Abstract
A novel peptide, conorfamide-Sr2 (CNF-Sr2), was purified from the venom extract of Conus spurius, collected in the Caribbean Sea off the Yucatan Peninsula. Its primary structure was determined by automated Edman degradation and amino acid analysis, and confirmed by electrospray ionization mass spectrometry. Conorfamide-Sr2 contains 12 amino acids and no Cys residues, and it is only the second FMRFamide-related peptide isolated from a venom. Its primary structure GPMγDPLγIIRI-nh2, (γ, gamma-carboxyglutamate;-nh2, amidated C-terminus; calculated monoisotopic mass, 1,468.72 Da; experimental monoisotopic mass, 1,468.70 Da) shows two features that are unusual among FMRFamide-related peptides (FaRPs, also known as RFamide peptides), namely the novel presence of gamma-carboxyglutamate, and a rather uncommon C-terminal residue, Ile. CNF-Sr2 exhibits paralytic activity in the limpet Patella opea and causes hyperactivity in the freshwater snail Pomacea paludosa and in the mouse. The sequence similarities of CNF-Sr2 with FaRPs from marine and freshwater mollusks and mice might explain its biological effects in these organisms. It also resembles FaRPs from polychaetes (the prey of C. spurius), which suggests a natural biological role. Based on these similarities, CNF-Sr2 might interact with receptors of these three distinct types of FaRPs, G-protein-coupled receptors, Na+ channels activated by FMRFamide (FaNaCs), and acid-sensing ion channels (ASICs). The biological activities of CNF-Sr2 in mollusks and mice make it a potential tool to study molecular targets in these and other organisms.
Keywords: Conoidea, cone snail, Conus spurius, conorfamide, gamma-carboxyglutamate, FaRP
1. Introduction
Some of the most effective and complex marine venoms are those produced by snails of the genus Conus (the cone snails, or cones), most of which inhabit the Indopacific Ocean [28]. There are approximately 500 known species of cones, and all are equipped with a highly specialized venom apparatus that is primarily used to paralyze and capture their prey [52]. Cones can be divided into three main groups according to the type of prey on which they feed: fish-hunting, mollusk-hunting, and worm-hunting [27]. Each Conus species produces more than 100 different, physiologically active venom peptides [40] that are injected into the prey through a disposable, hollow radular tooth which, in the case of the piscivorous Conus catus, has been shown to be propelled by a high-speed ballistic mechanism [58]. Conus venoms are extremely potent, and cone snails are thought to have caused approximately 30 human fatalities [21].
The peptide toxins in Conus venoms generally have 7–40 amino acid residues and often contain diverse post-translational modifications [6]. They may be divided into two groups: those that contain zero or only one disulfide bond (the “conopeptides”), and those containing two or more disulfide bonds (the “conotoxins”). Both types of toxins may bind membrane proteins such as voltage-gated ion channels, ligand-gated ion channels, and G-protein-coupled receptors, causing disruption of neuromuscular transmission and consequently paralysis in their prey [39]. In general, conopeptides and conotoxins bind their molecular targets with high affinity and specificity, and some of them have been used as molecular tools for studies of ion channels and receptors [3] and as potential pharmaceuticals [20, 32, 60]; furthermore, a conotoxin that blocks N-type calcium channels (ω-MVIIA) was recently approved for the treatment of chronic pain [61].
Conotoxins and conopeptides are generated by proteolytic processing of precursors with the general structure: signal peptide - “pro” region - mature toxin. Based on the sequence of the signal peptide of their precursors, conotoxins are classified into gene superfamilies, whereas their biological activities and molecular targets define pharmacological families [39]. So far, nine superfamilies of conotoxins (T, A, J, O, M, P, I, S, L) have been identified [26, 39, 43], and there is a correlation between the superfamily and the arrangement of Cys residues within the sequences of the mature conotoxins belonging to it. Each gene superfamily includes from 1 to 4 pharmacological families. Based on the same criterion, five superfamilies of conopeptides have been proposed: two of them (the conopressins and the contryphans) contain one disulfide bridge, whereas the other three (the contulakins, the conantokins, and the conorfamides) do not have Cys residues [39]. The group of the conorfamides was defined by conorfamide-Sr1, a peptide that belongs to the family of the FMRFamide-related peptides (FaRPs) [35].
Here we report the biochemical and preliminary biological characterization of the second member of the conorfamide family, conorfamide-Sr2 (CNF-Sr2), isolated from the venom of Conus spurius. CNF-Sr2 shows a novel feature within FaRPs, the presence of gamma-carboxyglutamate, and it has biological activity in mollusks and mice.
2. Materials and methods
2.1. Collection and storage of specimens
The specimens of Conus spurius were collected by shrimp-fishing vessels along the coasts of the Caribbean Sea off the Yucatan Peninsula, Mexico, from depths of 37 m to 100 m. The captured organisms were frozen at −20°C and, upon arrival at the laboratory, they were stored at −70°C until used.
2.2. Crude venom extraction
The shells of three specimens were broken mechanically to release the organisms, and the dissections of the venom ducts were made with the help of clamps and scissors while keeping the body of the snail on a Petri dish on ice. The venom ducts were homogenized in extraction solution (2% (v/v) trifluoroacetic acid (TFA) in 40% (v/v) aqueous acetonitrile (MeCN)). The mixture was centrifuged at 14,000 × g for 15 min at 4°C, the supernatant was saved, and the process was repeated four times. The supernatants (crude venom) were combined and taken to dryness (Speed Vac Plus SC110A; Savant, Holbrook NY). The crude venom was stored at −20°C until purified.
2.3. Crude venom fractionation and peptide purification
The crude venom was dissolved in 1 ml of a mixture (7:3 v/v) of solution A (0.1% (v/v) aqueous TFA) and solution B (0.1% (v/v) TFA in 90% (v/v) aqueous MeCN). It was then centrifuged at 11,130 × g for 15 min at room temperature. The supernatant was fractionated at room temperature by Reversed Phase-High Performance Liquid Chromatography using a C18 semipreparative column (Vydac; 218TP510, 10 × 250 mm, 5 μm particle diameter; 300 Å pore size) equipped with a C18 guard column (Vydac; 218GCC1210, 10 × 10 mm, 12 μm particle diameter, 300 Å pore size), filter (Alltech; 28689, 4 mm, 2 μm pore size), and a 2-ml sample-loading loop. The elution was conducted at 2.5 ml/min, employing solutions A and B. A linear gradient from 5 to 100% solution B was developed over 95 min.
Further purification of a major peak was achieved on a semipreparative C8 column (YMC; OC12S05-2510WT, 250 × 10 mm, 5 μm particle diameter, 120 Å pore size) with the corresponding C8 precolumn (YMC; OC12S05G304WTA, 4 mm × 23 mm, 5 μm particle diameter, 120 Å pore size) and a 2-ml sample-loading loop. The elution was performed at 1 ml/min, employing the same solutions as above. After an isocratic step at 24% solution B for 15 min, a linear gradient from 24 to 45% solution B was developed over 63 min.
Final purification was carried out on an analytical C8 column (Agilent; Eclipse XDB-C8, 4.6 × 150 mm, 5 μm particle diameter, 80 Å pore size) with a C8 precolumn (Varian, 310-793-2300; 4.6 × 15 mm, 5 μm particle size, 100 Å pore size) and a 200-μl sample-loading loop. The elution was conducted at 1 ml/min, employing the same solution A as above, and a different solution B containing 0.085% (v/v) TFA in 60% (v/v) aqueous MeCN. A linear gradient from 45 to 55% solution B over 40 min was employed. In all cases, the effluents were monitored at 220 nm.
2.4. Primary structure analysis
Sequence analysis and peptide quantitation were performed by automated Edman degradation in a Procise 491 Protein Sequencing System (Applied Biosystems, Foster City CA), using the Pulsed-Liquid Method.
2.5. Molecular mass determination
Electrospray ionization mass spectrum (ESI-MS) was measured using a Micromass Quattro II Triple Quadrupole Mass Spectrometer with MassLynx operating system. The sample (approximately 100 pmol) was dissolved in 100 μl of 50% (v/v) aqueous methanol and automatically infused with a flow rate of 0.05 ml/min in the same solvent. The sample was scanned over the m/z range 50–2000, with capillary voltage of 2.7 KV and a cone voltage of 50 V. The resulting data were analyzed using MassLynx software. For samples with MW ~1000, the accuracy was estimated to be within 100 ppm (0.01%) or 0.1 Da.
2.6. Amino acid composition
Amino acid analysis of the purified peptide (500 pmol) was performed by vapor-phase acid hydrolysis followed by derivatization with phenylisothiocyanate and quantitation of the resulting phenylthiocarbamyl (PTC) amino acids by RP-HPLC [5]. An amino acid standard solution (AA-S-18; Sigma, St. Louis MO) was hydrolyzed and derivatized in parallel with the peptide sample. Acid hydrolysis was conducted for 72 h in vacuo, in a Reaction Vial Assembly (WAT007363; Waters, Milford MA) containing 6 ml of 1% (v/v) liquified phenol in 30% (w/v) hydrochloric acid (Suprapur; Merck, Darmstadt, Germany). Quantitation of the PTC-derivatives was performed on a Nova-Pak column (WAT011695; 3.9 mm × 300 mm, 4 μm particle diameter, 60 Å pore size; Waters). Solution A was 0.05% (v/v) triethylamine in 0.14 M sodium acetate, adjusted to pH 5.00 with glacial acetic acid; solution B was 60% (v/v) aqueous MeCN. A convex gradient (curve 5, Waters) was developed from 10 to 51% solution B during 20 min at 1 ml/min at 45°C, employing a Waters chromatograph (2487 Dual λ Absorbance Detector, 600 Controller, 600 Pump; Waters). The effluent was monitored at 254 nm. The amino acid composition was calculated from the molar ratio, assuming one Arg residue per molecule, as indicated by automated sequencing.
2.7. Biological activity assays
The peptide was dissolved in normal saline solution (0.9% NaCl). Biological activity was tested in snails, fish, limpets, and mice. Snails and fish were purchased from local venders at Querétaro, Qro.; limpets were collected from the Pacific coast of Mazatlan, Sinaloa, Mexico, and maintained in laboratory aquaria for at least 24 h before use; mice were supplied by the animal facility of the Institute.
Fresh water snails Pomacea paludosa (15.5–20.1 g body weight) were injected in the rear part of the foot using 1-ml insulin syringes provided with 29-gauge hypodermic needles. Test snails (n = 3) were injected with 30 μl of the peptide sample (250 pmol). Control animals (n = 3) were injected with 30 μl of normal saline solution. A positive response was defined as the animal’s loss of ability to stay attached to the vertical glass surface of the aquarium and subsequent contraction of the foot [18]. The snails were observed for 1 h.
Limpets (Patella opea) of 170–230 mg body weight were used. Control limpets (n = 3) were injected with 10 μl of normal saline solution at the base of the foot pedal of an upturned animal. Ten-microliter aliquots (50 and 250 pmol) of the peptide sample were injected in test limpets (n = 3). The injected animals were subsequently placed in their normal position in a Petri dish containing fresh aerated sea water. Paralysis was determined 5 min post-injection. A paralyzed limpet shows an extreme contraction of the foot pedal, resulting in an obvious hardening of the foot and loss of the animal’s ability to cling to a flat surface [19].
Fresh water fish Lebistes reticulatus (200–230 mg body weight) were injected below the rear part of the dorsal fin. Ten microliter aliquots of normal saline solution or peptide sample (250 pmol) were injected into control (n = 3) and test (n = 3) animals, respectively. Fish were observed for up to 2 hours after injection [19].
Male mice (strain CD-1; 21 or 25 days old) were intracranially injected with 30 μl of normal saline solution (control animals, n = 3) or with the same volume containing 250 (n = 3) or 1000 (n = 3) pmol of peptide. After the injection, the mice were placed in a cage for observation for 1 h [41].
3. Results
3.1. Peptide purification
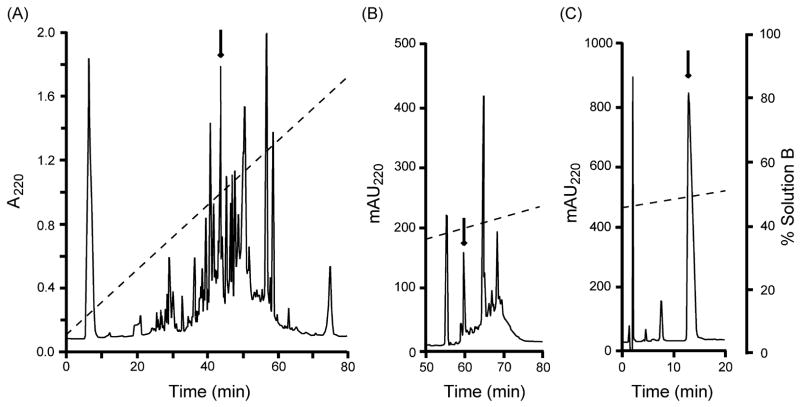
One major peak from the fractionation of the crude venom of Conus spurius on a semipreparative C18 column (arrow in Fig. 1A) was chosen for study. This peak displayed several components after chromatography on a semipreparative C8 column, one of which (arrow in Fig. 1B) was further purified on an analytical C8 column, yielding an apparently pure component (arrow in Fig. 1C).
Figure 1.
Purification of conorfamide-Sr2. (A) Fractionation of the crude extract from the venom ducts of Conus spurius by a semipreparative Vydac C18 column. The elution was conducted at room temperature with a linear gradient from 5 to 100% solution B over 95 min (dashed line) at 2.5 ml/min. HPLC solutions were: 0.1% (v/v) TFA in water (solution A), and 0.085% (v/v) TFA in 90% (v/v) aqueous MeCN (solution B). The absorbance was monitored at 220 nm. (B) The peak highlighted by the arrow in (A) was further purified on a semipreparative Vydac C8 column using a linear gradient from 24% to 45% solution B over 63 min after an isocratic step at 24% solution B for 15 min (dashed line), at 1 ml/min. (C) The peak highlighted by the arrow in (B) was rechromatographed on an analytical Agilent C8 column using a linear gradient from 45% to 55% solution B (0.1% (v/v) TFA in 60% (v/v) aqueous MeCN) over 40 min (dashed line), at 1 ml/min. The gradients are displayed at the same scale (right side).
3.2. Peptide sequencing
The automated Edman sequencing (14 cycles) of the purified peptide showed only one sequence: GPMγDPLγIIR (γ, gamma-carboxyglutamate). At positions 4 and 8 small amounts of PTH-Glu were observed; in both cases, the peaks of PTH-Glu were much smaller than both the preceding and the following residues, which indicates the presence of γ residues at these positions [11]. This phenomenon is thought to result from a small degree of decarboxylation when the peptide is heated under acidic conditions during the cleavage steps of the Edman degradation; although most of the γ residues are stable under these conditions, the anilinothiazolinone derivative of γ is too polar to be extracted [11], and only the PTH-derivative of the small amount of Glu (from the decarboxylation of γ) is observed. No increase of any PTH-derivative was observed at cycles 12–14. Thus, the 11-residue sequence obtained was GPMγDPLγIIR, with a calculated monoisotopic mass of 1,356.62 Da.
3.3. Molecular mass determination
Figure 2 shows the electrospray ionization (ESI) mass spectrum of conorfamide-Sr2, with several monoisotopic m/z signals corresponding to distinct [Mn + H]1+ species. These signals indicate molecular masses of 1,380.7 Da ([M1 + H]1+), 1,424.7 Da ([M2 + H]1+), 1,440.7 Da ([M3 + H]1+), 1,468.7 Da ([M4 + H]1+), and 1,484.6 Da ([M5 + H]1+).
Figure 2.
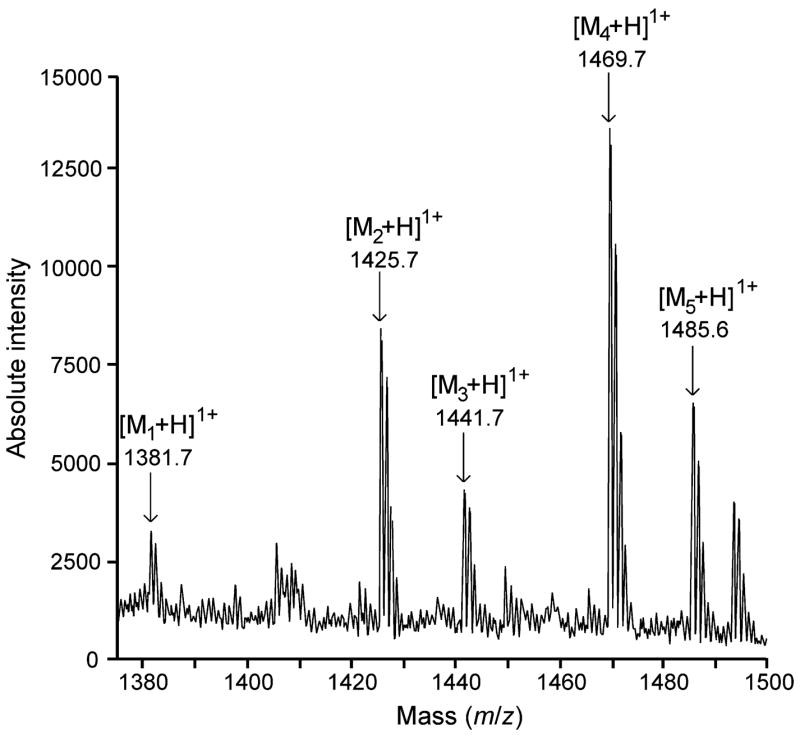
Molecular mass of conorfamide-Sr2. ESI-mass spectrum: the m/z signal at 1,469.7 indicates a monoisotopic mass of 1,468.70 Da for the native peptide. The signals at 1,425.7 and 1,381.7 correspond, respectively, to the monodecarboxylated and the didecarboxylated species generated during the mass spectrometry analysis. The signals at 1,485.6 and 1,441.7 correspond, respectively, to nondecarboxylated and monodecarboxylated species in which oxidation of Met-3 occurred during the purification process.
The difference of 44 Da between the pairs [M1 + H]1+ - [M2 + H]1+ and [M2 + H]1+ - [M4 + H]1+ equals the mass of a carboxyl moiety (monoisotopic mass, 44.01 Da) and indicates distinct degrees of decarboxylation produced during the analysis [11]. Thus, [M4 + H]1+ corresponds to the nondecarboxylated molecule (1,468.7 Da), whereas [M2 + H]1+ (1,424.7 Da) and [M1 + H]1+ (1,380.7 Da) correspond to the monodecarboxylated and the didecarboxylated species, respectively.
The difference of ~16 Da between the pairs [M2 + H]1+ - [M3 + H]1+ and [M4 + H]1+ - [M5 + H]1+ equals the mass of an oxygen atom (monoisotopic mass, 15.99 Da). Given that the purified peptide contains an easily oxidizable residue at position 3 (Met), the most probable explanation for the presence of the species [M3 + H]1+ and [M5 + H]1+ is the oxidation of this residue [11]. Thus, [M5 + H]1+ (1,484.6 Da) corresponds to the oxidized, nondecarboxylated molecule, whereas [M3 + H]1+ (1,440.7 Da) corresponds to the oxidized, monodecarboxylated species.
However, the mass of the nonoxidized nondecarboxylated species (1,468.7 Da) is 112.08 Da higher than the calculated mass (1,356.62 Da) for the 11-residue sequence determined chemically. This difference indicates that one or more C-terminal residues were not detected during the automated sequencing. In order to identify the missing residue(s) we determined the amino acid composition of the purified peptide (see next section).
3.4. Amino acid analysis
Given that only about 50% hydrolysis of the Ile-Ile bond (positions 9 and 10 of conorfamide-Sr2) occurs under standard conditions for acid hydrolysis (6 M HCl plus 0.1% (w/v) phenol at 110°C for 24 h in vacuo) [2], we employed a 72-h hydrolysis. Under these conditions, hydroxy-Pro was not detected, and only one (0.92) Leu and one (1.13) Gly residue per molecule were determined, consistent with the chemical sequencing (Table 1). However, three (2.88) Ile residues per molecule were obtained instead of the two detected during the automated Edman degradation (Table 1); thus, we concluded that the missing residue is Ile.
Table 1.
Amino acid composition of conorfamide-Sr2
| Amino acid | Yielda (pmol) | Residues per moleculeb | Chemical sequence | Proposed sequence |
|---|---|---|---|---|
| Asx c | 15.65 | 0.97 | 1 | 1 |
| Glxd | 32.14 | 1.99 | 2 | 2 |
| Ser | 0 | 0 | 0 | 0 |
| Gly | 18.26 | 1.13 | 1 | 1 |
| His | 0 | 0 | 0 | 0 |
| Arg | 16.19 | 1.00 | 1 | 1 |
| Thr | 0 | 0 | 0 | 0 |
| Ala | 0 | 0 | 0 | 0 |
| Pro | 32.32 | 2.00 | 2 | 2 |
| Tyr | 0 | 0 | 0 | 0 |
| Val | 0 | 0 | 0 | 0 |
| Met | 7.36 | 0.45e | 1 | 1 |
| Cys | N.D.f | N.D.f | 0 | 0 |
| Ile | 46.60 | 2.88 | 2 | 3 |
| Leu | 14.90 | 0.92 | 1 | 1 |
| Phe | 0 | 0 | 0 | 0 |
| Trp | N.D.f | N.D.f | 0 | 0 |
| Lys | 0 | 0 | 0 | 0 |
Average values (n = 3) obtained after acid hydrolysis in vacuo at 110°C for 72 h.
Molar ratio: Arg = 1.00.
Asx = Asp + Asn.
Glx = Glu + Gln + γ-carboxyglutamic acid.
Partial destruction of Met residues occurs under these conditions.
N.D., not determined.
However, the calculated mass (1,469.70 Da) for the putative 12-residue sequence containing Ile at the C-terminus (GPMγDPLγIIRI) is greater than the experimental mass (1,468.7 Da) by 1.00 Da. This difference is higher than the expected error (0.01%, or ~0.1 Da for peptides of ~1,000 Da) of ESI mass spectrometry [57]. Thus, we concluded that the C-terminus is amidated, which would decrease its monoisotopic mass by 0.98 Da, based on the difference between an amidated carboxyl group (44.01 Da) and a free carboxyl group (44.99 Da). The calculated monoisotopic mass for the proposed 12-residue sequence, GPMγDPLγIIRI-nh2 (-nh2, amidated C-terminus) (Figure 3), is 1,468.72 Da, in excellent agreement (Δ = 0.02 Da) with the experimental monoisotopic mass (1,468.70 Da).
Figure 3.

Primary structure of conorfamide-Sr2 and comparison with conorfamide-Sr1 and FMRFamide. The one-letter code is employed for standard amino acids; for post-translational modifications: γ, gamma-carboxyglutamate;-nh2, amidated C-terminus. Amino acid identities (*) or conservative substitutions (:) are indicated between FMRFamide and conorfamide-Sr1, and between conorfamide-Sr1 and conorfamide-Sr2.
3.5. Biological activity
As shown in Table 2, the purified peptide elicited effects in mice, snails, and limpets, but it had no effect in fish.
Table 2.
Biological activity of conorfamide-Sr2
| Organism | Dose (pmol) | Effect |
|---|---|---|
| Mouse | 250 | Brief clonus of limbsa; mild hyperactivityb |
| 1000 | Long clonusc of limbs; strong hyperactivityd | |
| Pomacea paludosa (freshwater snail) | 250 | Hyperactivitye |
| Patella opea (limpet) | 50 | Mild paralysisf |
| 250 | Strong paralysisg | |
| Lebistes reticulates (freshwater fish) | 50, 250 | No effect |
~ 10 s.
Continuous wandering.
~ 1 min.
Continuous wandering and jumping.
Continuous displacement around the aquarium.
Partial loss of attachment to the Petri dish; no reaction after being touched.
Complete stillness; loss of attachment to the Petri dish; no reaction after being touched.
Control mice showed transient symptoms associated with the injection (stillness for 2 to 9 min), followed by a period of alternating displacement and exploration in the cage, stillness, and grooming behavior for 15 to 18 min after the injection; then the mice became still and fell asleep. The mice injected with 250 pmol did not display the period of stillness observed in the control animals; they started to displace immediately and then showed a brief (~ 10 s) clonus of their limbs; after ~ 2 min they again started to displace and to explore the cage for ~ 3 min, after which they remained still for 4 min; then they resumed the displacement and, after ~ 3 min, they displayed contractions of the body. Finally, after ~ 5 min, they continued, with brief periods of stillness, the exploratory and grooming behaviors up to 43 to 53 min post-injection, after which they fell asleep. At 1000 pmol, the clonus was immediate and longer (~ 1 min), and two mice started to walk around the cage; one of them continued this behavior for 12 min and fell asleep after 45 min, whereas the other walked and jumped for 30 min. The third mouse stayed still for 18 min; then it started to move slowly for 5 min; after 10 min it started to jump, trying to grab the cover of the cage, for 19 min; after a brief period it started to jump again and continued doing this until the end of the period of observation (60 min).
Both control and test snails were attached with their foot to an aquarium glass. Immediately upon injection (250 pmol), one test snail dropped off, and its foot muscle shrank; the effect lasted less than 2 min; then, it began to move actively around the bottom of the aquarium for 22 min. Another snail started to move just after the injection, and continued doing so for 5 min; during this period it turned its head and extended its siphon for 10 s. The third snail did not show any apparent change upon the injection. By contrast, after the injection, the three control snails remained attached to the aquarium glass, without displacement.
Injection of the peptide (50 pmol) at the base of the limpets’ foot pedals caused them to lose their ability to cling to a flat substrate with normal strength; also, they did not react (contract) if touched. These symptoms developed within seconds and apparently were enhanced when the dose was increased (250 pmol). Control limpets kept their ability to attach to the Petri dish and reacted (contracted) immediately when touched.
Neither control (10 μl of normal saline solution) nor test fish (50 pmol/10 μl, and 250 pmol/10 μl) showed any apparent behavioral change upon injection.
Evidently, these results are preliminary, and do not yield information on the molecular target of the peptide; however, they indicate that it is active on the vertebrate nervous system, and on the molluskan muscle.
4. Discussion
The peptide characterized in this work, from the venom of the worm-hunting snail C. spurius, was named conorfamide-Sr2 (CNF-Sr2), because it has 33% sequence identity with conorfamide-Sr1 (CNF-Sr1) from the same species [35]; they share the first three N-terminal residues, and an Arg residue adjacent to the C-terminus; in addition, both peptides have hydrophobic residues at positions 9 and 12, and amidated C-termini (Figure 3).
CNF-Sr1, the first conorfamide isolated from any venom [35], is considered to belong to the group of the FMRFamide-related peptides (FaRPs, also known as RFamide peptides) because of its high local sequence identity (75%) with FMRFamide, the prototype of this class of peptides [46]. This sequence identity includes the C-terminal amide and the Arg residue (Figure 3) that are critical for the activity of FMRFamide on molluskan muscle preparations [42]; in addition, CNF-Sr1 conforms to the common C-terminal motif (F/Y)XRFamide (where X is a hydrophobic residue) that characterizes FaRPs [16]. There is high sequence identity between the four C-terminal residues of CNF-Sr1 and FMRFamide, and both produced a hyperactivity syndrome when injected intracranially into mice [35]. Although CNF-Sr1 is threefold as long as FMRFamide, it has been shown that N-terminal extensions do not necessarily alter the activity of FMRFamide analogs [42].
CNF-Sr2 also produces hyperactivity when injected intracranially into mice, among other symptoms (Table 2). Although the sequence identity of the four C-terminal residues of CNF-Sr2 with FMRFamide is only 25% (the Arg residue), CNF-Sr2 has other similarities with FMRFamide: conservative substitutions for the Phe-1 and the Met residues (which are not critical for activity of FMRFamide analogs [42]); a conservative substitution at the C-terminus (Figure 3), and the C-terminal amidation. Thus, CNF-Sr2 and FMRFamide share not only local sequence similarity (including identities and conservative substitutions), but they also cause similar biological effects in mice. Although FMRFamide is not present naturally in mammals, it has been shown to have a variety of effects on cardiovascular, behavioral, gastrointestinal, and endocrine functions of rats and mice (for review and references see [51]).
CNF-Sr2 has sequence similarity with FaRPs expressed in the CNS of the mouse. Table 3A displays the sequences of some mouse FaRPs, taking the sequence similarity between CNF-Sr2 and FMRFamide as the minimum for inclusion. In the rat, mRNAs encoding the precursor of the homologs of RFamided-related peptides RFRP-1 and RFRP-3 (Table 3A) are expressed in several hypothalamic regions, and the intracerebroventricular (i.c.v.) injection of the human homolog of RFRP-1 increases prolactin secretion; in addition, the i.c.v. administration of the rat homolog of RFRP-1 blocks or reduces morphine-induced anti-nociception in models of acute and persistent pain, respectively. The human homolog of RFRP-1 (hRFRP-1) has been shown to bind G-protein-coupled receptors OT7TO22 (also known as NPFF1) and HLWAR77 (also known as NPFF2) with high affinity. Other mammalian FaRPs with less similarity to CNF-Sr2 (not shown in Table 2) have also been demonstrated to have functions in the central nervous system: neuropeptide FF (NPFF) reduces morphine-induced analgesia and stimulates prolactin secretion; prolactin-releasing peptide (PrRP) is involved in the control of food intake, increases corticotrophin-releasing hormone and gonadotrophin secretion, and induces anti-nociception; kisspeptin (metastin) is involved in the control of reproductive function; peptide 26RFa stimulates food consumption. All these FaRPs bind G-protein-coupled receptors, such as OT7TO22 (NPFF1), HLWAR77 (NPFF2), GPR10, GPR54, and GPR103 (also referred to as SP9155 and AQ27) (for review and references see [9]). Thus, given its sequence similarity with these neuropeptides, the behavioral changes in mice reported in Table 2 might be the result of CNF-Sr2 acting on one or more of these receptors. Another possibility is that CNF-Sr2 affects acid-sensing ion channels (ASICs), which have been shown to be expressed in the mammalian central and peripheral nervous system and to be potentiated by distinct FaRPs; ASICs appear to be involved in normal and abnormal physiological processes such as learning and memory, acquired fear-related behavior, pain sensation in ischemia and inflammation, taste perception, hearing function, and mechanoperception, among others (for review and references see [31]).
Table 3.
Comparison of conorfamide-Sr2 with some FaRPs from mouse (A) and mollusks (B)
| Sequence | Organism or Mouse peptidea | Reference | |
|---|---|---|---|
| GPMγDPLγIIRI* | Conus spurius | This work | |
| (A) | |||
| SVSFQELKDWGAKNVIKMSPAPANKVPHSAANLPLRF* | RFRP-1 | [24] | |
| VNMEAGTRSHFPSLPQRF* | RFRP-3 | [24] | |
| (B) | |||
| FMRF* | Macrocallista nimbosa | [46] | |
| FLRF* | Pomacea paludosa | [45] | |
| GDPFLRF* | Siphonaria pectinata | [48] | |
| EFLRI* | Lymnaea stagnalis | [55] | |
| ZNYLAFPRM* | [44] | ||
| ZFLRI* | [55] | ||
| SGYLAFPRM* | [44] | ||
| QFYRI* | [55] | ||
| ZFYRI* | [55] | ||
| ALSGDAFLRF* | [33] | ||
| (G/S)DPFLRF* | [15] | ||
| (Z/Q/G)GSLFRF* | [25] | ||
| (G/N)TLLRF* | [25] | ||
| HDYMRF* | [54] | ||
| TLFRF* | [25] | ||
| FMRF* | [15] | ||
| LSSFVRI* | Fusinus ferrugineus | [29] | |
| (G/S)SLFRF* | [29] | ||
| MNYLAFPRM* | Helix aspersa | [49] | |
| SGYLAFPRM* | [49] | ||
| (S/N)DPFLRF* | [49] | ||
| ZDPFLRF* | [47] | ||
| ZFYRF* | [50] | ||
| FMRF* | [49] | ||
| FLRF* | [49] | ||
| PMSMLRL* | Aplysia californica | [12] | |
| MNYLAFPRM* | [49] | ||
| ARPGYLAFPRM* | [12] | ||
| GGALFRF* | [13] | ||
| STLFRF* | [13] | ||
| GSLFRF* | [22] | ||
| FMRF* | [56] | ||
| MNYLAFPRM* | Aplysia brasiliana | [37] | |
| FMRF* | [30] | ||
| GDPFLRF* | Helisoma trivolvis | [34] | |
| FMRF* | [34] | ||
| FLRF* | [34] | ||
Names have been taken from the original references. The one-letter code is employed for standard amino acids. For post-translational modifications: γ, gamma-carboxyglutamate; Z, pyroglutamic acid;
, amidated C-terminus. Bold-faced, double-underlined residues indicate identity with conorfamide-Sr2; bold-faced, underlined residues indicate conservative substitutions with conorfamide-Sr2. Some peptides differing only at the N-terminus have been grouped in the same line, indicating the distinct N-termini separated by slashes and between parentheses.
CNF-Sr2 also has biological activity in freshwater and marine mollusks (Table 2). This feature is not unexpected in view of its sequence similarity with FaRPs from these organisms (Table 3B). In mollusks, a variety of distinct FaRPs have been found [48]; they are active in cardiac tissue, noncardiac muscle, and nerve, and are involved in physiological processes such as cardioexcitation [38], neurotransmission, and neuromodulation [54]. Table 3B shows the sequences of molluskan FaRPs that have sequence similarities with CNF-Sr2 that equal or exceed that of the latter with FMRFamide; we have included only gastropod species (with the exception of the bivalve Macrocalista nimbosa), but some of these peptides are also present in other types of mollusks. We comment briefly on some of the known physiological roles of several peptides that have the highest levels of sequence similarity with CNF-Sr2: 1) PMSMLRLamide (myomodulin) potentiates contractions of the accessory radula closer muscle in Aplysia californica [12]; 2) EFLRIamide produces biphasic (inhibitory and excitatory) effects on the heartbeat rate of L. stagnalis [62]; 3) MNYLAFPRMamide (small cardioactive peptide B) enhances the defensive siphon- and gill-withdrawal reflex [1] and causes body wall contractions [10] in addition to its excitatory effects on the isolated heart of Aplysia [7]; 4) ZFYRIamide produces biphasic (inhibitory and excitatory) effects on the heartbeat rate of L. stagnalis [62]; 5) FMRFamide has a variety of effects on molluskan cardiorespiration, feeding, reproduction, and screening-pigment migration (for review and references see [14]); also, it inhibits the output of the siphon-withdrawal reflex circuit [4], produces a brief contraction followed by a prolonged relaxation of the isolated body wall of Aplysia [10], and increases the amplitude of the contraction of the pharyngeal retractor muscle of H. pomatia [23]; 6) FLRFamide has essentially identical activities with FMRFamide on diverse molluskan tissues [45]. Thus, in gastropods, these peptides have numerous and important physiological actions that might be altered by an exogenous analog, such as the peptide characterized in this work; although C. spurius is a vermivorous species, CNF-Sr2 might function as a defensive peptide against other predatory mollusks. FMRFamide and several FaRPs have been identified as ligands of: 1) G-protein-coupled receptors [8, 36], and 2) the invertebrate FMRFamide-gated Na+ channel (FaNaC), a member of the epithelial amiloride-sensitive Na+ channel and degenerin (ENaC/DEG) family of ion channels; this peptide-gated channel is expressed in neurons and to a lower extent in the pedal muscle, and is considered a candidate for peptidergic neurotransmission (for review and references see [31]). Thus, because of its sequence similarity with FMRFamide and other molluskan FaRPs, injection of CNF-Sr2 into mollusks might produce the behavioral changes described in Table 2 by acting on this Na+-selective channel.
CNF-Sr2 displays sequence similarity with several FaRPs from polychaete worms, the prey of C. spurius: 1) AMGMLRMamide (Pev-myomodulin) elicits a strong contraction of the esophagus of the polychaete annelid Perinereis vancaurica [59]; 2) FMRFamide causes relaxation of the longitudinal muscle of the body wall of the sedentary polychaete Sabellastarte magnifica; also, it can control heartbeat and the contraction of longitudinal muscles, and has an antidiuretic action in leeches [17] (for review and references see [53]). Thus, in polychaetes, these peptides have important physiological actions, which might be altered by exogenous analogs. Therefore, it seems probable that the venom peptide characterized in this work, CNF-Sr2, might function as an offensive agent to facilitate the capture of the prey (for example, by interfering with the movements of an escaping worm).
Although the molecular target of CNF-Sr2 has not yet been identified, its biological activities in mice and mollusks make it a potential tool to study molecular targets in these and other organisms. Furthermore, the similarities of CNF-Sr2 with distinct FaRPs suggest that it might interact with G-protein-coupled receptors, Na+ channels activated by FMRFamide (FaNaCs), and/or acid-sensing ion channels (ASICs).
Acknowledgments
This investigation was supported by Grant GM 48677 from The National Institute of General Medical Sciences, to B.M.O., and by Grants: 41477-Q (E.P.H.C.) and 43754-Q (M.B.A.) from Consejo Nacional de Ciencia y Tecnología, México (CONACYT), and IN-204403 from the Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica, Universidad Nacional Autónoma de México (E.P.H.C.). M.M. was supported by a fellowship from Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICYT, Venezuela). We thank Dr. Anthony G. Craig (The Clayton Foundation Laboratories for Peptide Biology, The Salk Institute, San Diego CA, USA) and the Mass Spectrometry Facility of the Chemistry Department, University of Utah, Salt Lake City (supported by the National Science Foundation [Grant CHE-9708413] and the University of Utah Institutional Funds Committee) for the mass analysis. We gratefully acknowledge Dr. Dorothy D. Pless for revising the manuscript. We also thank Lic. Pilar Galarza for retrieving bibliographic references and M.V.Z. Martín García Servín for providing the mice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abrams TW, Castellucci VF, Camargo JS, Kandel ER, Lloyd PE. Two endogenous neuropeptides modulate the gill and siphon withdrawal reflex in Aplysia by presynaptic facilitation involving cAMP-dependent closure of a serotonin-sensitive potassium channel. Proc Natl Acad Sci U S A. 1984;81:7956–60. doi: 10.1073/pnas.81.24.7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen G. In: Sequencing of proteins and peptides. Work TS, Burdon RH, editors. Amsterdam: Elsevier/north Holland; 1981. [Google Scholar]
- 3.Armishaw CJ, Alewood PF. Conotoxins as research tools and drug leads. Curr Protein Pept Sci. 2005;6:221–40. doi: 10.2174/1389203054065437. [DOI] [PubMed] [Google Scholar]
- 4.Belkin KJ, Abrams TW. FMRFamide produces biphasic modulation of the LFS motor neurons in the neural circuit of the siphon withdrawal reflex of Aplysia by activating Na+ and K+ currents. J Neurosci. 1993;13:5139–52. doi: 10.1523/JNEUROSCI.13-12-05139.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bidlingmeyer BA, Cohen SA, Tarvin TL. Rapid analysis of amino acids using pre-column derivatization. J Chromatogr. 1984;336:93–104. doi: 10.1016/s0378-4347(00)85133-6. [DOI] [PubMed] [Google Scholar]
- 6.Buczek O, Bulaj G, Olivera BM. Conotoxins and the posttranslational modification of secreted gene products. Cell Mol Life Sci. 2005;62:3067–79. doi: 10.1007/s00018-005-5283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cawthorpe DR, Rosenberg J, Colmers WF, Lukowiak K, Drummond GI. The effects of small cardioactive peptide B on the isolated heart and gill of Aplysia californica. Can J Physiol Pharmacol. 1985;63:918–24. doi: 10.1139/y85-152. [DOI] [PubMed] [Google Scholar]
- 8.Cazzamali G, Grimmelikhuijzen CJ. Molecular cloning and functional expression of the first insect FMRFamide receptor. Proc Natl Acad Sci U S A. 2002;99:12073–8. doi: 10.1073/pnas.192442799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chartrel N, Tsutsui K, Costentin J, Vaudry H. The Rfamide-related peptides. In: Kastin AJ, editor. Handbook of biologically active peptides. Burlington MA: Academic Press; 2006. pp. 779–86. [Google Scholar]
- 10.Cooper BF, Krontiris-Litowitz JK, Walters ET. Humoral factors released during trauma of Aplysia body wall. II. Effects of possible mediators. J Comp Physiol B. 1989;159:225–35. doi: 10.1007/BF00691743. [DOI] [PubMed] [Google Scholar]
- 11.Craig AG. The characterization of conotoxins. J Toxicol Toxin Rev. 2000;19:53–93. [Google Scholar]
- 12.Cropper EC, Tenenbaum R, Kolks MA, Kupfermann I, Weiss KR. Myomodulin: a bioactive neuropeptide present in an identified cholinergic buccal motor neuron of Aplysia. Proc Natl Acad Sci U S A. 1987;84:5483–6. doi: 10.1073/pnas.84.15.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cropper EC, Brezina V, Vilim FS, Harish O, Price DA, Rosen S, et al. FRF peptides in the ARC neuromuscular system of Aplysia: purification and physiological actions. J Neurophysiol. 1994;72:2181–95. doi: 10.1152/jn.1994.72.5.2181. [DOI] [PubMed] [Google Scholar]
- 14.Di Cosmo A, Di Cristo C. Molluscan bioactive peptides. In: Kastin AJ, editor. Handbook of biologically active peptides. Burlington MA: Academic Press; 2006. pp. 235–39. [Google Scholar]
- 15.Ebberink RH, Price DA, van Loenhout H, Doble KE, Riehm JP, Geraerts WP, et al. The brain of Lymnaea contains a family of FMRFamide-like peptides. Peptides. 1987;8:515–22. doi: 10.1016/0196-9781(87)90018-0. [DOI] [PubMed] [Google Scholar]
- 16.Espinosa E, Carrigan M, Thomas SG, Shaw G, Edison AS. A statistical view of FMRFamide neuropeptide diversity. Mol Neurobiol. 2000;21:35–56. doi: 10.1385/MN:21:1-2:035. [DOI] [PubMed] [Google Scholar]
- 17.Evans BD, Pohl J, Kartsonis NA, Calabrese RL. Identification of RFamide neuropeptides in the medicinal leech. Peptides. 1991;12:897–908. doi: 10.1016/0196-9781(91)90035-n. [DOI] [PubMed] [Google Scholar]
- 18.Fainzilber M, Zlotkin E. A new bioassay reveals mollusk-specificity in molluscivorous Conus venoms. Toxicon. 1992;30:464–9. doi: 10.1016/0041-0101(92)90543-e. [DOI] [PubMed] [Google Scholar]
- 19.Fainzilber M, Gordon D, Hasson A, Spira ME, Zlotkin E. Mollusc-specific toxins from the venom of Conus textile neovicarius. Eur J Biochem. 1991;202:589–95. doi: 10.1111/j.1432-1033.1991.tb16412.x. [DOI] [PubMed] [Google Scholar]
- 20.Grant MA, Morelli XJ, Rigby AC. Conotoxins and structural biology: a prospective paradigm for drug discovery. Curr Protein Pept Sci. 2004;5:235–48. doi: 10.2174/1389203043379710. [DOI] [PubMed] [Google Scholar]
- 21.Gray WR, Olivera BM, Cruz LJ. Peptide toxins from venomous Conus snails. Annu Rev Biochem. 1988;57:665–700. doi: 10.1146/annurev.bi.57.070188.003313. [DOI] [PubMed] [Google Scholar]
- 22.Greenberg MJ, Price DA. Relationships among the FMRFamide-like peptides. Prog Brain Res. 1992;92:25–37. doi: 10.1016/s0079-6123(08)61162-0. [DOI] [PubMed] [Google Scholar]
- 23.Hernadi L, Vehovszky A, Hiripi L, Gyori J, Walker RJ, Elekes K. Neuroanatomical, immunocytochemical, and physiological studies of the pharyngeal retractor muscle and its putative regulatory neurons playing a role in withdrawal and feeding in the snail, Helix pomatia. Cell Tissue Res. 2005;321:257–71. doi: 10.1007/s00441-005-1144-2. [DOI] [PubMed] [Google Scholar]
- 24.Hinuma S, Shintani Y, Fukusumi S, Iijima N, Matsumoto Y, Hosoya M, et al. New neuropeptides containing carboxy-terminal RFamide and their receptor in mammals. Nature Cell Biol. 2000;2:703–8. doi: 10.1038/35036326. [DOI] [PubMed] [Google Scholar]
- 25.Hoek RM, Li KW, van Minnen J, Lodder JC, de Jong-Brink M, Smit AB, et al. LFRFamides: a novel family of parasitation-induced RFamide neuropeptides that inhibit the activity of neuroendocrine cells in Lymnaea stagnalis. J Neurochem. 2005;92:1073–80. doi: 10.1111/j.1471-4159.2004.02927.x. [DOI] [PubMed] [Google Scholar]
- 26.Imperial JS, Bansal PS, Alewood PF, Daly NL, Craik DJ, Sporning A, et al. A novel conotoxin inhibitor of Kv1.6 channel and nAChR subtypes defines a new superfamily of conotoxins. Biochemistry. 2006;45:8331–40. doi: 10.1021/bi060263r. [DOI] [PubMed] [Google Scholar]
- 27.Kohn AJ. The ecology of Conus in Hawaii. Ecol Monogr. 1959;29:47–90. [Google Scholar]
- 28.Kohn AJ, Saunders PR, Weiner S. Preliminary studies of the venom of the marine snail Conus. Annu NY Acad Sci. 1960;90:706–25. doi: 10.1111/j.1749-6632.1960.tb26416.x. [DOI] [PubMed] [Google Scholar]
- 29.Kuroki Y, Kanda T, Kubota I, Ikeda T, Fujisawa Y, Minakata H, et al. FMRFamide-related peptides isolated from the prosobranch mollusk Fusinus ferrugineus. Acta Biol Hung. 1993;44:41–4. [PubMed] [Google Scholar]
- 30.Lehman HK, Price DA, Greenberg MJ. The FMRFamide-like neuropeptide of Aplysia is FMRFamide. Biol Bull. 1984;167:460–6. doi: 10.2307/1541290. [DOI] [PubMed] [Google Scholar]
- 31.Lingueglia E, Deval E, Lazdunski M. FMRFamide-gated sodium channel and ASIC channels: a new class of ionotropic receptors for FMRFamide and related peptides. Peptides. 2006;27:1138–52. doi: 10.1016/j.peptides.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 32.Livett BG, Gayler KR, Khalil Z. Drugs from the sea: conopeptides as potential therapeutics. Curr Med Chem. 2004;11:1715–23. doi: 10.2174/0929867043364928. [DOI] [PubMed] [Google Scholar]
- 33.Loi PK, Tublitz N. Molecular analysis of FMRFamide and FMRFamide-related peptides (FaRPs) in the cuttlefish Sepia officinalis. J Exp Biol. 1997;200:1483–9. doi: 10.1242/jeb.200.10.1483. [DOI] [PubMed] [Google Scholar]
- 34.Madrid KP, Price DA, Greenberg MJ, Khan HR, Saleuddin AS. FMRFamide-related peptides from the kidney of the snail, Helisoma trivolvis. Peptides. 1994;15:31–6. doi: 10.1016/0196-9781(94)90166-x. [DOI] [PubMed] [Google Scholar]
- 35.Maillo M, Aguilar MB, López-Vera E, Craig AG, Bulaj G, Olivera BM, et al. Conorfamide, a Conus venom peptide belonging to the Rfamide family of neuropeptides. Toxicon. 2002;40:401–7. doi: 10.1016/s0041-0101(01)00176-3. [DOI] [PubMed] [Google Scholar]
- 36.Meeusen T, Mertens I, Clynen E, Baggerman G, Nichols R, Nachman RJ, et al. Identification in Drosophila melanogaster of the invertebrate G protein-coupled FMRFamide receptor. Proc Natl Acad Sci U S A. 2002;99:15363–8. doi: 10.1073/pnas.252339599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morris HR, Oanico M, Karplus A, Lloyd PE, Riniker B. Elucidation by FAB-MS of the structure of a new cardioactive peptide from Aplysia. Nature. 1982;300:643–5. doi: 10.1038/300643a0. [DOI] [PubMed] [Google Scholar]
- 38.Moulis A. RFamide neuropeptide actions on the molluscan heart. Acta Biol Hung. 2004;55:335–41. doi: 10.1556/ABiol.55.2004.1-4.39. [DOI] [PubMed] [Google Scholar]
- 39.Norton RS, Olivera BM. Conotoxins down under. Toxicon. 2006;48:780–98. doi: 10.1016/j.toxicon.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 40.Olivera BM, Rivier J, Scott JK, Hillyard DR, Cruz LJ. Conotoxins. J Biol Chem. 1991;266:22067–79. [PubMed] [Google Scholar]
- 41.Olivera BM, Cruz LJ, Yoshikami D. Effects of Conus peptides on the behavior of mice. Curr Opin Neurobiol. 1999;9:772–7. doi: 10.1016/s0959-4388(99)00033-1. [DOI] [PubMed] [Google Scholar]
- 42.Painter SD, Morley JS, Price DA. Structure-activity relations of the molluscan neuropeptide FMRFamide on some molluscan muscles. Life Sci. 1982;31:2471–8. doi: 10.1016/0024-3205(82)90752-4. [DOI] [PubMed] [Google Scholar]
- 43.Peng C, Tang S, Pi C, Liu J, Wang F, Wang L, et al. Discovery of a novel class of conotoxin from Conus litteratus, lt14a, with a unique cysteine pattern. Peptides. 2006;27:2174–81. doi: 10.1016/j.peptides.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 44.Perry SJ, Dobbins AC, Schoefield MG, Piper MR, Benjamin PR. Small cardioactive peptide gene: structure, expression and mass spectrometric analysis reveals a complex pattern of co-transmitters in a snail feeding neuron. Eur J Neurosci. 1999;11:655–62. doi: 10.1046/j.1460-9568.1999.00472.x. [DOI] [PubMed] [Google Scholar]
- 45.Price DA. Evolution of a molluscan cardioregulatory neuropeptide. Am Zool. 1986;26:1007–15. [Google Scholar]
- 46.Price DA, Greenberg MJ. Structure of a molluscan cardio-excitatory neuropeptide. Science. 1977;197:670–1. doi: 10.1126/science.877582. [DOI] [PubMed] [Google Scholar]
- 47.Price GA, Cottrell KE, Doble MJ, Greenberg W, Jorenby W, Lehman HK, et al. A novel FMRFamide-related peptide in Helix: pQDPFLRFamide. Biol Bull. 1985;169:256–66. [Google Scholar]
- 48.Price DA, Davies NW, Doble KE, Greenberg MJ. The variety and distribution of the FMRFamide-related peptides in molluscs. Zool Sci. 1987;4:395–410. [Google Scholar]
- 49.Price DA, Lesser W, Lee TD, Doble KE, Greenberg MJ. Seven FMRFamide-related and two SCP-related cardioactive peptides from Helix. J Exp Biol. 1990;154:421–37. doi: 10.1242/jeb.154.1.421. [DOI] [PubMed] [Google Scholar]
- 50.Price DA, Doble KE, Lesser W, Greenberg MJ, Swiderek KM, Lee TD, et al. The peptide pQFYRFamide is encoded on the FMRFamide precursor of the snail Helix aspersa but does not activate the FMRFamide-gated sodium current. Biol Bull. 1996;191:341–52. doi: 10.2307/1543007. [DOI] [PubMed] [Google Scholar]
- 51.Raffa RB. The action of FMRFamide (Phe-Met-Arg-Phe-NH2) and related peptides on mammals. Peptides. 1988;9:915–22. doi: 10.1016/0196-9781(88)90141-6. [DOI] [PubMed] [Google Scholar]
- 52.Rockel D, Korn W, Kohn AJ. Manual of the living Conidae. Wiesbaden, Germany: Verlag Christa Hemmen; 1995. [Google Scholar]
- 53.Salzet M, Stefano GB. Biochemical evidence for an annelid neuroendocrine system: evolutionary conserved molecular mechanisms. Placebo. 2001;3:54–72. [Google Scholar]
- 54.Santama N, Benjamin PR. Gene expression and function of FMRFamide-related neuropeptides in the snail Lymnaea. Microsc Res Tech. 2000;49:547–56. doi: 10.1002/1097-0029(20000615)49:6<547::AID-JEMT5>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 55.Santama N, Wheeler CH, Skingsley DR, Yeoman MS, Bright K, Kaye I, et al. Identification, distribution and physiological activity of three novel neuropeptides of Lymnaea: EFLRIamide and pQFYRIamide encoded by the FMRFamide gene, and a related peptide. Eur J Neurosci. 1995;7:234–46. doi: 10.1111/j.1460-9568.1995.tb01059.x. [DOI] [PubMed] [Google Scholar]
- 56.Schaefer M, Picciotto MR, Kreiner T, Kaldani RR, Taussig R, Scheller RH. Aplysia neurons express a gene encoding multiple FMRFamide neuropeptides. Cell. 1985;41:457–67. doi: 10.1016/s0092-8674(85)80019-2. [DOI] [PubMed] [Google Scholar]
- 57.Scoble HA, Vath JE, Yu W, Martin SA. Mass spectrometric strategies for the structural characterization of proteins. In: Matsudaira P, editor. A practical guide to protein and peptide purification for microsequencing. 2. San Diego CA: Academic Press, Inc; 1993. pp. 125–53. [Google Scholar]
- 58.Schulz JR, Norton AG, Gilly WF. The projectile tooth of a fish-hunting cone snail: Conus catus injects venom into fish prey using a high-speed ballistic mechanism. Biol Bull. 2004;207:77–9. doi: 10.2307/1543581. [DOI] [PubMed] [Google Scholar]
- 59.Takahashi T, Matsushima O, Morishita F, Fujimoto M, Ikeda T, Minakata H, et al. A myomodulin-CARP-related peptide isolated from a polychaete annelid, Perinereis vancaurica. Zoolog Sci. 1994;11:33–8. [PubMed] [Google Scholar]
- 60.Wang CZ, Chi CW. Conus peptides - a rich pharmaceutical treasure. Acta Biochim Biophys Sin (Shanghai) 2004;36:713–23. doi: 10.1093/abbs/36.11.713. [DOI] [PubMed] [Google Scholar]
- 61.Wermeling DP. Ziconotide, an intrathecally administered N-type calcium channel antagonist for the treatment of chronic pain. Pharmacotherapy. 2005;25:1084–94. doi: 10.1592/phco.2005.25.8.1084. [DOI] [PubMed] [Google Scholar]
- 62.Willoughby D, Yeoman MS, Benjamin PR. Cyclic AMP is involved in cardioregulation by multiple neuropeptides encoded on the FMRFamide gene. J Exp Biol. 1999;202:2595–607. doi: 10.1242/jeb.202.19.2595. [DOI] [PubMed] [Google Scholar]