Abstract
The release of the genome sequences of two strains of Aspergillus niger has allowed systems-level investigations of this important microbial cell factory. To this end, tools for doing data integration of multi-ome data are necessary, and especially interesting in the context of metabolism. On the basis of an A. niger bibliome survey, we present the largest model reconstruction of a metabolic network reported for a fungal species. The reconstructed gapless metabolic network is based on the reportings of 371 articles and comprises 1190 biochemically unique reactions and 871 ORFs. Inclusion of isoenzymes increases the total number of reactions to 2240. A graphical map of the metabolic network is presented. All levels of the reconstruction process were based on manual curation. From the reconstructed metabolic network, a mathematical model was constructed and validated with data on yields, fluxes and transcription. The presented metabolic network and map are useful tools for examining systemwide data in a metabolic context. Results from the validated model show a great potential for expanding the use of A. niger as a high-yield production platform.
Keywords: Aspergillus niger, bibliome, genome, metabolic network, metabolome
Introduction
It is difficult to think of a filamentous fungus where the metabolic capabilities are of greater interest than Aspergillus niger. As a widely used industrial workhorse, the commercial applications of A. niger range from the high-yield production of citric and gluconic acid, through production of a range of industrial enzymes in high titers and the production of heterologous proteins such as chymosin or human interferon (van Brunt, 1986; Dunn-Coleman et al, 1991; Punt et al, 2002; Karaffa and Kubicek, 2003; Singh and Kumar, 2007). With such an impressive diversity of high-yield products, A. niger holds the potential to become an even more versatile cell factory platform in the future. The recent publishing of the genome sequence of A. niger CBS 513.88, has made it apparent that this fungus holds a genetic diversity that is a potential trove of new products (Pel et al, 2007). As commented by Cullen (2007), there is now a great potential for increasing the yield of both known and novel products by directed metabolic engineering. However, data integration on a systemic level is necessary to fully understand and exploit the potential of the metabolism.
To increase the understanding of central metabolism in A. niger, a number of models and overviews have been presented (Röhr and Kubicek, 1981; Torres, 1994a, 1994b; Schmidt et al, 1999; Pedersen et al, 2000b; David et al, 2003; Karaffa and Kubicek, 2003; Gheshlaghi et al, 2007). While these models describe specific aspects of metabolism well, no model has ever integrated a full genome-scale metabolic network with genomic annotation. A complete and accurate model of this type has several uses, e.g. (a) identification of targets for metabolic engineering (Patil et al, 2005); (b) interpretation of transcription data and identification of regulatory features (David et al, 2006); (c) metabolic flux analysis (Christensen and Nielsen, 1999) and (d) evaluation and improvement of gene annotation (Pel et al, 2007). Reconstruction of genome-scale metabolic networks has often been based on an identification of enzymatic activities from the genome annotation (Förster et al, 2003; Duarte et al, 2004b; Borodina et al, 2005; David et al, 2006). While this is a fast way of getting an initial list of reactions, the approach does have a number of innate weaknesses, as discussed by Borodina et al (2005). One major concern is that errors in the annotation cause false prediction of the presence of enzymes, which can affect the modeling results greatly. Some alleviation of this problem is often achieved with extensive curation afterwards (Borodina et al, 2005). An improvement of this is seen in the recent model for human metabolism by Duarte et al (2007), who used a combination of genomic and bibliomic information, thus basing the network on peer-reviewed features. An additional advantage of including an extensive literature survey in the metabolic reconstruction is that it allows inclusion of characteristics, which do not have identified genomic features and are unique for the studied organism. Additionally, integrating data from the literature reveals the inadequacies in our knowledge on metabolism, i.e. cases where specific steps in pathways have not been investigated. Finally, this approach creates linkages between the metabolic network of the specific organism and literature on the individual reactions, thereby providing a convenient topic-based index and direct reference to the relevant literature.
On the basis of manual curation of an extensive bibliomic survey of the literature on A. niger, we have provided a comprehensive catalog of all reported intracellular enzymatic activities in A. niger and thus established the entire metabolic network. It was compiled based on manual curation of an extensive bibliomic survey of the A. niger literature. On the basis of this, a stoichiometric metabolic model has been constructed and besides being useful for model simulation and data analysis as mentioned above, the model provides a link between reactions, genes and scientific papers and hence represents a comprehensive database on most of the currently available information on the metabolism of A. niger.
Results
Reconstruction of the metabolic network
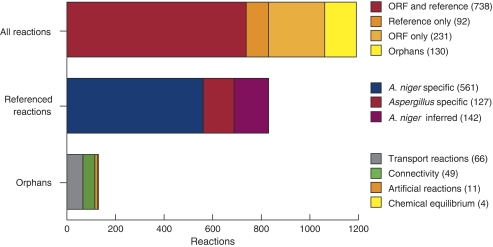
The reconstructed metabolic network of A. niger comprises 1190 biochemically unique reactions with no gaps. If reactions for isoenzymes are added based on the gene annotation, the total number is 2240 reactions. In total, 52 enzyme complexes were identified in the network. Overall, 371 cited articles and 871 unique ORFs are associated with the included reactions. The network comprises a total of 1045 metabolites distributed across three compartments; extracellular, cytosolic and mitochondrial. A summary of the support for the included reactions is shown in Figure 1. The full list of reactions with bibliography of the supporting articles is available in Supplementary Table I and the list of metabolites is given in Supplementary Table II. An SBML-formatted version of the model is supplied as Supplementary Information.
Figure 1.
Statistics of the reconstructed A. niger metabolic network. Top bar shows the support for the 1190 unique reactions. Middle bar shows the distribution of the references according to specificity. The bottom bar shows the reactions constituting the ‘orphans'—reactions with no supporting ORF or literature.
Biomass composition and growth energetics
A key element of the model is to have a correct biomass composition as this defines the drain of metabolites into the biomass pool. Here, the biomass composition for A. niger was calculated based solely on reported measurements. The overall biomass composition can be found in Table I. A more detailed description of the composition of the biomass components can be found in Supplementary Tables III–VIII. For modeling growth, key energetic parameters mATP, YxATP and the P/O ratios (see the Materials and methods section for details) were calculated from published data. mATP was calculated to be 1.9 (mmol ATP)/(g DW h). This is in agreement with the value found in the A. niger model by David et al (2003). YxATP was calculated to be 61 (mmol ATP)/(g DW). The P/O ratios were set to 2.64 for mitochondrial NADH, and 1.64 for succinate and cytosolic NADH.
Table 1.
Biomass composition of A. niger
| Biomass component | Mass (g)/(g DW) | References |
|---|---|---|
| Protein | 0.263 | Terroine and Bonnet (1927); Rockwell and O'Flaherty (1931); Smirnov and Chubova (1965); Christias et al (1975); Imshenetskii et al (1981) |
| DNA | 0.00244 | Imshenetskii et al (1981) |
| RNA | 0.01814 | Imshenetskii et al (1981) |
| Lipids | 0.10899 | Brennan et al (1974); Byrne and Brennan (1976); Chattopadhyay et al (1985); Morozova et al (2002); Nemec and Jernejc (2002) |
| Cell wall | 0.38 | Rockwell and O'Flaherty (1931); Smirnov and Chubova (1965); Imshenetskii et al (1981) |
| Small molecules | 0.131 | (Fuhrer et al (1980); Promper et al (1993); Witteveen and Visser (1995) |
| Ash | 0.075 | Nielsen et al (2003) |
| Sum | 0.97857 |
Overview of the metabolism
The reaction network of A. niger comprises central carbon metabolism, catabolic pathways for 115 different carbon sources and 23 different nitrogen sources and anabolic pathways for the components of the biomass. Additionally, reactions for the production of glucoamylase and alpha-amylase have been added to allow calculation of the theoretical maximum yields and the energetic drain caused by producing these enzymes in high yields.
The 115 different carbon sources can be divided into 49 carbon sources found in the literature to support growth (Supplementary Table IX), and 66 additional carbon sources that can support growth of the model (Supplementary Table X). Included in these 115 carbon sources are more than 20 xenobiotic compounds. Of the 23 different compounds the model can use as a sole nitrogen source, 16 of these are described in the literature (Supplementary Table XI).
A number of pathways were assembled de novo for this model based on the available literature. For these, the pathways in the model represent a review of the present knowledge of fungal metabolism supplemented with inferred reactions. Some of the catabolic pathways for xenobiotic compounds hold examples of these inferred reactions, since for many of these a complete pathway has not yet been hypothesized in fungi. Another example of this is the steroid biosynthesis, which has not been fully elucidated in fungi with all its cofactors. On the basis of a combination of reviews on other aspergilli (Ferreira et al, 2005), fungi in general (Chopra and Khuller, 1984) and the SGD and KEGG pathway databases (Cherry et al, 1998; Kanehisa et al, 2002), a model for the biosynthesis of lanosterol, zymosterol and ergosterol has been constructed and ORFs putatively assigned to the steps of the pathway.
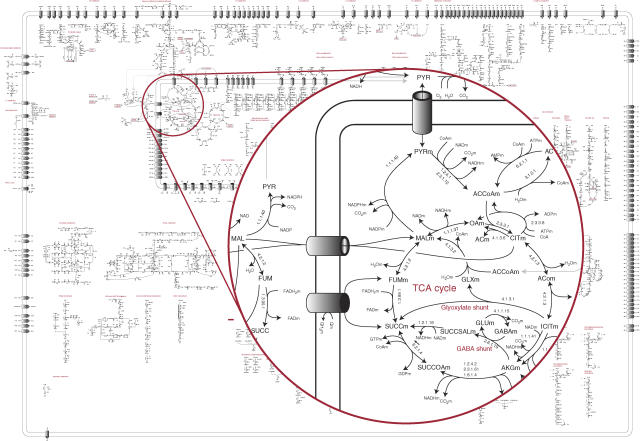
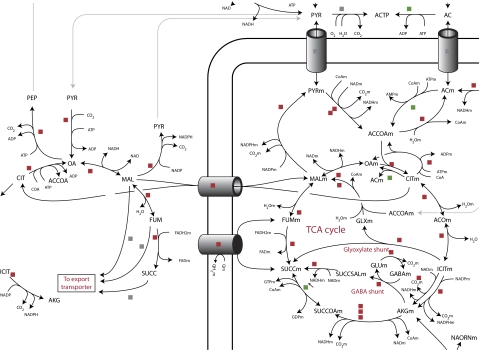
To provide an overview of the metabolism, a metabolic map of the 1190 reactions in the model was drawn (Figure 2; full map in Supplementary Figure 1).
Figure 2.
Metabolic map. The metabolic map of A. niger (Supplementary Figure 1) is showed in reduced size. An example of the level of detail is shown for the citric acid cycle.
Supplementing reactions and metabolites
A small number of reactions reported in the literature were not included in the metabolic network (65 additional unique reactions). These reactions are activities that were not possible to connect to the rest of the metabolic model, e.g. catalysis of complex processes (such as multiple steps of biopolymer degradation of a single enzyme), or single enzymatic activities with no role in normal metabolism (such as bio-catalysis reactions). Additionally, 56 pathways were reported in the literature, but not included. These are pathways for the biosynthesis of complicated secondary metabolites and catabolic pathways for complex aromatic substrates and share the feature that the end product was reported in the literature, but no information on the intermediate step(s) was available. However, as this catalog of information still holds relevant information on the metabolic capabilities of A. niger it is included as Supplementary Tables XII and XIII. References to the 87 articles describing these pathways and activities are included as well. Inclusion of these references brings the total number of citations up to 409.
Assignment of ORFs
Upon manual inspection of the ORFs assigned from the annotation of the CBS 513.88 genome sequence, some irregularities were found (a) only one inorganic diphosphatase (EC 3.6.1.1) was found in the genome sequence. For this reason, this ORF was assigned to both the cytosol and the mitochondrion; (b) the same seven ORFs were assigned to the K+ and the Ca+ transporting ATPase, as these are difficult to separate based on sequence. The Na+, Mg2+ and other cation transporters may possibly be found within this group as well (MIPS category 67.04.01.02); (c) seven amine oxidases were found (four putative and three characterized). As the available literature does not correlate the reported activities to sequence or enzyme, all seven were assigned to the same reactions and (d) for several general enzymes, a large number was found. These were alcohol dehydrogenases (20), amidases (14) and acid phosphatases (7). These were not possible to separate into specific functions based on sequence, but it is unlikely that they are all catalyzing the same reaction, as the annotation may indicate.
To further expand the applications of the reconstructed network, ORFs from the genome sequence of A. niger ATCC 1015 (Baker, 2006) were assigned to the model as well. Of the 871 ORFs from the CBS 513.88 genome assigned to the model, 792 had direct candidates in the A. niger ATCC 1015 genome sequence (more than 90% identity and e-values below 10−75. Another 18 ORFs had candidates with identity between 80 and 90% and e-values below 10−75. Three more short proteins had an identity between 80 and 90%. To improve the assignment, the results were examined manually, and an additional 31 ORFs were paired up. A total of 30 putative enzymes (3.5%) in the A. niger CBS 513.88 genome were not found in the A. niger ATCC 1015 protein sequences. Twenty-one of these were found at the nucleotide level using blastn (McGinnis and Madden, 2004), but were not present in the best gene predictions. The assignment of ORFs from both genome sequences to reactions is shown in Supplementary Table I.
Comparison of fungal metabolic networks
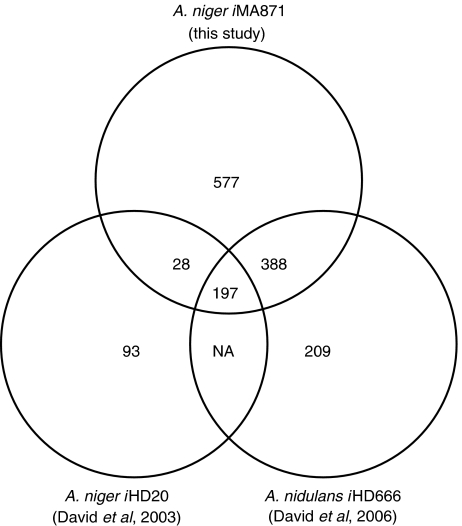
The key statistics of the A. niger metabolic network were compared to that of other fungal stoichiometric models (Table II). To aid the reader, we adopt the naming convention proposed for Escherichia coli metabolic models by Reed et al (2003). Much like a strain number, the name of the organism is added as a designation for the model with the following syntax: iXXxxx. i is for in silico, distinguishing it as a computer model and not a strain. This is followed by the initials (XX) of the person who developed the model, in this case iMA, and the number of ORFs (xxx) included in the metabolic network. Henceforth, the model presented in this study will be denoted as A. niger iMA871, or iMA871 as a shorthand form. To evaluate the overlap of the metabolic network of the aspergilli, the reactions in A. niger iMA871 were compared to a model of A. niger central metabolism (David et al, 2003) (A. niger iHD20) and a genome-scale model of A. nidulans (David et al, 2006) (A. nidulans iHD666). The results are shown in Figure 3.
Table 2.
Comparison of fungal models
| Model | References | Genes | Metabolitesa | Unique reactionsb |
|---|---|---|---|---|
| S. cerevisiae iFF708 | Förster et al (2003) | 708 | 584 | 842 |
| S. cerevisiae iND750 | Duarte et al (2004a) | 750 | 646 | 1149 |
| A. nidulans iHD666 | David et al (2006) | 666 | 551 | 794 |
| A. niger iHD20 | David et al (2003) | 20c | 244 | 318 |
| A. niger iMA871 | This study | 871 | 782 | 1190 |
Comparison of key statistics of selected fungal stoichiometric models.
aNumber of chemically distinct metabolites, not counting presence in multiple compartments.
bUnique reactions are defined as reactions being biochemically unique in their own compartment or transport reactions. Isoenzymes are thus not included in this number.
cReconstructed prior to the release of an A. niger genome sequence.
Figure 3.
Venn diagram of reaction statistics for three Aspergillus models. The diagram shows the number of unique reactions shared and specific for the three models. Overlap between A. nidulans iHD666 and A. niger iHD20 was not investigated.
In total, 93 reactions were found exclusively in A. niger iHD20. These are for the largest part lumped reactions and artificial reactions producing biomass. The remaining difference is mostly due to divergences in compartmentalization, with the enzyme being present in both models, but the subcellular localization differing. A. nidulans iHD666 contains 209 reactions uniquely found in that network. These are to a large extent reactions relating to lipid synthesis, as this has been completely redesigned for A. niger iMA871. Additionally, a number of pathways found in A. nidulans, but not in A. niger, are included in these 209 reactions (e.g. aflatoxin, statin and penicillin biosynthesis). Most interestingly, the reactions found only in A. niger iMA871 are found all through metabolism, with the exception of the very central metabolism, thus suggesting that the bibliomic survey has been thorough, and that the model captures many activities unique for A. niger. The largest part of these reactions is found in the lipid biosynthesis and xenobiotic catabolism.
Model validation
To evaluate the predictive capabilities of A. niger iMA871 and assess the inference of the modeling results, experimental results from articles were compared to the predictions of simulations of A. niger iMA871. The prediction of yields, the intracellular distribution of carbon fluxes and physiological responses were examined. Additionally, transcription data were used to assess the network topology and the validity of the gene annotation.
Prediction of yields
A number of studies have examined the yields of different products and biomass on substrates for cultivations of A. niger. We have compared these to the predictions of A. niger iMA871: van de Merbel (1994) reports production of 13 g/l oxalic acid from a mixture of 10 g/l glucose and 10 g/l fructose in 70 h (van de Merbel et al, 1994). iMA871 predicts a theoretical maximum of a conversion of 100% of carbon, giving 30 g/l oxalic acid. A review by Karaffa and Kubicek (2003) states that industrial citric acid producer strains can produce as much as 95% citrate from sugar on a weight basis. iMA871 predicts a theoretical maximum of 98% molar yield (approximately 101% on a weight basis). The molar yield of gluconic acid on glucose has been reported by Singh and Kumar (2007) to be as high as 98%. Predictions by iMA871 give a theoretical maximum of 100%. On the basis of the data presented by Schrickx et al (1993), an examination of the prediction of glucoamylase overproduction was performed. The study examines the growth of A. niger N402 and a glucoamylase overproducing mutant in continuous cultures and calculates the yields of biomass on glucose and oxygen for the strains. Simulations with A. niger iMA871 give values that are 5–10% higher than the yields reported by Schrickx et al, but well within the given confidence intervals (Table III). The slight over-prediction of the yields may be partially explained by the fact that the data in the article only accounts for 94–97% of the carbon.
Table 3.
Comparison of biomass and oxygen yields for model and A. niger N402
| N402 | iMR871 | N402 (glaA) | iMR871 (glaA) | |
|---|---|---|---|---|
| Yxs (g DW/mol glucose) | 100 (88.14–114.6) | 105.5 | 96.29 (85.94–109.5) | 105.1 |
| Yxo (g DW/mol O2) | 54.68 (46.08–67.25) | 60.6 | 54.14 (42.59–74.29) | 60.6 |
| Carbon (%) | 94.4 | 100 | 97.1 |
Comparison of biomass and oxygen yields for A. niger N402 (Schrickx et al, 1993) and iMR871. ‘glaA' denotes a mutant producing more glucoamylase. 95% confidence intervals are given in parentheses. The ‘carbon' row shows the amount of carbon accounted for.
Modeling flux distributions
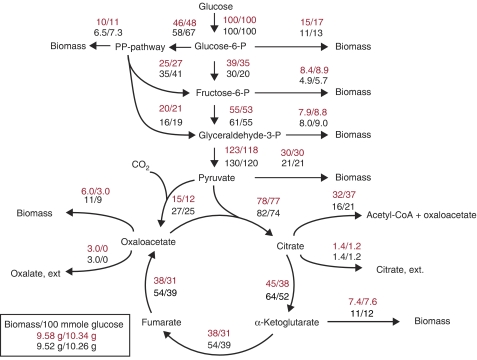
The study of Pedersen et al (2000b) reports the effect of deleting the gene encoding oxaloacetate hydrolase (oah) on the metabolic fluxes in central metabolism using 13C labeling and a simplified model of the central carbon metabolism (40 reactions). To evaluate the predictive powers of A. niger iMA871 for flux modeling, it was used to re-model the fluxes measured by Pedersen et al Modeling was carried out both for the wild-type results and the fluxes measured in the oah deletion mutant. In both cases, the model was optimized for growth, with the measured values for citrate and oxalate from Pedersen et al set as constraints.
Initial modeling indicated that only 20% of the glucose uptake was sent to the pentose phosphate pathway, which is low for a filamentous fungus, where values as high as 61% has been reported for Penicillium chrysogenum (Henriksen et al, 1996). This underestimation was due to the high activity of cytosolic NADP+ isocitrate dehydrogenase, generating the necessary NADPH for synthesis of biomass components. The presence of this enzyme is established—the in vitro activity of NADP+ isocitrate dehydrogenase was described by Muller (1975). The enzyme is described as present in both the cytosol and the mitochondria in the publication of the A. niger CBS 513.88 genome sequence (Pel et al, 2007), but it seems only slightly active in the cytosol under the conditions examined by Pedersen et al. For this reason, a new round of modeling was conducted with the cytosolic activity of this enzyme limited to a low value, giving the flux distribution of Figure 4. Examining the results of Figure 4, it is evident that the flux to the PP pathway is still lower than the measured values, but in the same order of magnitude. For all measured fluxes, A. niger iMA871 predicts the direction of the response to the oah deletion accurately, and the numerical size of the response in the simulation (i.e. the difference between the flux in the wild type and the mutant) generally approximates to the measured values. This is especially true for the fluxes toward biomass. iMA871 predicts the exact size of the perturbation in almost all cases, as well as the increase in total biomass yield by the deletion of oah. When calculating the yield of NADPH on glucose, A. niger iMA871 accurately predicts a production of 100–120 mol NADPH/100 mol glucose, which is the yield reported by Pedersen et al.
Figure 4.
Comparison of measured oah deletion fluxes to predicted values. Values in black (lower) are the calculated fluxes based on 13C labeling, wt/Δoah. Red values (upper) are predicted with A. niger iMA871. Parts of the figure in black are from Pedersen et al (2000b).
Modeling of physiological responses
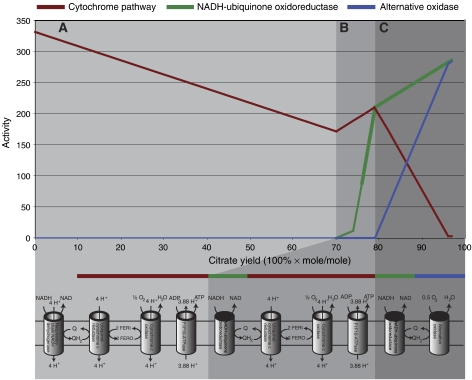
One of the best-studied physiological responses of A. niger is respiration during high-yield citrate production. A number of studies and reviews (Promper et al, 1993; Kirimura et al, 1999, 2006; Karaffa and Kubicek, 2003) have reported activity of both the cytochrome pathway, and the alternative oxidase during citrate production. Additionally, it has been reported, that when the alternative oxidase is inhibited with salicylhydroxamic acid, citric acid production is drastically decreased (Kirimura et al, 2006). Furthermore, it has been hypothesized that the alternative oxidase is present to remove excess NADH without production of further ATP (Karaffa and Kubicek, 2003). To study this response with A. niger iMA871, the fluxes through the oxidative pathways on different citrate yields were simulated (Figure 5), and the results do indeed show activity of the non-proton-pumping NADH-ubiquinone oxidoreductase and the alternative oxidase, when intermediate to high yields of citrate are obtained. However, at carbon yields above 98%, the model predicts the use of other means to reduce the NADH excess. As yields this high have never been reported, the use of these may be an artifact caused by the true natural limitation being reached.
Figure 5.
Predicted activities of respiratory pathways in citric acid fermentations. In phase A, the standard respiratory pathway is the sole source of NADH turnover. In phase B, the proton-pumping NADH dehydrogenase is substituted for the non-proton-pumping NADH-ubiquinone oxidoreductase, resulting in a pathway that is a meld of the alternative oxidase pathway and the standard oxidative pathway. In phase C, the cytochrome pathway is substituted for the alternative oxidase pathway. The net result is a decreasing amount of ATP produced per metabolized NADH as the citric acid yield increases, resulting in a changing P/O ratio.
ORF assignment and network topology
To assess the validity of the reconstructed metabolic network of iMA871 and the ORFs assigned to the enzyme activities, the network was compared to transcription data published with the genome sequencing of A. niger CBS 513.88 (Pel et al, 2007). The transcription data are from the third day of a fed-batch cultivation with glucose and ammonium as carbon and nitrogen sources, respectively. The presence/absence of mRNA from the relevant ORFs was plotted onto the metabolic map and visually inspected (full map in Supplementary Figure 2, central metabolism in Figure 6). As seen from Figure 6, the succinate-CoA ligase complex is absent. Neither α- nor β-chain of the complex is present, which suggests that an alternative pathway such as the GABA shunt is active. This is in coherence with reportings of Kumar et al (2000), showing the GABA shunt to be active during citric acid production.
Figure 6.
Genes expressed in central carbon metabolism when A. niger CBS 513.88 is grown on glucose. Red box: at least one isoenzyme of this process is present; green box: all absent; gray box: no ORF has been assigned to the process.
Upon examining the remaining part of metabolism, it was found that the expressed genes correlated nicely with the pathways essential for biomass production. In no case were transcripts from an entire essential pathway found to be absent. Expression of genes involved in polyol synthesis for the biomass was also found to be active. All enzymes of the pathways producing coenzyme A, biotin, NADH/NADPH and nucleoside phosphates were found to be present. All pathways not necessary for growth on glucose were found to be absent, with the only exception of the pathway from L-iditol to D-fructose. This includes catabolism of C6 substrates other than glucose, C5, C4, C3 and C2 substrates and the urea and nitrate assimilation pathways. A few enzymes in the middle of linear pathways were found to be absent, while the up- and downstream ones were present. These pathways were phosphocholine, cardiolipin, folate, lanosterol and ergosterol biosynthesis. This may be due to poor annotation of these genes, as there are a limited number of studies of these pathways in aspergilli.
Assessing the metabolic capabilities of A. niger
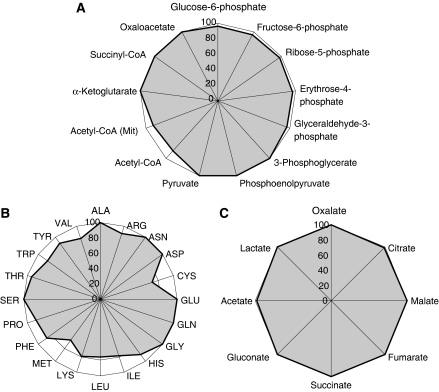
A. niger is widely used in industrial processes due to its innate ability to produce organic acids and enzymes in high yields. Using A. niger iMA871, we have examined the theoretical maximum yields of different metabolic precursors and acids from glucose (Figure 7). Panel A of Figure 7 shows that A. niger is highly efficient at producing all 12 essential precursor molecules—as defined by Stephanopoulos et al (1998). The conversion of these into amino acids is, as panel B shows, not completely efficient for all of the amino acids, but given the number of reduction equivalents necessary for synthesis of many of the amino acids, this is not surprising. It does, however, suggest that enzyme production yields may be increased by supplementing the medium with amino acids, especially the branched and the sulfur-containing amino acids. The plot of panel C demonstrates that A. niger is theoretically capable of converting glucose to a number of organic acids at 100% efficiency, as has experimentally been demonstrated for several of the acids in the plot.
Figure 7.
Radar plots of the theoretical maximum yields of selected metabolites from glucose and ammonium. Value axis shows percentage of C moles of the metabolite from glucose; 100% denotes a full conversion of glucose to the product. Plot (A) shows the twelve essential precursor metabolites as defined in (Stephanopoulos et al, 1998). Plot (B) shows the 20 common amino acids. Plot (C) shows selected organic acids.
In addition to the compounds discussed above, the theoretical maximum yields of α-amylase and glucoamylase were calculated, giving 0.61 g α-amylase and 0.62 g glucoamylase/g glucose, respectively. Glucoamylase yields of 0.023 g/g have been observed in lab-scale fed-batch by Pedersen et al (2000a), but yields of glucoamylase in commercial fed-batch fermentations are estimated to be 0.33–0.66 g/g, under the assumption that such fermentations are economically feasible (L Lasure, personal communications). This is in accordance with the model predictions.
Discussion
The metabolic network of A. niger has been reconstructed using a multi-omic approach. The reconstruction method was based on labor-intensive approaches of a bibliome survey combined with extensive manual curation in every step of the process. A reconstruction of the A. niger metabolic network presented by Sun et al (2007) was based on automated techniques with no curation or basis in features reported in the literature. This study reports a network of approximately the same size as the network of Sun et al's study (998 unique EC numbers, 2443 reactions in total). Since the network of our study includes 1190 unique reactions and 2240 reactions in total, it is evident that manual reconstruction of the network can provide a validated result that is at least as comprehensive as the result of an automated approach, but with additional advantages such as the inclusion of citations. However, some caution should be taken in this comparison. The network presented by Sun et al (2007) was constructed for comparison of metabolic features between fungi, and is thus not a metabolic model, does not contain any compartmentalization and has a large number of reactions that are not connected to central metabolism. The network reconstructions thus have different scopes, and a comparison based solely on numerical statistics may not do full justice to the unique features of each of the reconstructions.
The assignment of ORFs to reactions was based on the manual annotation of the A. niger CBS 513.88 genome sequence. While this is a conservative way of assigning ORFs to enzymatic functions this may still overestimate the number of genes assigned to a specific reaction. In the case of general enzymes such as alcohol dehydrogenase (EC 1.1.1.1), it was not possible to differentiate between enzymatic specificities of the individual genes. Thus, all genes were assigned to the same reactions. This leads to an overestimation, meaning that the total number of 2240 reactions described above may be closer to an upper limit than the actual number. For this reason, we do not find that this is a useful statistic for comparing individual models.
For the purpose of communication of the presented model, and an easy overview of the model, we have prepared a map of the metabolic network. However, since the map is prepared with a notation similar to the one known from biochemical textbooks, one may easily confuse it with these well-described pathways. It is important to bear in mind that the map is only a representation of the model, and a number of reactions are added for connectivity and are not based on reported experiments. One should always examine the relevant citations before drawing conclusions based on the metabolic map. With this in mind, we believe it to be an easily accessible reference tool for Aspergillus metabolism and future efforts in metabolic engineering.
The constructed model (A. niger iMA871) was based on the metabolic network and validated by several methods. A comparison of predicted yields of selected products compared to those yields reported in the literature showed the model predictions to be in good accordance with experimental values. One exception was the prediction of the theoretical maximum yield of oxalate. However, since no strain optimization for oxalic acid production has ever been reported, the discrepancy between experimental and predictive yields is not surprising. Oxalic acid is produced industrially through chemical processes (Wallace, 1926), and in the selection of most fermentation strains, it is sought to eliminate oxalic acid as a by-product (Hjort and Pedersen, 2000).
The prediction of flux distributions was examined by modeling the results of Pedersen et al (2000b). While the overall prediction of biomass yields and perturbation effects was in good accordance with the measured values, in some cases, the numerical size of the fluxes was not. The sizes of the fluxes through mitochondrial fumarate and α-ketoglutarate are noticeably lower in the modeled results than in the measured results, while the flux from citrate to oxaloacetate+acetyl-CoA through a cytosolic pathway is higher in the prediction. Since the same amount of flux to cytosolic oxaloacetate is approximately too high as the flux to α-ketoglutarate is too low, it seems plausible that the problem is the flux distribution between TCA in the cytosol and mitochondria between the modeled and the measured values. This may be caused by the compartmentalization of reactions used by Pedersen et al, as it is a lot less sophisticated than the one of A. niger iMA871. However, this is compensated for by Pedersen et al by the measurement of the labeling pattern in amino acids, which can estimate the carbon fluxes through the individual compartments. Another explanation might be that the difference in fluxes is an artifact of the A. niger iMA871 reconstruction process. In the compartmentalization of iMA871 reactions, a pathway was only placed in the mitochondria, if evidence suggested that a part of it is located there. This means that the number of reactions assigned to the mitochondria of A. niger iMA871 is most likely lower than the actual number, thus decreasing mitochondrial carbon fluxes, and increasing cytosolic fluxes. While this artifact influences the flux distribution inside the cell, the nature of the model ensures that it does not affect the prediction of the ‘phenotype' of A. niger iMA871, such as product or biomass yields.
Using the example of oxidative phosphorylation during production of citric acid, we examined the prediction of physiological responses. Predictions were in good accordance with the known response of the different oxidative pathways (Promper et al, 1993; Kirimura et al, 1999, 2006; Karaffa and Kubicek, 2003). Interestingly, we find three modes of operation (phases A–C of Figure 5) that allow a flexible regulation of the P/O ratio to dispose of excess mitochondrial NADH produced during high-yield citrate production. When examining these results, it is important to bear in mind that the results are very much dependent on the estimation of the P/O ratio of mitochondrial NADH included in the model parameters. The exact points of change from one phase to another, and the lengths of the phases, are not robust to changes in this value. One should therefore be careful in concluding at which yields a specific pathway is needed. However, the pattern of the phases, the order of them and the physiological response are robust toward changes in this value, and we thus believe that these systems are indeed in effect in A. niger during high-yield production of citrate.
In conclusion, we present an integration of the genome, reactome, metabolome and bibliome of A. niger through reconstruction and modeling of the metabolic network. With 70% of the 1190 included reactions backed by literature from 371 articles, and above 80% of the reactions associated with one or more of 871 assigned ORFs from the A. niger CBS 513.88 and the ATCC 1015 genome sequences (Baker, 2006; Pel et al, 2007), the network represents a comprehensive knowledge base for the metabolism of A. niger. A map providing a unique overview of the reconstructed metabolism in a format easily accessible to most readers has been made. Stoichiometric modeling based on the metabolic network was validated with a wide array of data types. The systematization of data into the presented metabolic network and map has produced useful tools for examining systemwide data in a metabolic context. The included example of the network-aided interpretation of transcription data validates the gene assignments and demonstrates the potential of using the network for visualization and analysis of transcriptomic data. The presented modeling results produced new information on physiological traits of A. niger as well as showing that A. niger has tremendous potential for being a versatile cell factory in the current development toward a bio-based economy.
Materials and methods
Model reconstruction
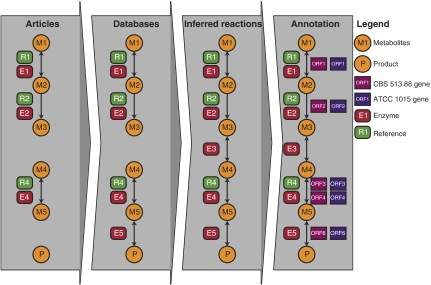
The reaction list for A. nidulans iHD666 was purged of isoenzymes and used as a reaction template. An overview of the reconstruction procedure is shown in Figure 8. Initially, the A. niger bibliome was searched for references on metabolism, the bibliome being all articles concerning A. niger found on PubMed (http://www.ncbi.nlm.nih.gov/entrez) and on the ISI Web of Science. Information on enzymes/biochemical reactions, pathways, metabolites or biomass was manually extracted from the articles. The article references were added to the metabolic network reaction list. In addition to this, all literature references for A. niger enzymes found in BRENDA (http://www.brenda-enzymes.info/), the ExPASy enzyme database (http://www.expasy.ch/enzyme) and Swiss-Prot (http://www.expasy.ch/sprot) were examined, and added to the list of reactions as necessary. In some cases, reactions leading to the anabolism/catabolism of a given metabolite were inferred from A. niger literature based on indirect observations (an example of this is the degradation of ferulic acid from the reportings of Milstein et al (1988)).
Figure 8.
Overview of the model reconstruction process. Pathway addition was based on the reported presence of products and/or enzyme activities reported for A. niger. This was supplemented with enzymes reported present in other aspergilli. Pathway databases (KEGG) were used where possible to fill gaps in pathways. If necessary, reactions were inferred to close the remaining gaps. Finally, ORFs were connected to reactions using the CBS 513.88 sequence annotation. No new reactions were added based solely on sequence information.
Upon completion of the A. niger references from the bibliome and databases, the assembled information, representing lists of the reactome and metabolome, was examined for missing steps in pathways. If information on a given enzyme necessary for connectivity was lacking from A. niger literature, references to the reaction from other aspergilli were added if available. After this step, information from KEGG (http://www.genome.jp/kegg/kegg2.html) was used to fill as many of the remaining gaps as possible. Gaps remaining after this were closed with the simplest possible reaction(s) (i.e. a dehydrogenation step or an elimination of water) resulting in a final network without gaps. Unless information existed stating otherwise, all reactions were defined as reversible. For modeling, some reactions were defined as irreversible due to artificial transhydrogenation cycles allowing NADH/NAPDH conversions. After the last reaction was added, extensive curation of the reaction list was performed. Reactions from the template A. nidulans iHD666 reaction list were removed, if no evidence existed for them in A. niger. The correctness of the individual reactions was checked by ensuring that C-, N-, O-, P- and S-balances hold for all pathways and production of all biomass components. After the metabolic network was compiled, ORFs were assigned to the included reactions.
ORF assignment
EC numbers in the annotation of the A. niger CBS 513.88 genome (Pel et al, 2007) were used to assign ORFs to the reactions of the metabolic network. No new reactions were added based solely on sequence information. This was supplemented by a manual examination of the ORF reaction pairs. Homologs of the genes from the A. niger CBS 513.88 genome were found in the A. niger ATCC 1015 genome sequence (version 1.0) using blastp (McGinnis and Madden, 2004). The names of the ORFs from both genome sequences were added to the reaction list.
Biomass composition and growth energetics
To model growth, a biomass equation was added to the list of reactions. This equation acts as a drain of the component molecules used in growth. The biomass composition of A. niger was determined through a bibliome survey. A metabolite was added to the biomass equation if it was reported present in A. niger and the specific content quantified. The following parameters were estimated: the ATP cost of growth-associated maintenance (YxATP), the ATP requirement for non-growth-associated activities (maintenance) (mATP) and the operational P/O ratios. mATP was estimated using the relation between mATP, substrate consumption rate (qs) and the specific growth rate (μ) described for continuous cultures of A. niger by Schrickx et al (1993). YxATP was calculated using the biomass yield on glucose in continuous cultures of A. niger from Pedersen et al (2000b). The P/O ratios for the model were taken from the survey carried out by David et al (2003).
Enzyme complexes
Enzymes that are part of a complex were identified by interrogating the A. niger CBS 513.88 annotation of the included proteins for the words ‘subunit', ‘chain' or ‘complex'. This information was added to the reaction list and used in the calculation of isoenzymes.
Compartmentalization and transport
The model has three compartments: extracellular space, cytosol and mitochondrion. Unless otherwise reported in the literature, the enzymatic reactions were assumed to be cytosolic. When an enzyme was reported to have isoenzymes in both mitochondrion and the cytosol, the compartmentalization predictions of the A. niger CBS 513.88 annotation were used to assign each ORFs to a compartment. Most transport reactions had to be inferred based on the knowledge of reactions or pathways found in a specific compartment. To reduce the number of unsupported transport reactions, pathways have as a rule been assigned to the mitochondrion if one or more enzymes in the pathway were reported to be present there. Enzymes reported to be present in the cell wall were placed in the extracellular compartment. As there is little information in the literature on peroxisomal localization for A. niger enzymes, this compartment was not included as a part of the model. Enzymes normally found in the peroxisome are therefore placed in the cytosol. One exception is the degradation of fatty acids, which in most fungi takes place in the peroxisome and the mitochondrion, with an ill-defined distribution between the two. This has in the model been placed in the mitochondrion, along with the glyoxylate shunt.
Modeling
The quantification of metabolic fluxes was carried out using the flux balance analysis (Stephanopoulos et al, 1998; Schilling et al, 1999; Nielsen et al, 2003). The compiled set of metabolic reactions is converted to a stoichiometric matrix S of dimensions m × n, where m is the number of metabolites and n is the number of reactions or fluxes to be computed. Under the assumption that the metabolite concentrations are in a pseudo-steady-state and the dilution effects from growth are negligible, the model can be represented as:
where vector v represents the flux activity for each metabolic reaction. As the number of reactions (n) is greater than m (the number of metabolites), the equation system is underdetermined. The system was solved using linear programming by formulating a specific objective function (z). In the case of a maximization problem, the linear programming problem can be formulated as:
where c is a row vector containing the influence of the individual fluxes on the objective function z. The individual fluxes are constrained to the interval of [αi,βi]. For irreversible reactions, either αi or βi are set to 0. In case of reactions defining the limiting substrate, αi and βi are set to the same positive value. Unless otherwise stated in the text, the optimization parameter was maximizing flux through the biomass equation. Transport fluxes for phosphate, sulfate, ammonia/nitrate and oxygen were not limited. For metabolites not present in the medium, the uptake rates (vi) were set to 0. Secretion of all major metabolic products (organic acids, alcohols, amino acids, H2O, CO2, etc.) was allowed (vi⩽0). All calculations were performed using the commercially available software Lindo (Lindo Systems Inc.).
Supplementary Material
Supplementary Information
Supplementary Figure 1
Supplementary Figure 2
Supplementary Information
Acknowledgments
We thank Gerald Hofmann for valuable feedback during the reconstruction process, Renata Usaite, Gaëlle Lettier and José Manuel Otero for help with translation of articles, Jette Thykjær for scientific discussions on 13C labeling, DSM Food Specialities for giving early access to the genome sequence and genome annotation of A. niger CBS 513.88 and the US Department of Energy—Joint Genome Institute for making the A. niger ATCC 1015 sequence available. MRA has been funded by the Danish Research Agency for Technology and Production.
References
- Baker SE (2006) Aspergillus niger genomics: past, present and into the future. Med Mycol 44: S17–S21 [DOI] [PubMed] [Google Scholar]
- Borodina I, Krabben P, Nielsen J (2005) Genome-scale analysis of Streptomyces coelicolor A3(2) metabolism. Genome Res 15: 820–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan PJ, Griffin PFS, Lösel DM, Tyrrell D (1974) The lipids of fungi. Prog Chem Fats Other Lipids 14: 49–89 [DOI] [PubMed] [Google Scholar]
- Byrne PF, Brennan PJ (1976) Isolation and characterization of inositol-containing glycosphingolipids from Aspergillus niger. Biochem Soc Trans 4: 893–895 [DOI] [PubMed] [Google Scholar]
- Chattopadhyay P, Banerjee SK, Sen K, Chakrabarti P (1985) Lipid profiles of Aspergillus niger and its unsaturated fatty acid auxotroph, UFA2. Can J Microbiol 31: 352–355 [DOI] [PubMed] [Google Scholar]
- Cherry JM, Adler C, Ball C, Chervitz SA, Dwight SS, Hester ET, Jia Y, Juvik G, Roe T, Schroeder M, Weng S, Botstein D (1998) SGD: Saccharomyces genome database. Nucleic Acids Res 26: 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra A, Khuller GK (1984) Lipid metabolism in fungi. CRC Crit Rev Microbiol 11: 209–271 [DOI] [PubMed] [Google Scholar]
- Christensen B, Nielsen J (1999) Metabolic network analysis: a powerful tool in metabolic engineering. Adv Biochem Eng Biotechnol 66: 209–231 [PubMed] [Google Scholar]
- Christias C, Couvaraki C, Georgopoulos SG, Vomvoyanni V (1975) Protein content and amino acid composition of certain fungi evaluated for microbial protein production. Appl Microbiol 29: 250–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen D (2007) The genome of an industrial workhorse. Nat Biotech 25: 189–190 [DOI] [PubMed] [Google Scholar]
- David H, Åkesson M, Nielsen J (2003) Reconstruction of the central carbon metabolism of Aspergillus niger. Eur J Biochem 270: 4243–4253 [DOI] [PubMed] [Google Scholar]
- David H, Hofmann G, Oliveira AP, Jarmer H, Nielsen J (2006) Metabolic network driven analysis of genome-wide transcription data from Aspergillus nidulans. Genome Biol 7: R108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte NC, Becker SA, Jamshidi N, Thiele I, Mo ML, Vo TD, Srivas R, Palsson B (2007) Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci USA 104: 1777–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte NC, Herrgård MJ, Palsson B (2004a) Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model. Genome Res 14: 1298–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte NC, Palsson BØ, Fu P (2004b) Integrated analysis of metabolic phenotypes in Saccharomyces cerevisiae. BMC Genomics 5: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn-Coleman NS, Bloebaum P, Berka RM, Bodie E, Robinson N, Armstrong G, Ward M, Przetak M, Carter GL, LaCost R, Wilson LJ, Kodama KH, Baliu EF, Bower B, Lamsa M, Heinsohn H (1991) Commercial levels of chymosin production by Aspergillus. Biotechnology (NY) 9: 976–981 [DOI] [PubMed] [Google Scholar]
- Ferreira ME, Colombo AL, Paulsen I, Ren Q, Wortman J, Huang J, Goldman MH, Goldman GH (2005) The ergosterol biosynthesis pathway, transporter genes, azole resistance in Aspergillus fumigatus. Med Mycol 43: S313–S319 [DOI] [PubMed] [Google Scholar]
- Förster J, Famili I, Fu P, Palsson BØ, Nielsen J (2003) Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res 13: 244–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrer L, Kubicek CP, Röhr M (1980) Pyridine nucleotide levels and ratios in Aspergillus niger. Can J Microbiol 26: 405–408 [DOI] [PubMed] [Google Scholar]
- Gheshlaghi R, Scharer JM, Moo-Young M, Douglas PL (2007) Metabolic flux analysis for optimizing the specific growth rate of recombinant Aspergillus niger. Bioprocess Biosyst Eng 30: 397–418 [DOI] [PubMed] [Google Scholar]
- Henriksen CM, Christensen LH, Nielsen J, Villadsen J (1996) Growth energetics and metabolic fluxes in continuous cultures of Penicillium chrysogenum. J Biotechnol 45: 149–164 [DOI] [PubMed] [Google Scholar]
- Hjort C, Pedersen H (2000) Oxaloacetate hydrolase deficient fungal host cells. International Patent WO 00/50576
- Imshenetskii AA, Lysenko SV, Demina NS (1981) Comparative physiological and biochemical characteristics of Aspergillus niger isolated from the mesosphere and of its mutant. Mikrobiologiia 50: 1042–1045 [PubMed] [Google Scholar]
- Kanehisa M, Guto S, Kawashima S, Nakaya A (2002) The KEGG databases at GenomeNet. Nucleic Acids Res 30: 42–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaffa L, Kubicek CP (2003) Aspergillus niger citric acid accumulation: do we understand this well working black box? Appl Microbiol Biotechnol 61: 189–196 [DOI] [PubMed] [Google Scholar]
- Kirimura K, Ogawa S, Hattori T, Kino K (2006) Expression analysis of alternative oxidase gene (aox1) with enhanced green fluorescent protein as marker in citric acid-producing Aspergillus niger. J Biosci Bioeng 102: 210–214 [DOI] [PubMed] [Google Scholar]
- Kirimura K, Yoda M, Usami S (1999) Cloning and expression of the cDNA encoding an alternative oxidase gene from Aspergillus niger WU-2223L. Curr Genet 34: 472–477 [DOI] [PubMed] [Google Scholar]
- Kumar S, Punekar NS, Satyanarayan V, Venkatesh KV (2000) Metabolic fate of glutamate and evaluation of flux through the 4-aminobutyrate (GABA) shunt in Aspergillus niger. Biotechnol Bioeng 67: 575–584 [PubMed] [Google Scholar]
- McGinnis S, Madden TL (2004) BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res 32: W20–W25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein O, Trojanowski J, Huttermann A, Gressel J (1988) Catabolism of single ring aromatic acids by four Aspergillus species. Microbios 55: 7–16 [PubMed] [Google Scholar]
- Morozova EV, Kozlov VP, Tereshina VM, Memorskaya AS, Feofilova EP (2002) Changes in lipid composition and carbohydrate composition of Aspergillus niger conidia during germination. Appl Biochem Microbiol 38: 129–133 [PubMed] [Google Scholar]
- Muller HM (1975) Oxalate accumulation from citrate by Aspergillus niger. II. Involvement of the tricarboxylic acid cycle. Arch Microbiol 104: 159–162 [DOI] [PubMed] [Google Scholar]
- Nemec T, Jernejc K (2002) Influence of Tween 80 on lipid metabolism of an Aspergillus niger strain. Appl Biochem Biotechnol 101: 229–238 [DOI] [PubMed] [Google Scholar]
- Nielsen J, Villadsen J, Lidén G (2003) Bioreaction Engineering Principles, 2nd edn. New York: Kluwer Academic/Plenum Publishers [Google Scholar]
- Patil KR, Rocha I, Förster J, Nielsen J (2005) Evolutionary programming as a platform for in silico metabolic engineering. BMC Bioinformatics 23: 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen H, Beyer M, Nielsen J (2000a) Glucoamylase production in batch, chemostat and fed-batch cultivations by an industrial strain of Aspergillus niger. Appl Microbiol Biotechnol 53: 272–277 [DOI] [PubMed] [Google Scholar]
- Pedersen H, Christensen B, Hjort C, Nielsen J (2000b) Construction and characterization of an oxalic acid nonproducing strain of Aspergillus niger. Metab Eng 2: 34–41 [DOI] [PubMed] [Google Scholar]
- Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JA, van den Berg M, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EG et al. (2007) Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol 25: 221–231 [DOI] [PubMed] [Google Scholar]
- Promper C, Schneider R, Weiss H (1993) The role of the proton-pumping and alternative respiratory chain NADH:ubiquinone oxidoreductases in overflow catabolism of Aspergillus niger. Eur J Biochem 216: 223–230 [DOI] [PubMed] [Google Scholar]
- Punt PJ, van Biezen N, Conesa A, Albers A, Mangnus J, van den Hondel C (2002) Filamentous fungi as cell factories for heterologous protein production. Trends Biotechnol 20: 200–206 [DOI] [PubMed] [Google Scholar]
- Reed JL, Vo TD, Schilling CH, Palsson BO (2003) An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GSR). Genome Biol 4: R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell GE, O'Flaherty F (1931) Studies in the physiology of molds. II. Chemical composition and culture of molds. J Am Leather Chemists Assoc 26: 216–222 [Google Scholar]
- Röhr M, Kubicek CP (1981) Regulatory aspects of citric acid fermentation by Aspergillus niger. Process Biochem June/July: 34–44 [Google Scholar]
- Schilling CH, Schuster S, Palsson BO, Heinrich R (1999) Metabolic pathway analysis: basic concepts and scientific applications in the post-genomic era. Biotechnol Prog 15: 296–303 [DOI] [PubMed] [Google Scholar]
- Schmidt K, Nørregaard LC, Pedersen B, Meissner A, Duus JØ, Nielsen JØ, Villadsen J (1999) Quantification of intracellular metabolic fluxes from fractional enrichment and 13C–13C coupling constraints on the isotopomer distribution in labeled biomass components. Metab Eng 1: 166–179 [DOI] [PubMed] [Google Scholar]
- Schrickx JM, Krave AS, Verdoes JC, van den Hondel CA, Stouthamer AH, van Verseveld HW (1993) Growth and product formation in chemostat and recycling cultures by Aspergillus niger N402 and a glucoamylase overproducing transformant, provided with multiple copies of the glaA gene. J Gen Microbiol 139: 2801–2810 [DOI] [PubMed] [Google Scholar]
- Singh OV, Kumar R (2007) Biotechnological production of gluconic acid: future implications. Appl Microbiol Biotechnol 75: 713–722 [DOI] [PubMed] [Google Scholar]
- Smirnov VI, Chubova VP (1965) Biochemical indexes of mycelium of Aspergillus niger. Mikrobiologicheskie Protessy v Pochvakh Moldavii 2: 88–91 [Google Scholar]
- Stephanopoulos GN, Aristidou AA, Nielsen J (1998) Metabolic Engineering—Principles and Methodologies. New York (Orlando, FL/London/San Diego, CA): Academic Press [Google Scholar]
- Sun J, Lu X, Rinas U, Zeng AP (2007) Metabolic peculiarities of Aspergillus niger disclosed by comparative metabolic genomics. Genome Biol 8: R182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terroine E-F, Bonnet R (1927) L'energie de croissance. X. Formation des matiéres grasses aux dépens des glucides chez les microorganisms. Bull Soc Chim Biol (Paris) 9: 588–596 [Google Scholar]
- Torres NV (1994a) Modeling approach to control of carbohydrate metabolism during citric acid accumulation by Aspergillus niger: I. Model definition and stability of the steady state. Biotechnol Bioeng 44: 104–111 [DOI] [PubMed] [Google Scholar]
- Torres NV (1994b) Modeling approach to control of carbohydrate metabolism during citric acid accumulation by Aspergillus niger: II. Sensitivity analysis. Biotechnol Bioeng 44: 112–118 [DOI] [PubMed] [Google Scholar]
- van Brunt J (1986) Fungi: the perfect hosts? Biotechnology (NY) 4: 1057–1062 [Google Scholar]
- van de Merbel NC, Ruijter GJG, Lingeman H, Brinkman UAT, Visser J (1994) An automated monitoring system using on-line ultrafiltration and column liquid chromatography for Aspergillus niger fermentations. Appl Microbiol Biotechnol 41: 658–663 [Google Scholar]
- Wallace W (1926) Manufacture of oxalates and oxalic acid. United States Patent US 1 602 802
- Witteveen CFB, Visser J (1995) Polyol pools in Aspergillus niger. FEMS Microbiol Lett 134: 57–62 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Figure 1
Supplementary Figure 2
Supplementary Information