Abstract
As part of continuing studies of the venom components present in Conus austini (syn.: Conus cancellatus), a vermivorous cone snail collected in the western Gulf of Mexico, Mexico, two major peptides, as14a and as14b, were purified and characterized. Their amino acid sequences were determined by automatic Edman sequencing after reduction and alkylation. Their molecular masses, established by matrix-assisted laser desorption ionization time-of-flight mass spectrometry, confirmed the chemical analyses and indicated that as14a and as14b have free C-termini. Each peptide contains four Cys residues arranged in a pattern (C-C-C-C, framework 14). The primary structure of as14a is GGVGRCIYNCMNSGGGLNFIQCKTMCY (experimental monoisotopic mass 2,883.92 Da; calculated monoisotopic mass 2,884.20 Da), whereas that of as14b is RWDVDQCIYYCLNGVVGYSYTECQTMCT (experimental monoisotopic mass 3,308.63 Da; calculated monoisotopic mass 3,308.34 Da). Both purified peptides elicited scratching and grooming activity in mice, and as14b also caused body and rear limb extension and tail curling immediately upon injection. The high sequence similarity of peptide as14a with peptide vil14a from the vermivorous C. villepinii suggests that the former might block K+ channels.
Keywords: Conoidea, Conidae, cone snail, Conus austini, conotoxins, vermivorous, worm-hunting, four Cys, framework 14
1. Introduction
The venoms of numerous aquatic animals, such as jellyfish, anemones, and cone snails, have been shown to contain a variety of proteinaceous compounds that facilitate immobilization and digestion of prey and are also used for defense [13]. These compounds often take the form of enzymes that promote tissue invasion, and of peptide toxins that interact with specific cell surface receptors and ion channels. In Conus, the toxins are primarily used to stun, immobilize, and kill prey [27]. It has been estimated that the venom of each Conus species has between 50 and 200 biologically active peptides, named conotoxins, which are mostly cysteine-rich peptides of 7–40 amino acids. Conotoxins typically contain 1 to 4 disulfide bridges, and cysteine residues are separated by 0 to 6 amino acids [25]. The cysteines are arranged in only a few patterns, each of which is characteristic for a given gene superfamily and is associated with limited group of pharmacological targets [26]. Most conotoxins affect the function of ion channels and receptors in a highly potent and specific way; thus, some of them are widely used as pharmacological reagents in neuroscience, and several are being developed as diagnostic and therapeutic agents [12, 19]. Recently, a conotoxin from Conus magus was approved for the treatment of chronic pain [36].
Although the vast majority of Conus species (~75%) prey exclusively on polychaete worms, most studies have been done on conotoxins from fish- and molluskeating species; in fact, only ~10–15% of the species envenomate fish, and a similar percentage preys on other gastropods [17]. Up to now, almost all the cone snails studied were collected in coral reefs of the Indo-Pacific region [17, 31].
Here we describe the isolation and sequencing of two novel conotoxins from a vermivorous cone, Conus austini, collected in the Gulf of Mexico. These peptides, named as14a and as14b, are major components of C. austini venom, and both elicit behavioral changes when injected intracranially into mice. Both peptides show sequence similarity with peptides vil14a and flf14a-c from C. villepinii and C. floridanus floridensis, respectively [23]. Based on the high structural similarity of peptide as14a with peptide vil14a, we suggest that it might block K+ channels.
2. Materials and methods
2.1. Materials
Octadecyl- (C18) and octyl- (C8) Reversed-Phase High Performance Liquid Chromatography (RP-HPLC) columns were from Vydac (Hesperia CA). Reagents for N-terminal sequencing were from Applied Biosystems (Foster City CA). HPLC-grade acetonitrile (ACN) was purchased from Caledon (Georgetown, Ontario, Canada); 4-vinylpyridine from Sigma (St. Louis MO) was distilled under vacuum; trifluoroacetic acid (TFA) was from Fluka (Buchs, Switzerland).
2.2 Specimen collection
Adult C. austini specimens were collected by shrimping vessels and the research vessel (R/V) Justo Sierra at depths of 60–80 m in muddy areas along the coast of Tamaulipas, Mexico.
2.3. Venom separation and fractionation
Venom ducts were dissected from the animals. Crude venom extract was obtained by homogenizing 10 venom ducts in 5 ml of extraction buffer solution (40% ACN containing 0.1% TFA) at 4 °C. The homogenate was centrifuged at 10,000 × g at 4 °C for 20 min, and the supernatant was lyophilized and stored at −20 °C. Lyophilized whole venom was dissolved in deionized water containing 0.1 % of TFA and then centrifuged at 10,100 × g at 4 °C for 20 min. Total protein was quantified by the Bradford method [3] using bovine serum albumin as standard (Protein Assay Kit; Bio-Rad, Hercules CA). For isolation of the peptides from the crude venom and all subsequent purification steps, solution A consisted of 0.085% of TFA in water, and solution B was 0.10% TFA in 90% ACN. Venom was loaded ~1 mg at a time onto an analytical RP-HPLC C18 column (Vydac 218TP54; 4.6 × 250 mm, 5 μm particle size) provided with a C18 guard column (Vydac 218GK54; 4.6 × 10 mm, 5 μm particle size). Components were eluted at room temperature, first isocratically (5% solution B for 10 min), and then by a linear gradient (5 to 55% of solution B over 100 min) at a flow rate of 1 ml/min. The absorbance was monitored at 220 nm.
2.4. Toxin purification
Two fractions, as14a and as14b, were further purified at room temperature. The first step used the same analytical C18 column employed for the fractionation of the venom, using an isocratic step (20% solution B for 10 min) followed by a gradient of 20 to 35% solution B over 60 min, at a flow rate of 1 ml/min. The second purification step involved an analytical C8 column (Vydac 208TP54; 4.6 × 250 mm, 5 μm particle size) provided with a MetaGuard Nucleosil C8 column (4.6 × 10 mm, 5 μm particle size) (Varian 0120-MG; Torrance CA), using the same elution conditions as above.
2.5. Molecular mass characterization
Samples of the native peptides (~100 pmol) were subjected to matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry on a Voyager DE Mass Spectrometer (Applied Biosystems) equipped with delayed ion extraction. Spectra were obtained in positive reflector mode using sinapinic acid as matrix.
2.6. Sequence determination
Because of the probable presence of disulfide bonds in the peptides, samples of as14a and as14b were subjected to reduction and alkylation before sequencing. Each peptide was dissolved in 100 μl of 0.1 M Tris-HCl, pH 8.0, and 100 mg of guanidine hydrochloride (final concentration, 6 M) was added and dissolved. After addition of 45 μl of 50 mM dithiothreitol (final concentration, 10 mM), the mixture was incubated at 65 °C for 25 min under nitrogen. Subsequently, 4 μl of 4-vinylpyridine (final concentration, 157 mM) was added, and the solution was incubated at room temperature for 16 h under nitrogen [34].
Each sample was desalted at room temperature using an analytical C18 column eluted with an isocratic step (10% solution B for 20 min) followed by a linear gradient (10 to 40% solution B over 30 min), with a flow rate of 1 ml/min.
Amino acid sequencing of pyridylethylated peptides was performed by automated Edman degradation using a Procise 491 Protein Sequencing System (Applied Biosystems).
2.7. Biological assay
Four-week-old male mice (strain CD-1, 24–27 g body weight) were used. The samples of as14a and as14b were dissolved in 0.9% normal saline solution. Mice were injected intracerebrally with 30 μl of the peptide sample (120 pmol/mouse, n = 3). The animals were observed to detect any behavioral change or death, for up to 3 h post-injection. Control animals were injected with 30 μl of 0.9 % normal saline solution (n = 3) [4].
2.8. Sequence alignment
In order to make sequence comparisons CLUSTAL W (1.83), was employed using the default settings (http://www.ebi.ac.uk/clustalw/) [35].
3. Results
3.1. Venom fractionation and toxin purification
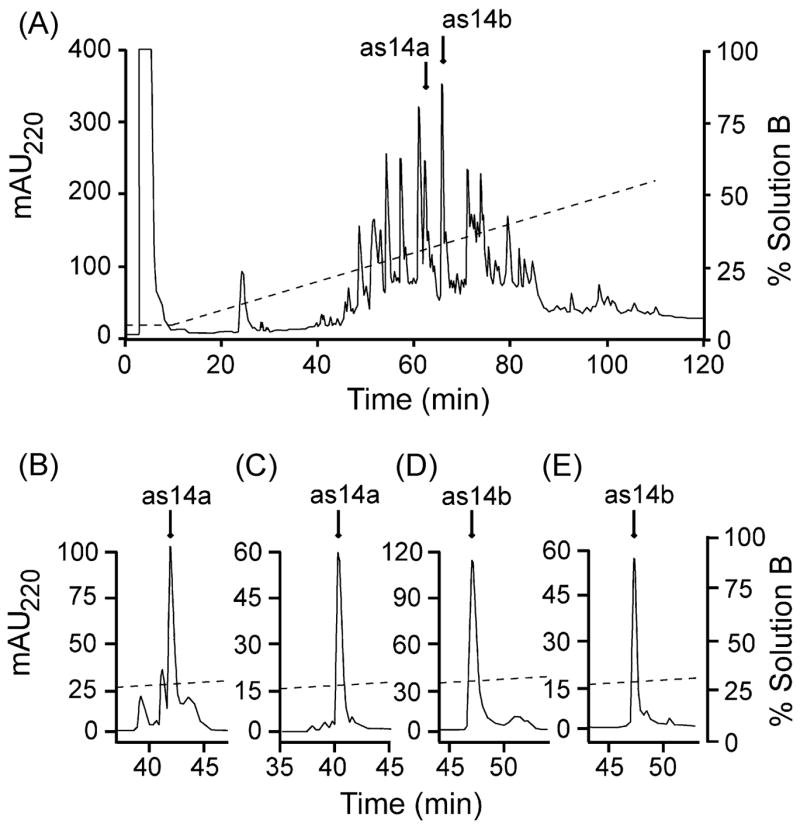
We continued to characterize the components of the venom of C. austini [37] by isolating two prominent peaks, named as14a and as14b, from the RP-HPLC of the venom crude extract (Figure 1A). These peptides were further purified, and their elution positions are shown in Figure 1B–E (see legend).
Figure 1.
Purification of peptides as14a and as14b. Panel A) Fractionation of the venom extract (~1 mg total protein) by an analytical C18 column eluted with an isocratic step at 5% solution B for 10 min followed by a linear gradient from 5–55% solution B over 100 min at 1 ml/min. HPLC solutions were: 0.1% (v/v) TFA in water (solution A), and 0.085% (v/v) TFA in 90% aqueous ACN (solution B). The absorbance was monitored at 220 nm. The peaks highlighted by arrows in panel A were further purified on the same column, using an isocratic step (20% solution B for 10 min) and then increasing solution B by 0.25%/min at 1 ml/min (as14a, panel B; as14b, panel D). The peaks highlighted by arrows in panels B (as14a) and D (as14b) were further purified on an analytical C8 column using the same elution procedure as above (as14a, panel C; as14b, panel E).
3.2. Mass spectrometry
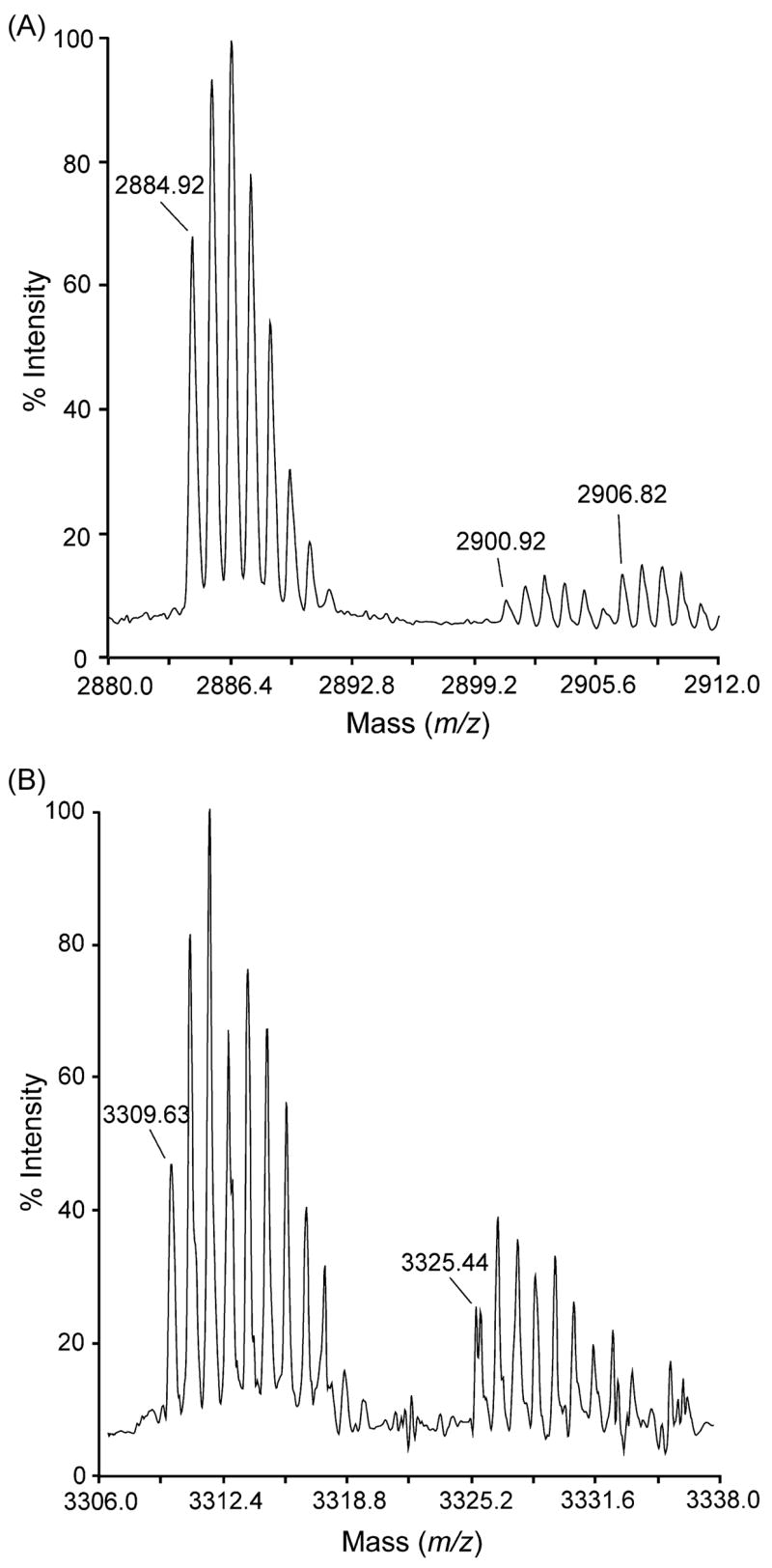
The MALDI-TOF mass spectra of peptides as14a and as14b are shown in Figure 2. The m/z signals at 2,884.92 (Figure 2A) and 3,309.63 (Figure 2B) correspond to monoisotopic masses of 2,883.92 Da (as14a) and 3,308.63 Da (as14b). In both cases, there were clusters of minor signals with individual m/z values ~16 units higher than those of the corresponding signal in the major cluster. These minor signals are probably due to molecules containing one oxidized Met residue. In the case of as14a, a second cluster of minor signals with individual m/z values ~22 units higher than those of the corresponding signal in the major cluster was also observed; these minor signals are probably derived from a small fraction of sodium adducts.
Figure 2.
MALDI-TOF mass spectra of conotoxins as14a and as14b. (A) The m/z signal obtained at 2884.92 ([M + H]+) for as14a indicates a monoisotopic mass of 2883.92 Da. (B) The m/z signal at 3309.63 ([M + H]+) for as14b corresponds to a monoisotopic mass of 3308.63 Da. Clusters of minor signals on the right correspond to molecules with oxidized Met residues (panels A and B) and sodium adducts (panel A) (see text).
3.3. Determination of amino acid sequence
Automatic microsequencing after reduction and pyridylethylation gave single sequences with 27 unambiguous amino acid residues for as14a and as14b (Figure 3A). On the basis of the chemical sequences and assuming two disulfide bridges, the theoretical monoisotopic masses for as14a and as14b are, respectively, 2,884.20 Da and 3,207.29 Da for a free C-terminus, and 2,883.20 Da and 3,206.28 Da for an amidated C-terminus. Thus, the mass spectrometry analysis of peptide as14a (2,883.92 Da) confirmed its chemical sequence and indicates that Cys residues are present as disulfides; in addition, this result suggests that this peptide has a free C-terminus. The oxidation of one Met residue at position 11 or 25 in as14a may account for the cluster of minor signals in the mass spectrum noted above. In the case of peptide as14b the mass analysis (3,308.63 Da) revealed that the chemical sequence was incomplete by 102.35 Da if an amidated C-terminal residue is proposed, and by 101.34 Da if a free C- terminus is considered. This latter value is very close to the monoisotopic mass of a Thr residue (101.05 Da). If one assumes that a free Thr residue is the C-terminus of as14b, its theoretical monosisotopic mass would be 3,308.34 Da, which is in very good agreement with the experimental mass (3,308.63 Da). Thus, we propose that as14b is a 28-residue peptide with 2 disulfide bonds and a free C-terminus. The oxidation of one Met residue at position 26 in as14b may account for the cluster of minor signals in the mass spectrum.
Figure 3.

A) Sequences of peptides as14a and as14b from C. austini and their comparison by a CLUSTAL W alignment. B) Alignment of peptides as14a and as14b with other framework 14-conotoxins (either purified or cloned) from C. planorbis (pl14a, pl14.1-3), C. ferrugineus (fe14.1-2), C. geographus (Con14), C. villepinii (vil14a), C. floridanus floridensis (flf14a-c), and C. litteratus (lt14a). The one-letter code is employed for standard amino acids. For post-translational modifications: γ, γ-carboxyglutamate; &, amidated C-terminus. Identical residues (*), conserved substitutions (:), and semiconserved substitutions (.) are shown for some pairs of peptides (intermediate lines) and for all the peptides (bottom line). Residues potentially involved in functional Lys/Tyr dyads in vil14a and as14a are in bold type and doubly underlined. C) Comparison of intercysteine spacings of the framework 14-conotoxins known so far.
3.4. Biological effects of the purified peptides
The intracranial injection of peptides as14a and as14b (120 pmol/mouse) in 28-day-old mice caused increased scratching and grooming activities as compared to the control mice. The biological activity of as14b was much more apparent and lasted 20 min longer than that of as14a, and in addition included an initial extension of the body and rear limbs and curling of the tail (Table 1).
Table 1.
Estimated potencies (EC50 ± 95% confidence intervals) and maximal bioluminescence response of all the peptides tested on tick (BmLK3) and mosquito (E10) receptor transfected cell lines. EC50 are an estimate of the concentration required to induce a half-maximal response.
| Peptides | Tick receptor, (BmLK3 cell line) | Mosquito receptor, (E10 cell line) | ||
|---|---|---|---|---|
| EC50 ± C I | Maximal Bioluminescence response at 1μM | EC50 ± C I | Maximal Bioluminescence response at 1μM | |
| AFSWGa | 100 ± 70 | 70 ± 30 | ||
| FASWGa | 586 ± 40nM | 5600 ± 300 | 400 ± 50 | |
| FFAWGa | 64 ± 15nM | 12800 ± 900 | 621 ± 70nM | 3050 ± 150 |
| FFSAGa | 200 ± 20 | 30 ± 20 | ||
| FFSWAa | 417 ± 40nM | 10600 ± 1000 | 2.8 ± 0.2μM | 1830 ± 170 |
| FFSWGa | 590 ± 60 nM | 10800 ± 400 | 525 ± 100 | |
| FFWGa | 100 ± 90 | 30 ± 20 | ||
| FFFWa | 50 ± 40 | 50 ± 30 | ||
| FFFWG-OH | 300 ± 50 | 70 ± 40 | ||
| FFFSWGa | 259 ± 50nM | 13000 ± 900 | 562 ± 70nM | 10000 ± 800 |
| FF[Aib]WGa | 29 ± 10nM | 12700 ± 900 | 445 ± 110nM | 9300 ± 900 |
4. Discussion
The present study reports the purification and the biochemical and biological characterization of two peptides, as14a and as14b, isolated from the venom of Conus austini, a vermivorous species found in the Gulf of Mexico [37]. These peptides have identical amino acids at 11 positions (40.7% sequence identity), including 4 Cys residues (Fig. 3A); thus, they might belong to the same gene superfamily and pharmacological family, which remain to be determined.
The Cys residues of peptides as14a-b are arranged in the pattern C-C-C-C (where “-“ represents one or more non-Cys residues), which was found for the first time in Con14, a “scratcher” peptide from the piscivorous C. geographus [5, 28]. Recently, this arrangement of Cys residues, named framework 14 [23], has also been found in peptides from vermivorous species: peptides flf14a-c, from C. floridanus floridensis and peptide vil14a from C. villepinii [23]; peptide pl14a and clones pl14.1-3, from C. planorbis, and clones fe14.1-2, from C. ferrugineus [14]; and clone lt14a, from C. litteratus [30] (Fig. 3B). Despite having the same Cys framework, these peptides have four distinct inter-Cys spacings (Fig. 3C) and two different disulfide connectivities; furthermore, some have been shown to belong to distinct gene superfamilies: the precursors of the peptides from C. planorbis and C. ferrugineus from the Philippines (Cys spacing, CX3CX10CX1C; disulfide bonds, I–III, II–IV) have very similar signal peptides and have defined the J-superfamily of conotoxins [14], whereas the precursor of the peptide from C. litteratus from the South China Sea (Cys spacing, CX3CX3CX2C; disulfide bonds, I–III, II–IV) has a very different signal sequence and has defined the L-superfamily [30]. The sequences of the precursors (and therefore, the types of gene superfamilies) of the peptide from C. geographus (Cys spacing, CX3CX8CX1C; disulfide bonds, unknown), a piscivorous species from the Philippines, and of the peptides with the spacing CX3CX11CX3C, from C. floridanus floridensis, C. villepinii (disulfide bonds, I–IV, II–III) and C. austini (disulfide bonds, unknown), all of which are vermivorous species from the Western Atlantic, remain to be determined. A phylogenetic analysis indicated that, within framework 14-conotoxins, the peptides from the vermivorous species of the Western Atlantic (C. floridanus floridensis and C. villepinii) form a separate branch from the J-conotoxins of the vermivorous species found in the Philippines (C. planorbis and C. ferrugineus) [14], which suggests the existence of a third framework-14 gene superfamily. Thus, these data indicate that distinct types of framework 14-conotoxins are produced by: 1) species of the same feeding type inhabiting different geographic locations, and 2) species from the same habitat having different feeding types. Indeed, this finding is not surprising given that the species harboring the peptides with each of the four different Cys spacings are believed to belong to four distinct taxonomic clades [8, 9].
When peptide as14a was compared (by a CLUSTAL W sequence alignment) with framework 14-conotoxins from the other species, sequence identities ranging from 12% to 92% were obtained. Peptide as14a has the highest sequence identity with peptide vil14a (from C. villepinii, a vermivorous species from the Western Atlantic), which blocks potassium channels in PC12 cells [23]. The 3D structure of this latter peptide, obtained by nanoNMR analysis and comparative modeling based on the structure of -κhefutoxin-1 from the scorpion Heterometrus fulvipes [33], shows that residues Lys-23 and Tyr-8, and Lys-23 and Tyr-27 are separated by ~ 6A, forming two potential functional Lys/Tyr dyads, a motif that has been found in diverse toxins that block potassium channels from scorpions, sea anemones [7], and Conus snails [32]. Peptide as14a has Lys and Tyr residues at the same positions (Fig. 3B), and therefore it might also contain two Lys/Tyr dyads for binding and blocking potassium channels; however, these possibilities need to be confirmed, respectively, by the determination of the 3D structure, and by electrophysiological assays.
The comparison of peptide as14b with the other framework 14-conotoxins yielded sequence identities from 8% to 74%. Peptide as14b has the highest sequence identity with peptide flf14b (from C. floridanus floridensis, a vermivorous species from the Western Atlantic) [23]. The high sequence identity between these two peptides suggests that both might have the same pharmacological site of action, which remains to be identified.
Thus, the peptides characterized in this work are most related to peptides from other vermivorous species from the Western Atlantic. It is interesting that C. austini expresses two classes of framework 14-conotoxins so far found separately in C. villepinii and C. floridanus floridensis [23]. These findings are consistent with an earlier proposal that C. austini, C. villepinii, and C. floridanus floridensis belong to the same taxonomical clade of vermivorous cones [9].
The biological activities of many conotoxins have been evaluated by direct injection of a venom fraction into the central nervous system (CNS) of mice, and this has proven to be an effective method for identifying components active on vertebrates [15, 18, 29], even when peptides were isolated from vermivorous and molluscivorous cones, whose prey are invertebrates. Examples of conotoxins from vermivorous Conus that are active on mice are: α-ImI and α-ImII, from C. imperialis (seizures and tremors; target nicotinic ACh receptors) [22]; contryphan-Vn, of C. ventricosus (stiff tail syndrome; target Ca2+-dependent K+ channels) [21]; and conorfamide-Sr1, of C. spurius (hyperactivity syndrome; target unknown) [20]. In addition, several “scratcher” peptides have been isolated from the venom of vermivorous Conus species: Lys-conopressin-G of C. imperialis, which acts on vasopressin receptors [24], and QcIIIA, QcIIIB, and QcVIA of C. quercinus, with unknown pharmacological targets [1].
The intracranial injection in mice of peptides as14a and as14b elicited a symptomatology characterized by increased scratching and grooming behavior (Table 1). The biological activity of these peptides from C. austini (whose natural prey are polychaete worms) [37] in the mouse CNS might be explained by the moderate conservation of the amino acid sequence and structure of ion channels across the evolution of the distinct eukaryotic taxa, from invertebrates to vertebrates [2]. The activity of peptides as14a and as14b is similar to that produced by the injection of peptide Con14 in the same organism [5, 28]; this result, which suggests that the three peptides might have the same, as yet unidentified, type of molecular target in the mouse CNS, is rather surprising given the low degrees of sequence identity between peptide Con14 and peptides as14a and as14b; according to the multiple CLUSTAL W alignment, only 2 non-Cys residues are shared, by each of these peptides and peptide Con14 (Fig. 3B). Undoubtedly, it is important to identify the molecular targets of these peptides in order to understand the correlation between their structures and the behaviors they elicit.
Conotoxins with a broad range of phylogenetic specificity, such as the two novel “scratcher” peptides as14a and as14b from C. austini described in this work, might become useful tools to study the structure, function, and evolution of ion channels or receptors that determine the physiology of the nervous system of invertebrates and vertebrates [6]. It is interesting that peptide as14b contains sequences (-YY- and -VVG-) that are present in peptides that produce effects in the CNS, such as neuropeptides Y and YY [16], bovine neuropeptide B [11], and some deltorphins [10]; these structural elements of peptide as14b could contribute to the biological activity observed in the CNS of mice.
Acknowledgments
This work was supported by Grants: GM 48677, from The National Institute of General Medical Sciences (B.M.O.); 41477-Q, from Consejo Nacional de Ciencia y Tecnología-México (CONACYT), and IN-204403, from the Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica, Universidad Nacional Autónoma de México (E.P.H.C.); 43754 from CONACYT (M.B.A.). A.Z.-C. was supported by fellowships from CONACYT (144622), from Dirección General de Estudios de Posgrado, Universidad Nacional Autónoma de México. We thank the officers and crew of the RV Justo Sierra during the campaign SIGSBEE V, under the direction of Dr. Elva Escobar. We thank the Mass Spectrometry Facility of the Chemistry Department, University of Utah, Salt Lake City (supported by the National Science Foundation [Grant CHE-9708413] and the University of Utah Institutional Funds Committee) for the mass analysis. We gratefully acknowledge the assistance of María Eugenia Ramos Aguilar for the collection of C. austini specimens, of Carmen Frías Castañeda for bioassay on mice, and of Dr. Dorothy D. Pless for revision of the manuscript. We also thank Lic. Pilar Galarza for the retrieval of bibliographic references.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abogadie FC, Ramilo CA, Corpuz GP, Cruz LJ. Biologically active peptides from Conus quercinus, a worm-hunting species. Trans Natl Acad Sci Tech (Philippines) 1990;12:219–32. [Google Scholar]
- 2.Anderson PAV, Greenberg RM. Phylogeny of ion channels: clues to structure and function. Comp Biochem Physiol B. 2001;129:17–28. doi: 10.1016/s1096-4959(01)00376-1. [DOI] [PubMed] [Google Scholar]
- 3.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Clark C, Olivera BM, Cruz LJ. A toxin from Conus geographus venom which acts on the vertebrate central nervous system. Toxicon. 1981;19:691–9. doi: 10.1016/0041-0101(81)90106-9. [DOI] [PubMed] [Google Scholar]
- 5.Craig AG. The characterization of conotoxins. J Toxicol-Toxin Rev. 2000;19:53–93. [Google Scholar]
- 6.Cruz LJ, Ramilo CA, Corpuz GP, Olivera BM. Conus peptides: phylogenetic range of biological activity. Biol Bull. 1992;183:159–64. doi: 10.2307/1542418. [DOI] [PubMed] [Google Scholar]
- 7.Dauplais M, Lecoq A, Song JX, Cotton J, Jamin N, Gilquin B, et al. On the convergent evolution of animal toxins: conservation of a dyad of functional residues in potassium channel-blocking toxins with unrelated structures. J Biol Chem. 1997;272:4302–9. doi: 10.1074/jbc.272.7.4302. [DOI] [PubMed] [Google Scholar]
- 8.Duda TF, Jr, Kohn AJ. Species-level phylogeography and evolutionary history of the hyperdiverse marine gastropod genus Conus. Mol Phylogenet Evol. 2005;34:257–72. doi: 10.1016/j.ympev.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 9.Duda TF, Jr, Rolan E. Explosive radiation of Cape Verde Conus, a marine species flock. Mol Ecol. 2005;14:267–72. doi: 10.1111/j.1365-294X.2004.02397.x. [DOI] [PubMed] [Google Scholar]
- 10.Erspamer V. The opioid peptides of the amphibian skin. Int J Dev Neurosci. 1992;10:3–30. doi: 10.1016/0736-5748(92)90003-i. [DOI] [PubMed] [Google Scholar]
- 11.Fujii R, Yoshida H, Fukusumi S, Habata Y, Hosoya M, Kawamata Y, et al. Identification of a neuropeptide modified with bromine as an endogenous ligand for GPR7. J Biol Chem. 2002;277:34010–16. doi: 10.1074/jbc.M205883200. [DOI] [PubMed] [Google Scholar]
- 12.Grant MA, Morelli XJ, Rigby AC. Conotoxins and structural biology: a prospective paradigm for drug discovery. Curr Protein Pept Sci. 2004;5:235–48. doi: 10.2174/1389203043379710. [DOI] [PubMed] [Google Scholar]
- 13.Halstead BW. Poisonous and venomous marine animals of the world. New Jersey: The Darwin Press, Inc; 1988. [Google Scholar]
- 14.Imperial JS, Bansal PS, Alewood PF, Daly NL, Craik DJ, Sporning A, et al. A novel conotoxin inhibitor of Kv1. 6 channel and nAChR subtypes defines a new superfamily of conotoxins. Biochemistry. 2006;45:8331–40. doi: 10.1021/bi060263r. [DOI] [PubMed] [Google Scholar]
- 15.Jimenez EC, Shetty RP, Lirazan M, Rivier J, Walker C, Abogadie FC, et al. Novel excitatory Conus peptides define a new conotoxin superfamily. J Neurochem. 2003;85:610–21. doi: 10.1046/j.1471-4159.2003.01685.x. [DOI] [PubMed] [Google Scholar]
- 16.Keire DA, Bowers CW, Solomon TE, Reeve JR., Jr Structure and receptor binding of PYY analogs. Peptides. 2002;23:305–21. doi: 10.1016/s0196-9781(01)00602-7. [DOI] [PubMed] [Google Scholar]
- 17.Kohn AJ, Perron FE. Life history and biogeography: patterns in Conus. Oxford: Oxford University Press; 1994. [Google Scholar]
- 18.Lirazan MB, Hooper D, Corpuz GP, Ramilo CA, Bandyopadhyay P, Cruz LJ, et al. The spasmodic peptide defines a new conotoxin superfamily. Biochemistry. 2000;39:1583–8. doi: 10.1021/bi9923712. [DOI] [PubMed] [Google Scholar]
- 19.Livett BG, Gayler KR, Khalil Z. Drugs from the sea: conopeptides as potential therapeutics. Curr Med Chem. 2004;11:1715–23. doi: 10.2174/0929867043364928. [DOI] [PubMed] [Google Scholar]
- 20.Maillo M, Aguilar MB, López-Vera E, Craig A, Bulag G, Olivera BM, et al. Conorfamide Sr1, a Conus venom peptide belonging to the FMRFamide-like family of neuropeptides. Toxicon. 2002;40:401–17. doi: 10.1016/s0041-0101(01)00176-3. [DOI] [PubMed] [Google Scholar]
- 21.Massilia GR, Schinina ME, Ascenzi P, Polticelli F. Contryphan-Vn: a novel peptide from the venom of the Mediterranean snail Conus ventricosus. Biochem Biophys Res Commun. 2001;288:908–13. doi: 10.1006/bbrc.2001.5833. [DOI] [PubMed] [Google Scholar]
- 22.McIntosh JM, Yoshikami D, Mahe E, Nielsen DB, Rivier JE, Gray WR, et al. A nicotinic acetylcholine receptor ligand of unique specificity. α-Conotoxin ImI. J Biol Chem. 1994;269:16733–9. [PubMed] [Google Scholar]
- 23.Möller C, Rahmankhah S, Lauer-Fields J, Bubis J, Fields GB, Marí F. A novel conotoxin framework with a helix-loop-helix (Cs α/α) fold. Biochemistry. 2005;44:15986–96. doi: 10.1021/bi0511181. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen DB, Dykert J, Rivier JE, McIntosh JM. Isolation of Lys-conopressin-G from the venom of the worm-hunting snail Conus imperialis. Toxicon. 1994;32:845–8. doi: 10.1016/0041-0101(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 25.Norton RS, Olivera BM. Conotoxins down under. Toxicon. 2006;48:780–98. doi: 10.1016/j.toxicon.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 26.Olivera BM. Conus venom peptides, receptor and ion channel targets and drug design: 50 million years of neuropharmacology. Mol Biol Cell. 1997:2101–9. doi: 10.1091/mbc.8.11.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olivera BM. Conus venom peptides: reflections from the biology of clades and species. Annu Rev Ecol Syst. 2002;33:25–42. [Google Scholar]
- 28.Olivera BM, River J, Clark C, Ramilo CA, Corpuz GP, Abogadie FC, et al. Diversity of Conus neuropeptides. Science. 1990;249:257–63. doi: 10.1126/science.2165278. [DOI] [PubMed] [Google Scholar]
- 29.Olivera BM, Cruz LJ, Yoshikami D. Effects of Conus peptides on the behavior of mice. Curr Op Neurobiol. 1999;9:772–7. doi: 10.1016/s0959-4388(99)00033-1. [DOI] [PubMed] [Google Scholar]
- 30.Peng C, Tang S, Pi C, Liu J, Wang F, Wang L, et al. Discovery of a novel class of conotoxin from Conus litteratus, lt14a, with a unique cysteine pattern. Peptides. 2006;27:2174–81. doi: 10.1016/j.peptides.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 31.Rockel D, Korn W, Kohn AJ. Manual of the living Conidae. Vol. 1. Indo-Pacific. Wiesbaden: Verlag Christa Hemmen; 1995. [Google Scholar]
- 32.Savarin P, Guenneugues M, Gilquin B, Lamthanh H, Gasparini S, Zinn-Justin S, et al. Three-dimensional structure of κ-conotoxin PVIIA, a novel potassium channel-blocking toxin from cone snails. Biochemistry. 1998;37:5407–16. doi: 10.1021/bi9730341. [DOI] [PubMed] [Google Scholar]
- 33.Srinivasan KN, Sivaraja V, Huys I, Sasaki T, Cheng B, Kumar TK, et al. κ-Hefutoxin1, a novel toxin from the scorpion Heterometrus fulvipes with unique structure and function. Importance of the functional dyad in potassium channel selectivity. J Biol Chem. 2002;277:30040–7. doi: 10.1074/jbc.M111258200. [DOI] [PubMed] [Google Scholar]
- 34.Tarr GE. Manual Edman sequencing system. In: Shively JE, editor. Methods of Protein Microcharacterization. Clifton: Humana Press; 1986. pp. 155–194. [Google Scholar]
- 35.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment though sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wermeling DP. Ziconotide, an intrathecally administered N-type calcium channel antagonist for the treatment of chronic pain. Pharmacotherapy. 2005;25:1084–94. doi: 10.1592/phco.2005.25.8.1084. [DOI] [PubMed] [Google Scholar]
- 37.Zugasti-Cruz A, Maillo M, López-Vera E, Falcón A, Heimer de la Cotera EP, Olivera BM, et al. Amino acid sequence and biological activity of a γ-conotoxin-like peptide from the worm-hunting snail Conus austini. Peptides. 2006;27:506–11. doi: 10.1016/j.peptides.2005.07.021. [DOI] [PubMed] [Google Scholar]


