables I and II).
Discussion
In this paper, we investigated the contributions of various actin nucleation pathways to lamellipod and filopod protrusion in tumor cells. WAVE2 is the major regulator of lamellipod formation, whereas N-WASP does not affect lamellipod protrusion. WAVE2 KD increases filopod formation but N-WASP KD had no effect on filopod formation. The absence of both WAVE2 and N-WASP reveals an underlying pathway for polymerization and protrusion of a lamellar structure involving mDia1. The activation of mDia1 is correlated with increased RhoA activity when both N-WASP and WAVE2 are simultaneously removed, suggesting a mechanism for the activation of mDia1. Hence, the appearance of mDia1-dependent protrusions occurs when signaling through both WAVE2 and N-WASP is interrupted, suggesting that lamellipod protrusion and lamellar extensions are coordinately regulated (Fig. 10).
Figure 10.
Summary of cell phenotypes resulting from inhibition of WASP family members. (Top) Control. All pathways are intact and regulated. (Top, N-WASP KD) Lamellipod extension containing dendritic network is normal. The only difference is the absence of ventral protrusions called invadopods (Yamaguchi et al., 2005). (Top, WAVE2 KD) Inhibition of lamellipods and loss of dendritic network but not invadopods. Lamellar structures are more prominent. (Top, N-WASP/WAVE2 KD) Inhibition of both N-WASP and WAVE2 results in loss of lamellipods and invadopods and activation of the mDia1 pathway, leading to the formation of a prominent lamella, filopods, and jagged protrusions. (Bottom, 1) The N-WASP-Arp2/3 complex is necessary for invadopod formation (Yamaguchi et al., 2005). (Bottom, 2) WAVE2 regulates lamellipod formation, not filopod formation. (Bottom, 3) Inhibition of both WAVE2 and N-WASP up-regulates RhoA and mDia1 activities, resulting in lamellar but not lamellipodial extension.
WAVE2 is the major regulator of lamellipod formation
The original proposal for the function of WAVE2 and N-WASP was that WAVE2 regulates lamellipod formation, whereas N-WASP regulates filopod formation (Miki and Takenawa, 1998). A study of cell protrusion in D. melanogaster demonstrated that WAVE depletion resulted in inhibition of both lamellipod and filopod formation, whereas WASP depletion had no affect (Biyasheva et al., 2004). These results led to the proposal of a new model in which WAVEs are required for lamellipod formation and filopods arise from the lamellipod in response to additional signals and convergent elongation of actin filaments resulting from their cross-linking by bundling proteins (Biyasheva et al., 2004). Furthermore, fibroblasts from N-WASP–deficient mice still extend some filopods, suggesting that N-WASP is not the sole regulator of filopod formation (Snapper et al., 2001) and additionally suggesting that the original model may not be correct.
We investigated the role of WAVE2 on actin protrusions in invasive and metastatic cancer cells. We found that WAVE2, but neither N-WASP nor WAVE1, is required for lamellipod formation. The absence of WAVE2 protein inhibited lamellipod formation. However, in contrast to the model in which WAVEs are required for filopods in D melanogaster (Biyasheva et al., 2004), more filopods were produced in the absence of WAVE2 in tumor cells, suggesting that additional protrusion activities are evident when lamellipod protrusion is inhibited. These results also indicate that the precise regulation of protrusive activity may vary with cell type, consistent with the different physiological functions of the different types of migratory cells.
N-WASP localization to the leading edge after EGF stimulation in tumor cells suggests a role for N-WASP in lamellipod formation (Lorenz et al., 2004; Sukumvanich P. et al., 2004). However, N-WASP KD in tumor cells did not affect either filopod or lamellipod formation. This result does not fit the model put forward by Miki and Takenawa (1998) and suggests other functions for N-WASP in the lamellipod. N-WASP activity is present in the lamellipods and invadopods of tumor cells (Lorenz et al., 2004) and is required for invadopod formation (Yamaguchi et al., 2005) but not lamellipod formation as shown here. Because invadopod formation initiates at the cell periphery in the lamellipods of tumor cells followed by migration of invadopods to the cell center (Yamaguchi et al., 2005), N-WASP may function in the lamellipod in the initiation of invadopods in this peripheral location.
Simultaneous inhibition of both N-WASP and WAVE2 activates mDia1-dependent protrusions
Formins have been shown to nucleate actin filament assembly (Pruyne et al., 2002), produce Rho-induced stress fibers (Watanabe et al., 1999; Tominaga et al., 2000), and form filopods (Peng et al., 2003; Pellegrin and Mellor, 2005; Schirenbeck et al., 2005). When N-WASP and WAVE2 were simultaneously knocked down in tumor cells we found a significant increase in filopod formation and stress fibers. Consistent with the increase in filopods was an increase in the number of BEs over the levels seen in WAVE2 KD cells. This suggested that the formin pathway was being activated in N-WASP/WAVE2 KD cells. A similar phenotype has been seen in cells microinjected with tropomyosin, resulting in an increase of filopod formation and actin polymerization from BEs at the leading edge that was independent of the Arp2/3 and ADF/cofilin mechanisms (Gupton et al., 2005). In N-WASP/WAVE2 KD cells, we also saw a decrease in the Arp2/3 complex localization at the leading edge and an increase in tropomyosin localization at the leading edge (Fig. S3), which indicated that the protrusions being formed in MTLn3 cells under these conditions are lamella (Gupton et al., 2005). Furthermore, the protrusions formed by the N-WASP/WAVE2 KD cells required mDia1 expression and could be induced by expression of active mDia1. Therefore, the removal of both N-WASP and WAVE2 generated a protrusive phenotype similar to that described for lamella, in which the activation of mDia1 is responsible for actin polymerization and protrusion.
The role for formins in filopod assembly
The observation that mDia1 has an essential role for filopodia formation in MTLn3 cells is surprising. Previously, mDia2 had been implicated in filopod formation in mouse embryonic stem cells (Peng et al., 2003). In contrast to the previous study, we show here that mDia2 expression did not appear to be altered by mDia1 KD and that mDia1 is sufficient for filopod formation. The lack of change in mDia2 expression is likely because of the different approaches used by the two different studies.
In the initial study, Drf1 (mDia1)-null cells were generated over a long selection process. In this context, the extended selection likely drove cells to increase mDia2 expression to compensate for loss of mDia1 expression (Peng et al., 2003). Collectively, the results from Peng et al. (2003) and this study indicate redundant roles for mDia1 and 2 in the assembly of F-actin in protrusive structures such as filopods.
RhoA activation is up-regulated in the absence of N-WASP and WAVE2
The regulatory connection between the simultaneous inhibition of N-WASP and WAVE2 and the activation of mDia1 was investigated by looking at the activation status of RhoA, the upstream regulator of mDia1 activity (Alberts et al., 1998; Watanabe et al., 1999; Alberts, 2001). We found that RhoA activity is up-regulated in N-WASP/WAVE2 KD cells. We showed that the mDia1-dependent protrusions in the N-WASP/WAVE2 KD cells are directly dependent on RhoA activation. EGF stimulation further activates RhoA and amplifies the production of filopods and lamellar protrusions. Furthermore, increasing RhoA activity further increased the formation of these protrusions.
In particular, our work demonstrates a previously undetected interaction between the regulation of protrusion formation by WAVE2 and N-WASP and mDia1. Our results here, and those of previous studies (Yamaguchi et al. 2005), indicate that the formation of lamellipods, invadopods, and filopods is coordinated by the WAVE2, N-WASP, and mDia1 pathways, respectively, and that the relative amount of each type of protrusion is determined by cross talk between these pathways (Fig. 10). In addition, our work suggests that the relative balance between the protrusion of lamellipods and the extension of the lamella is determined by the cross talk between N-WASP, WAVE2, and mDia1. Our work also supports the speculation by Gupton et al. (2005) that an Arp2/3-independent generation of BEs is responsible for extension of the lamellum. Further work will be required to understand the consequences of this cross talk in determining the invasive and metastatic phenotype of tumor cells.
Materials and methods
Cell cultures
MTLn3 cells (rat mammary adenocarcinoma cell line) were maintained, starved, and stimulated as described previously (DesMarais et al., 2004). For light microscopy experiments, cells were plated on glass-bottom dishes (MatTek Corporation) that had been treated with 1 M HCl for 10 min, followed by one wash with 75% ethanol and then one wash with PBS. Before each experiment, cells were starved in L15 medium (Invitrogen) supplemented with 0.35% BSA (starvation medium) for 3–4 h. For stimulation, MTLn3 cells were treated at 37°C with a bath application of 5 nM EGF (Invitrogen) for various times. Previous work has shown that serum stimulation of MTLn3 protrusion activity, cell motility, RhoA activity, and Cdc42 activity is mediated by the EGF receptor (Wyckoff et al., 2004; El-Sibai et al., 2007, 2008). Therefore, we used EGF stimulation in place of serum to examine the motility pathways in MTLn3 cells after stimulation.
Antibodies
Primary antibodies were purchased from Santa Cruz Biotechnology, Inc. or supplied by T. Takenawa (Tokyo University, Tokyo, Japan) and A. Alberts (Van Andel Research Institute, Grand Rapids, MI). WAVE2 (C-14), a goat polyclonal Ig antibody, was used to stain against WAVE2 in immunofluorescence at a concentration of 20 μg/ml (Santa Cruz Biotechnology, Inc.). Arp3 (H-110), a rabbit polyclonal antibody, was used to blot against the Arp2/3 complex at 1:300 dilution (Santa Cruz Biotechnology, Inc.) Anti-mDia1 rabbit polyclonal was used against mDia1 at a concentration of 1 μg/ml. Westerns blots were probed with N-WASP–specific monoclonal Ig rabbit antibodies (donated by T. Takenawa) at a dilution of 1:3,000. Immunoblots for WAVE2 were probed by WAVE2-specific monoclonal Ig rabbit antibodies (provided by T. Takenawa) at a dilution of 1:500. Immunoblots for Abi1 were probed for Abi1-specific Ig goat antibodies (T-15; Santa Cruz Biotechnology, Inc.) at a dilution of 1:300. mDia1 was stained using an anti-mDia1 (C-terminal) rabbit polyclonal antibody (provided by A. Alberts) at a 1 μg/ml dilution for Western blots and a 0.2-μg/ml dilution for immunofluorescence. LC24, a mouse monoclonal IgG specific to TM4, was provided by J. Lin (University of Iowa, Iowa City, IA; DesMarais et al., 2002). For BEs, the Cy5-conjugated antibiotin (Jackson ImmunoResearch Laboratories) was used at a 1:50 dilution.
Constructs
Expression plasmid DN-mDia1 (mDia1 F2ΔN1 YFP) encodes N-terminal modifications of the FH2 region (codons 752–1182) of mDia1 (Copeland and Treisman, 2002) fused to pEYFP-C1 expression vectors (Clontech Laboratories, Inc.; Wallar et al., 2007). DA-mDia1:GFP-ΔGBD-mDia1 (543–1,255) fusion constructs were based on the pEGFP-C1 expression vectors (Clontech Laboratories, Inc.; Wallar et al., 2007). CFP-RhoA-G14V fusion constructs were fused to pECFP-C1 expression vector (Clontech Laboratories, Inc.). The EYFP and ECFP plasmids were originally altered (A206K) to eliminate artifactual dimerization of fluorescent proteins (Wallar et al., 2007). Vectors were transfected into cells using Lipofectamine 2000 (Invitrogen). Full-length human WAVE2 was expressed in pBabe puro retroviral vector (Morgenstern and Land, 1990).
Protein
Recombinant TAT-C3 protein was a gift from E. Sahai (Research UK London Research Institute, London, UK). TAT protein transduction domain was added to recombinant C3 exoenzyme. A pGEX-KG MYC-C3 plasmid was modified to include the nucleotide sequence 5′-GGAGGATACGGCCGAAAGAAGCGACGACAGCGACGCCGTGGAGGA-3′ at a position 5′ to the C3 coding sequence. The C3 sequence corresponds to nucleotides 1–780 of EMBL accession X51464 (Sahai and Olson, 2006). Cells were treated with a final concentration of 5.3 μg/ml TAT-C3 in full media for 12 h.
siRNA
The WAVE2, N-WASP, WAVE1, mDia1, mDia2, and P34 siRNA duplexes were designed against the following gene sequences: WAVE2, 5′-AAACCTATAACAGTGTGACG-3′; Scrambled, 5′-AAGGAATCGGATCTCGTAAGG-3′; N-WASP, 5′-AAGACGAGATGCTCCAAATGG-3′; WAVE1, 5′-AAGTGCAGGGGCTGCCTCCGC-3′; P34, 5′-AAGGAACTTCAGGCACACGGA-3′ (Ambion); mDia1, 5′-AAGCAGGAGCTTCGAGAGATTTT-3′; and mDia2, 5′-GCAGGAGCTTCGAGAGATTTT-3′ (Dharmacon). Abi1 siRNA was predesigned (QIAGEN). MTLn3 cells were transfected with the siRNA duplex in the presence of oligofectamine (Invitrogen). The transfection was terminated after 4 h by using 2× serum-containing media.
Quantitative RT-PCR method
RNA was extracted from MTLn3 cells using the TRIzol reagent (Invitrogen), as per the manufacturer's instructions. A further round of RNA purification was performed using RNeasy (QIAGEN) for RNA clean up, as per the manufacturer's instructions. RNA was diluted to a concentration of 2 μg/5 μl for the reverse transcription reaction. The reverse transcription reaction included 2 μg RNA, 25 mM MgCl2, deoxyribonucleotide triphosphates (each at a final concentration of 10 mM), random hexamers (N808-0127), RNase inhibitor (N808-0119), and Multiscribe reverse transcription (N808-0118; all reagents were obtained from Applied Biosystems). Thermal cycling was done at 25°C for 10 min, 48°C for 30 min, and a final incubation at 95°C for 5 min. Target mDia1, mDia2, and mDia3 sequences were developed using Primer Express software (version 1.5; Applied Biosystems). The sequences include the following: mDia1 forward (agg atg cac agg aac agt ata aca aac), mDia1 reverse (acg aag tag tca cct agc tcc tta tag ag), mDia2 forward (ttt gaa gag cag gtg aac aac atc), mDia2 reverse (tga tct cct cgc agg cag tac), mDia3 forward (aga gac cgc aga aag cga att), mDia3 reverse (gcg aga gcg aga tct ttc ca), WAVE1 forward (cca ctc cca caa ggt gaa gt), WAVE1 reverse (agg cag gac ttg tga agg tg), WAVE2 forward (ctt ttc cgt cgt ccc tgt aa), WAVE2 reverse (tca ctc agc tag atc cca agg), N-WASP forward (tgg tga cca tca agt tcc ag), N-WASP reverse (ggt tgg tgg tgt gga ctc tt), P34 forward (gaa caa ccg cat cat cga g), and P34 reverse (aag gac gcc atc aaa atc tg). These primers were used with internal probe sequences that had FAM dye incorporation for accurate measurement in real time. A total of 200 ng cDNA was added to the Universal PCR mastermix (Applied Biosystems) with 100 μM of forward primer, 100 μM of reverse primer, and 100 μM of probe. The thermal cycling was at 50°C for 2 min, 95°C for 10 min, and 95°C for 15 s, cycling 40 times, followed by a final incubation at 58°C for 1 min. The reactions were all performed in an ABI 7700 (Applied Biosystems) machine. WASP family member mRNA levels were analyzed by quantitative RT-PCR as previously described (Wang et al., 2002).
Pulldown assay and Western blots
Activation of Rho was assayed using GST-RBD. Activation of Cdc42 and Rac was assayed using GST–Cdc42-Rac interactive binding domain (CRIB). GST-RBD and GST-CRIB were expressed in Escherichia coli and purified using glutathione-agarose. The cells were lysed in buffer A (25 mM Hepes, 1% Igepal, 150 mM NaCl, 10 mM MgCl2, 10% glycerol, 1 mM EDTA, 1 mM NaVO4, 20 mM NaF, 1 mM PMSF, 100 μg/ml aprotinin, and 5 μM leupeptin). Lysates were cleared by centrifugation and incubated with 20 μg GST-RBD protein for 30 min at 4°C while rotating. The beads were washed three times with buffer A and boiled in Laemmli sample buffer. Rho was detected by Western blotting and by incubating the membrane with an antibody against RhoA using the rabbit monoclonal anti-Rho antibody (clone 3L74). For detecting Cdc42, we used a rabbit polyclonal antibody (Millipore) and for Rac1 we used a mouse monoclonal antibody (Millipore). Immunoblots were incubated with the appropriate secondary antibody that were either HRP- or fluorescently conjugated and quantified by densitometry using the Image analysis software (version 1.62; National Institutes of Health).
Immunofluorescence
Cells were plated at low density on glass-bottom plates (MatTek Corporation) as described in Cell cultures. Cells were untreated or stimulated with EGF. They were fixed with 3.7% paraformaldehyde (Electron Microscopy Sciences) in cytoskeleton fix buffer (5 mM KCl, 137 mM NaCl, 4 mM NaHCO3, 1.1 mM Na2HPO4, 0.4 mM KH2PO4, 2 mM MgCl2, 5 mM Pipes, 2 mM EGTA, and 5.5 mM glucose, pH 7.2) at 37°C for 5 min. Cells were then permeabilized with 0.5% Triton X-100 in cytoskeleton fix buffer for 20 min. Next, the cells were rinsed once and then incubated for 10 min with 0.1 M glycine in cytoskeleton fix buffer. Then cells were washed with TBS (20 mM Tris and154 mM NaCl, pH 8) five times for 5 min each time. Cells were blocked/stabilized using TBS with 1% BSA, 1% FBS, and 5 μM rhodamine phalloidin (Invitrogen). Next, the cells were incubated for 1 h with primary antibody diluted in TBS containing 1% BSA at room temperature. Cells were rinsed five times for 5 min with TBS containing 1% BSA after a 1-h incubation at room temperature with a FITC-conjugated rabbit anti–goat secondary antibody (Sigma-Aldrich). After five final washes with TBS (1% BSA) for 5 min each time, cells were mounted with 50% glycerol, 6 mg/ml N-propyl gallate, and 0.02% sodium azide in TBS.
Live-cell imaging
GFP-actin–expressing MTLn3 cells were used as described previously (Lorenz et al., 2004). In brief, for EGF stimulation, time-lapse series were recorded using a 60× NA 1.4 infinity-corrected optics on a microscope (IX170; Olympus) supplemented with a computer-driven cooled charge-coupled device camera (Sensicam QE; Cooke Corporation) and operated by IPLab Spectrum software (VayTek). Time lapse was recorded at a rate of 1 frame/10 s and analyzed in Image J. Cells were initially transfected on 100-mm culture dishes and split after 24 h onto glass-bottom dishes (MatTek Corporation). 44 h after transfection, cells were then starved with Leibovitz's L15 medium (Invitrogen) supplemented with 0.35% BSA for 3–4 h. Cells were then stimulated with a bath application of 5 nM EGF (Invitrogen) treated at 37°C. Cells were imaged for 1 min before and 6 min after EGF stimulation.
Electron microscopy: preparation of samples for rapid freezing, freeze drying, and rotary shadowing (FDS)
MTLn3 siRNA–treated cells were grown on 5-mm glass coverslips, starved for 3 h, and then stimulated with EGF for 3 min. After the postfixation step, coverslips were rinsed three times in water and processed for FDS based on the procedure described by Hartwig (1992). In brief, fixed coverslips were washed with two changes of distilled water. They were placed on a specimen mount of the rapid freezing apparatus (CF100; Life Cell) and frozen by slamming them into a liquid nitrogen–cooled copper block. Freezing tabs containing the frozen coverslips were transferred to a liquid nitrogen–cooled stage of a freeze fracture apparatus (CFE-50; Cressington), the stage temperature was raised to −90°C for 30 min, and the rotary was shadowed at a 45° angle with 1.2–1.3 nm tantalum-tungsten followed by 2.5 nm carbon at 90°. Replicas were separated from the coverslip with 25% hydrofluoric acid, washed in distilled water, and picked up on formvar-coated copper grids. The samples were observed using a transmission electron microscope (100CX; JEOL) at 100 kV. By convention, images were viewed as negatives.
BE assay
BE assays were visualized using a previously described protocol (Chan et al., 1998) with slight modifications. In brief, cells grown on Mattek dishes were stimulated with EGF at 37°C, and permeabilized in the presence of 0.45 μM biotin-labeled actin in buffer C (138 mM KCl, 10 mM Pipes, pH 6.9, 0.1 mM ATP, 3 mM EGTA, pH 6.9, and 4 mM MgCl2) with 0.025% saponin and 1% BSA for 1 min. After a brief rinse in buffer C, cells were fixed in 3.7% formaldehyde in cytoskeleton stabilization buffer for 5 min, followed by a 10-min incubation in 0.1 M glycine in cytoskeleton stabilization buffer. After a rinse in TBS, samples were incubated with 5 μM phalloidin for 20 min in TBS/BSA/FCS, pH 8.1, and washed five times for 5 min with TBS/BSA. Samples were incubated with 1:50 CY5-conjugated mouse anti-biotin antibody in TBS/BSA for 1 h, and then washed five times for 5 min with TBS/BSA and mounted in 50% glycerol in TBS, pH 8.1, and 6 mg/ml N-propyl gallate. Quantification of fluorescence intensity (see Microscopy and fluorescence quantification) versus distance from the cell periphery was used to determine the number of free BEs in the leading edge. The mean intensities, which correspond to the zone between 0 and 0.66 μm inside the cell, plotted separately versus time reveal that the generation of free BEs in response to EGF stimulation follows two transients with the two peaks corresponding to 1 and 3 min after addition of EGF. siRNA and plasmid transfections were performed as described in Constructs and siRNA.
Microscopy and fluorescence quantification
Pictures were taken using either a 20× NA 0.75 or 60× NA 1.4 infinity-corrected optics on a microscope (IX170) supplemented with a computer-driven cooled charge-coupled device camera and operated by IPLab Spectrum software.
For BE, tropomyosin, and WAVE2 localization analysis, cells were fixed as described in Immunofluorescence. Images were analyzed using a macro for Image designed to measure cell edge fluorescence intensity, developed by the Analytical Imaging Facility of the Albert Einstein College of Medicine. Cells were traced and the macro automated the collection of pixel intensity in a perimeter of the cell starting at 1.98 μm outside the cell and extending to 4.18 μm inside the cell in 0.11-μm steps (background was automatically subtracted from the measured mean fluorescence intensity). The mean intensity corresponds to the zone between 0 and 0.66 μm inside the cell, plotted separately versus time. For area time-lapse analysis, Image J was used to quantitate cell area at each time point. For all fixed cells, image acquisition was done at room temperature.
Online supplemental material
Fig. S1 shows that WAVE1 KD has no affect on lamellipod formation. Fig. S2 shows quantitation of protein expression of siRNA KD cells. Fig. S3 shows that N-WASP/WAVE2 KD has a more prominent tropomyosin compartment at the cell periphery and decreased Arp2/3 localization and that mDia1 KD inhibits tropomyosin at the cell periphery. Fig. S4 shows that KD of either mDia1 or 2 does not affect lamellipod protrusion at the leading edge. Fig. S5 shows Cdc42 and Rac activity in N-WASP/WAVE2 KD cells. Video 1 shows that upon EGF stimulation, control MTLn3 cells exhibit lamellipod extension and accumulation of F-actin at the leading edge of the lamellipod. Video 2 shows that N-WASP/WAVE2 siRNA KD cells exhibit jagged protrusions and increased filopod formation upon EGF stimulation. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200708123/DC1.
Supplemental Material
Acknowledgments
This work was supported by grants GM38511 (J. Condeelis), CA10023 (J.M. Backer), and ACS RSG-05-033-01-CSM (A. Alberts).
Abbreviations used in this paper: Arp, actin-related protein; BE, barbed end; CRIB, Cdc42/Rac interactive binding domain; DN, dominant negative; FH2, formin homology 2; KD, knockdown; N-WASP, neural WASP; RBD, Rho binding domain; WASP, Wiskott-Aldrich syndrome protein.
References
- Alberts, A.S. 2001. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J. Biol. Chem. 276:2824–2830. [DOI] [PubMed] [Google Scholar]
- Alberts, A.S., N. Bouquin, L.H. Johnston, and R. Treisman. 1998. Analysis of RhoA-binding proteins reveals an interaction domain conserved in heterotrimeric G protein beta subunits and the yeast response regulator protein Skn7. J. Biol. Chem. 273:8616–8622. [DOI] [PubMed] [Google Scholar]
- Bailly, M., I. Ichetovkin, W. Grant, N. Zebda, L.M. Machesky, J.E. Segall, and J. Condeelis. 2001. The F-actin side binding activity of the Arp2/3 complex is essential for actin nucleation and lamellipod extension. Curr. Biol. 11:620–625. [DOI] [PubMed] [Google Scholar]
- Biyasheva, A., T. Svitkina, P. Kunda, B. Baum, and G. Borisy. 2004. Cascade pathway of filopodia formation downstream of SCAR. J. Cell Sci. 117:837–848. [DOI] [PubMed] [Google Scholar]
- Chan, A.Y., S. Raft, M. Bailly, J.B. Wyckoff, J.E. Segall, and J.S. Condeelis. 1998. EGF stimulates an increase in actin nucleation and filament number at the leading edge of the lamellipod in mammary adenocarcinoma cells. J. Cell Sci. 111:199–211. [DOI] [PubMed] [Google Scholar]
- Condeelis, J., and J.E. Segall. 2003. Intravital imaging of cell movement in tumours. Nat. Rev. Cancer. 3:921–930. [DOI] [PubMed] [Google Scholar]
- Condeelis, J., R.H. Singer, and J.E. Segall. 2005. The great escape: when cancer cells hijack the genes for chemotaxis and motility. Annu. Rev. Cell Dev. Biol. 21:695–718. [DOI] [PubMed] [Google Scholar]
- Copeland, J.W., and R. Treisman. 2002. The Diaphanous-related formin mDia1 controls serum response factor activity through its effects on actin polymerization. Mol. Biol. Cell. 13:4088–4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland, J.W., S.J. Copeland, and R. Treisman. 2004. Homo-oligomerization is essential for F-actin assembly by the formin family FH2 domain. J. Biol. Chem. 279:50250–50256. [DOI] [PubMed] [Google Scholar]
- DesMarais, V., I. Ichetovkin, J. Condeelis, and S.E. Hitckcock-DeGregori. 2002. Spatial regulation of actin dynamics: a tropomyosin-free, actin-rich compartment at the leading edge. J. Cell Sci. 115:4649–4660. [DOI] [PubMed] [Google Scholar]
- DesMarais, V., F. Macaluso, J. Condeelis, and M. Bailly. 2004. Synergistic interaction between the Arp2/3 complex and cofilin drives stimulated lamellipod extension. J. Cell Sci. 117:3499–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sibai, M., P. Nalbant, H. Pang, R.J. Flinn, C. Sarmiento, F. Macaluso, M. Cammer, J.S. Condeelis, K.M. Hahn, and J.M. Backer. 2007. Cdc42 is required for EGF-stimulated protrusion and motility in MTLn3 carcinoma cells. J. Cell Sci. 120:3465–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sibai, M., O. Pertz, H. Pang, S.C. Yip, M. Lorenz, M. Symons, J.S. Condeelis, K.M. Hahn, and J.M. Backer. 2008. RhoA/ROCK-mediated switching between Cdc42- and Rac1-dependent protrusion in MTLn3 carcinoma cells. Exp. Cell Res. 10.1016/j.yexcr.2008.01.016. [DOI] [PMC free article] [PubMed]
- Fukuoka, M., S. Suetsugu, H. Miki, K. Fukami, T. Endo, and T. Takenawa. 2001. A novel neural Wiskott-Aldrich syndrome protein (N-WASP) binding protein, WISH, induces Arp2/3 complex activation independent of Cdc42. J. Cell Biol. 152:471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton, S.L., K.L. Anderson, T.P. Kole, R.S. Fischer, A. Ponti, S.E. Hitchcock-DeGregori, G. Danuser, V.M. Fowler, D. Wirtz, D. Hanein, and C.M. Waterman-Storer. 2005. Cell migration without a lamellipodium: translation of actin dynamics into cell movement mediated by tropomyosin. J. Cell Biol. 168:619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, E.S., and H.N. Higgs. 2004. Actin cytoskeleton: formins lead the way. Curr. Biol. 14:R520–R522. [DOI] [PubMed] [Google Scholar]
- Hartwig, J.H. 1992. Mechanisms of actin rearrangements mediating platelet activation. J. Cell Biol. 118:1421–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs, H.N. 2005. Formin proteins: a domain-based approach. Trends Biochem. Sci. 30:342–353. [DOI] [PubMed] [Google Scholar]
- Innocenti, M., A. Zucconi, A. Disanza, E. Frittoli, L.B. Areces, A. Steffen, T.E.B. Stradal, P.P.D. Fiore, M.-F. Carlier, and G. Scita. 2004. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat. Cell Biol. 6:319–327. [DOI] [PubMed] [Google Scholar]
- Kempiak, S.J., and J.E. Segall. 2004. Stimulation of cells using EGF-coated magnetic beads. Sci. STKE. 2004:pl1. [DOI] [PubMed] [Google Scholar]
- Kheir, W.A., J.C. Gevrey, H. Yamaguchi, B. Isaac, and D. Cox. 2005. A WAVE2-Abi1 complex mediates CSF-1-induced F-actin-rich membrane protrusions and migration in macrophages. J. Cell Sci. 118:5369–5379. [DOI] [PubMed] [Google Scholar]
- Kovar, D.R. 2006. Molecular details of formin-mediated actin assembly. Curr. Opin. Cell Biol. 18:11–17. [DOI] [PubMed] [Google Scholar]
- Lorenz, M., V. DesMarais, F. Macaluso, R.H. Singer, and J. Condeelis. 2004. Measurement of barbed ends, actin polymerization, and motility in live carcinoma cells after growth factor stimulation. Cell Motil. Cytoskeleton. 57:207–217. [DOI] [PubMed] [Google Scholar]
- Machesky, L.M., R.D. Mullins, H.N. Higgs, D.A. Kaiser, L. Blanchoin, R.C. May, M.E. Hall, and T.D. Pollard. 1999. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl. Acad. Sci. USA. 96:3739–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki, H., and T. Takenawa. 1998. Direct binding of the verprolin-homology domain in N-WASP to actin is essential for cytoskeletal reorganization. Biochem. Biophys. Res. Commun. 243:73–78. [DOI] [PubMed] [Google Scholar]
- Miki, H., K. Miura, and T. Takenawa. 1996. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. EMBO J. 15:5326–5335. [PMC free article] [PubMed] [Google Scholar]
- Miki, H., T. Sasaki, Y. Takai, and T. Takenawa. 1998. a. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature. 391:93–96. [DOI] [PubMed] [Google Scholar]
- Miki, H., S. Suetsugu, and T. Takenawa. 1998. b. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 17:6932–6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki, H., H. Yamaguchi, S. Suetsugu, and T. Takenawa. 2000. IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature. 408:732–735. [DOI] [PubMed] [Google Scholar]
- Mogilner, A., and G. Oster. 2003. Force generation by actin polymerization II: the elastic ratchet and tethered filaments. Biophys. J. 84:1591–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern, J.P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins, R.D., W.F. Stafford, and T.D. Pollard. 1997. Structure, subunit topology, and actin-binding activity of the Arp2/3 complex from Acanthamoeba. J. Cell Biol. 136:331–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrin, S., and H. Mellor. 2005. The Rho family GTPase Rif induces filopodia through mDia2. Curr. Biol. 15:129–133. [DOI] [PubMed] [Google Scholar]
- Peng, J., B.J. Wallar, A. Flanders, P.J. Swiatek, and A.S. Alberts. 2003. Disruption of the diaphanous-related formin Drf1 gene encoding mDia1 reveals a role for Drf3 as an effector for Cdc42. Curr. Biol. 13:534–545. [DOI] [PubMed] [Google Scholar]
- Pruyne, D., M. Evangelista, C. Yang, E. Bi, S. Zigmond, A. Bretscher, and C. Boone. 2002. Role of formins in actin assembly: nucleation and barbed-end association. Science. 297:612–615. [DOI] [PubMed] [Google Scholar]
- Rohatgi, R., L. Ma, H. Miki, M. Lopez, T. Kirchhausen, T. Takenawa, and M.W. Kirschner. 1999. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 97:221–231. [DOI] [PubMed] [Google Scholar]
- Sahai, E., and M.F. Olson. 2006. Purification of TAT-C3 exoenzyme. Methods Enzymol. 406:128–140. [DOI] [PubMed] [Google Scholar]
- Schirenbeck, A., T. Bretschneider, R. Arasada, M. Schleicher, and J. Faix. 2005. The Diaphanous-related formin dDia2 is required for the formation and maintenance of filopodia. Nat. Cell Biol. 7:619–625. [DOI] [PubMed] [Google Scholar]
- Sidani, M., D. Wessels, G. Mouneimne, M. Ghosh, S. Goswami, C. Sarmiento, W. Wang, S. Kuhl, M. El-Sibai, J.M. Backer, et al. 2007. Cofilin determines the migration behavior and turning frequency of metastatic cancer cells. J. Cell Biol. 179:777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapper, S.B., F. Takeshima, I. Anton, C.-H. Liu, S.M. Thomas, D. Nguyen, D. Dudley, H. Fraser, D. Purich, M. Lopez-Ilasaca, et al. 2001. N-WASP deficiency reveals distinct pathways for cell surface projections and microbial actin-based motility. Nat. Cell Biol. 3:897–904. [DOI] [PubMed] [Google Scholar]
- Steffen, A., K. Rottner, J. Ehinger, M. Innocenti, G. Scita, J. Wehland, and T.E. Stradal. 2004. Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J. 23:749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu, S., H. Miki, and T. Takenawa. 1999. Identification of two human WAVE/SCAR homologues as general actin regulatory molecules which associate with the Arp2/3 complex. Biochem. Biophys. Res. Commun. 260:296–302. [DOI] [PubMed] [Google Scholar]
- Suetsugu, S., H. Miki, H. Yamaguchi, T. Obinata, and T. Takenawa. 2001. Enhancement of branching efficiency by the actin filament-binding activity of N-WASP/WAVE2. J. Cell Sci. 114:4533–4542. [DOI] [PubMed] [Google Scholar]
- Sukumvanich, P., V. DesMarais, C.V. Sarmiento, Y. Wang, I. Ichetovkin, G. Mouneimne, S. Almo, and J. Condeelis. 2004. Cellular localization of activated N-WASP using a conformation-sensitive antibody. Cell Motil. Cytoskeleton. 59:141–152. [DOI] [PubMed] [Google Scholar]
- Takenawa, T. 2001. WASP and WAVE family proteins: key molecules for rapid rearrangement of cortical actin filaments and cell movement. J. Cell Sci. 114:1801–1809. [DOI] [PubMed] [Google Scholar]
- Tominaga, T., E. Sahai, P. Chardin, F. McCormick, S.A. Courtneidge, and A.S. Alberts. 2000. Diaphanous-related formins bridge Rho GTPase and Src tyrosine kinase signaling. Mol. Cell. 5:13–25. [DOI] [PubMed] [Google Scholar]
- Wallar, B.J., and A.S. Alberts. 2003. The formins: active scaffolds that remodel the cytoskeleton. Trends Cell Biol. 13:435–446. [DOI] [PubMed] [Google Scholar]
- Wallar, B.J., A.D. DeWard, J.H. Resau, and A.S. Alberts. 2007. RhoB and the mammalian Diaphanous-related formin mDia2 in endosome trafficking. Exp. Cell Res. 313:560–571. [DOI] [PubMed] [Google Scholar]
- Wang, W., J.B. Wyckoff, V.C. Frohlich, Y. Oleynikov, S. Huttelmaier, J. Zavadil, L. Cermak, E.P. Bottinger, R.H. Singer, J.G. White, et al. 2002. Single cell behavior in metastatic primary mammary tumors correlated with gene expression patterns revealed by molecular profiling. Cancer Res. 62:6278–6288. [PubMed] [Google Scholar]
- Wang, W., S. Goswami, K. Lapidus, A.L. Wells, J.B. Wyckoff, E. Sahai, R.H. Singer, J.E. Segall, and J.S. Condeelis. 2004. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res. 64:8585–8594. [DOI] [PubMed] [Google Scholar]
- Watanabe, N., T. Kato, A. Fujita, T. Ishizaki, and S. Narumiya. 1999. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat. Cell Biol. 1:136–143. [DOI] [PubMed] [Google Scholar]
- Wyckoff, J., W. Wang, E.Y. Lin, Y. Wang, F. Pixley, E.R. Stanley, T. Graf, J.W. Pollard, J. Segall, and J. Condeelis. 2004. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 64:7022–7029. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, H., M. Lorenz, S. Kempiak, C. Sarmiento, S. Coniglio, M. Symons, J. Segall, R. Eddy, H. Miki, T. Takenawa, and J. Condeelis. 2005. Molecular mechanisms of invadopodium formation: the role of the N-WASP–Arp2/3 complex pathway and cofilin. J. Cell Biol. 168:441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip, S.C., M. El-Sibai, S.J. Coniglio, G. Mouneimne, R.J. Eddy, B.E. Drees, P.O. Neilsen, S. Goswami, M. Symons, J.S. Condeelis, and J.M. Backer. 2007. The distinct roles of Ras and Rac in PI 3-kinase-dependent protrusion during EGF-stimulated cell migration. J. Cell Sci. 120:3138–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.