Abstract
The biological community is now awash in high-throughput data sets and is grappling with the challenge of integrating disparate data sets. Such integration has taken the form of statistical analysis of large data sets, or through the bottom–up reconstruction of reaction networks. While progress has been made with statistical and structural methods, large-scale systems have remained refractory to dynamic model building by traditional approaches. The availability of annotated genomes enabled the reconstruction of genome-scale networks, and now the availability of high-throughput metabolomic and fluxomic data along with thermodynamic information opens the possibility to build genome-scale kinetic models. We describe here a framework for building and analyzing such models. The mathematical analysis challenges are reflected in four foundational properties, (i) the decomposition of the Jacobian matrix into chemical, kinetic and thermodynamic information, (ii) the structural similarity between the stoichiometric matrix and the transpose of the gradient matrix, (iii) the duality transformations enabling either fluxes or concentrations to serve as the independent variables and (iv) the timescale hierarchy in biological networks. Recognition and appreciation of these properties highlight notable and challenging new in silico analysis issues.
Keywords: biochemical network, duality, gradient, hierachical analysis, thermodynamics
Introduction
In the past decade, we have witnessed significant advances in the development of statistical analysis of genome-scale networks (Slonim, 2002), which has been propelled by the availability of genome-scale high-throughput data sets and the successes of constraint-based modeling approaches (Pharkya et al, 2004; Price et al, 2004; Kummel et al, 2006; Palsson, 2006; Reed et al, 2006). The foundation of such genome-scale analysis is built on the stoichiometric matrix, S, which describes all the biochemical transformations in a network in a self-consistent and chemically accurate matrix format. Much progress has been made with the genome-scale network reconstruction process and a growing number of genome-scale metabolic reconstructions are now available (Reed et al, 2006; Feist et al, 2007; Jamshidi and Palsson, 2007; Oh et al, 2007; Resendis-Antonio et al, 2007).
Reconstructions of genome-scale biochemical reaction networks (Reed et al, 2006) have been analyzed by topological- (Barabasi and Oltvai, 2004) and constraints-based (Price et al, 2004) methods, but dynamic models, at this scale, still need development. It turns out that S is not only a requisite for dynamic models but also a major determinant in their properties and thus it is important to have well-curated reconstructions available. The growing availability of metabolomic and fluxomic data sets (Goodacre et al, 2004; Hollywood et al, 2006; Sauer, 2006; Breitling et al, 2008) and methods to estimate the thermodynamic properties (Mavrovouniotis, 1991; Henry et al, 2006, 2007) of biochemical reactions has opened up the possibility to formulate large-scale kinetic models.
The structure of the workflow that leads to large-scale dynamic models is emerging and so are the associated data and mathematical challenges. Here, we will propose a framework for the data integration and mathematical challenges that come with the construction of genome-scale kinetic models. We proceed in four steps. First, we briefly describe the governing equations for dynamic analysis of genome-scale networks. Second, we will outline a proposed workflow for the formulation of large-scale kinetic models. Third, we describe the matrix format of the basic data and the information that it entails. Fourth, we outline the four newly identified key mathematical properties of the resulting models. These four properties are fundamental and the modeling community is now faced with the challenge of studying them in detail. We illustrate some of these properties with examples.
The basic equations used for dynamic analysis
The dynamic mass balances
The reconstruction of a genome-scale reaction network requires the identification of all its chemical components and the chemical transformations that they participate in. This process primarily relies on annotated genomes and detailed bibliomic assessment (Reed et al, 2006). The result of this process is the stoichiometric matrix, S, that is used in the dynamic mass balances
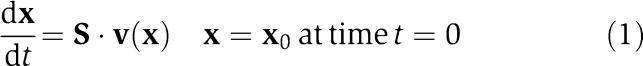 |
that are the basis of all kinetic models. Here d(.)/dt denotes the time derivative, x is the vector of the concentrations of the compounds in the network and v(x) is the vector of the reaction rates. All biochemical transformations are fundamentally uni- or bi-molecular. Such reactions can be represented by mass action kinetics, or generalizations thereof (Segel, 1975). The net reaction rate for every elementary reaction in a network can be represented by the difference between a forward and reverse flux (e.g. see Figure 1).
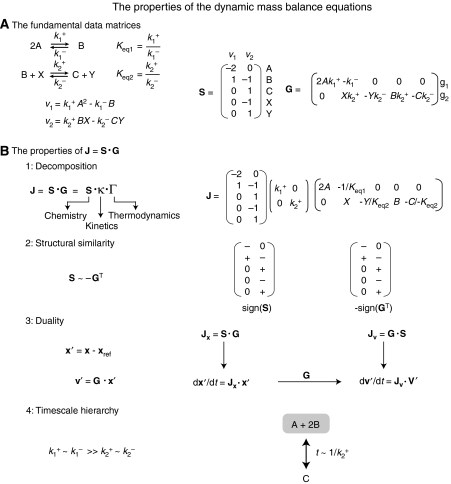
Figure 1.
(A) The fundamental matrices describing the dynamic states of biological networks: the stoichiometric matrix S and the gradient matrix G. The corresponding stoichiometric and gradient matrices (for mass action kinetics) are shown. (B) The decomposition of the Jacobian matrix into the stoichiometric and gradient matrices looks similar to other mathematical factorizations; however, this approach is biologically meaningful and driven by the underlying chemistry, kinetics and thermodynamics. There are different ways to factor G into Γ and the diagonal κ matrix. In this example, the forward rate constant is factored out of G. The structural similarity between S and GT is highlighted by the fact that their row-reduced forms are equivalent and illustrated for a simple example involving two reactions. A duality exists between the concentrations and fluxes. The practical significance of this duality is that in the linear regimen, the relationship between fluxes and concentrations can be determined independently of the specific rate law formulation, if one can approximate the gradient matrix. When certain reactions occur much faster than others, the related metabolites pool together into aggregate variables. For the example in (A), when the forward and reverse rate constants of the first reaction (2A↔B; dimerization reaction) are much faster than the rate constants for the second reactions (B+X↔C+Y; cofactor exchange reaction), then the substrate and products of the first reaction form an aggregate pool. Since B is a ‘dimer' of A, the pool will consist of A and B in a 1:2 ratio. In this example, the ratios between the metabolites can also be found in the left null space. Furthermore, since the first reaction occurs much more quickly than the second, the interaction between the A+2B pool and the C (X and Y are considered cofactors in this example) is determined by the rate constant(s) for the second reaction.
This commonly used formulation is based on several well-known assumptions, such as constant temperature, volume and homogeneity of the medium. If S, v(x) and the initial conditions (x0) are known, then these ordinary differential equations can be numerically solved for a set of conditions of interest.
Linear form
The characterization of the dynamic states of networks can be studied through numerical simulation or through using mathematical analysis. A simulation is context dependent and represents a case study. Mathematical methods for the analysis of model characteristics typically rely on studying the properties of the transformation between the concentrations and fluxes. The analysis of such fundamental properties normally relies on the linearization of the governing equations at a defined condition. The linearization of the dynamic mass balance equations comes down to the linearization of the reaction rate vector to form the gradient matrix
 |
and then forming the Jacobian matrix at a reference state xref:
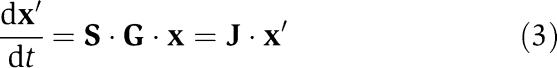 |
where x′=x−xref and J is the well-known Jacobian matrix. Analysis of the characteristics of Jacobian matrix is standard procedure in mathematical analysis of system dynamics (e.g. Strogatz, 1994). The application of these methods to biochemical networks has been carried out for decades (Heinrich et al, 1977, 1991; Heinrich and Sonntag, 1982; Palsson et al, 1987) and in recent years there has been renewed interest and recently further developments in the dynamic analysis of the properties of J have appeared (Teusink et al, 2000; Kauffman et al, 2002; Famili et al, 2005; Bruggeman et al, 2006; Steuer et al, 2006; Grimbs et al, 2007).
The Jacobian matrix for biochemical reaction networks is the product of two data matrices. Prior to looking at the fundamental properties of J, we consider the workflows and data properties that relate to S and G.
The workflow associated with constructing large-scale dynamic models
The equations used to describe the dynamic states of networks and outlined above are fairly well known, with the exception of the explicit representation of the gradient matrix. This factorization of the Jacobian matrix turns out to be important in formulating the workflows and in the analysis of the properties of these basic equations.
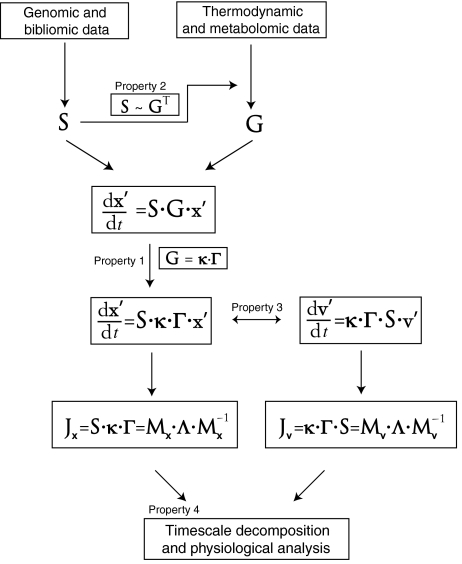
The integration of genomic, bibliomic, fluxomic and metabolomic data with thermodynamic information into dynamic models of metabolism is illustrated in Figure 2. The process of reconstructing S is now well developed (Palsson, 2006). The formulation of G can now be performed based on metabolomic data and methods to estimate thermodynamic properties. As discussed below the chemical equations that make up S determine the location of the non-zero elements in G and hence its structure.
Figure 2.
The workflow for constructing approximations to genome-scale kinetic networks and how the four properties discussed in this article and highlighted in Figure 1 enable such reconstructions.
This workflow brings up two important issues. First, the representation and the properties of the data that the two key matrices are made up of. Second, the mathematical challenges associated with analyzing the resulting equations. The matrices represent data, and the equations represent physical laws. Thus, these two issues basically represent data analysis challenges under the constraints of the governing physical laws. We now discuss both of these issues.
Key data and fundamental scientific considerations are found in a matrix format
Factoring J into the S and G matrices is not simply a mathematical exercise, but represents a decomposition of J into two fundamental factors each with its own relevance (Table I). Comparing the properties of S and G further highlights the contributions of each matrix to characteristics of biological networks. For example, the formation of pools and reaction co-sets is determined by S (Heinrich and Schuster, 1996; Papin et al, 2004; Jamshidi and Palsson, 2006), whereas timescale separation is in the realm of G. This decomposition factors the various underlying data needed for the formulation of genome-scale kinetic models. Furthermore, it illustrates the various underlying disciplinary interests that need to be considered and integrated to successfully achieve the analysis of dynamic states of genome-scale networks.
Table 1.
A comparison of the properties of the stoichiometric and gradient matrices
| Properties | Stoichiometric matrix | Gradient matrix |
|---|---|---|
| Biological | Species differences Distal causation | Individual differences Proximal causation |
| Genetic | Genomic characteristics represents a species | Genetic characteristics represents an individual |
| Informatic | Annotated genome Bibliomic Comparative genomics | Kinetic data Metabolomics Fluxomics |
| Physico-chemical | Chemistry Conservation laws | Kinetics Thermodynamics |
| Mathematical | Integer entries Knowable matrix | Real numbers Entries have errors |
| Numerical | Sparse Well conditioned Non-stiff | Sparse Ill conditioned Leads to stiffness |
| Systemic | Pool formation Network structure | Timescale separation Dynamic function |
Bioinformatic considerations
S is primarily derived from an annotated genomic sequence fortified with any direct bibliomic data on the organisms' gene products. The construction of G will rely on fluxomic and metabolmic data, in addition to direct kinetic characterization of individual reactions and assessment of thermodynamic properties.
Physico-chemical considerations
S represents chemistry (i.e. stoichiometry of reactions), while G represents kinetics and thermodynamics. The chemical information is relatively easy to get, the thermodynamics information harder but possible (Henry et al, 2007), and the kinetic information is the hardest to acquire. The former two represent hard physico-chemical constraints, while the third represents biologically manipulable numbers; i.e. reaction rates are accelerated by enzymes.
Biological and genetic considerations
The matrix S is reconstructed based on the content of a genome and is a property of a species. It has thus been used productively for the analysis of distal causation (Ibarra et al, 2002; Fong and Palsson, 2004; Pal et al, 2006; Harrison et al, 2007). Distal (or ultimate) causation results from (genomic) changes that occur from generation to generation, in contrast to proximal (or proximate) causation that occurs against a fixed genetic background (i.e. an individual) (Mayr, 1961). G is genetically derived and can represent the variations among individuals in a biopopulation. It is important in studying proximal causation and the differences in phenotypes of individuals in a biopopulation. For example, many disease states in higher order organisms result from disruptions or deficiencies in enzyme kinetics (Jamshidi et al, 2002). These changes are reflected in G since this contains the information about kinetics, consequently the analysis of disease states, inter-individual differences and transitions from a healthy to disease state in a particular individual will in general focus on G.
Mathematical and numerical considerations
Whereas S is a ‘perfect' matrix comprised of integers (i.e. digital), G is an analog matrix whose entries are real numbers and we may only know within an order of magnitude. From a numerical and mathematical standpoint, S is a well-conditioned matrix comprised of integers (−2, −1, 0, 1, 2), whereas G is an ill-conditioned matrix of real numbers that can differ up to 10 orders of magnitude in their numerical values. This property leads to timescale separation. Both matrices are sparse, that is, most of their elements are zero.
The four fundamental properties
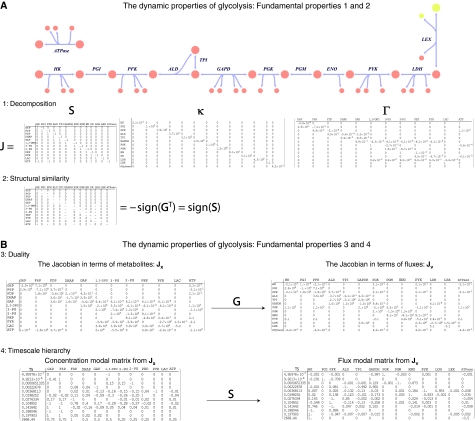
Study of the result from the linearization of the dynamic mass balance equations yields four properties that are of fundamental importance (Figure 1). These properties are illustrated using the familiar glycolytic pathway (Figure 3A).
Figure 3.
(A) An example of the first two properties for the glycolytic pathway. A reaction map of the glycolytic pathway is shown. The decomposition of the Jacobian (Jx) into the stoichiometric, κ, and Γ matrices follows below (1−norm used for the factorization of Γ). The negative transpose of the gradient matrix is shown directly below the stoichiometric matrix, demonstrating the structural similarity. (B) Explicit illustration of the third and fourth properties via the resulting data matrices. The Jacobian duals are shown; they are related by the gradient matrix. Hierarchical analysis can be carried out of the network in terms of metabolites or fluxes. The resultant modal matrices can be related to one another via the stoichiometric matrix. As illustrated in Figure 4, sometimes it is convenient to think about the hierarchical structure in terms of metabolites and sometimes it is more intuitive to think in terms of the fluxes. The network was constructed using equilibrium constants and concentrations from a kinetic red cell model, which has been validated in the literature. Network dynamics were then described using mass action kinetics for a particular steady state. Abbreviations: ALD, fructose-bisphosphate aldolase; ATPase, ATP hydrolysis (demand/utilization); ENO, enolase; GAPD, glyceraldehyde phosphate dehydrogenase; HK, hexokinase; LDH, lactate dehydrogenase; LEX, lactate transporter; PFK, phosphofructokinase; PGI, phosphoglucoisomerase; PGK, phosphoglucokinase; PGM, phosphoglucomutase; PYK, pyruvate kinase; TPI, triose-phosphate isomerase; TS, timescales in hours.
Property 1. Fundamental structure of the Jacobian
The wide differences in the numerical values of the entries of G lead to its factorization (G=κ·Γ) by scaling it by the length of its rows to yield a factorization of J into three matrices:
where κ is a diagonal matrix of the norms of the rows in G (Figure 1B1). We emphasize that ith column of S contains the stoichiometric coefficients of the ith reaction in the network and the ith row in G contains the forward and reverse rate constants of that same reaction. Thus, the reciprocal of the diagonal entries, 1/κii, corresponds to characteristic time constants of the corresponding reactions. Their numerical values will differ significantly.
The factorization of the Jacobian in equation (4) shows that the study of the dynamic properties of biochemical networks can be formally decomposed into chemistry (represented by S), kinetics (represented by κ) and thermodynamic driving forces (represented by Γ). The effects of each can thus be formally determined. Chemistry and thermodynamics are physico-chemical properties, whereas the kinetic constants are biologically set through a natural selection process. The particular numerical values (chosen through the selection process) lead to the formation of biologically meaningful dynamic properties of the network. These biological features of the network can be assessed through timescale decomposition, see property 4 below.
Glycolysis
We use the familiar glycolytic pathway (Figure 3A) to illustrate this and the properties below (See Supplementary Information for the data matrices). A full kinetic model of red cell metabolism (Jamshidi et al, 2001) is available and the stoichiometric and gradient matrices are readily obtained for its glycolytic pathway. G can be factored into κ and (Figure 3A). We see that the elements in κ are spread over approximately 10 (log[κmax/κmin]=9.7) orders of magnitude. The matrix S is universal and so are the elements of G for a given set of physico-chemical conditions, such as temperature.
Property 2. The structural relationship between the stoichiometric and gradient matrices
It can be readily shown from equation (2) that if sij=0 then gji is also zero, that is, if a compound does not participate in a reaction it has no kinetic effect on it. Conversely, if sij≠0 then gji is also not zero. When elementary rate law formulations are used, this relationship holds for allosteric regulation as well, for net reactions. Further inspection reveals the property that S is structurally similar to −GT as illustrated in Figure 1B2. Thus, the non-zero entries in S have corresponding non-zero elements in −GT, but with a different numerical value. This fundamental feature shows that the topology of the network as reflected in S has a dominant effect on its dynamic features, providing another example of the biological principle that structure has a dominant effect on function.
Glycolysis
The structural similarity between the stoichiometric matrix and negative of the transpose of the gradient matrix for glycolysis is immediately apparent (Figure 3A1).
Property 3. Duality—either fluxes or concentrations can be used as the independent variables
A flux deviation variable, v′ can be defined such that v′=G·x′, from which it follows that
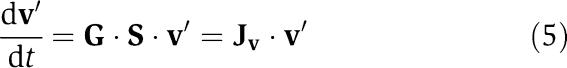 |
This transformation illustrates the switch from concentrations to fluxes as the independent variables. While concentrations have historically been used as the independent variables, the use of fluxes has grown in recent years as they tie together the multiple parts of a network to form its overall functions. Furthermore, the ability to relate the fluxes and concentrations independently of a specific rate law formulation, if the elements of G can be approximated, has significant implications for the construction and analysis of kinetic networks.
The two Jacobian matrices, G·S and S·G, are similar and share eigenvalues. Equation (5) is the ‘dynamic' flux balance equation since the variables in it are the fluxes, v′. One can thus analyze network properties either in terms of concentrations or fluxes as illustrated within Figure 1B3. The fluxes are ‘network' variables, as they tie all the components together, while the concentrations are ‘component' variables. Note that since Jv has not been fully recognized and studied in this field, when not otherwise specified, J is the metabolite Jacobian, or Jx.
Glycolysis
The duality between fluxes and concentrations highlights a deep relationship in network dynamics. The gradient matrix can be used to convert a set of differential equations in terms of metabolites into a set of differential equations in terms of fluxes. Consequently, the Jacobian can be defined in terms of metabolites or fluxes (Figure 3B3).
Property 4. Multi-timescale analysis of network dynamics
The properties of the Jacobian matrix that determine the characteristics of the network dynamics are its eigen properties. The eigenvalues are network-based time constants (in contrast to the reaction-based time constants in κ). Formally, the standard eigen analysis is performed by the diagonalization of the Jacobian matrix as:
where M is the matrix of eigenvectors, Λ is a diagonal matrix of the eigenvalues and M−1 is the matrix of eigenrows.
If the decomposition of equation (7) is introduced into equation (3), we obtain differential equations for the modes (m=M−1·x′)
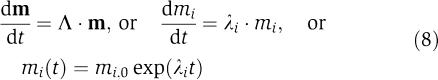 |
or a set of completely decoupled dynamic variables, that is, each mi moves on its own timescale defined by λi independent of all the other mj. The eigenrows give lumped or aggregate variables that move independently on each timescale since m is a set of dynamically independent variables. The eigenvectors form a natural coordinate system to describe the dynamic motion of the modes. We note that this decomposition can be applied to Jx or Jv (Figure 3B4). The eigenvalues will be the same whereas the eigenvectors and eigenrows will differ since the variable sets (concentrations versus fluxes) will not be the same.
The distribution of the numerical values of the eigenvalues is the basis for timescale separation. Timescale separation forms the basis for decomposition of biochemical reaction networks in time and the interpretation of the biochemical events that take place on the various time constants. Timescale separation has been analyzed in the context of biological networks and shown to lend insight and enable the simplification of these networks (Reich and Selkov, 1981; Heinrich and Sonntag, 1982; Palsson et al, 1987). Glycolysis provides an example.
Glycolysis
Pooling of variables (metabolites or fluxes) refers to the formation of aggregate groups of variables, in which the group of variables move together in a concerted manner. Pools that form on very fast timescales reflect the formation of chemical equilibrium pools, whereas pooling that occurs on the slower timescales reflects physiologically relevant interactions. Moving from the very fast timescales to the slower ones (Figure 4), one observes the well-known examples of pool formation between hexose phosphates (HPs) and phosphoglycerates (PGs). With successive removal of modes on the slower timescales, more and more of the metabolites begin to form aggregate variables and move together in a concerted fashion at fixed ratios. For glycolysis alone, the successive aggregation of chemical moieties (i.e. HP, PG) culminates in, on the slowest timescale, the formation of a physiologically meaningful pool that represents the sum of high-energy phosphate bonds found in the glycolytic intermediates (i.e. their ATP equivalents) (Figures 3.4 and 4). The last row of M−1 for Jv shows that this pool is moved by hexokinase as the input and ATPase as the output (Figure 3B4).
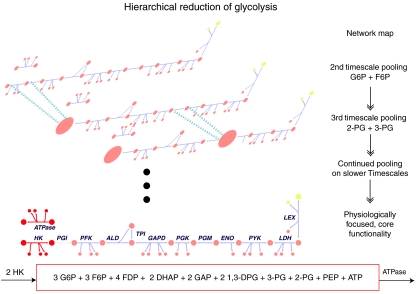
Figure 4.
Illustration of pooling for glycolysis. The pools that form on the fastest timescales reflect achievement of chemical equilibria between particular metabolites, such as the hexose phosphates (HPs) on the second timescale and the partial phosphoglycerate (PG) pool on the third timescale. Pools that form on the slower timescales reflect more biologically relevant interactions. For this example of glycolysis, the slowest timescale reflects the sum total of the high-energy phosphate bonds, which can be viewed in terms of the metabolites or fluxes (see Figure 3B3 and 3B4). From the flux point of view, the net balance can be described by the hexokinase flux and the ATPase (in a ratio of 2:1). On this very slow timescale, all of the other glycolytic metabolites have pooled together and the physiological function of the pathway emerges; namely, the sum of the high-energy phosphate bonds found in the glycolytic intermediates. The bottom illustration and corresponding equation detail the metabolites and fluxes comprising the final mode, in the fixed ratios. Metabolite abbreviations: ATP, adenosine triphosphate; DHAP, dihydroxyacetone phosphate; 1,3-DPG, 1,3-bis-phosphoglycerate; F6P, fructose-6-phosphate; FDP, fructose-2,6-phosphate; G6P, glucose-6-phosphate; GAP, glyceraldehyde-3-phosphate; LAC, lactate; PEP, phosphoenolpyruvate; 2-PG, 2-phosphoglycerate; 3-PG, 3-phosphoglycerate; PYR, pyruvate.
Recapitulation
Once recognized, these four properties will require further study.
The first property is a result of the explicit reconstruction process and the incorporation of different data types. The properties, completeness and accuracy of the data can be explicitly traced to dynamic properties. This decomposition will not only tie the models directly back to the data but also explicitly gives us the parts of the model that are under biological control and subject to change with adaptation or evolution. Measurement uncertainties are primarily in κ and are subject to evolutionary changes. These ‘biological' design parameters will likely need to be dealt with through the use of methods that bracket the range of values within uncertainty limitations.
The second property is a result of the full delineation of the chemical equations that make up a network and ideally their representation as net combinations of elementary reactions (i.e. vnet=v+−v−). In this format, we not only determine the structure of the gradient matrix but also make integration of multiple networks possible and enable the explicit analysis of the effects of regulatory molecules. Furthermore, it explicitly recognizes the underlying bilinear kinetic nature of network dynamics, as all chemical reactions are combinations of bilinear interactions, including regulatory processes (Segel, 1975).
The third property is helpful now that we have systems biological thinking developing fast in the community. The systems biology paradigm of ‘components → networks → computational models → physiological states' naturally leads to the use of fluxes as variables to characterize the functional states of a network. Fluxomic data tie components in a network together and are interpreted through network models, whereas concentration data are component data. Fluxes have been widely used for steady-state analysis and can now be used to study dynamic states as well.
The fourth property has been known and studied for several decades (Heinrich et al, 1977; Reich and Selkov, 1981; Heinrich and Sonntag, 1982; Palsson et al, 1987). Such studies have primarily been performed for small-scale models today, but their conceptual foundation has been established. At larger scales new issues will arise. These are likely to include, data sensitivity, course graining and modularization of physiological functions in time. New methods to study the bases vectors in M and M−1 that directly relate them to biochemistry and physiological functions need to be established. The promise of the elucidation of (dynamic) structure–(physiological) function relationships (Palsson et al, 1987) may now grow to large-scale networks.
Example of a cell-scale kinetic model
With fluxomic, metabolomic and thermodynamic data, we can anticipate the ability to generate large-scale kinetic models where more complicated structures of chemical and physiological pool formation will be found. Currently, human red cell is the only cell-scale kinetic model available, whose formulation followed a 30-year history of iterative model building (summarized in Joshi and Palsson, 1989, 1990). Analysis of the dynamic structure of this model resulted in the simplification of network dynamics and the description of the cellular functions in terms of physiologically meaningful pools of metabolites.
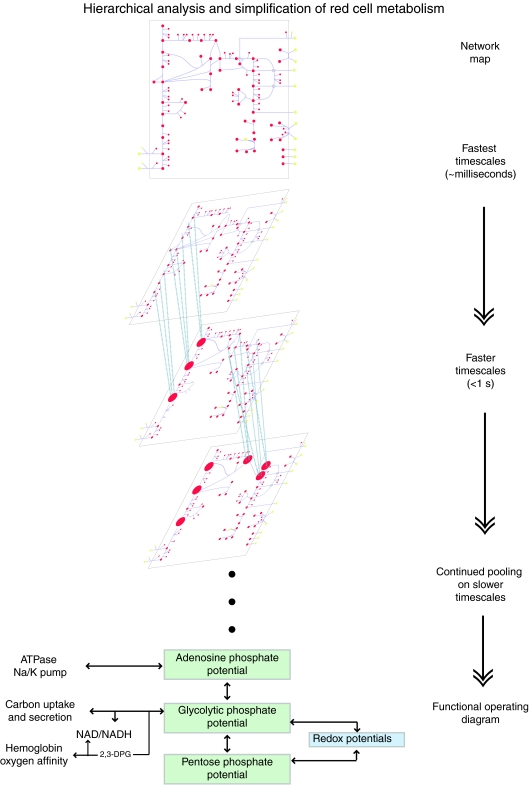
Analysis of the hierarchical dynamics of the human red cell model resulted in a richer and more complex physiological pool formation (Kauffman et al, 2002) that is detailed above for glycolysis alone. An overview of the hierarchical reduction of the network into a functional diagram is schematized in Figure 4. For example, the adenosine phosphate potential is defined analogously to Atkinson's energy charge (Atkinson, 1968). As originally elaborated by Reich and Selkov (1981), this ‘potential' is the ratio of the number of energy-rich phosphate bonds and the ability or capacity to carry such bonds. The different pools of metabolites in the red cell contribute to phosphate potentials and/or oxidation/reduction potentials. The result of the pool formation is an ‘operating diagram' (bottom of Figure 5) that describes the function of the metabolic network in the red cell slower timescales (minutes to hours).
Figure 5.
Hierarchical simplification of metabolic dynamics in the human red cell. Illustration of the reduction of biological networks through the formation of aggregate pools on progressively slower timescales, for a full kinetic model of the human red cell. Pool formation, as observed for the simple glycolytic pathway in Figure 4, occurs in a more complicated form in the red cell metabolism. A map for the complete kinetic model of human red cell metabolism is illustrated as pools form on progressive timescales. Ultimately, the function of the red cell can be reduced to redox and adenosine ‘potentials' in a similar spirit to Atkinson's energy charge (Atkinson, 1968). The adenosine phosphate potential drives the sodium potassium ATPase pump, the redox potential reflects redox state of the cell and determines the oxidative loads that it can withstand. The glycolytic phosphate potential interacts with the redox and adenosine and pentose phosphate potentials. It also affects the redox state of hemoglobin and the oxygen-binding affinity via NADH and 2,3-DPG. This ‘functional block diagram' describes the principle functionalities of red cell metabolism.
This example shows how physiologically meaningful dynamic structures form as a result of the particular numerical values in G and how they overlay on the network structure given by S. Not all sets of numerical values give this dynamic structure. It has been shown that genetic variation, as represented by sequence polymorphism in particular enzymes (pyruvate kinase and glucose-6-phosphate dehydrogenase), can disrupt this dynamic structure and lead to pathological states (Jamshidi et al, 2002).
Concluding remarks
Large-scale kinetic models have not been successfully constructed to date, with the human red blood cell being an exception (Heinrich et al, 1977; Heinrich, 1985; Joshi and Palsson, 1989, 1990; Mulquiney and Kuchel, 1999; Jamshidi et al, 2001). The chief reason for this lack of success is the large number of kinetic parameters required to define the system that is confounded by the fact that in vitro measurements of kinetic constants may not be representative of their numerical values in vivo (e.g. for a recent example, see Teusink et al, 2000). Thus, the probability of achieving other cell-scale models using these approaches appears to be very low. Recently, there have been efforts by investigators to develop methods to fill the gap between constraint-based models and kinetic models (Famili et al, 2005; Smallbone et al, 2007; Ishii et al, 2008).
As metabolomic data become available and drives toward genome-scale coverage (Brauer et al, 2006; Wishart et al, 2007) and approaches for approximating thermodynamic quantities using computational approaches (Mavrovouniotis, 1991) are being realized on the genome-scale (Henry et al, 2007), the data needed to build large-scale kinetic models will become available. In anticipation of the completion of these developments, we present here a workflow for the formulation of large-scale dynamic models and identification of four fundamental properties of the governing equations that genome-scale dynamic analysis will be based on. By focusing on the key structural and dynamic properties of networks and the inherent relationships between fluxes and concentrations, it will become possible to achieve dynamic descriptions of genome scale models, as illustrated in Figure 2.
Supplementary Material
Supplementary Information
References
- Atkinson DE (1968) The energy charge of the adenylate pool as a regulatory parameter. Interaction with feedback modifiers. Biochemistry 7: 4030–4034 [DOI] [PubMed] [Google Scholar]
- Barabasi AL, Oltvai ZN (2004) Network biology: understanding the cell's functional organization. Nat Rev Genet 5: 101–113 [DOI] [PubMed] [Google Scholar]
- Brauer MJ, Yuan J, Bennett BD, Lu W, Kimball E, Botstein D, Rabinowitz JD (2006) Conservation of the metabolomic response to starvation across two divergent microbes. Proc Natl Acad Sci USA 103: 19302–19307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling R, Vitkup D, Barrett MP (2008) New surveyor tools for charting microbial metabolic maps. Nat Rev Microbiol 6: 156–161 [DOI] [PubMed] [Google Scholar]
- Bruggeman FJ, de Haan J, Hardin H, Bouwman J, Rossell S, van Eunen K, Bakker BM, Westerhoff HV (2006) Time-dependent hierarchical regulation analysis: deciphering cellular adaptation. Syst Biol (Stevenage) 153: 318–322 [DOI] [PubMed] [Google Scholar]
- Famili I, Mahadevan R, Palsson BO (2005) k-Cone analysis: determining all candidate values for kinetic parameters on a network scale. Biophys J 88: 1616–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist AM, Henry CS, Reed JL, Krummenacker M, Joyce AR, Karp PD, Broadbelt LJ, Hatzimanikatis V, Palsson BO (2007) A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol 3: 121 10.1038/msb4100155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong SS, Palsson BO (2004) Metabolic gene-deletion strains of Escherichia coli evolve to computationally predicted growth phenotypes. Nat Genet 36: 1056–1058 [DOI] [PubMed] [Google Scholar]
- Goodacre R, Vaidyanathan S, Dunn WB, Harrigan GG, Kell DB (2004) Metabolomics by numbers: acquiring and understanding global metabolite data. Trends Biotechnol 22: 245–252 [DOI] [PubMed] [Google Scholar]
- Grimbs S, Selbig J, Bulik S, Holzhutter HG, Steuer R (2007) The stability and robustness of metabolic states: identifying stabilizing sites in metabolic networks. Mol Syst Biol 3: 146 10.1038/msb4100186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison R, Papp B, Pal C, Oliver SG, Delneri D (2007) Plasticity of genetic interactions in metabolic networks of yeast. Proc Natl Acad Sci USA 104: 2307–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich R (1985) Mathematical models of metabolic systems: general principles and control of glycolysis and membrane transport in erythrocytes. Biomed Biochim Acta 44: 913–927 [PubMed] [Google Scholar]
- Heinrich R, Rapoport SM, Rapoport TA (1977) Metabolic regulation and mathematical models. Prog Biophys Mol Biol 32: 1–82 [PubMed] [Google Scholar]
- Heinrich R, Schuster S (1996) The Regulation of Cellular Systems. Berlin (Heidelberg/New York): Springer [Google Scholar]
- Heinrich R, Schuster S, Holzhutter HG (1991) Mathematical analysis of enzymic reaction systems using optimization principles. Eur J Biochem 201: 1–21 [DOI] [PubMed] [Google Scholar]
- Heinrich R, Sonntag I (1982) Dynamics of non-linear biochemical systems and the evolutionary significance of time hierarchy. Biosystems 15: 301–316 [DOI] [PubMed] [Google Scholar]
- Henry CS, Broadbelt LJ, Hatzimanikatis V (2007) Thermodynamics-based metabolic flux analysis. Biophys J 92: 1792–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry CS, Jankowski MD, Broadbelt LJ, Hatzimanikatis V (2006) Genome-scale thermodynamic analysis of Escherichia coli metabolism. Biophys J 90: 1453–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollywood K, Brison DR, Goodacre R (2006) Metabolomics: current technologies and future trends. Proteomics 6: 4716–4723 [DOI] [PubMed] [Google Scholar]
- Ibarra RU, Edwards JS, Palsson BO (2002) Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature 420: 186–189 [DOI] [PubMed] [Google Scholar]
- Ishii K, Nakamura S, Morohashi M, Sugimoto M, Ohashi Y, Kikuchi S, Tomita M (2008) Comparison of metabolite production capability indices generated by network analysis methods. Biosystems 91: 166–170 [DOI] [PubMed] [Google Scholar]
- Jamshidi N, Edwards JS, Fahland T, Church GM, Palsson BO (2001) Dynamic simulation of the human red blood cell metabolic network. Bioinformatics 17: 286–287 [DOI] [PubMed] [Google Scholar]
- Jamshidi N, Palsson BO (2006) Systems biology of SNPs. Mol Syst Biol 2: 38 10.1038/msb4100077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamshidi N, Palsson BO (2007) Investigating the metabolic capabilities of Mycobacterium tuberculosis H37Rv using the in silico strain iNJ661 and proposing alternative drug targets. BMC Syst Biol 1: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamshidi N, Wiback SJ, Palsson B (2002) In silico model-driven assessment of the effects of single nucleotide polymorphisms (SNPs) on human red blood cell metabolism. Genome Res 12: 1687–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A, Palsson BO (1989) Metabolic dynamics in the human red cell. Part I—A comprehensive kinetic model. J Theor Biol 141: 515–528 [DOI] [PubMed] [Google Scholar]
- Joshi A, Palsson BO (1990) Metabolic dynamics in the human red cell. Part III––Metabolic reaction rates. J Theor Biol 142: 41–68 [DOI] [PubMed] [Google Scholar]
- Kauffman KJ, Pajerowski JD, Jamshidi N, Palsson BO, Edwards JS (2002) Description and analysis of metabolic connectivity and dynamics in the human red blood cell. Biophys J 83: 646–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummel A, Panke S, Heinemann M (2006) Putative regulatory sites unraveled by network-embedded thermodynamic analysis of metabolome data. Mol Syst Biol 2: 2006.0034 10.1038/msb4100074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrovouniotis ML (1991) Estimation of standard Gibbs energy changes of biotransformations. J Biol Chem 266: 14440–14445 [PubMed] [Google Scholar]
- Mayr E (1961) Cause and effect in biology. Science 134: 1501–1506 [DOI] [PubMed] [Google Scholar]
- Mulquiney PJ, Kuchel PW (1999) Model of 2,3-bisphosphoglycerate metabolism in the human erythrocyte based on detailed enzyme kinetic equations: equations and parameter refinement. Biochem J 342 (Pt 3): 581–596 [PMC free article] [PubMed] [Google Scholar]
- Oh YK, Palsson BO, Park SM, Schilling CH, Mahadevan R (2007) Genome-scale reconstruction of metabolic network in Bacillus subtilis based on high-throughput phenotyping and gene essentiality data. J Biol Chem 282: 28791–28799 [DOI] [PubMed] [Google Scholar]
- Pal C, Papp B, Lercher MJ, Csermely P, Oliver SG, Hurst LD (2006) Chance and necessity in the evolution of minimal metabolic networks. Nature 440: 667–670 [DOI] [PubMed] [Google Scholar]
- Palsson BO (2006) Systems Biology: Determining the Capabilities of Reconstructed Networks. Cambridge, London/New York: Cambridge University Press [Google Scholar]
- Palsson BO, Joshi A, Ozturk SS (1987) Reducing complexity in metabolic networks: making metabolic meshes manageable. Fed Proc 46: 2485–2489 [PubMed] [Google Scholar]
- Papin JA, Reed JL, Palsson BO (2004) Hierarchical thinking in network biology: the unbiased modularization of biochemical networks. Trends Biochem Sci 29: 641–647 [DOI] [PubMed] [Google Scholar]
- Pharkya P, Burgard AP, Maranas CD (2004) OptStrain: a computational framework for redesign of microbial production systems. Genome Res 14: 2367–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price ND, Reed JL, Palsson BO (2004) Genome-scale models of microbial cells: evaluating the consequences of constraints. Nat Rev Microbiol 2: 886–897 [DOI] [PubMed] [Google Scholar]
- Reed JL, Famili I, Thiele I, Palsson BO (2006) Towards multidimensional genome annotation. Nat Rev Genet 7: 130–141 [DOI] [PubMed] [Google Scholar]
- Reich J, Selkov E (1981) Energy Metabolism of the Cell: a Theoretical Treatise. New York (Orlando, FL/London/San Diego, CA): Academic Press [Google Scholar]
- Resendis-Antonio O, Reed JL, Encarnacion S, Collado-Vides J, Palsson BO (2007) Metabolic reconstruction and modeling of nitrogen fixation in Rhizobium etli. PLoS Comput Biol 3: 1887–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer U (2006) Metabolic networks in motion: 13C-based flux analysis. Mol Syst Biol 2: 62. 10.1038/msb4100109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segel I (1975) Enzyme Kinetics. New York: John Wiley & Sons [Google Scholar]
- Slonim DK (2002) From patterns to pathways: gene expression data analysis comes of age. Nat Genet 32 (Suppl): 502–508 [DOI] [PubMed] [Google Scholar]
- Smallbone K, Simeonidis E, Broomhead DS, Kell DB (2007) Something from nothing: bridging the gap between constraint-based and kinetic modelling. FEBS J 274: 5576–5585 [DOI] [PubMed] [Google Scholar]
- Steuer R, Gross T, Selbig J, Blasius B (2006) Structural kinetic modeling of metabolic networks. Proc Natl Acad Sci USA 103: 11868–11873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strogatz S (1994) Nonlinear Dynamics and Chaos: with Applications to Physics, Biology, Chemistry, and Engineering. Cambridge: Perseus [Google Scholar]
- Teusink B, Passarge J, Reijenga CA, Esgalhado E, van der Weijden CC, Schepper M, Walsh MC, Bakker BM, van Dam K, Westerhoff HV, Snoep JL (2000) Can yeast glycolysis be understood in terms of in vitro kinetics of the constituent enzymes? Testing biochemistry. Eur J Biochem 267: 5313–5329 [DOI] [PubMed] [Google Scholar]
- Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, Nikolai L, Lewis M, Coutouly MA, Forsythe I, Tang P, Shrivastava S, Jeroncic K, Stothard P, Amegbey G et al. (2007) HMDB: the human metabolome database. Nucleic Acids Res 35: D521–D526 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information