Abstract
Recent studies have emphasized the value of including structural information into the topological analysis of protein networks. Here, we utilized structural information to investigate the role of intrinsic disorder in these networks. Hub proteins tend to be more disordered than other proteins (i.e. the proteome average); however, we find this only true for those with one or two binding interfaces (‘single'-interface hubs). In contrast, the distribution of disordered residues in multi-interface hubs is indistinguishable from the overall proteome. Surprisingly, we find that the binding interfaces in single-interface hubs are highly structured, as is the case for multi-interface hubs. However, the binding partners of single-interface hubs tend to have a higher level of disorder than the proteome average, suggesting that their binding promiscuity is related to the disorder of their binding partners. In turn, the higher level of disorder of single-interface hubs can be partly explained by their tendency to bind to each other in a cascade. A good illustration of this trend can be found in signaling pathways and, more specifically, in kinase cascades. Finally, our findings have implications for the current controversy related to party and date-hubs.
Keywords: hubs, intrinsic disorder, structural networks
Introduction
There have been many advances in the study of protein interaction networks enabled by the advent of high-throughput technology (Barabasi and Oltvai, 2004). Recent studies have started to put these networks into the context of 3D protein structures (Aloy and Russell, 2006; Kim et al, 2006). Many genomic properties that had been previously linked to topological properties were shown to be better described by structural quantities. In particular, the notion of network hubs was refined to two different kinds of hubs, ‘single' (or singlish)-interface and multi-interface hubs (Kim et al, 2006). The former have only few interaction interfaces (two at most) and tend to be enriched in signaling proteins, whereas the latter correspond to central members of larger protein complexes.
In contrast to the classical view of structured proteins, the concept of intrinsically disordered regions has recently emerged (Dunker et al, 2002; Linding et al, 2003; Iakoucheva et al, 2004; Radivojac et al, 2007). Disordered regions are segments of a protein that does not completely fold and remains flexible and unordered. Computational predictions of disordered regions have found that, although proteomes of archaea and bacteria comprise only a small fraction of intrinsically disordered proteins (about 2–4%), eukaryotic proteomes include a large fraction (about 33%) of long regions that are natively disordered and thus do not adopt a fixed structure (Ward et al, 2004b). The functions of disordered regions have been classified into four categories: molecular recognition, molecular assembly, protein modification, and entropic chain activity (Wright and Dyson, 1999; Sugase et al, 2007). Disordered regions of proteins have been shown to have key physiological roles, for example, are involved as communicators in many cellular signaling pathways. In particular, the target sites of both protein kinases and many modular protein domains (such as SH3, PDZ, SH2 etc.) generally lie in disordered regions (Iakoucheva et al, 2004; Beltrao and Serrano, 2005; Fuxreiter et al, 2007), presumably because disordered regions are more prone to present the short linear motifs that these domains and kinases bind to.
The initial studies on structural networks did not examine the role of disorder (Kim et al, 2006; Beltrao et al, 2007; Devos and Russell, 2007). In this work, we make the first rigorous investigation of disorder in structural networks and its role for many cellular properties.
Results and discussion
Singlish-interface hubs have a higher propensity for disorder, whereas multi-interface hubs have the same propensity as normal proteins
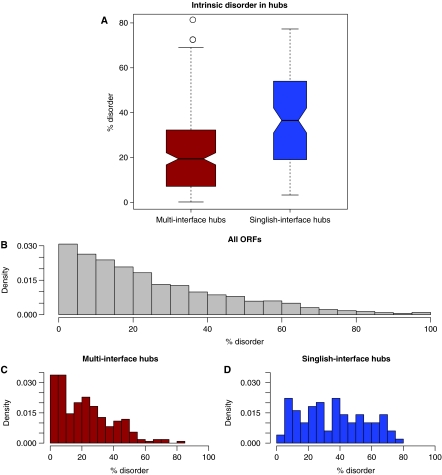
It has been pointed out before that hubs, that is, proteins with a large number of interaction partners, have a higher average number of disordered residues (Dunker et al, 2005; Haynes et al, 2006; Patil and Nakamura, 2006; Singh et al, 2007). This result may be surprising, as one might assume that interactions would constrain the protein towards ordered regions. Indeed, a recent study has disagreed with the previous finding (Schnell et al, 2007). Here, we seek to clarify this result by putting it in the context of structural interaction networks. Surprisingly, we find that in the Structural Interaction Network (SIN v2.0) (Kim et al, 2006), singlish-interface hubs have a much higher fraction of disordered residues than multi-interface hubs (Figure 1A). The reason for the higher disorder of singlish-interface versus multi-interface hubs seems obvious: multi-interface hubs tend to be much more constrained than singlish-interface hubs. Hence, we expect multi-interface hubs to have a significantly reduced level of disorder than non-hub proteins, whereas singlish-interface hubs would be at approximately the same level. However, when we compare both types of hubs to all other proteins, we find that multi-interface hubs have about the same propensity for disorder as other proteins, whereas singlish-interface hubs have a much higher propensity than other proteins (Figure 1B–D). Hence, the difference in degree of disorder between the two types of hubs is unlikely to be the result of structural constraints on multi-interface hubs, as the other proteins would also have a similar absence of these constraints.
Figure 1.
(A) Disorder in singlish-interface hubs versus multi-interface hubs. (Wilcoxon ranksum test, P=1.0e−8). (B) Distribution of disordered residues in the yeast proteins. As can be seen, most yeast proteins have a relatively low level of disorder; however, almost all have some fraction of disordered residues. (C) Distribution of disorder in multi-interface hubs. The distribution does not significantly deviate from the distribution of the yeast proteome (Kolmogorov-Smirnov test, P-value=0.11). In other words, multi-interface hubs do not have different levels of disorder than normal proteins. (D) Distribution of disorder in singlish-interface hubs. Singlish-interface hubs show a different distribution in terms of disorder (Kolmogorov–Smirnov test, P=2.0e−6).
Disordered regions in proteins tend to be under less evolutionary constraints contributing to the faster evolutionary rate of singlish-interface hubs
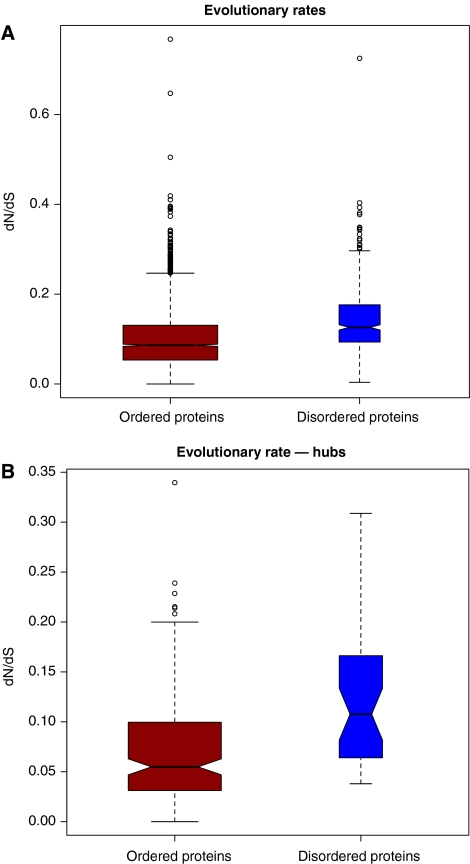
Previous studies have found that singlish-interface hubs have a significantly higher evolutionary rate than multi-interface hubs, presumably due to stronger constraints of the multiple interfaces (Kim et al, 2006). However, other studies have suggested that this difference is due to a difference in protein abundance (Batada et al, 2007). We hypothesized that the higher level of disorder would be related to this higher evolutionary rate. Indeed, it has been suggested that disordered proteins evolve faster than structured ones (Brown et al, 2002). We find here that in a genome-wide analysis, disordered proteins have a significantly higher evolutionary rate than structured proteins (Figure 2A and B). As disordered proteins also tend to be expressed at a lower rate than structured ones (Supplementary Table S3), the causality is unclear. Hence, we looked at the evolutionary rate on a residue-by-residue basis, independent of any bias at the gene level. We find that disordered regions in proteins tend to evolve much faster than the other regions (Supplementary Table S4). Although structural factors only partly determine the evolutionary rate of proteins (Bloom et al, 2006), a difference in disorder is likely to be a contributing factor.
Figure 2.
dN/dS ratio of ordered and disordered proteins, and hubs. (A) All yeast proteins, split by order/disorder (Wilcoxon rank sum test, P-value <2.2e−16); (B) Hubs only (Wilcoxon rank sum test, P=1.5e−6).
Binding interfaces are structured
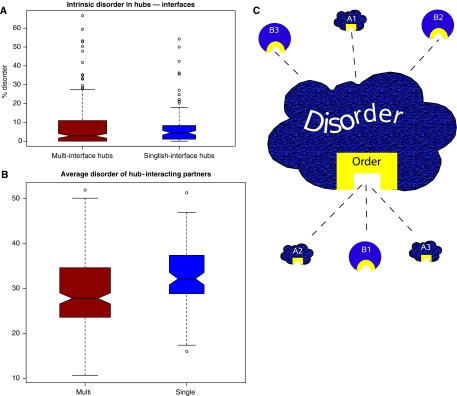
Disordered regions have been implicated in mediating promiscuous binding (Dunker et al, 2005; Patil and Nakamura, 2006), thus enabling a protein to functionally bind to many diverse interacting partners. Also, singlish-interface hubs are known to be promiscuous binders and their interfaces presumably interact with many different partners. Hence, it seems reasonable to assume that the heightened level of disorder in singlish-interface hubs is due to their interfaces being involved in promiscuous binding (Singh et al, 2007). Therefore, their binding interfaces should be highly disordered. However, when we examine the binding interfaces of singlish-interface hubs, we find them to be largely structured. Moreover, we do not find a significant difference in level of disorder between interfaces of singlish-interface and multi-interface hubs (Figure 3A).
Figure 3.
(A) Disorder in the interface regions of singlish- and multi-interface hubs. (Wilcoxon ranksum test, P=0.4). (B) Disorder of the binding partner of singlish- and multi-interface hubs (Wilcoxon ranksum test, P=4.5e−5). BIOGRID data are reported in the boxplot. Similar results were found for Kim et al (2006) and Batada et al (2006) data sets (Supplementary Figure S5). (C) Schematic representation of disorder in singlish-interface hubs. Singlish-interface hubs have large portions of disordered regions (painted in gray). However, the interface is itself is highly structured (painted in black). One reason for the disorder in the bulk of the protein is the fact that singlish-interface hubs often are targeted by kinases. On the other hand, they tend to be kinases themselves and target disordered regions in other proteins.
This leaves us with two questions: (1) with structured interfaces, how is the promiscuous binding of singlish-interface hubs mediated? (2) What leads to their higher level of disorder, if not promiscuous binding at the binding interface?
Higher disorder in interacting partners of singlish-interface hubs
We first turn to the question of how the binding promiscuity of singlish-interface hubs is mediated. We hypothesized that if the interface of the singlish-interface hub itself is structured, perhaps the binding partners would be disordered, thus leading to promiscuous binding. This case of a disordered-structured promiscuous interaction has been described recently (Dunker et al, 2005). Indeed, when examining the binding partners of singlish-interface hubs for disorder, we find that they are significantly more disordered than the binding partners of multi-interface hubs, as well as more disordered than other proteins (Figure 3B). Hence, promiscuous binding is partly mediated by disorder, but not in the interface in the singlish-interface hub itself, rather in the interacting partners.
Enrichment of disordered regions in singlish-interface hubs can be rationalized by their cascading nature as is illustrated in their involvement in signaling pathways
We hypothesized that the higher propensity of disorder in interacting partners of singlish-interface hubs may be related to their own higher level of intrinsic disorder. That is, if singlish-interface hubs had a tendency to interact with each other in a cascade fashion, it would lead to a separate region in the singlish-interface hub: a highly structured binding interface (that binds disordered regions in other proteins) and a disordered region, which in turn is bound by other singlish-interface hubs (Figure 3C). A recent study listed a number of examples of proteins with just this layout (Xie et al, 2007). We find here for the particular case of singlish-interface hubs, a higher tendency to interact with each other (on average, 68% of singlish-interface hub partners are singlish-interface hubs). This cascading property is well illustrated in signaling pathways. Signaling pathways are thought to have evolved through a mix-and-match principle, consistent with the cascading nature that we observed in singlish-interface hubs (Pawson and Nash, 2003). That is, repeated duplications of these signaling genes must have occurred during evolution. Furthermore, it is known that signaling pathways tend to be enriched in disordered proteins (Iakoucheva et al, 2002, 2004). We find here that disordered proteins in particular have significantly higher numbers of paralogs than other proteins, suggesting that they have been duplicated more often (Supplementary Table S2; Supplementary Figure S4), and are perhaps less dosage sensitive.
Kinases serve as a good illustrative example of signaling pathways. Indeed, there is a significant enrichment for protein kinases among singlish-interface hubs: about 34% of singlish-interface hubs are kinases (hypergeometric test, P-value=1e−33). Furthermore, it is known that the binding sites of both protein kinases and modular protein domains tend to lie in disordered regions (Iakoucheva et al, 2004; Beltrao and Serrano, 2005). When checking for the likely targets of protein kinases, we find a significant enrichment in singlish-interface hubs (Table I). Likewise, and in agreement with previous results, we find that disordered proteins are much more likely to be kinase targets (Supplementary Table S1). Hence, for these proteins, some of the heightened level of disorder may be due to the fact that they present kinase target sites.
Table 1.
Kinase targets
| Hubs |
||
|---|---|---|
| Multi-interface | Singlish-interface | |
| Non-kinase targets | 165 | 56 |
| Kinase targets | 54 | 43 |
Contingency table of kinase targets versus hub interface. Singlish-interface hubs are enriched for being kinase targets (Fisher's exact test, P=0.001).
Furthermore, the concept of distal docking motifs for kinase targeting has recently been proposed (Remenyi et al, 2006; Ubersax and Ferrell, 2007). This notion fits in very well with our results. In the simplest case, a kinase has a structured catalytic region and a second disordered region, which could harbor a distal docking motif.
Implications for different types of hubs in other networks
A related concept to singlish-interface and multi-interface hubs is the notion of party and date hubs (Han et al, 2004), and it has been shown that there is some correspondence of the two (Kim et al, 2006). Indeed, we find, consistent with earlier results (Ekman et al, 2006; Singh et al, 2007), that date hubs tend to have a higher degree of disorder than party hubs (in two versions of the FYI (Bertin et al, 2007), Supplementary Figure S2a–b). However, there has been some controversy about the notion of date and party hubs (Batada et al, 2006, 2007) and potential biases in different data sets. Indeed, when examining the date-party hubs as defined by Batada and co-workers, we do not find a difference in the level of disorder (Supplementary Figure S2c). From this, one may conclude that the differences in disorder we observed are strongly dependent on data set choice and gene expression data sets tend to be confounded by noise. Hence, we examined the HCI data set by Batada and co-workers more closely and used an approximate inference of which hubs would be singlish-interface and which would be multi-interface in their data set (see Materials and methods). We believe that this inference of number of interfaces may be somewhat more robust, as it is related to a real biophysical property of proteins. Now we observe a significant difference in disorder between the two hub classes (Supplementary Figure S3). In summary, we find evidence for the notion that some hubs have few binding interfaces (hence interact with their partners at different times), whereas others have many and that both groups have distinct properties, such as a different level of disorder. This suggests that the notion of date and party hubs, since related, also reflects two distinct groups of proteins.
Conclusions
We have presented evidence here that intrinsic disorder is an important feature in protein networks. Specifically, it further distinguishes two types of hubs, multi- and singlish-interface, and is important in mediating promiscuous binding. However, the disordered regions do not seem to be enriched at the interface regions of singlish-interface hubs, but are rather enriched in their binding targets, presumably due to their central role in signaling pathways. Furthermore, the feature of protein disorder brings further evidence to the difference in evolutionary constraints of protein hubs.
Materials and methods
A number of different sources were utilized in this study. Hereafter, a description of the data sets and the analysis methods is reported.
Disorder prediction
We used DISOPRED (Ward et al, 2004a) to obtain disorder predictions of 6714 ORFs of Saccharomyces cerevisiae (including many dubious ORFs). This software tool provides both a score and a disorder classification for each residue. DISOPRED is among the top-ranking disorder prediction tools evaluated at the ‘Critical Assessment of Techniques for Protein Structure Prediction (CASP) conference (Moult et al, 2007). The percentage of disordered residues is computed by dividing the number of disordered residues by ORF length. ORFs with a percentage of disordered residues greater than 50% were considered disordered.
Similarly, we computed the percentage of disorder of interacting interfaces by dividing the number of disordered residues in the interface by the interface length.
Structural interaction network version 2
The definition of singlish- and multi-interface hubs is reported by Kim et al (2006). We used an updated version of the SIN (SIN version 2.0). Among 316 hubs, 98 are singlish-interface and 218 are multi-interface hubs.
Party-hubs and date-hubs
Information about party- and date- hubs derives from three data sets: Han et al (2004), Bertin et al (2007), and Batada et al (2007). In Han et als' data set, 108 party- and 91 date-hubs are included. In Bertin et als' data set, there are 306 date- and 240 party-hubs.
Concerning the data set by Batada and co-workers, we determined party- and date-hubs by first selecting the ORFs with more than 10 interacting partners. Then, party (date) have an average correlation with their corresponding interacting partners greater than (less than) 0.25. This resulted in 175 date- and 33 party-hubs. Coexpression correlation was computed based on the compendium data by Hughes et al (2000).
Pfam interacting domains
Pfam interacting domains were obtained from PFAM repository (Bateman et al, 2002). To analyze the disorder of interfaces, the working hypothesis is that interacting domains confer binding capability to protein regions. The following cutoff values were used for domain assignment: (1) e-value of alignment <1e−4; (2) matched sequence length >80% of domain length; (3) domain length >12 residues. When using these constraints, we have 1342 ORFs with at least one interacting domain.
In addition, this data set was employed to infer which of date- and party-hubs by Batada and co-workers are multi- or singlish-interface hubs. In this case, more stringent criteria to assign a domain to an ORF were used: (1) e-value of alignment <1e−7; (2) matched sequence length >95% pfam domain length; (3) domain length >5 residues. Accordingly, 1738 ORFs have at least one pfam domain: divided in 1441 singlish-domain ORFs and 327 multi-domain ORFs. Among those 1738, we only consider hubs (defined as having more than 10 interacting partners); resulting in 73 singlish-domain and 24 multi-domain hubs.
Kinase target data
We used the phosphorylome data set (Ptacek et al, 2005) to obtain the list of kinase interaction partners. It contains 1325 ORFs known as targets for kinases.
Interaction data
Interaction data derives from several sources: BIOGRID (Stark et al, 2006), Batada et al (2006), and Kim et al (2006). Each data set provides a list with the interacting ORFs. Considering BIOGRID, we included interactions determined by Affinity Capture-MS, Affinity Capture-RNA, Affinity Capture-Western, biochemical activity, co-crystal structure, Far Western, FRET, Protein-peptide, Protein-RNA, Reconstituted Complex, and Two-hybrid. Above-mentioned sources contain 61 634, 28 915, and 4080 interactions, respectively.
We computed the average disorder of the interacting partners for each hub and assessed whether a difference between the partners of singlish- and multi-interface hubs is present by means of the Wilcoxon rank sum test. As singlish-interface hubs have other singlish-interface hubs as interacting partners, which are more disordered, we therefore repeated the same analysis by excluding other singlish-interface hubs partners. The difference between multi- and singlish-interface hubs partners is still significant (Supplementary Figure S5).
Biases in the interaction network may affect our results. Indeed, the SIN is smaller than other interaction networks, and as it is based on proteins with solved crystal structures, it may be depleted in disordered proteins. However, we find contrasting evidence: the average percentage of disordered residues in the SIN is about the same as the genomic average: 26% (25% is the genomic average—Wilcoxon rank sum test, P=0.08 (Supplementary Figure S1).
Orthologs/paralogs
Orthologs and paralogs information was computed from the Clusters of Orthologous Groups (COGs) (Tatusov et al, 2003). Cluster information was used to determine the number of paralogs for each ORF.
Evolutionary rates
Sequence alignment between S. cervisiae and S. bayanus was performed through BLAST. Each residue is then labeled as mutated or non-mutated. Disorder analysis was then computed residue-by-residue.
Computational analysis
We used R 2.5 to perform the statistical analysis (R Development Core Team, 2007).
All data sets used and the updated version of the SIN along with detailed description and statistics are available at http://sin.gersteinlab.org.
Supplementary Material
Supplementary Information 1
Supplementary Information 2
Supplementary Information 3
Supplementary Information 4
Acknowledgments
We thank Hunter Fraser for helpful discussions and careful reading of the manuscript. We thank the ‘Yale University Biomedical High Performance Computing Center' (NIH grant: RR19895) for providing the computational support. This work was supported by the NIH.
References
- Aloy P, Russell RB (2006) Structural systems biology: modelling protein interactions. Nat Rev Mol Cell Biol 7: 188–197 [DOI] [PubMed] [Google Scholar]
- Barabasi AL, Oltvai ZN (2004) Network biology: understanding the cell's functional organization. Nat Rev Genet 5: 101–113 [DOI] [PubMed] [Google Scholar]
- Batada NN, Reguly T, Breitkreutz A, Boucher L, Breitkreutz BJ, Hurst LD, Tyers M (2006) Stratus not altocumulus: a new view of the yeast protein interaction network. PLoS Biol 4: e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batada NN, Reguly T, Breitkreutz A, Boucher L, Breitkreutz BJ, Hurst LD, Tyers M (2007) Still stratus not altocumulus: further evidence against the date/party hub distinction. PLoS Biol 5: e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL (2002) The Pfam protein families database. Nucleic Acids Res 30: 276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrao P, Kiel C, Serrano L (2007) Structures in systems biology. Curr Opin Struct Biol 17: 378–384 [DOI] [PubMed] [Google Scholar]
- Beltrao P, Serrano L (2005) Comparative genomics and disorder prediction identify biologically relevant SH3 protein interactions. PLoS Comput Biol 1: e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin N, Simonis N, Dupuy D, Cusick ME, Han JD, Fraser HB, Roth FP, Vidal M (2007) Confirmation of organized modularity in the yeast interactome. PLoS Biol 5: e153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JD, Drummond DA, Arnold FH, Wilke CO (2006) Structural determinants of the rate of protein evolution in yeast. Mol Biol Evol 23: 1751–1761 [DOI] [PubMed] [Google Scholar]
- Brown CJ, Takayama S, Campen AM, Vise P, Marshall TW, Oldfield CJ, Williams CJ, Dunker AK (2002) Evolutionary rate heterogeneity in proteins with long disordered regions. J Mol Evol 55: 104–110 [DOI] [PubMed] [Google Scholar]
- Devos D, Russell RB (2007) A more complete, complexed and structured interactome. Curr Opin Struct Biol 17: 370–377 [DOI] [PubMed] [Google Scholar]
- Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z (2002) Intrinsic disorder and protein function. Biochemistry 41: 6573–6582 [DOI] [PubMed] [Google Scholar]
- Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN (2005) Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J 272: 5129–5148 [DOI] [PubMed] [Google Scholar]
- Ekman D, Light S, Bjorklund AK, Elofsson A (2006) What properties characterize the hub proteins of the protein-protein interaction network of Saccharomyces cerevisiae? Genome Biol 7: R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxreiter M, Tompa P, Simon I (2007) Local structural disorder imparts plasticity on linear motifs. Bioinformatics 23: 950–956 [DOI] [PubMed] [Google Scholar]
- Han JD, Bertin N, Hao T, Goldberg DS, Berriz GF, Zhang LV, Dupuy D, Walhout AJ, Cusick ME, Roth FP, Vidal M (2004) Evidence for dynamically organized modularity in the yeast protein–protein interaction network. Nature 430: 88–93 [DOI] [PubMed] [Google Scholar]
- Haynes C, Oldfield CJ, Ji F, Klitgord N, Cusick ME, Radivojac P, Uversky VN, Vidal M, Iakoucheva LM (2006) Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput Biol 2: e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai HY, He YDD, Kidd MJ, King AM, Meyer MR, Slade D, Lum PY, Stepaniants SB, Shoemaker DD, Gachotte D, Chakraburtty K, Simon J et al. (2000) Functional discovery via a compendium of expression profiles. Cell 102: 109–126 [DOI] [PubMed] [Google Scholar]
- Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK (2002) Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol 323: 573–584 [DOI] [PubMed] [Google Scholar]
- Iakoucheva LM, Radivojac P, Brown CJ, O'Connor TR, Sikes JG, Obradovic Z, Dunker AK (2004) The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res 32: 1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PM, Lu L, Xia Y, Gerstein M (2006) Relating 3D structures to protein networks provides evolutionary insight. Science 314: 1938–1941 [DOI] [PubMed] [Google Scholar]
- Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB (2003) Protein disorder prediction: implications for structural proteomics. Structure 11: 1453–1459 [DOI] [PubMed] [Google Scholar]
- Moult J, Fidelis K, Kryshtafovych A, Rost B, Hubbard T, Tramontano A (2007) Critical assessment of methods of protein structure prediction—Round VII. Proteins Struct Funct Bioinform 69 (S8): 3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil A, Nakamura H (2006) Disordered domains and high surface charge confer hubs with the ability to interact with multiple proteins in interaction networks. FEBS Lett 580: 2041–2045 [DOI] [PubMed] [Google Scholar]
- Pawson T, Nash P (2003) Assembly of cell regulatory systems through protein interaction domains. Science 300: 445–452 [DOI] [PubMed] [Google Scholar]
- Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, McCartney RR, Schmidt MC, Rachidi N, Lee SJ, Mah AS, Meng L, Stark MJ, Stern DF, De Virgilio C, Tyers M et al. (2005) Global analysis of protein phosphorylation in yeast. Nature 438: 679–684 [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2007) R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK (2007) Intrinsic disorder and functional proteomics. Biophys J 92: 1439–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remenyi A, Good MC, Lim WA (2006) Docking interactions in protein kinase and phosphatase networks. Curr Opin Struct Biol 16: 676–685 [DOI] [PubMed] [Google Scholar]
- Schnell S, Fortunato S, Roy S (2007) Is the intrinsic disorder of proteins the cause of the scale-free architecture of protein-protein interaction networks? Proteomics 7: 961–964 [DOI] [PubMed] [Google Scholar]
- Singh GP, Ganapathi M, Dash D (2007) Role of intrinsic disorder in transient interactions of hub proteins. Proteins 66: 761–765 [DOI] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M (2006) BioGRID: a general repository for interaction datasets. Nucleic Acids Res 34: D535–D539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugase K, Dyson HJ, Wright PE (2007) Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature 447: 1021–1025 [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax JA, Ferrell JE Jr (2007) Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol 8: 530–541 [DOI] [PubMed] [Google Scholar]
- Ward JJ, McGuffin LJ, Bryson K, Buxton BF, Jones DT (2004a) The DISOPRED server for the prediction of protein disorder. Bioinformatics 20: 2138–2139 [DOI] [PubMed] [Google Scholar]
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT (2004b) Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol 337: 635–645 [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ (1999) Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol 293: 321–331 [DOI] [PubMed] [Google Scholar]
- Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Uversky VN, Obradovic Z (2007) Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J Proteome Res 6: 1882–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information 1
Supplementary Information 2
Supplementary Information 3
Supplementary Information 4