Abstract
We present, here, a detailed and curated map of molecular interactions taking place in the regulation of the cell cycle by the retinoblastoma protein (RB/RB1). Deregulations and/or mutations in this pathway are observed in most human cancers. The map was created using Systems Biology Graphical Notation language with the help of CellDesigner 3.5 software and converted into BioPAX 2.0 pathway description format. In the current state the map contains 78 proteins, 176 genes, 99 protein complexes, 208 distinct chemical species and 165 chemical reactions. Overall, the map recapitulates biological facts from approximately 350 publications annotated in the diagram. The network contains more details about RB/E2F interaction network than existing large-scale pathway databases. Structural analysis of the interaction network revealed a modular organization of the network, which was used to elaborate a more summarized, higher-level representation of RB/E2F network. The simplification of complex networks opens the road for creating realistic computational models of this regulatory pathway.
Keywords: cell-cycle regulation, E2F, RB pathway, RB1, systems-biology standards
Introduction
The cell cycle is the succession of four phases called G1, S, G2 and M. In dividing cells, DNA replication (S phase) and mitosis (M phase) alternate (Alberts et al, 1994), and are separated by two gap phases, G1 and G2 phases. In quiescent cells, the cells are considered to be in G0 phase. When they receive external signals, such as growth factors, a series of activations push the cell from a G0 to a G1 state and enters the cell cycle. The whole process of cell division is mainly orchestrated by complexes composed of two subunits, a kinase and a cyclin partner. These complexes phosphorylate a certain number of proteins, either activating or inhibiting them. Among them, the retinoblastoma tumour suppressor protein RB (RB1) is a key regulator in cell-cycle entry (transition G1/S). It sequesters a family of transcription factors, the E2Fs, responsible for the transcription of many genes involved in cell-cycle regulation, DNA replication and other functions like the activation of the apoptotic pathway (Muller et al, 2001). RB functions as a brake in the cell cycle, which is released when external signals trigger S-phase entry. The main targets of the external signals are the G1 cyclin/CDK complexes. Once active, the complexes, among them CycD1/CDK4,6, act as starters of the cell cycle (Novak et al, 2007) and phosphorylate RB, which then releases E2F (DeGregori, 2004).
RB is a member of a family of proteins called the pocket proteins (Knudsen and Wang, 1997). These proteins RB, p107 and p130, share sequence similarities, especially in the ‘pocket domain' (Stevaux and Dyson, 2002), which is responsible for their repressor function. RB protein contains domains where the binding sites for co-repressors (E2F proteins and viral oncoproteins) are situated. These sites are subjected to most mutations.
RB is a tumour suppressor gene. Because of its implication in so many, if not all, cancers (Sherr and McCormick, 2002), the study of RB regulation requires a special attention.
More specifically, the RB/E2F pathway is commonly deregulated in cancer through genetic or epigenetic mechanisms, resulting in E2F activation. Several common oncogenes (involved in many cancer types) are the activators of the pathway, whereas several common tumour suppressor genes are inhibitors of the pathway. For example, cyclin D1 (CCND1), E2F3 and the two cyclin-dependent kinases CDK4 and CDK6 can be activated by translocation, amplification or mutation, whereas RB (RB1) and the cyclin-dependent kinase inhibitors p16INK4a (CNKN2A) and p15INK4b (CDKN2B) can be inactivated by point mutation, homozygous deletion or DNA methylation. In addition, RB can be inactivated by several oncogenic viral proteins including E7 from human papillomavirus, which is responsible for more than 90% of cervical carcinomas (Munger et al, 2001). Tumour suppressor gene inactivation is found not only in sporadic tumours but also in tumour-prone families. Germline mutations of RB1 results in retinoblastoma with a high penetrance early in young individuals and late in life in sarcomas and lung and bladder carcinomas (Knudson, 1971; Nevins, 2001; Giacinti and Giordano, 2006). Germinal mutations of p16INK4a results in melanomas (Hussussian et al, 1994). Finally, the importance of the RB/E2F pathway in tumour formation based on genetic or epigenetic arguments, has been confirmed by in vivo experiments using transgenic mice (Classon and Harlow, 2002).
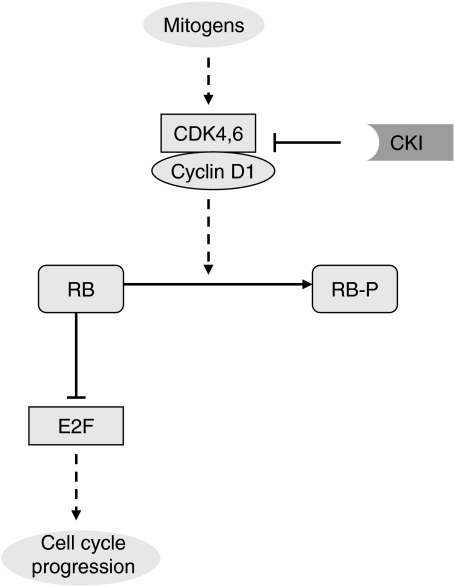
Often, textbooks present a simplified picture of RB regulation (Figure 1): mitogens activate CycD1/CDK complexes, which phosphorylate RB, and which, in turn, releases the hold on E2F transcription factors. E2F activity controls cell-cycle progression.
Figure 1.
Textbook view of the RB/E2F pathway. When CycD1/CDK4,6 is activated, it phosphorylates RB, which in turn is inactivated and releases E2F transcription factors leading to cell proliferation.
However, the real picture of interactions around RB is much more complex due to combinatorial complexity and the number of important regulators, which are not taken into account in the simple pathway view. There are not only several E2F transcription factors but also several proteins besides RB with which the E2F proteins bind to be inactivated. Moreover, the RB-related proteins not only can sequester the E2Fs but also can actively repress transcription. There are 10 known E2F proteins—7 of which bind to dimerization partners (DP1 and DP2) to become active—that can associate with the three pocket proteins RB, p107 and p130, and proteins from the polycomb group. These E2F proteins can be (1) activators of transcription (E2F1, E2F2 and E2F3a) or (2) repressors of transcription (E2F3b, E2F4, E2F5 and E2F6). The last E2F family proteins, E2F7a, E2F7b and E2F8, form homodimers, do not bind to pocket proteins and seem to repress transcription.
We believe that this complexity has to be taken into account in any realistic consideration of the RB network (such as the creation of a computational model). Thus, we stated our problem as a very careful and focused consideration of the whole corpus of available biological facts published in high-level biological literature to reconstruct a detailed network of interactions around RB protein.
We started from the simplified picture (Figure 1) and added several tens of proteins involved in cell-cycle regulation, carefully connecting them to the rest of the network by means of known biochemical transformations. There were several questions that had to be answered during this reconstruction:
What should the language be for such a detailed description? It should be standard and expressive enough to readily incorporate available biological facts. A choice was made to use Systems Biology Graphical Notation (SBGN) standard partially implemented in CellDesigner software (Kitano et al, 2005). This choice also defined for us the level of details that we should keep in the description.
How to deliver the collected information to the public? Together with CellDesigner diagram, we chose to use the BioPAX format for storing pathway information. An automatic conversion tool from CellDesigner (Funahashi et al, 2003) to BioPAX was developed (Zinovyev et al, 2007).
How to define the ‘borders' of RB pathway? RB interacts with more than 100 proteins (Morris and Dyson, 2001). We chose to concentrate mainly on genes involved in cell-cycle regulation and in apoptosis entry.
How to make the resulting detailed network usable and understandable? We performed a structural analysis of the resulting network, and it was found that the network has a modular structure. A module was defined as a cluster of relevant cycles in the reaction graph. A special software, BiNoM, plugin of Cytoscape (Zinovyev et al, 2007), was developed to perform such analysis (see Materials and methods).
How to maintain and expand the pathway? A web-page is associated with the pathway (http://bioinfo-out.curie.fr/projects/rbpathway/) and will be regularly updated as some new papers appear and with the input we expect from the RB experts worldwide (listed in ‘Contributors' section on our web-page).
One purpose behind the construction of this diagram, once validated, is to provide a map of RB/E2F pathway that can become not only a reference while studying different cancers and mutations but also a tool to analyse formally the pathway and predict its behaviour in response to different types of deregulations (Nevins, 2001).
Table I recapitulates all current names for genes used in this article and their corresponding HUGO names. RB, p107 and p130 are the common names for the proteins referenced as RB1, RBL1 and RBL2 (HUGO names), respectively.
Table 1.
List of protein names used in the diagram and the corresponding HUGO names
| Cell designer name | Used name | HUGO name |
|---|---|---|
| AKT1 | AKT1 | AKT1 |
| APC | Anaphase-promoting complex | |
| ATM | ATM | ATM |
| ATR | ATR | ATR |
| BAT8 | G9a | BAT8 |
| BMI1* | BMI1 | PCGF4 |
| CDC14A | CDC14A | CDC14A |
| CDC2 | CDK1 | CDC2 |
| CDC20 | CDC20 | CDC20 |
| CDC25C | CDC25C | CDC25C |
| CDC37 | CDC37 | CDC37 |
| CDH1* | CDH1 | FZR1 |
| CDK2 | CDK2 | CDK2 |
| CDK3 | CDK3 | CDK3 |
| CDK4 | CDK4 | CDK4 |
| CDK6 | CDK6 | CDK6 |
| CDK7 | CDK7 | CDK7 |
| CDKN3 | KAP1 | CDKN3 |
| CHEK1 | Chk1 | CHEK1 |
| CHEK2 | Chk2 | CHEK2 |
| CREBBP | CBP | CREBBP |
| cyclin A2* | cyclin A2, CycA2 | CCNA2 |
| cyclin B1* | cyclin B1, CycB1 | CCNB1 |
| cyclin C* | cyclin C, CycC | CCNC |
| cyclin D1* | cyclin D1, CycD1 | CCND1 |
| cyclin E1* | cyclin E1, CycE1 | CCNE1 |
| cyclin H* | cyclin H, CycH | CCNH |
| DP1* | DP1 | TFDP1 |
| DP2* | DP2 | TFDP2 |
| E2F1 | E2F1 | E2F1 |
| E2F2 | E2F2 | E2F2 |
| E2F3 | E2F3 | E2F3 |
| E2F4 | E2F4 | E2F4 |
| E2F5 | E2F5 | E2F5 |
| E2F6 | E2F6 | E2F6 |
| E2F7 | E2F7 | E2F7 |
| E2F8 | E2F8 | E2F8 |
| EED | EED | EED |
| EHMT1 | Eu-HMT1 | EHMT1 |
| EP300 | p300 | EP300 |
| EPC1 | EPC1 | EPC1 |
| EZH2 | EZH2 | EZH2 |
| GSK3B | GSK3B | GSK3B |
| HDAC1 | HDAC1 | HDAC1 |
| HP1gamma* | HP1gamma | CBX3 |
| HSP90 | HSP90 family | |
| MAX | MAX | MAX |
| MDM2 | MDM2 | MDM2 |
| MEL-18* | MEL-18 | PCGF2 |
| MGA | MGA | MGA |
| MNAT1 | MAT1 | MNAT1 |
| NBS1 | NBS1 | NBS1 |
| p107* | p107 | RBL1 |
| p130* | p130 | RBL2 |
| p14ARF* | p14ARF | CDKN2A |
| p15INK4b* | p15INK4b | CDKN2B |
| p16INK4a* | p16INK4a | CDKN2A |
| p18INK4c* | p18INK4c | CDKN2C |
| p19INK4d* | p19INK4d | CDKN2D |
| p21Cip1* | p21Cip1 | CDKN1A |
| p27Kip1* | p27Kip1 | CDKN1B |
| p53* | p53 | TP53 |
| p57Kip2* | p57Kip2 | CDKN1C |
| PCAF | P/CAF | PCAF |
| PCNA | PCNA | PCNA |
| PHC1 | Mph1 | PHC1 |
| PKMYT1 | Myt1 | PKMYT1 |
| PP1 | PP1 | PP1 |
| pRB* | RB, pRB | RB1 |
| RING1 | RING1 | RING1 |
| RYBP | RYBP | RYBP |
| SERTAD1 | SERTAD1 | SERTAD1 |
| SIN3B | SIN3B | SIN3B |
| SUV39H1 | SUV39H1 | SUV39H1 |
| SWI/SNF | SWI/SNF | SWI/SNF |
| TFIIH* | TFIIH | |
| TOPBP1 | TOPBP1 | TOPBP1 |
| WEE1 | WEE1 | WEE1 |
Asterisks are added to the CellDesigner names each time the name used does not correspond to the HUGO name.
A comprehensive map of RB/E2F pathway
Map of the pathway in CellDesigner and BioPAX representations
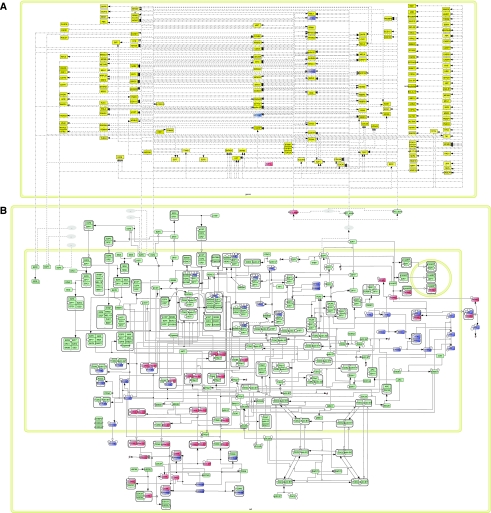
Figure 2 shows the resulting comprehensive map of RB pathway. The diagram utilizes SBGN system (http://www.sbgn.org) to represent proteins and their specific modifications, protein complexes and genes, as well as various protein transformations (binding, unbinding, phosphorylation, acetylation, transport and so on) and their effects of activation or inhibition of chemical reactions, including transcriptional activation or inhibition.
Figure 2.
The textbook pathway of RB has been expanded by integrating data from the literature. The E2F transcription factors (represented here by single proteins in the nuclear compartment) are connected by activation and inhibition arrows to their gene targets. (A) Map of target genes of E2F transcription factors. Each E2F associates with different cofactors to activate or inhibit the transcription of many genes; pointed arrows mean activation and flat arrows mean inhibitions (B) Map of protein–protein interaction network. Each icon on the diagram represents distinct chemical species. See Kitano and co-workers' description of CellDesigner's standard notation (Kitano et al, 2005) for a detailed meaning of shapes. When the information is available (from Atlas Oncology web-page: http://www.atlasgeneticsoncology.org/), tumour suppressor genes and the corresponding proteins are coloured in blue and oncogenes in red, the other proteins are in green. To read and navigate through the map, visit our webpage: http://bioinfo-out.curie.fr/projects/rbpathway/. The map is clickable and allows easy access to all included information (such as literature references or standard protein ids) and hyperlinked to other databases.
In the general organization of the diagram, there is a part representing the set of genes regulated by the E2F family of transcription factors (2A) and another one representing the location of the different proteins in the three different cellular compartments (the nucleus, nucleolus and cytoplasm) (2B).
The resulting map has a total of 78 proteins, which are represented in 208 distinct chemical species (158 of them are located in the nucleus, 47 in the cytoplasm and 3 in the nucleolus), 526 reactions and regulations (among them, 57 protein associations, 13 protein dissociations, 68 post-translational modifications, 361 transcriptional regulations and 27 transport pseudo-reactions), 176 genes, and compiles experimental results from more than 350 publications (301 research papers and 56 reviews).
Once compiled in CellDesigner software, the pathway information was translated into BioPAX format using BiNoM Cytoscape plugin (see Materials and methods). All the files in SBML and BioPAX formats are available in the Supplementary information. An interactive version of the map can also be found on the web site.
We compared our RB/E2F map to existing pathway databases such as Reactome (Joshi-Tope et al, 2005) or Transpath (Krull et al, 2003) and concluded that we provide a more systematic description of RB/E2F network than that contained in the general purpose pathway databases (details at http://bioinfo-out.curie.fr/projects/rbpathway/Comparison.html).
Modular decomposition of the pathway
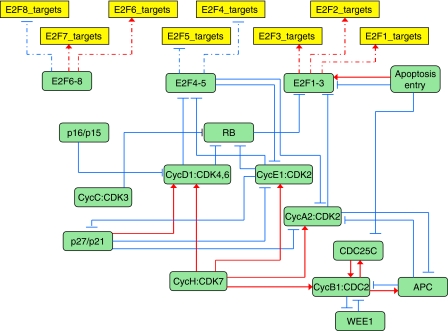
Using clustering of relevant cycles in the reaction graph (see Materials and methods for details) and consequent manual curation, the detailed RB pathway was decomposed into 16 network modules. As a result, almost every module can be thought as a detailed sequence of events that occur with a particular protein or protein complex whose name designates the whole module. Thus, we identified RB, E2F1-3, E2F4-5, E2F6-8, CycC/CDK3, CycH/CDK7, CycD1/CDK4,6, CycE1/CDK2, CycA2/CDK2, CycB1/CDC2, p16INK4a/p15INK4b, p27KIP1/p21CIP1, CDC25C and Wee1, APC and Apoptosis entry modules. Eight additional modules were considered representing the transcriptional targets of each of the E2Fs described in the RB/E2F map.
Identification of these modules allowed us to compile a modular RB pathway view (Figure 3), where network modules are connected by ‘activation' and ‘inhibition' relations. The information about these relations is derived from the detailed diagram. For example, in the detailed map, E2F1 is phosphorylated by CycA2/CDK2 and is subsequently recognized for degradation, which is translated in the modular map by CycA2/CDK2 module inhibiting E2F1-3 module.
Figure 3.
Modular view of the comprehensive map presented in Figure 2. The comprehensive map has been simplified using curated structural analysis techniques to divide the graph in modules. The links between the nodes represent the influence that a module has on the others. There are 16 protein modules (green rounded rectangles) and 8 E2F target gene modules (yellow rectangles).
Because of the space limitation, the module descriptions and figures are provided below only for E2Fs and RB modules (Figure 4). However, the ‘clickable' modular view leading to a description and the corresponding figures of each of the modules can be found in the Supplementary information (http://bioinfo-out.curie.fr/projects/rbpathway/Modules.html).
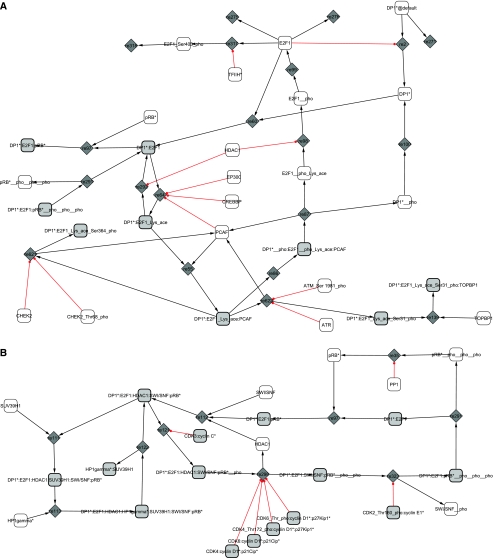
Figure 4.
Cytoscape view of (A) E2F1-3 module and (B) RB module.
The RB module
Two cycles appear in this module revealing two roles of RB. (1) A transcriptional repressor: HDAC1 is a marker of transcriptional repression and seems to require the chromatin remodeler SWI-SNF to form a repressor complex with DP1/E2F1/RB (Frolov and Dyson, 2004). The complex recruits both HP1-γ and SUV39H1 and continues to prevent transcription (Nielsen et al, 2001). (2) A repressor of E2F1/DP1: RB binds to E2F1/DP1 and blocks its transcriptional activity. The affinity between E2F1/DP1 and RB is decreased through sequential phosphorylations by the Cyclin/CDK complexes. The first phosphorylations by CycC/CDK3 favours the passage from G0 to G1 phase, then CycD1/CDK4,6 modifies its conformation and releases HDAC1 (Zhang et al, 2000), revealing a new site of phosphorylation targeted by CycE1/CDK2 (Vidal and Koff, 2000; Muchardt and Yaniv, 2001). The phosphorylation by CycD1/CDK4,6 already allows some genes to be transcribed (such as Cyclin E).
The complexes CycD1/CDK4 and CycD1/CDK6 act as sensors of growth factors. When activated, they precipitate the cells into S phase by further phosphorylating RB (Weinberg, 1995; Planas-Silva and Weinberg, 1997).
When RB is hyperphosphorylated, the complex dissociates and E2F1/DP1 is released from the inactive complex. Later, RB is dephosphorylated by the phosphatase PP1 towards the end of M phase and able to repress E2Fs again (Vidal and Koff, 2000).
The E2F transcription factors modules
The pocket proteins RB, p130 and p107 inhibit a family of transcription factors, the E2F, through association and can also act as active repressors by recruiting other partners.
We already mentioned three subgroups of E2F transcription factors. The first group contains activators of transcription: E2F1, E2F2 and E2F3a, which bind to RB; the second one regroups inhibitors of transcription: E2F3b, E2F4 and E2F5, which bind to either p107 or p130 and to some extent RB; and the third group includes E2F6, E2F7 and E2F8, which do not need to bind to pocket proteins to become active repressors of transcription. These three groups correspond to three different modules that are described below.
Activator E2F1-3 module
The E2F1-3 module includes the E2F activator family of transcription E2F1, E2F2 and E2F3a. Even though the proteins slightly differ in their cell-cycle role, we chose to describe the three E2F transcription factors as one, as they share a lot of functional similarities. In a future version of the pathway, we plan to differentiate the activity of the three transcription activators.
Experiments show that dimerization between E2F1 and its partner DP1 is stable and that E2F1 stimulates nuclear localization of DP1 (Magae et al, 1996). E2F1/DP1 is acetylated by three acetyltransferases—PCAF, CREB binding protein and p300—to stabilize the E2F1 protein (Frolov and Dyson, 2004). The acetylated complex is capable of binding to PCAF to form an active dimer. The complex ability of binding to DNA on the promoter sites of its target genes along with its transcriptional activity is increased during the G1–S transition.
At G2, the complex is phosphorylated by CycA2/CDK2 (He and Cress, 2002). The affinity between E2F1 and DP1 is then diminished leading to the dissociation of the complex (Tsantoulis and Gorgoulis, 2005) and the release of PCAF. The proteins undergo further modifications before degradation: E2F1 is deacetylated by HDAC1 (Martinez-Balbas et al, 2000), dephosphorylated and phosphorylated de novo during S phase by TFIIH kinase for rapid degradation (Ianari et al, 2004).
On DNA damage, the complex PCAF/E2F1/DP1 can be phosphorylated and stabilized either by CHEK1 and CHEK2 through phosphorylation at Ser-364 or by ATM and ATR (Dimova and Dyson, 2005; Powers et al, 2004), preventing E2F1 ubiquitination (Wang et al, 2004). E2F1 mediates the transcription of many genes involved in apoptosis. However, E2F1 transcriptional activity can also be inhibited when bound to the topoisomerase TopBP1 to give time to the cell to repair the damage (Liu et al, 2003).
Repressor E2F4–5 module
E2F4 associates successively with two different pocket proteins: p130 in quiescent cells and p107 in proliferating cells. E2F4 is initially found in the cytoplasm. When in complex, E2F4 is translocated in the nucleus, where it acts as a repressor of transcription during G0 and G1 phases (Verona et al, 1997). Once in the nucleus, the complex binds to DNA and recruits some co-repressors of transcription: the chromatin remodeler Sin3B, the deacetylase histone HDAC1 and the methyltransferase histones SUV39H1 (Rayman et al, 2002; Liu et al, 2005).
At the G0–G1 transition, when the quiescent cells receive signals from growth factors, p130 starts to be phosphorylated and gets dissociated from the complex it was forming with E2F4/DP2. Later, p130 can also be phosphorylated by CycD1/CDK4,6 and CycE1/CDK2 when present and degraded in late G1. It is then replaced by p107 (Farkas et al, 2002). When E2F4/DP2 is in complex with p107, it continues to repress transcription of target genes until it is phosphorylated by CycD1/CDK4,6. E2F4 is then translocated to the cytoplasm, where it can no longer repress transcription, whereas both p130 and p107 are able to inhibit CycE1/CDK2 and CycA2/CDK2 activities (Litovchick et al, 2004).
In this module, E2F3b and E2F5 should also be considered. Their repressive role in the pathway is not described yet but will be detailed in future versions of the pathway.
Repressor E2F6–8 module
E2F6 seems to play a role in S-phase entry. It binds to both DP partners, DP1 and DP2, in the cytoplasm, and the complexes are then translocated in the nucleus. As opposed to other E2F family members, E2F6 does not associate with pocket proteins but rather recruits some proteins from the polycomb group (PcG) to repress transcription.
The first polycomb group with which E2F6 is involved includes BMI1, Mel-18, PHC1, RING1 and RYBP and acts as a repressor of transcription (Trimarchi et al, 2001; Sánchez-Beato et al, 2004). Similarly, E2F6, Max and HP1-γ bind to form a complex that has also revealed a repressive transcription role in quiescent cells (Ogawa et al, 2002). However, E2F6 seems to intervene in other transitions of the cell cycle than only that of G0–G1. Another complex involving EPC1 and EZH2 has been found in proliferating cells: E2F6/DP1 is repressing transcription when in complex with EPC1 alone or with both EPC1 and EZH2 (Attwooll et al, 2005). E2F6/DP1 binds consecutively to EPC1 and then to both EZH2 and EED. The roles of the different complexes are not clearly established yet and more experiments will be needed to confidently describe their specific actions in proliferating and quiescent cells.
E2F7 and E2F8 regulations have not been carefully defined in our pathway yet. Both are known to inhibit transcription although (de Bruin et al, 2003; Logan et al, 2005). The details will be added as more publications on their role in RB pathway appear.
RB pathway transcriptional activity modules
The eight ‘E2F*_targets' modules correspond to the gene targets of the eight E2F transcription factors. The three transcription factors E2F1, E2F2 and E2F3, although not having well-documented differences at the level of protein–protein interactions and represented by a generic entity in our diagram (Figure 2B), show their specificities at the transcription level. For this reason, the generic entity is decomposed into the three individual components in the upper part of the diagram (Figure 2A).
According to our diagram, RB/E2F pathway is a self-regulating molecular mechanism, as there exist multiple positive and negative feedbacks through transcription: among the 78 proteins described in the diagram, 23 are targets of the E2Fs transcription factors, both activators and inhibitors. The structure of these feedbacks is detailed in Supplementary Figure S1 and Supplementary Table S1. E2F1 is a target gene for all E2Fs with the exception of E2F5. In turn, E2F1 regulates expression of E2F1, E2F2, E2F6, E2F7 and E2F8 genes resulting in several negative and positive feedback control circuits.
To analyse the differences between the E2Fs and their roles in other contexts, we calculated the significance of the overlaps of all E2F1-8 transcriptional targets with other known pathways such as MSigDB database (BROAD, MIT) (Subramanian et al, 2005). The results are provided in Supplementary Table S2. As expected, these lists have highly significant overlaps with cell-cycle-, G1–S transition-, Rb/E2F- or p21-related pathways. Some other overlaps highlight the involvement of RB/E2F in some differentiation-related pathways and reciprocally. However, only E2F1 target list is linked to various apoptosis-related pathways (APOPTOSIS_GENMAP, APOPTOSIS_KEGG, DEATH_PATHWAY and others). In particular, E2F1, but not the other E2Fs, targets p53 tumour suppressor gene in RB/E2F pathway. This confirms the recent findings that E2F1 is the only specific inducer of apoptosis among the E2F transcription factors, even though its level can be regulated by other E2Fs (Lazzerini Denchi and Helin, 2005).
Case study of bladder tumour data
As an example of the potentialities of the map we have assembled, we performed a study on 55 bladder tumours (Stransky et al, 2006). This study exemplified how the map and its modular decomposition can be used to explain the differences between two different tumour progression pathways and/or different stages of cancer progression.
More specifically, we analysed transcriptome and comparative genomics hybridization data collected for 55 patients with bladder cancers. We verified which groups of genes (modules) are up- or downregulated in invasive cancers and identified two different paths that both lead to invasive aggressive cancers. This study confirmed known facts about bladder cancer, for example overexpression of CCND1 in low-grade tumours, and led to new observations, for example downregulation of E2F4-5, E2F6-8, Wee1, APC modules in invasive cancers.
The approach and our map can similarly be used for comparing other biological contexts. Indeed, further analyses will be developed to obtain more insights into molecular mechanisms of cancer progression. The details and results of this preliminary study can be found on our webpage: http://bioinfo-out.curie.fr/projects/rbpathway/case_study.html.
Discussion
In this paper, we present a comprehensive representation of the molecular interactions regulating RB activity in cell-cycle-related events. We were able to integrate an important amount of information and represent it in a hierarchical manner, with both a detailed and a summarized and readable representation. This map reflects our understanding of the numerous publications we used to build the pathway. Our study opens perspectives for understanding functioning of RB/E2F pathway and for integrating this information into realistic computational models of mammalian cell cycle.
Pathway databases have rapidly grown during the last years. There exists a number of publicly available or commercial databases: Reactome (Joshi-Tope et al, 2005), KEGG (Kanehisa and Goto, 2000), Transpath (Krull et al, 2006), Ingenuity (www.ingenuity.com), BioCyc (Krummenacker et al, 2005), and so on. They implement different data models, represent molecular interactions at different level of biological details and specialize in different aspects of cellular interactome description. These databases provide an important source of information; however, due to large-scale effort undertaken for their creation, it is difficult to guarantee that the interaction information collected for some specific part of the cellular network is exhaustive. Moreover, biological publications often contain ambiguous statements or even contradictions to other publications, such that for some specific biological pathway, normal level of expertise of a pathway database curator may not be sufficient to resolve these issues in a systematic and self-consistent way.
When a structural model of a specific complex molecular process is created, the pathway databases can be used to build a model draft that needs to be carefully curated by experts specialized in this particular field. The resulting diagram presents a consensus point of view of the experts in which ambiguities and contradictions are resolved according to their opinion. As examples of such focused studies, we can mention reconstruction of human cell-cycle events by Kohn (Kohn, 1999), comprehensive maps of EGFR pathway (Oda et al, 2005) or Toll-like receptor signalling pathway (Oda and Kitano, 2006) and so on.
Our RB pathway reconstruction enlarges this collection. Detailed knowledge of this pathway is necessary for understanding deregulation of mammalian cell cycle in human cancers where the RB network is very frequently affected by genetic and epigenetic alterations. Using automated querying of Reactome database, we confirmed that the reconstruction of RB pathway we present in this paper is more systematic and comprehensive.
Standardization of pathway knowledge representation is of outmost importance in the process of pathway curation (Hucka et al, 2003). In recent years, knowledge representation standards have emerged in Systems Biology and gained momentum in the community (Klipp et al, 2007; Le Novere et al, 2005). SBGN standard partially implemented in CellDesigner software showed good performance in large-scale efforts such as PANTHER database (Mi et al, 2007) and was adopted in this study. It was confirmed that this tool presents a practical compromise between readability of the resulting diagram and the exhaustiveness of the interaction representation. Biologists who are familiar with it can use CellDesigner as an ‘input' device for entering experimental facts in a well-defined framework.
To facilitate the manipulation and analysis of the big pathway diagram, we developed BiNoM software (Zinovyev et al, 2007) which, among many other features, imports information from CellDesigner to Cytoscape (Shannon et al, 2003). More specifically, methods of pathway structure analysis that we implemented in BiNoM allowed to define a modular structure of RB pathway and create its higher-level modular view (see Materials and methods for details). Importantly, this higher-level pathway representation is fully based on the underlying detailed map and helps navigate through it. If necessary, it is always possible to refer to detailed mechanisms of the individual module's functioning. Similar approach to modular pathway modelling is implemented in ProMoT system (Saez-Rodriguez et al, 2006).
The most up-to-date and comprehensive description of the pathway, CellDesigner and BioPAX files are available on the web-page with Supplementary information. The interactive online version of the pathway diagram is accessible at: http://bioinfo-out.curie.fr/projects/rbpathway/complete_network.htm. Such functionalities as zooming or centering the screen on a molecule, complex or reaction, isolating parts of the diagram (modules) in both Cytoscape and CellDesigner views facilitate the navigation through the network and the extraction of specific information. The CellDesigner map file was used to automatically generate a navigable web site, allowing to access the pathway information details, including literature references and connection to some other databases, in few clicks. Using BiNoM software, such interactive online representations of the pathway diagrams can be automatically created for other large CellDesigner diagrams.
Further directions we preview for this study include (1) creating a computational qualitative model of the regulation of mammalian cell cycle by RB; (2) superimposing this model with available data on genetic/epigenetic alteration status of key proteins in tumour samples of certain cancers (such as breast or bladder cancers); and (3) providing permanent update of the information collected in the pathway.
Materials and methods
The pathway is available in SBML format from the BioModels database (http://www.ebi.ac.uk/biomodels) with the accession number MODEL4132046015.
BiNoM
CellDesigner 3.5 version (Funahashi et al, 2003) was used to enter biological facts from a carefully studied selection of papers (see the whole bibliography on the web site with Supplementary information). Whenever the details of a biological fact could not be naturally expressed with CellDesigner standard notations, it was fixed and some solution was proposed. For example, we added a notation (co-factor) to describe all the components intervening in the transcription of genes mediated by the E2F family proteins. To perform reaction network structural analysis, we also developed BiNoM Cytoscape plugin (Zinovyev et al, 2007) available at http://bioinfo-out.curie.fr/projects/binom/.
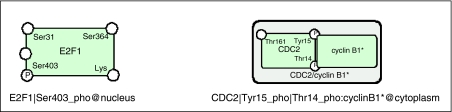
When importing CellDesigner information into Cytoscape, the graphical notation used to describe composition of complexes or modification status in CellDesigner was substituted by textual description, such that the label of a chemical species would be sufficient to identify the species in a unique way. The general template of the species label was the following:
Here, ‘:' delimitates components of a complex if the species has several components. Optional suffixes ‘active' or ‘hm' describe active state of the chemical species or N-homodimer state. Several examples of such conversion are presented in Figure 5.
Figure 5.
Examples of textual representation of species structure. In CellDesigner, information about sites of phosphorylation or acetylation, and so on can be showed on the protein itself. A complex is represented as a black box surrounding the proteins composing the complex.
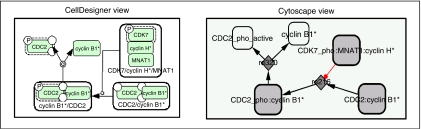
The reaction graph of CellDesigner is represented as a bipartite reaction graph in Cytoscape. Two types of vertices are used in this graph: reaction vertices and species vertices. An example of a little network is provided in Figure 6.
Figure 6.
Example of a simple network converted from CellDesigner to Cytoscape. In Cytoscape, single proteins are represented by white squares and complexes by grey squares.
Methods for extracting modules in BiNoM
A step-by-step method for creating the modular view of the RB/E2F pathway is provided on the webpage. The directed bipartite graph, representing the RB reaction network in Cytoscape was analysed using the following steps:
All strongly connected components were extracted using the standard Tarjan's algorithm, implemented in BiNoM, and used in further analysis. A strongly connected component is a subgraph, in which there exists a directed path from any graph vertex to any vertex.
Every strongly connected component was decomposed into relevant cycles, using modification of Vismara's algorithm (Vismara, 1997), implemented in BiNoM. A relevant cycle is a cycle that can not be further decomposed into smaller cycles (Gleiss et al, 2001). A set of relevant cycles is by definition the union of all minimum cyclic bases. Therefore, it is a minimum unique cyclic graph decomposition (Vismara, 1997). A minimal cycle basis of a graph is a set of all independent cycles with minimum summary length. In general, this set is not unique.
For the collection of cycles obtained at the previous step, a simple agglomerative clustering algorithm with asymmetric similarity (proportion of common nodes) was applied: if a subgraph is included in another subgraph in P>50% of its vertices, it will be merged with that subgraph. If a subgraph could be merged with several other subgraphs, it would be merged with the one with which it had the biggest intersection. If the intersection is the same, the subgraph will be merged with the biggest subgraph. This process was repeated until there was no possibility to merge subgraphs.
For every cycle union, the ‘majority rule' (Ma et al, 2004) was applied to classify all reactions from noncyclic part of the network: all incoming and outgoing linear and branching pathways were included in the cluster of cycles to which most of them were initially attached. This gave the initial definition of modules.
Step 4 was repeated with some range of values of P (from 30 to 70%) to ensure that it gives a robust decomposition of the network. The clustering in this range of the parameter always gave the same number of modules.
All resulting modules were carefully studied and named according to the name of the protein (complex) with major participation. In the process of manual curation, some important reactions and regulations were re-assigned, and some modules were split.
For extracting modules in RB reaction graph, we defined a cycle in the reaction graph as an elementary functional unit. Tightly coupled cycles form the module core. There are two types of cycles in the RB/E2F network: (1) cycles of mass flow and (2) cycles of information (perturbation) flow. In the first case, a cycle of reactions corresponds to an elementary mode in Stoichiometric Network Analysis (Schilling et al, 2000), but applied to the pseudo-monomolecular approximation of the real reaction network, when the reaction A+B → C is considered as A → C and B → C with kinetic constants dependent on the other reaction participant concentrations. This approximation is valid when, for example concentration of A is much higher than that of the B. Then, A in the reaction A+B → C is considered as a relatively slowly changing environment.
As it is argued in Gorban and Radulescu (2007) (http://arxiv.org/abs/physics/0703278v2), in a complex network dynamics, at a given moment of time, it might be that only a small subset of reactions functions in a truly nonlinear mode (however, this subset changes with time), and the others can be approximated as pseudo-monomolecular. This way, the combinatorial problem of finding true elementary modes is simplified. Similar decoupling is applied by Klamt et al (2006) for studying both signal and mass flows in the logical framework.
Numerous approaches for module extraction in reaction networks have been developed during the past years (Ravasz et al, 2002; Stelling et al, 2002). An approach based on T-invariants in Petri Nets (a notion similar to elementary mode) was used by Sackmann et al (2006) to define modules in metabolic networks. Agglomerative (Ma et al, 2004) and divisive (Holme et al, 2003) hierarchical clusterings of reactions of a metabolic network were also proposed based on the analysis of the shortest paths. Our method is different, as it uses clustering subgraphs (relevant cycles) rather than individual reactions.
Linking RB/E2F pathway to other signalling pathways
To estimate the significance of the overlap among E2F's target lists and MSigDB database, we applied the following classical hypergeometric test to calculate the P-value:
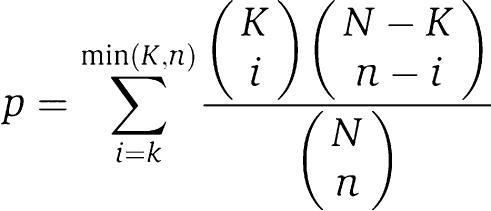 |
where N is the total number of different genes in MSigDB signatures, K is the number of genes in a pathway from MSigDB, n is the number of genes in one of the lists of E2Fx transcriptional targets. Having in mind multiple hypotheses testing, we considered only the overlaps with P⩽10−6 as significant.
Supplementary Material
Supplementary Information
Supplementary Materials
Acknowledgments
This project was partly funded by the EC contract ESBIC-D (LSHG-CT-2005-518192), the PIC Rétinoblastome from Institut Curie, the PIC Bioinformatique et Biostatistiques from Institut Curie and the Research Networks Program in Bioinformatics from the High Council for Scientific and Technological Cooperation between France and Israël (Ministère des Affaires Etrangères, Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche). LC, AG, AZ and EB are members of the team “Systems Biology of Cancer,” Equipe labellisée par la Ligue Nationale Contre le Cancer. FR and AG are members of the team “Oncologie Moléculaire,” Equipe labellisée par la Ligue Nationale Contre le Cancer. We thank Nicolas Stransky, Aurélie Hérault and Yves Allory for providing bladder tumour data and Simon Saule for his valuable suggestions for the RB pathway map. We also thank the anonymous reviewers for their useful and stimulating comments.
References
- Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J (eds). (1994) Molecular Biology of the Cell. Garland Publishing, New York, USA [Google Scholar]
- Attwooll C, Oddi S, Cartwright P, Prosperini E, Agger K, Steensgaard P, Wagener C, Sardet C, Moroni MC, Helin K (2005) A novel repressive E2F6 complex containing the polycomb group protein, EPC1, that interacts with EZH2 in a proliferation-specific manner. J Biol Chem 280: 1199–1208 [DOI] [PubMed] [Google Scholar]
- de Bruin A, Maiti B, Jakoi L, Timmers C, Buerki R, Leone G (2003) identification and characterization of E2F7, a novel mammalian E2F family member capable of blocking cellular proliferation. J Biol Chem 278: 42041–42049 [DOI] [PubMed] [Google Scholar]
- Classon M, Harlow E (2002) The retinoblastoma tumour suppressor in development and cancer. Nat Rev Cancer 2: 910–917 [DOI] [PubMed] [Google Scholar]
- DeGregori J (2004) The Rb network. J Cell Sci 117: 3411–3413 [DOI] [PubMed] [Google Scholar]
- Dimova DK, Dyson NJ (2005) The E2F transcriptional network: old acquaintances with new faces. Oncogene 24: 2810–2826 [DOI] [PubMed] [Google Scholar]
- Farkas T, Hansen K, Holm K, Lukas J, Bartek J (2002) Distinct phosphorylation events regulate p130- and p107-mediated repression of E2F-4. J Biol Chem 277: 26741–26752 [DOI] [PubMed] [Google Scholar]
- Frolov MV, Dyson NJ (2004) Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J Cell Sci 117: 2173–2181 [DOI] [PubMed] [Google Scholar]
- Funahashi A, Tanimura N, Morohashi M, Kitano H (2003) CellDesigner: a process diagram editor for gene-regulatory and biochemical networks. BIOSILICO 1: 159–162 [Google Scholar]
- Giacinti C, Giordano A (2006) RB and cell cycle progression. Oncogene 25: 5220–5227 [DOI] [PubMed] [Google Scholar]
- Gleiss P, Stadler P, Wagner A, Fell DA (2001) Relevant cycles in chemical reaction networks. Adv Complex Systems 4: 207–226 [Google Scholar]
- He Y, Cress WD (2002) E2F-3B is a physiological target of cyclin A. J Biol Chem 277: 23493–23499 [DOI] [PubMed] [Google Scholar]
- Holme P, Huss M, Jeong H (2003) Subnetwork hierarchies of biochemical pathways. Bioinformatics 19: 532–538 [DOI] [PubMed] [Google Scholar]
- Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, the rest of the SF, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, Cuellar AA, Dronov S, Gilles ED, Ginkel M, Gor V, Goryanin II, Hedley WJ, Hodgman TC, Hofmeyr JH, , et al. , SBML Forum (2003) The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 19: 524–531 [DOI] [PubMed] [Google Scholar]
- Hussussian CJ, Struewing JP, Goldstein AM, Higgins PAT, Ally DS, Sheahan MD, Clark WH, Tucker MA, Dracopoli NC (1994) Germline p16 mutations in familial melanoma. Nat Genet 8: 15–21 [DOI] [PubMed] [Google Scholar]
- Ianari A, Gallo R, Palma M, Alesse E, Gulino A (2004) Specific role for p300/CREB-binding protein-associated factor activity in E2F1 stabilization in response to DNA damage. J Biol Chem 279: 30830–30835 [DOI] [PubMed] [Google Scholar]
- Joshi-Tope G, Gillespie M, Vastrik I, D'Eustachio P, Schmidt E, de Bono B, Jassal B, Gopinath GR, Wu GR, Matthews L, Lewis S, Birney E, Stein L (2005) Reactome: a knowledgebase of biological pathways. Nucl Acids Res 33: D428–D432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucl Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H, Funahashi A, Matsuoka Y, Oda K (2005) Using process diagrams for the graphical representation of biological networks. Nat Biotech 23: 961–966 [DOI] [PubMed] [Google Scholar]
- Klamt S, Saez-Rodriguez J, Lindquist JA, Simeoni L, Gilles ED (2006) A methodology for the structural and functional analysis of signaling and regulatory networks. BMC Bioinformatics 7: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klipp E, Liebermeister W, Helbig A, Kowald A, Schaber J (2007) Systems biology standards—the community speaks. Nat Biotech 25: 390–391 [DOI] [PubMed] [Google Scholar]
- Knudsen ES, Wang JY (1997) Dual mechanisms for the inhibition of E2F binding to RB by cyclin-dependent kinase-mediated RB phosphorylation. Mol Cell Biol 17: 5771–5783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson AJ (1971) Mutation and cancer: statistical study of retinoblastoma. PNAS 68: 820–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn KW (1999) Molecular interaction map of the mammalian cell cycle control and DNA repair systems. Mol Biol Cell 10: 2703–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull M, Pistor S, Voss N, Kel A, Reuter I, Kronenberg D, Michael H, Schwarzer K, Potapov A, Choi C, Kel-Margoulis O, Wingender E (2006) TRANSPATH: an information resource for storing and visualizing signaling pathways and their pathological aberrations. Nucleic Acids Res 34: D546–D551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull M, Voss N, Choi C, Pistor S, Potapov A, Wingender E (2003) TRANSPATH: an integrated database on signal transduction and a tool for array analysis. Nucleic Acids Res 31: 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krummenacker M, Paley S, Mueller L, Yan T, Karp PD (2005) Querying and computing with BioCyc databases. Bioinformatics 21: 3454–3455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzerini Denchi E, Helin K (2005) E2F1 is crucial for E2F-dependent apoptosis. EMBO Rep 6: 661–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Novere N, Finney A, Hucka M, Bhalla US, Campagne F, Collado-Vides J, Crampin EJ, Halstead M, Klipp E, Mendes P, Nielsen P, Sauro H, Shapiro B, Snoep JL, Spence HD, Wanner BL (2005) Minimum information requested in the annotation of biochemical models (MIRIAM). Nat Biotech 23: 1509–1515 [DOI] [PubMed] [Google Scholar]
- Litovchick L, Chestukhin A, DeCaprio JA (2004) Glycogen synthase kinase 3 phosphorylates RBL2/p130 during quiescence. Mol Cell Biol 24: 8970–8980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DX, Nath N, Chellappan SP, Greene LA (2005) Regulation of neuron survival and death by p130 and associated chromatin modifiers. Genes Dev 19: 719–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Lin F-T, Ruppert JM, Lin W-C (2003) Regulation of E2F1 by BRCT domain-containing protein TopBP1. Mol Cell Biol 23: 3287–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan N, Graham A, Zhao X, Fisher R, Maiti B, Leone G, Thangue NBL (2005) E2F-8: an E2F family member with a similar organization of DNA-binding domains to E2F-7. Oncogene 24: 5000–5004 [DOI] [PubMed] [Google Scholar]
- Ma HW, Zhao XM, Yuan YJ, Zeng AP (2004) Decomposition of metabolic network into functional modules based on the global connectivity structure of reaction graph. Bioinformatics 20: 1870–1876 [DOI] [PubMed] [Google Scholar]
- Magae J, Wu CL, Illenye S, Harlow E, Heintz NH (1996) Nuclear localization of DP and E2F transcription factors by heterodimeric partners and retinoblastoma protein family members. J Cell Sci 109: 1717–1726 [DOI] [PubMed] [Google Scholar]
- Martinez-Balbas MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T (2000) Regulation of E2F1 activity by acetylation. EMBO J 19: 662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Guo N, Kejariwal A, Thomas PD (2007) PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res 35: D247–D252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris E, Dyson N (2001) Retinoblastoma protein partners. Adv Cancer Res 82: 1–54 [DOI] [PubMed] [Google Scholar]
- Muchardt C, Yaniv M (2001) When the SWI/SNF complex remodels the cell cycle. Oncogene 20: 3067–3075 [DOI] [PubMed] [Google Scholar]
- Muller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner JD, Helin K (2001) E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev 15: 267–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger K, Basile JR, Duensing S, Eichten A, Gonzalez SL, Grace M, Zacny VL (2001) Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene 20: 7888–7898 [DOI] [PubMed] [Google Scholar]
- Nevins JR (2001) The Rb/E2F pathway and cancer. Hum Mol Genet 10: 699–703 [DOI] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer U-M, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, Kouzarides T (2001) Rb targets histone H3 methylation and HP1 to promoters. Nature 412: 561–565 [DOI] [PubMed] [Google Scholar]
- Novak B, Tyson JJ, Gyorffy B, Csikasz-Nagy A (2007) Irreversible cell-cycle transitions are due to systems-level feedback. Nat Cell Biol 9: 724–728 [DOI] [PubMed] [Google Scholar]
- Oda K, Kitano H (2006) A comprehensive map of the toll-like receptor signaling network. Mol Syst Biol 2: 2006.0015 10.1038/msb4100057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K, Matsuoka Y, Funahashi A, Kitano H (2005) A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Biol 1: 2005.0010. 10.1038/msb4100014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y (2002) A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296: 1132–1136 [DOI] [PubMed] [Google Scholar]
- Planas-Silva MD, Weinberg RA (1997) The restriction point and control of cell proliferation. Curr Opin Cell Biol 9: 768–772 [DOI] [PubMed] [Google Scholar]
- Powers JT, Hong S, Mayhew CN, Rogers PM, Knudsen ES, Johnson DG (2004) E2F1 uses the ATM signaling pathway to induce p53 and Chk2 phosphorylation and apoptosis. Mol Cancer Res 2: 203–214 [PubMed] [Google Scholar]
- Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi AL (2002) Hierarchical organization of modularity in metabolic networks. Science 297: 1551–1555 [DOI] [PubMed] [Google Scholar]
- Rayman JB, Takahashi Y, Indjeian VB, Dannenberg J-H, Catchpole S, Watson RJ, te Riele H, Dynlacht BD (2002) E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev 16: 933–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackmann A, Heiner M, Koch I (2006) Application of Petri net based analysis techniques to signal transduction pathways. BMC Bioinformatics 7: 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Rodriguez J, Mirschel S, Hemenway R, Klamt S, Gilles ED, Ginkel M (2006) Visual setup of logical models of signaling and regulatory networks with ProMoT. BMC Bioinformatics 7: 506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Beato M, Sánchez E, García JF, Pérez-Rosado A, Montoya MC, Fraga M, Artiga MJ, Navarrete M, Abraira V, Morente M, Esteller M, Koseki H, Vidal M, Piris MA (2004) Abnormal PcG protein expression in Hodgkin's lymphoma. Relation with E2F6 and NF? B transcription factors. J Pathol 204: 528–537 [DOI] [PubMed] [Google Scholar]
- Schilling CH, Letscher D, Palsson BO (2000) Theory for the systemic definition of metabolic pathways and their use in interpreting metabolic function from a pathway-oriented perspective. J Theor Biol 203: 229–248 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ, McCormick F (2002) The RB and p53 pathways in cancer. Cancer Cell 2: 103–112 [DOI] [PubMed] [Google Scholar]
- Stelling J, Klamt S, Bettenbrock K, Schuster S, Gilles ED (2002) Metabolic network structure determines key aspects of functionality and regulation. Nature 420: 190–193 [DOI] [PubMed] [Google Scholar]
- Stevaux O, Dyson NJ (2002) A revised picture of the E2F transcriptional network and RB function. Curr Opin Cell Biol 14: 684–691 [DOI] [PubMed] [Google Scholar]
- Stransky N, Vallot C, Reyal F, Bernard-Pierrot I, de Medina SG, Segraves R, de Rycke Y, Elvin P, Cassidy A, Spraggon C, Graham A, Southgate J, Asselain B, Allory Y, Abbou CC, Albertson DG, Thiery JP, Chopin DK, Pinkel D, Radvanyi F (2006) Regional copy number-independent deregulation of transcription in cancer. Nat Genet 38: 1386–1396 [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Fairchild B, Wen J, Lees JA (2001) The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc Natl Acad Sci USA 98: 1519–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsantoulis PK, Gorgoulis VG (2005) Involvement of E2F transcription factor family in cancer. Eur J Cancer 41: 2403–2414 [DOI] [PubMed] [Google Scholar]
- Verona R, Moberg K, Estes S, Starz M, Vernon JP, Lees JA (1997) E2F activity is regulated by cell cycle-dependent changes in subcellular localization. Mol Cell Biol 17: 7268–7282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal A, Koff A (2000) Cell-cycle inhibitors: three families united by a common cause. Gene 247: 1–15 [DOI] [PubMed] [Google Scholar]
- Vismara P (1997) Union of all the minimum cycle bases of a graph. Electr J Comb 4: 73–87 [Google Scholar]
- Wang B, Liu K, Lin F-T, Lin W-C (2004) A Role for 14-3-3{tau} in E2F1 stabilization a. J Biol Chem 279: 54140–54152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RA (1995) The retinoblastoma protein and cell cycle control. Cell 81: 323–330 [DOI] [PubMed] [Google Scholar]
- Zhang HS, Gavin M, Dahiya A, Postigo AA, Ma D, Luo RX, Harbour JW, Dean DC (2000) Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 101: 79–89 [DOI] [PubMed] [Google Scholar]
- Zinovyev A, Viara E, Calzone L, Barillot E (2007) BiNoM: a Cytoscape plugin for manipulating and analyzing biological networks. Bioinformatics; advance online publication 16 November 2007; doi:10.1093/bioinformatics/btm553 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Materials