Abstract
We have used a supervised classification approach to systematically mine a large microarray database derived from livers of compound-treated rats. Thirty-four distinct signatures (classifiers) for pharmacological and toxicological end points can be identified. Just 200 genes are sufficient to classify these end points. Signatures were enriched in xenobiotic and immune response genes and contain un-annotated genes, indicating that not all key genes in the liver xenobiotic responses have been characterized. Many signatures with equal classification capabilities but with no gene in common can be derived for the same phenotypic end point. The analysis of the union of all genes present in these signatures can reveal the underlying biology of that end point as illustrated here using liver fibrosis signatures. Our approach using the whole genome and a diverse set of compounds allows a comprehensive view of most pharmacological and toxicological questions and is applicable to other situations such as disease and development.
Keywords: biomarker, data mining, liver, toxicity, toxicology, xenobiotic
Introduction
To increase our understanding of liver biology and to aid preclinical drug characterization, we have identified many of the biological response programs to xenobiotics and drugs in the liver by measuring RNA abundance changes. Using these patterns, the similarities and differences in basic biological, pharmacological and toxicological responses to different classes of chemicals can be fully characterized.
To achieve these goals, we built a very large liver xenobiotic and pharmacological response data set. Liver RNA from rats treated using multiple doses and at several time points with 344 chemicals and drugs belonging to 70 pharmacologic activity classes and matching vehicle controls was hybridized to whole genome microarrays. The resulting data set is composed of 1695 individual animal studies and 5288 microarrays. Coupled to these data are clinical chemistry, hematology and hepatic histopathology end points selected to represent data typically collected in pharmacology and toxicology studies of drug candidates (Ganter et al, 2005).
Others have approached some of these issues using similar but much smaller gene expression data sets mostly designed to identify individual signatures, or biomarkers, of one or two types of phenotypic or pharmacologic end points (Waring and Ulrich, 2000; Hamadeh et al, 2002; Heinloth et al, 2004; Elrick et al, 2005; Nie et al, 2006; Slatter et al, 2006). Because of their limited coverage of different drugs, chemical structures and pharmacological responses, these studies do not provide a full description of the xenobiotic response of the liver, and may suffer from a lack of specificity due to inadequate representation of the diversity of drug responses.
This data set presents a data mining investigation of substantial complexity. Both unsupervised and supervised (Hastie et al, 2000, 2001; Quackenbush, 2001; Liu and Ringner, 2004) methods can be used for analysis. Because supervised methods provide a quantitative measure of similarity of new chemicals to known chemicals, we used them to develop gene expression signatures (classifiers), which are short, weighted probe lists used to assign a sample to one of two classes (El Ghaoui et al, 2003; Natsoulis et al, 2005).
The scope of our analysis allows us to derive general conclusions on the number of phenotypes resolvable by gene expression and the characteristics of genes and gene expression changes capable of classifying all resolvable phenotypes. In addition, these studies provide a method to identify all of the genes that are necessary and sufficient to form a classifier for a given phenotype. The list of necessary genes allows one to understand the biology of a phenotype in great detail, as illustrated using liver fibrosis as an example.
Results
Systematic mining of data set
We systematically divided each phenotype measurement by severity, dose or time, and attempted to find patterns of gene expression changes that were able to classify the phenotype and characterize its severity. The phenotypes measured include histopathological findings, clinical chemistry and hematology assay results, and other traditional measures of health such as body and organ weights. Other phenotypes include the pharmacological and chemical properties of the compounds. Samples were separated into two classes, those that share a given phenotype (positive class) and those that do not (negative class) (Table I).
Table 1.
Summary description of the database
| Characteristics of the liver xenobiotic and pharmacologic response data set | |
|---|---|
| Arrays (4941 treated+347 untreated controls) | 5288 |
| Treatments (biological triplicate) | 1695 |
| Compounds | 344 |
| Structure activity classes (SACs)a | 171 |
| Pharmacologic activity classes (ACs)b | 77 |
| Clinical chemistryc | 46 |
| Liver histopathology annotations | 57 |
aDefined by chemists as being distinct structural classes.
bSecond level of a two-step hierarchy. Several structurally distinct but related SACs are grouped into one AC if they share a target.
cIncludes blood chemistry and hematology assays.
A total of 2112 two-class classification questions were submitted to the sparse linear programming (SPLP) algorithm (Natsoulis et al, 2005), and the results were internally cross-validated using the split-sample cross-validation procedure (Table II). In total, 180 signatures met performance cutoffs for pharmacology-type and toxicity-type signatures (see Materials and methods); 41 pharmacology signatures had an average 65.4% sensitivity and 99.7% specificity and averaged 37 probes in length (range 7–70), while the 139 toxicity signatures had an average 52.6% sensitivity and 99.2% specificity and averaged 79 probes in length (range 27–167). Toxicity signatures were generally longer than pharmacology signatures, likely due to the complexity of capturing several distinct biological processes that converge to a common phenotype.
Table 2.
Summary description of the results of the systematic mining
| Signature type: | Candidates | Passing validity criteria |
|---|---|---|
| Body and organ weights | 25 | 5 |
| Clinical chemistrya | 477 | 61 |
| Histopathology | 317 | 65 |
| Therapeutic indication | 52 | |
| Pharmacology | 1241 | 49 |
| Total | 2112 | 180 |
aIncludes blood chemistry and hematology assays.
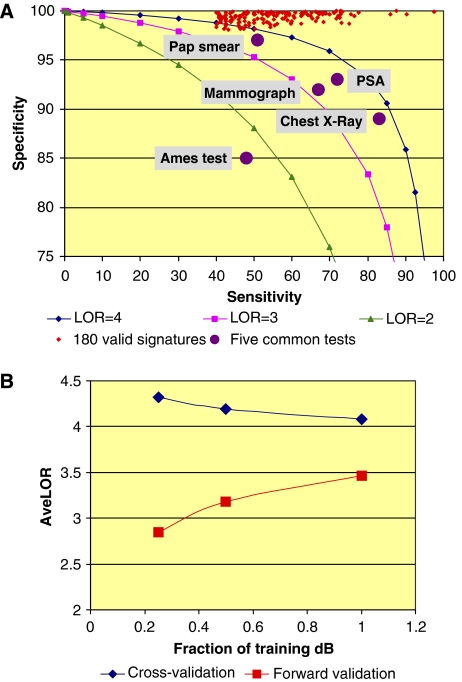
The quality of signatures derived from this systematic mining effort can be evaluated in several ways. First, each of the 180 signatures has a better log odds ratio (LOR) than any of five commonly used preclinical and clinical tests (Figure 1A) (Kim and Margolin, 1999; Mistry and Cable, 2003; Loy et al, 2004; http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=hstat1.table.7254). The increased performance is generally due to the intentional feature of the SPLP algorithm to create signatures with very high specificity (El Ghaoui et al, 2003; Natsoulis et al, 2005). The high specificity avoids false-positive calls, which, in drug development, could trigger a premature elimination of a compound or costly secondary testing. We have developed in addition a modified algorithm (adjusted SPLP, ASPLP) that weighs false positives and false negatives differently to obtain signatures with enhanced sensitivity and a slightly reduced specificity (GRG Lanckriet and G Natsoulis, unpublished results). Second, the performance metrics are estimates derived from split-sample cross-validation of the data and tend to overestimate forward validation performance obtained on independent data. Using a simulation based on our own results (Figure 1B), we show that the gap between these values decreases as the size of the training data set increases, illustrating that signatures derived from a large database are more predictive of future performance on independent data (Michiels et al, 2005). Finally, the convergence of the two curves (Figure 1B) suggests that our data set is sampling a substantial portion of the liver gene expression repertoire.
Figure 1.
(A) Comparing the 180 valid signatures with five commonly used tests. The sensitivity and specificity of the 180 valid signatures are compared for Pap smears, PSA, mammograph, chest X-ray and Ames test (http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=hstat1.table.7254; Kim and Margolin, 1999; Mistry and Cable, 2003; Loy et al, 2004). Iso-log odds curves for LOR equal to 2, 3 and 4 are shown for reference. (B) Modeling the average performance of signatures derived from databases of increasing size. The complete data set was split 20 times into a training data set and a forward validation data set. The training set was further split into half-sized and quarter-sized training sets. All splits were carried out at the compound level (i.e. all samples treated with the same compound are either included or not in a given set), thus modeling a growing toxicology database. The five signatures with the largest positive class were chosen for this study out of the set of 180 valid signatures. Each signature was re-derived and internally cross-validated as previously described from the 20 different quarter-size, half-size and full-size training sets. Each signature was also evaluated on the forward validation data set. The graph represents the average cross-validation and forward validation results for the five signatures.
Signature redundancy
Many of the classification sets were based on phenotypes that are biologically similar in interpretation, yet based on distinct end points (i.e. clinical chemistry versus histopathology), so it was of interest to determine how many of these 180 signatures are truly unique. We considered determining the overlap of signatures by comparing gene composition, class label similarity and similarity between rules defining the classes. None of these approaches was ideal as signatures measuring the same phenotype can have no gene in common (Natsoulis et al, 2005); looking at class labels does not take into account outliers that can drastically affect the signature characteristics and finally, some phenotypic end points that appear different are in fact similar when the functional annotations of the genes are compared. Thus, dissimilarities in class names can hide biological similarity.
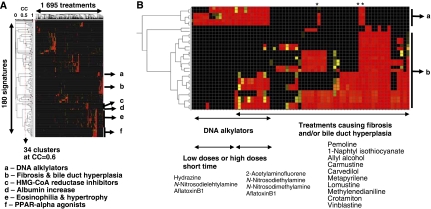
In view of these considerations, we chose a data-derived definition of what constitutes a unique signature. Signature matching scores (scalar products) for all 1695 treatments against all 180 valid signatures were clustered (Figure 2A). The number of unique signatures present in a set of high-performing signatures can be defined as the number of distinct clusters at a given threshold. At a correlation of 0.6, there are 34 clusters of signatures. Using a higher threshold (0.8) derives 55 groups that separate very closely related phenotypes. Using a lower threshold (0.4) derives 23 clusters. At that level events that are unique, as defined in the literature, are clustered together. A correlation of 0.6 ensures that most clusters are composed of signatures for either a single mode of action or a single pathological end point and divides the data into groups that follow both known and unexpected biological relationships. A specific list of unique non-redundant signatures was obtained by choosing as representative the signature with the highest positive predictive value from each cluster (Table III).
Figure 2.
Determination of a unique set of liver signatures. (A) A scalar product by treatment heat map, expressing the scores of all expression profiles against all signatures. (B) A blowup of regions a and b of (A) is shown. Negative values were set to zero and the resulting positive SP table was submitted to unweighted average-linkage clustering using an uncentered Pearson's correlation metric. Applying a correlation of 0.6 results in 34 clusters of signatures. The best representative signature per cluster is the signature with the highest positive predictive value (PPV). Color scale is continuous between black (SP=0), yellow (SP=0.5) and red (SP⩾1). Samples marked with a star are all treatments with methylenedianiline, a DNA alkylator whose profile match both DNA alkylator and fibrosis/bile duct hyperplasia groups of signatures.
Table 3.
Characteristics of 34 unique signatures
| Unique signature ID | Use | Type | Subtype | Class 1 size (no. of treatments) | Class −1 size (no. of treatments) | Class 1 compound count | Class −1 compound count | Class 1 AC count | Class −1 AC count | Number of signature in cluster | PPV | Sen- sitivity | Spec- ificity | Log odds ratio (LOR) | Signature length (no. of genes) | Number of signature cycles | Number of probes in NGS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SV0567082R5RU | T | CP | Absolute monocyte increase | 34 | 680 | 24 | 276 | 16 | 64 | 2 | 0.54 | 43.2 | 98.1 | 3.7 | 102 | 1 | 113 |
| SV0567098R5RU | T | CP | Creatinine increase | 38 | 757 | 18 | 308 | 16 | 66 | 3 | 0.62 | 47.8 | 98.5 | 4.1 | 141 | 1 | 127 |
| SV0567149R5RU | T | CP | Albumin increase | 33 | 604 | 17 | 273 | 11 | 65 | 2 | 0.71 | 52.5 | 98.8 | 4.5 | 105 | 2 | 186 |
| SV0562011R5RU | T | CP | Mean corpuscular hemoglobin concentration decrease (diagnostic, 3–7D time points) | 32 | 753 | 16 | 311 | 10 | 69 | 14 | 0.82 | 72.7 | 99.3 | 5.9 | 74 | 3 | 252 |
| SV0571010R5RU | T | CP | Mean corpuscular hemoglobin concentration. decrease (predictive, 0.25–1D time points) | 18 | 559 | 15 | 267 | 10 | 66 | 3 | 0.65 | 42.5 | 99.2 | 4.5 | 75 | 2 | 140 |
| SV0567088R5RU | T | CP | Glucose increase | 27 | 717 | 14 | 310 | 10 | 68 | 5 | 0.77 | 46.4 | 99.5 | 5.1 | 104 | 4 | 379 |
| SV0650093R5RU | T | H | Liver—centrilobular, inflammatory cell infiltrate, mixed cell | 37 | 676 | 16 | 264 | 9 | 66 | 2 | 0.6 | 47.0 | 98.3 | 3.9 | 111 | 1 | 131 |
| SV0567153R5RU | T | CP | Total protein increase | 28 | 612 | 14 | 295 | 9 | 66 | 2 | 0.67 | 48.8 | 98.8 | 4.4 | 91 | 2 | 181 |
| SV0635003R5RU | T | CP | Leukocyte count increase | 9 | 183 | 9 | 161 | 9 | 54 | 2 | 0.79 | 47.5 | 99.3 | 4.8 | 34 | 3 | 97 |
| SV0562020R5RU | T | CP | Hemoglobin decrease | 40 | 623 | 19 | 286 | 8 | 67 | 3 | 0.63 | 47.2 | 98.2 | 3.9 | 100 | 1 | 103 |
| SV0643003R5RU | T | BO | Relative liver weight decrease | 13 | 1158 | 11 | 310 | 7 | 68 | 1 | 0.77 | 48.3 | 99.8 | 6.2 | 68 | 10 | 840 |
| SV0562050R5RU | T | CP | Alkaline phosphatase decrease | 15 | 593 | 8 | 287 | 7 | 66 | 1 | 0.6 | 41.7 | 99.3 | 4.6 | 66 | 3 | 156 |
| SV0643002R5RU | T | BO | Relative spleen weight decrease | 21 | 651 | 14 | 294 | 6 | 67 | 9 | 0.97 | 73.3 | 99.9 | 8.1 | 29 | 21 | 918 |
| SV0562014R5RU | T | CP | Mean corpuscular hemoglobin decrease (diagnostic, 3–7D time points) | 14 | 739 | 7 | 310 | 6 | 70 | 4 | 0.86 | 53.3 | 99.8 | 6.5 | 60 | 16 | 1164 |
| SV0562026R5RU | T | CP | Leukocyte count decrease | 33 | 539 | 16 | 276 | 5 | 67 | 3 | 0.73 | 56.1 | 98.7 | 4.5 | 83 | 2 | 177 |
| SV0650033R5RU | T | H | Liver—periportal, hypertrophy | 22 | 699 | 11 | 270 | 5 | 68 | 3 | 0.57 | 46.1 | 98.9 | 4.3 | 71 | 1 | 78 |
| SV0567174R5RU | T | CP | Absolute basophil increase | 16 | 833 | 9 | 303 | 5 | 66 | 5 | 0.88 | 55.0 | 99.8 | 6.6 | 63 | 5 | 390 |
| SV0642001R5RU | T | BO | Relative liver weight increase | 11 | 604 | 9 | 282 | 5 | 63 | 2 | 0.76 | 47.0 | 99.7 | 5.6 | 38 | 11 | 492 |
| SV0651106R5RU | T | H | Liver—diffuse, cytoplasm, eosinophilia | 31 | 1273 | 8 | 277 | 5 | 68 | 27 | 0.84 | 53.1 | 99.7 | 6.0 | 97 | 14 | 1232 |
| SV0575020R5RU | T | CP | Lipase increase | 15 | 563 | 7 | 274 | 5 | 65 | 1 | 0.62 | 40.8 | 99.3 | 4.6 | 57 | 3 | 189 |
| SV0571053R5RU | T | CP | Absolute lymphocyte decrease | 16 | 429 | 11 | 201 | 4 | 55 | 2 | 0.75 | 46.4 | 99.4 | 4.9 | 60 | 2 | 125 |
| SV0650143R5RU | T | H | Liver—periportal, fibrosis | 12 | 754 | 5 | 284 | 4 | 68 | 25 | 0.85 | 71.0 | 99.8 | 7.0 | 42 | 25 | 1380 |
| SV0562116R5RU | T | CP | Glucose decrease | 16 | 754 | 5 | 317 | 3 | 67 | 2 | 0.61 | 42.1 | 99.4 | 4.7 | 69 | 6 | 434 |
| SV0650106R5RU | T | H | Liver—hepatocyte, periportal, lipid accumulation | 13 | 1063 | 4 | 248 | 3 | 66 | 5 | 0.85 | 55.0 | 99.9 | 6.7 | 52 | 16 | 1003 |
| SV0650121R5RU | T | H | Liver—hepatocyte, centrilobular, lipid accumulation, microvesicular | 18 | 1050 | 4 | 248 | 3 | 66 | 3 | 0.6 | 41.3 | 99.5 | 4.9 | 95 | 7 | 627 |
| SV0599196R5RU | P | SAC | GR-MR agonist | 13 | 655 | 7 | 318 | 1 | 67 | 8 | 0.9 | 71.2 | 99.8 | 7.0 | 34 | 6 | 203 |
| SV0614125R5RU | T | SAC | Toxicant, DNA alkylator | 15 | 856 | 6 | 325 | 1 | 69 | 6 | 0.89 | 51.9 | 99.9 | 6.8 | 59 | 4 | 216 |
| SV0614137R5RU | P | SAC | Estrogen receptor agonist, steroidal | 13 | 866 | 5 | 328 | 1 | 69 | 7 | 0.9 | 51.2 | 99.9 | 6.9 | 40 | 4 | 137 |
| SV0614148R5RU | P | SAC | PPAR α agonist, fibric acid | 14 | 861 | 5 | 328 | 1 | 69 | 15 | 0.97 | 72.8 | 100.0 | 8.6 | 20 | 19 | 659 |
| SV0599539R5RU | P | SAC | H+/K+ ATPase inhibitor | 17 | 1610 | 4 | 327 | 1 | 68 | 1 | 0.66 | 50.0 | 99.7 | 5.9 | 56 | 1 | 68 |
| SV0614270R5RU | P | SAC | PDE4 inhibitor | 14 | 1629 | 4 | 334 | 1 | 69 | 2 | 0.77 | 58.3 | 99.8 | 6.7 | 60 | 2 | 122 |
| SV0599291R5RU | T | SAC | Toxicant, heavy metal (3, 5 and 7D, other non-metal toxicants in negative class) | 12 | 877 | 3 | 333 | 1 | 70 | 1 | 0.8 | 50.0 | 99.9 | 6.5 | 50 | 6 | 353 |
| SV0614202R5RU | T | SAC | Toxicant, heavy metal (0.25–7D allowed, other toxicants not in negative class) | 12 | 1115 | 3 | 318 | 1 | 68 | 1 | 0.67 | 40.0 | 99.8 | 5.9 | 55 | 1 | 58 |
| SV0614084R5RU | P | SAC | HMG-CoA reductase inhibitors | 6 | 662 | 3 | 322 | 1 | 68 | 8 | 0.99 | 87.5 | 100.0 | 10.1 | 7 | 15 | 240 |
| Averages | 19.9 | 794 | 10 | 290 | 5.6 | 66.4 | 5.3 | 0.75 | 52.9 | 99.4 | 5.7 | 66.7 | 6.5 | 381 | |||
Class size and class composition (columns 5–10); classification performance and signature length (columns 11–16) and results of necessary gene set definition algorithm (columns 17 and 18).
Notes: The unique signature ID relates each signature to one of the 180 derivation rules, Figure 2. The use column indicates the toxicity-type signatures (T) or the pharmacology-type signatures (P) as discussed in the Materials and methods. There are four types of signatures presented, body and organ weight (BO), histopathology (H), clinical pathology (CP) which includes blood chemistry and blood hematology end points and structure activity class (SAC), which includes signatures of the SAC type and AC type as described in the Materials and methods.
In contrast to the supervised method described above, 2D hierarchical clustering of all 1695 liver experiments by all genes resolved only a few phenotypes such as HMG-CoA reductase inhibitors and acute phase responses, while PCA could resolve no phenotypes. PCA could only resolve phenotypes when a much smaller data set (<200 experiments) containing experiments with very distinct gene expression changes was analyzed (data not shown).
To verify that the patterns observed in Figure 2A do not occur by chance only, we randomized the class assignments (label permutation) of each of the 180 valid signatures 100 times each and derived and cross-validated a signature for each permuted label set. Even though the average LOR of the 100 randomized label sets is close to zero for each of the 180 signatures, the maximum LOR ranges between 1.2 and 5.7 and averages 2.9, well below the average LOR for the 180 valid signatures (5.7). The permuted label set signatures are also much longer (averaging 135 genes) than signatures derived from real class labels. We chose the signature with the highest LOR from each set of 100 randomizations and repeated the clustering experiment described in Figure 2A. Not a single cluster with more than one member is observed at a correlation of 0.6, while the highest correlation for a cluster with at least two members is 0.48 (Supplementary information S12).
Biological relationships between signatures
Beyond its practical use in defining a set of unique signatures, biologically interesting relationships are revealed between end points (Figure 2A). For instance, one large cluster consists of signatures for PPARα agonists, albumin increase, hepatic eosinophilia, hypertrophy, HMG-CoA reductase inhibitors and lipase increase. Both PPARα agonists and HMG-CoA reductase inhibitors are used to lower serum cholesterol and affect lipid metabolism. PPARα agonists are known to cause hepatomegaly and hepatocyte proliferation, and albumin output of the liver increases as a consequence of hepatomegaly (Peters et al, 1997). Finally, we commonly observed increased blood albumin concentration as well as hepatocellular eosinophilia and hypertrophy in both PPARα agonist- and HMG–CoA reductase inhibitor-treated animals (data not shown).
In another case, certain samples match both the toxicant, DNA alkylator and the fibrosis and bile duct hyperplasia signatures (Figure 2B), thus suggesting a biological relationship between cellular damage caused by DNA alkylators and liver remodeling, bile duct hyperplasia and fibrosis. In our experiments, most treatments that caused fibrosis also caused bile duct hyperplasia, and as a consequence, signatures for fibrosis were well correlated with bile duct hyperplasia signatures (r=0.68).
Characteristics of signature genes
A common assumption is that highly weighted signature genes in valid biomarkers have large amplitudes of regulation in the positive class. Our data indicate little correlation between the magnitude of the weight and the amplitude of gene regulation and between weights and average expression levels in untreated controls (Supplementary information S2). These findings suggest that small changes in expression play important roles in signatures and specific liver responses.
A liver xenobiotic response gene set
A total of 1704 probes (representing 1660 distinct genes) appear at least once in the 34 signatures. Less than 10% (150 probes) account for more than 50% of the sum of impacts (a measure of importance; see Materials and methods) across all 34 signatures, while about 25% (400 probes) account for 75% of the sum of impacts (Supplementary information S1). Thus a small number of genes contribute disproportionately to signature performance for all end points.
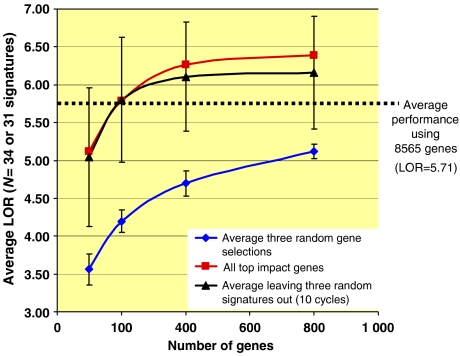
The average LOR across the 34 signatures is 5.7. Each of the 34 signatures was re-derived, with no additional feature reduction, using the top 100, 200, 400, 800 and 1600 probes by impact. A more impartial analysis was carried out by selecting gene sets from 31 randomly chosen signatures and evaluating the performance of the other three signatures on each of the gene sets. The procedure was repeated 10 times, evaluating the performance of a different set of three signatures each time. The two curves for average LOR, training on genes from all or training on genes from all but three signatures, are statistically indistinguishable (Figure 3) and form a rapidly rising curve which reaches the performance of the full set at 200 probes and exceeds the performance of all genes at 400 probes. The average performance of the 34 signatures derived from three separate random gene sets of equal size drawn from the array is lower. The entire procedure was repeated using permuted label sets (Supplementary information S12). In this case, the two impact-based curves do not separate from the curve based on random gene choice.
Figure 3.
Performance of signatures derived from gene sets of increasing size. Red curve, squares: gene sets of increasing size were chosen based on the sum of total impacts across all 34 signatures. Blue curve, diamonds: average of three random choices of gene sets of increasing size; performance was evaluated on all 34 signature training sets. Average and standard deviation of the results are reported. Black curve, triangles: gene sets were chosen based on the sum of total impacts across 31 randomly chosen signatures from the 34. The performance of the gene sets was evaluated on the three left out signatures. The procedure was repeated 10 times and the average and standard deviation of the result are reported. Black dotted line indicates the average LOR obtained when all 8565 probes are used without any pre-selection. See Supplementary information S12 for an equivalent study including permuted label sets.
Clearly, a subset of genes selected based on importance in signature performance performs better than the whole. The performance of any classification algorithm can decrease in the presence of a large excess of lower information content variables (genes), ultimately overwhelming the classification algorithm. SPLP is rather insensitive although not completely immune to this effect (Natsoulis et al, 2005). In addition, pre-selection of genes based on probe variance or highest signal intensity had little effect on the average LOR, whereas pre-selection based on the average log ratio across the positive class and statistical significance of fold change reduces signature performance (Supplementary information S2).
This analysis suggests that the genes selected by the impact metric are of general applicability, and just a small portion, by themselves, can characterize a large fraction of liver response to xenobiotics. We further characterized the utility of the small gene set to derive signatures for a biological event distinct from xenobiotic exposure, caloric restriction. The 200 gene set was far superior to a random selection of genes in classifying the response to caloric restriction (Supplementary information S10).
Gene composition of the xenobiotic response gene set
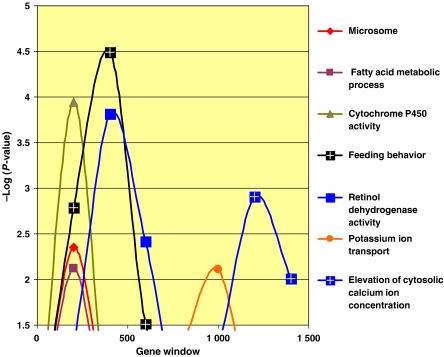
Given that just 200 probes out of the 1704 probes in the unique 34 signatures can create valid signatures for a diverse set of end points, this set would be expected to contain many genes involved in liver xenobiotic metabolism, pharmacology and response to tissue injury. The probes were ranked by the sum of their impact across all signatures and examined for enrichment in gene ontology (GO) terms (Figure 4 and Supplementary information S8). We sought to increase the sensitivity of detecting GO term enrichment by analyzing successive portions of that ranked list as some terms, only enriched in the very top of the list, might not appear significantly enriched in the entire list. GO terms associated with many of the liver's functions are enriched in the first windows of analysis (Figure 4), such as the terms cytochrome P450 and microsome, two partially overlapping GO terms (Supplementary information S9), reflecting the importance of cytochrome P450s in xenobiotic metabolism (Ioannides, 2002). The term fatty acid metabolism, comprising genes such as fatty acid synthase, enoyl-CoA hydratase, acyl-CoA synthetase among others is also enriched. The liver is a major site for fatty acid and lipid metabolism, and several major classes of compounds present in the database (statins, fibrates, glitazones, estrogen receptor modulators and others) affect lipid synthesis and degradation. Terms such as feeding behavior (including genes such as orexin and glucagon) and potassium ion transport and elevation of cytosolic calcium ion concentration suggest that genes belonging to these categories are also involved in xenobiotic responses but are less important than cytochrome P450 and fatty acid metabolism which peak earlier.
Figure 4.
Analysis of gene types enriched in the ranked list of 1660 genes appearing in 34 unique signatures. Genes were ranked by impact across all signatures. Overlapping windows of 400 genes were tested for enrichment in GO terms. Terms with enrichment exceeding −log(P-value)=2 are shown.
Identification of complete gene sets capable of forming a classifier
We have previously observed that several signatures for the same end point can be derived with no gene in common (Natsoulis et al, 2005). Therefore, it was of interest to determine how many different genes are capable, in various combinations, of yielding a signature with a performance exceeding a certain threshold for a given classification question. Such a gene set could be considered a necessary gene set (NGS) because no valid signature can be derived from the complement of the NGS (i.e. all genes present on the array minus the NGS). To determine the NGS for each of the 34 end points described above (Table III and Supplementary information S11), all genes were submitted to the signature-generating algorithm initially. If a valid signature was obtained, the highest impact signature probes, accounting for ⩾90% of total impact, were removed from the set of all probes and the resulting stripped set was resubmitted to the algorithm. The cycle was repeated until the performance of the signatures dropped below a chosen threshold (for this experiment LOR ⩾4); we call this procedure ‘stripping'. Some signatures, such as spleen weight decrease and periportal lipid accumulation, continue to yield valid signatures even after more than 20 cycles of stripping. The performance of other signatures such as monocyte increase and periportal hypertrophy rapidly decays after one or two cycles. Examination of the genes in these stripped signatures for each type suggests that in the former case, a large number of genes belonging to different pathways are sufficiently characteristic of the end point in question to yield a valid signature. In the latter case, just a few genes have the ability to diagnose the phenotype and once removed from consideration, no other gene can substitute.
Liver fibrosis
To illustrate the value of the NGS gene list formed by the stripping procedure, we chose the liver fibrosis signatures for detailed analysis (SV0650143R5RU, Table III). Chronic liver fibrosis can ultimately result in liver failure and is a significant risk factor for liver cancer, and remains difficult to prevent and treat (Takahara et al, 2006; Iredale, 2007). Better understanding of the biological responses during development of fibrosis has emerged via studies using multiple experimental model systems (Huang et al, 2004; Jiang et al, 2004; Utsunomiya et al, 2004; Takahara et al, 2006; Gnainsky et al, 2007; Iredale, 2007) and genome-wide characterization of gene expression changes as described here.
Hepatic fibrosis commonly occurs following injury from a variety of insults, including drugs and toxicants, and is accompanied by an inflammatory response triggered by Kupffer cells, resident monocytes and other types of immune cells. Hepatic stellate cells (HSCs) are normally quiescent; upon hepatocyte injury, however, HSCs are activated by inflammation and differentiate into myofibroblast-like cells that can proliferate and migrate. HSCs and periportal fibroblasts repair hepatic injury by secreting extracellular matrix proteins such as collagen, whose synthesis is promoted by the fibrogenic cytokine TGFβ (Ramm et al, 2000).
Gene-type enrichment in signatures reveals pathways and processes activated by pharmacology or pathology
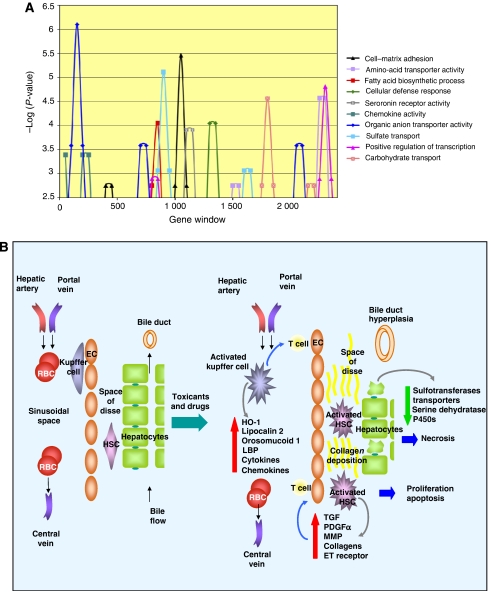
The 1380 unique probes (Supplementary information S11) that were present in all stripped fibrosis signatures that exceeded LOR=4 are statistically enriched (P-value <0.05) in GO terms, such as cell–matrix adhesion, amino-acid transporter activity, fatty acid biosynthetic process, cellular defense response, chemokine activity, organic anion transporter activity, sulfate transport, positive regulation of transcription and carbohydrate transport, most of which are affected during injury and subsequent fibrosis and bile duct hyperplasia (Figure 5A). Other terms such as serotonin receptor activity, sensory perception and brain development were enriched at P-values <0.001, indicating that local innervation and paracrine regulation of liver functions are remodeled during fibrosis. Many of these enrichments are not observed until the later signature cycles (cell–matrix adhesion and serotonin transporter activity, for example) and could be missed with more conventional methods of analysis.
Figure 5.
(A) Analysis of gene types enriched in the necessary gene set for the fibrosis signatures. Genes were ranked by cycle across all signatures. Overlapping windows of 50 genes were tested for enrichment in GO terms. Terms with enrichment exceeding −log(P-value)=2.5 are shown. (B) Model of gene expression events in liver fibrosis annotated with genes observed to be regulated by profibrotic xenobiotic treatments. The normal architecture of the liver (left) is remodeled following toxic exposure to compounds and drugs. Injury to hepatocytes from metabolites and reactive oxygen species activates resident macrophages, Kupffer cells, which produce acute phase response proteins, chemokines and cytokines. These proteins in turn attract other immune cells such as T cells, and activate quiescent hepatic stellate cells (HSCs). Both HSCs and endothelial cells (ECs) respond by proliferating and undergoing subsequent apoptosis. HSCs also produce growth factors that stimulate proliferation as well as collagens and other proteins involved in the deposition of collagen in the extracellular space.
Downregulation of a number of liver-specific genes may signal a loss of function of and/or an actual loss of the major parenchymal cells in the liver, hepatocytes, which comprise 80% of the normal liver cell population. Genes that are preferentially downregulated include those that are involved in amino-acid metabolism, organic anion and amino-acid transport and metabolism, and several sulfotransferases and cytochrome P450s (Figure 5B).
Genes that are preferentially upregulated in contrast include those involved in cell adhesion, cytoskeleton organization, cell–cell signaling, proliferation, xenobiotic metabolism and the immune response. Molecules that are upregulated induce or promote cell adhesion (PDGFα, endothelin 1, cd36, osteoblast-specific factor and procollagen C-endopeptidase enhancer) and remodeling of the actin cytoskeleton (Flna, Tekt1, Krt2-7 and Cappa1). Both PDGFα and endothelin 1 promote activation of HSCs and consequently fibrosis (Eng and Friedman, 2000; Iredale, 2007). In total, 84 of 137 probes that averaged twofold or more upregulation had low expression in the liver (average log signal intensity <−0.3; P-value=8.8 × 10−7). Many of these genes are those upregulated in rare cell types that are activated during liver injury and fibrosis, such as HSCs and Kupffer cells (Figure 5B).
TGFβ1, which is a strong promoter of collagen production and fibrosis, was itself unchanged on average. TGFβ1 must be proteolytically processed before becoming active (Zhu and Burgess, 2001) and thus there may be a change in conditions that favor processing of latent TGFβ1 during liver injury. Evidence that TGFβ1 is exerting an active influence includes the induction of both TGFβ1-induced transcript 1 and follistatin. Follistatin is an antagonist of activin, a growth factor that is a member of the TGF superfamily, and may modulate TGF action (Matzuk et al, 1995).
Activated HSCs produce collagen, which is deposited as part of the remodeling of the extracellular matrix during fibrosis. Expression of collagen at the sites of injury correlates with induction of fibrosis and scarring (Leveille and Arias, 1993; Alcolado et al, 1997; Gabele et al, 2003). Seven different genes encoding collagen molecules were part of the NGS, including procollagen type 1 α1 (upregulated about twofold on average across the positive class), the primary molecule in collagen I (data not shown).
The NGS analyses for all 34 unique signatures is summarized in Supplementary information S11. Similar insight into biology can be obtained through these analyses. For example, the HMG CoA reductase inhibitor signature NGS is enriched in cholesterol biosynthesis genes, reflecting the mechanism of action of these drugs. In addition, this signature was derived from high-dose treatments that caused liver injury. Enrichment of cell cycle and DNA replication terms were also observed, a possible reflection of the liver response to injury.
Discussion
The concept of the connectivity map was recently introduced, whereby a set of signatures is compared to a reference database of gene expression profiles obtained from compound-treated cell lines (Lamb et al, 2006). The authors show that close examination of gene expression profiles correlated and anticorrelated with a given signature provides insights into the multiple modes of action of certain compound classes and into the multiple biological mechanisms underpinning the phenotype of interest (Lamb et al, 2006). Here, we explore a much larger xenobiotic response database in an in vivo model. The breadth of the database and the systematic nature of the methodology allow us to derive a number of observations of general interest. (1) We have developed methods to identify biologically synonymous end points; these synonymous end points uncovered unexpected associations between apparently unrelated phenotypes. Using this method, we identify signatures (classifiers) for 34 distinct end points. (2) We show that signature genes are not appreciably enriched in genes showing large amplitude of regulation or high levels of expression; we also show that aggressive gene pre-selection by amplitude of expression change or statistical significance reduces classifier quality. (3) We show that a small number of genes (∼200) is sufficient to classify all unique phenotypic end points in the liver. (4) We show that this limited gene set involves many genes in the xenobiotic response repertoire. (5) Finally, we show that a large data set encompassing a wide variety of toxicological and pharmacological activities yields signatures with higher performance.
Our approach also identifies examples of very different signatures for a single end point. Similar results have been reported before and have often been regarded as problematic for the studies themselves or of the field in general (Michiels et al, 2005). Here, we describe a formal method to identify all the genes capable of defining a classifier for a given phenotype, at a chosen quality threshold. These lists of necessary genes form a ranked list of the genes involved in a particular phenotype and can be used to characterize the gene expression changes and thus infer biological changes that underlie a phenotype. Because the NGS includes all potential genes that could be used as part of a diagnostic test, a definition of the NGS for a particular phenotype or disease provides robust intellectual property and a barrier to entry for others attempting to build diagnostics for the same phenotype.
The liver fibrosis NGS provides insights into the biological pathways involved in the progression from liver injury to fibrosis. We show that xenobiotic insult leads to loss of certain gene expression apparently secondary to hepatocyte cell death through necrosis and apoptosis and leads to the upregulation of weakly expressed genes, probably due to activation and expansion of less abundant cell types, such as HSCs and Kupffer cells.
This study illustrates that a comprehensive approach can distill a complex and broad issue to a definable set of answers, increase our knowledge and develop useful signatures and diagnostics. Using a similar approach would better characterize other model systems and molecular phenotypes underlying disease processes and lead ultimately to clinically useful diagnostic markers.
Materials and methods
Rat liver xenobiotic and pharmacology database
The construction of the database was previously described in detail (Ganter et al, 2005). We focus here on the liver portion of the data set. In total, the data set was comprised of 5288 individual animal studies (arrays). The array data used in this study have been deposited in the Gene Expression Omnibus (GEO accession no. GSE8858). Microarray analysis was performed on liver mRNA using CodeLink Rat UniSet 1 Bioarrays (now provided by Applied Microarrays, Tempe, AZ, USA) with analysis restricted to 8565 probes and about 7700 individual genes. Coupled to these data are the blood clinical chemistry, hematology and histopathology findings typically measured in pharmacology and toxicology studies of new chemicals and drugs (Tables I and II and Supplementary information S3 describe the dimensions and contents of the data set). Taken together, the RNA abundance measurements and the phenotypic measurements constitute a uniform data set, which we have explored using supervised mining methods using classification rules established as described below.
Definitions
During the course of this study we employed several widely used concepts, and a few novel metrics, which are defined here to increase clarity. Treatment: a biological triplicate group of samples derived from animals treated with the same dose, for the same time and with the same compound. Upregulated, downregulated: indicate changes in the steady-state level of expression of a RNA in the liver, we note that tissues are composed of several cell types and changes may reflect multiplication, activation, inactivation or death of cell populations, in addition to selective RNA degradation, or changes in primary transcription rates. Orthogonal data: data describing samples that are not gene expression data; for example, clinical chemistry, hematology-, histopathology-, pharmacology- and literature-derived annotations. True positive (TP): samples for which the biomarker indicates the sample is positive and the orthogonal data indicate the sample is positive for the phenotype under investigation. True negative (TN): samples for which the biomarker indicates the sample is negative and the orthogonal data indicate the sample is negative for the phenotype under investigation. False negative (FN): samples for which the biomarker indicates the sample is negative and the orthogonal data indicate the sample is positive. False positive (FP): samples for which the biomarker indicates the sample is positive and the orthogonal data indicate the sample is negative. Log ratio: always refers to log10 ratio of mean signals of treated samples to vehicle treated controls for the gene in question. Sensitivity: sensitivity=TP/(TP+FN). Specificity: specificity=TN/(FP+TN). Positive predictive value (PPV): PPV=TP/(TP+FP). Log odds ratio (LOR): LOR=ln(((TP+0.5)(TN+0.5))/(FP+0.5)(FN+0.5)). The scalar product (SP): defined for a treatment as SP=Σwixi−b, where wi is the weight for gene i and xi is the log10 ratio for gene i; the sum is over all genes of the signature. Note that the list of genes and weights is the output of the SPLP algorithm. Impact: the impact of a gene in a signature is computed by multiplying the average log ratio x for that gene across the positive class defined in the signature definition (see below) by the weight w of that gene in the signature. The total impact of the gene is that value, minus the equivalent value calculated for the negative class. Gene list and GO analysis: or examinations of lists of genes for enrichment of various terms; enrichment is calculated by use of Fisher's exact test and often expressed as the P-value or −log10 of the P-value for the particular term(s).
Rule types for class definition
The rules were implemented using the SQL query language according to the following logical steps. First, the ‘universe' of profiles relevant to the two-class classification question was defined. The universe could be further restricted based on dose, time or both considerations. Profiles outside the universe were not considered further. Next the universe was split into three classes: the positive class, the negative class and the excluded class. The positive class was usually defined as the set of samples sharing a particular property, while the negative class was often defined as the remainder of the universe. A portion of the universe was sometimes assigned to an excluded class when the true phenotype might not be known for some samples because they were not assayed, or they were assayed but assay values were missing or uncertain. Alternatively, when classes were defined based on a continuous assay value, samples were often ranked, by fold change or P-value versus control, for example. The positive and negative classes were then defined as the extremes (top one-third and bottom one-third, for example) of this distribution and the intermediate samples were assigned to the excluded class. This had the advantage of training neither for nor against samples with intermediate values. Most of the clinical chemistry and hematology rules were structured in this manner since these values were continuous. In these cases, derivation of signatures along the variable distribution was often systematically explored. For example, signatures were systematically derived for a particular clinical value from treatments with fold changes of 100-, 30-, 10-, 3-, 1.5- and 1.1-fold; similar systematic schemes were applied as appropriate to ranked lists, P-values, ridit scores and other metrics indicating intensity. These systematic studies often revealed how the phenotype changed with intensity.
To allow cross-validation through split-sample procedure and to impose chemical diversity on the training set, thereby broadening the applicability, we imposed minimum class sizes. A minimum class size was imposed for both the positive and negative classes and for all rule types. We used split-sample cross-validation across 20 randomly selected splits to estimate the performance of the signature. We uniformly applied a split ratio of 60% training and 40% test, which in cases of a positive class size equal to 6 corresponded to 3.6 training and 2.4 test, or after rounding, 3 positive samples in the training set and 3 in the test set. Imposing a lower limit of six experiments in the positive class ensured that a minimum number of 6!/(3!*3!)=20 distinct splits of the positive class were possible. The minimum negative class size was set at 44 so that the sum of minimum sizes for both classes comprised at least 50 samples. The larger minimum class requirement for the negative class was intended to ensure diversity within the negative class, so that the resulting signature was capable of discriminating between the ‘phenotype of interest' and a large variety of other effects. A minimum of three distinct chemicals was also imposed on the positive class to ensure that the signatures recognized a general property of the class and not idiosyncratic characteristics of an individual compound. Additionally, for clinical chemistry and hematology and histopathology signatures, we required that the compounds used in the positive class treatments belong to three separate activity classes. Again, this was to ensure that the resulting signature is characteristic of the common pathology and not of an individual compound class. The logical steps described above were combined and automated to generate rule sets that could be categorized by the type of data defining the positive class as distinct from most of the other treatments within the database.
The systematic mining, using the restrictions and procedures mentioned, resulted in 2112 signature-derivation rules. The positive classes of these signatures averaged 36 treatments and included an average of 14 drugs from 11 classes per signature, and the negative class averaged 754 treatments and included an average of 270 drugs from 63 activity classes. Thus, the diversity of the treatments, drugs and activity classes was very large. Examples of the complete list of 2112 rules are presented by rule type for the activity class, structure activity class, pharmacology, clinical chemistry, hematology, histopathology rules and the body-and-organ weight rules (Supplementary information S4). The list of histopathology annotations and of the clinical chemistry and hematology assays on which the rules are based is presented (Supplementary information S3). Full Excel® files containing all of the rules also accompany this paper. The compound classification in terms of activity class and structure activity class as well as the class labels for the 34 unique signatures is shown in Table III and other characteristics of the set of 34 unique signatures can be found in Supplementary information S5, S6, S7 and S8.
Signature derivation and cross-validation
The classification algorithm used was the SPLP algorithm (El Ghaoui et al, 2003; Natsoulis et al, 2005). We note that this algorithm makes use of the mean log ratio and the standard error of the mean within each biological triplicate experiment, thus accounting for variability of measurements in classifier construction. In all cases, probes were eliminated for missing data (for the positive class if any data were missing; or for the negative class if >5% missing values). Missing probes occurred because of array technical failures, values below threshold and several other less common technical reasons. For computational speed consideration, probe pre-selection (feature reduction) by variance was used for those systematic mining signature derivation runs, which aimed to characterize all possible drug signatures, 2112 derivations in total (Tables I and II).
Split-sample cross-validation
In all cases, a 60/40 split-sample procedure was applied and the performance was reported as the average of the test results for 20 random partitions of the data (Simon et al, 2003; Allison et al, 2006; Varma and Simon, 2006). In cases where the sample class identities (labels) were set according to the properties of the compound (structure activity class, activity class and pharmacology signatures) the treatments were split by compounds. Splitting by compounds placed all dose–time combinations of treatments for a given compound either in the training or in the test set. This avoided situations in which a signature could be trained on samples treated with multiple dose–time combinations of a compound and evaluated on other dose–time combinations of the same compound. Evaluation was performed at the level of the treatment. We refer to this modified cross-validation procedure as split by compound, count by treatment. In cases where the labels were set according to the properties of the sample (e.g. signatures for histopathology and clinical chemistry end points where dose level and/or time point are critical to development of the phenotype), both the partitioning and the evaluation were carried out at the level of the sample; we refer to this cross-validation procedure as split by treatment, count by treatment.
Validity criteria
We defined two different validity cutoffs for signatures based on their anticipated use and the sensitivity of the expected user groups to false-positive or false-negative errors. A specificity ⩾95% and sensitivity ⩾50% was used for signatures assessing pharmacology, and specificity ⩾98% and sensitivity ⩾40% for signatures classifying toxicity end points.
Liver xenobiotic response gene set
An impact table for all genes appearing in the 34 unique signatures recomputed without gene pre-selection is presented (Supplementary information S8). Genes were sorted according to the sum of impacts across all signatures. This sorted list is referred to in the text as the impact-based list. The top 200 genes from this list are referred to as the liver xenobiotic response gene set.
Identification of NGSs
For a given end point, all available gene variables (i.e. no feature reduction) were submitted to the SPLP algorithm. If, upon cross-validation, the performance of the resulting signature exceeded LOR=4, the highest impact genes participating in the signature were set aside from the data set. The signature was re-derived and the procedure was repeated until the performance of the signature dropped below LOR=4. The union of all the set-aside gene sets was the NGS. We have observed that on average less than half the genes contribute more than 90% of the impact in any given signature. To focus the algorithm on the genes contributing most to the signature, for each cycle genes were ranked by impact, and the highest ranked genes, accounting for at least 90% of the total impact, were designated as the most important genes in the signature and set aside. GO analysis of the stripped gene set was performed as follows: genes were arranged in order of stripping cycles. GO analysis was performed on a series of overlapping windows of 50 genes with increments of 25. GO term enrichment was calculated for each window and for each GO term using Fisher's exact test.
Supplementary Material
Supplementary Information
Supplementary Material 1
Supplementary Material 2
References
- Alcolado R, Arthur MJ, Iredale JP (1997) Pathogenesis of liver fibrosis. Clin Sci (Lond) 92: 103–112 [DOI] [PubMed] [Google Scholar]
- Allison DB, Cui X, Page GP, Sabripour M (2006) Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet 7: 55–65 [DOI] [PubMed] [Google Scholar]
- El Ghaoui L, Lanckriet GRG, Natsoulis G (2003) Robust classifiers with interval data. Report no. UCB/CSD-03-1279 Computer Science Division (EECS), University of California, Berkeley, CA
- Elrick M, Kramer J, Alden C, Blomme E, Bunch R, Cabonce M, Curtiss S, Kier L, Kolaja K, Rodi C (2005) Differential display in rat livers treated for 13 weeks with phenobarbital implicates a role for metabolic and oxidative stress in nongenotoxic carcinogenicity. Toxicol Pathol 33: 118–126 [DOI] [PubMed] [Google Scholar]
- Eng FJ, Friedman SL (2000) Fibrogenesis I. New insights into hepatic stellate cell activation: the simple becomes complex. Am J Physiol Gastrointest Liver Physiol 279: G7–G11 [DOI] [PubMed] [Google Scholar]
- Gabele E, Brenner DA, Rippe RA (2003) Liver fibrosis: signals leading to the amplification of the fibrogenic hepatic stellate cell. Front Biosci 8: d69–d77 [DOI] [PubMed] [Google Scholar]
- Ganter B, Tugendreich S, Pearson CI, Ayanoglu E, Baumhueter S, Bostian KA, Brady L, Browne LJ, Calvin JT, Day GJ, Breckenridge N, Dunlea S, Eynon BP, Furness LM, Ferng J, Fielden MR, Fujimoto SY, Gong L, Hu C, Idury R et al. (2005) Development of a large-scale chemogenomics database to improve drug candidate selection and to understand mechanisms of chemical toxicity and action. J Biotechnol 119: 219–244 [DOI] [PubMed] [Google Scholar]
- Gnainsky Y, Kushnirsky Z, Bilu G, Hagai Y, Genina O, Volpin H, Bruck R, Spira G, Nagler A, Kawada N, Yoshizato K, Reinhardt DP, Libermann TA, Pines M (2007) Gene expression during chemically induced liver fibrosis: effect of halofuginone on TGF-beta signaling. Cell Tissue Res 328: 153–166 [DOI] [PubMed] [Google Scholar]
- Hamadeh H, Bushel P, Jayadev S, Martin K, DiSorbo O, Sieber S, Bennett L, Tennant R, Stoll R, Barrett J (2002) Gene expression analysis reveals chemical-specific profiles. Toxicol Sci 67: 219. [DOI] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Eisen MB, Alizadeh A, Levy R, Staudt L, Chan WC, Botstein D, Brown P (2000) ‘Gene shaving' as a method for identifying distinct sets of genes with similar expression patterns. Genome Biol 1: RESEARCH0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman J (2001) Elements of Statistical Learning: Data Mining, Inference and Prediction. New York, USA: Springer-Verlag [Google Scholar]
- Heinloth A, Irwin R, Boorman G, Nettesheim P, Fannin R, Sieber S, Snell M, Tucker C, Li L, Travlos G (2004) Gene expression profiling of rat livers reveals indicators of potential adverse effects. Toxicol Sci 80: 193. [DOI] [PubMed] [Google Scholar]
- Huang Q, Jin X, Gaillard ET, Knight BL, Pack FD, Stoltz JH, Jayadev S, Blanchard KT (2004) Gene expression profiling reveals multiple toxicity endpoints induced by hepatotoxicants. Mutat Res 549: 147–167 [DOI] [PubMed] [Google Scholar]
- Ioannides C (2002) Enzyme Systems that Metabolise Drugs and Other Xenobiotics. Chichester, New York, USA: John Wiley & Sons [Google Scholar]
- Iredale JP (2007) Models of liver fibrosis: exploring the dynamic nature of inflammation and repair in a solid organ. J Clin Invest 117: 539–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Liu J, Waalkes M, Kang YJ (2004) Changes in the gene expression associated with carbon tetrachloride-induced liver fibrosis persist after cessation of dosing in mice. Toxicol Sci 79: 404–410 [DOI] [PubMed] [Google Scholar]
- Kim BS, Margolin BH (1999) Prediction of rodent carcinogenicity utilizing a battery of in vitro and in vivo genotoxicity tests. Environ Mol Mutagen 34: 297–304 [PubMed] [Google Scholar]
- Lamb J, Crawford E, Peck D, Modell J, Blat I, Wrobel M, Lerner J, Brunet J, Subramanian A, Ross K (2006) The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313: 1929. [DOI] [PubMed] [Google Scholar]
- Leveille CR, Arias IM (1993) Pathophysiology and pharmacologic modulation of hepatic fibrosis. J Vet Intern Med 7: 73–84 [DOI] [PubMed] [Google Scholar]
- Liu Y, Ringner M (2004) Multiclass discovery in array data. BMC Bioinformatics 5: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy CT, Irwig L, Mistry K, Cable G, Kim BS, Margolin BH (2004) Accuracy of diagnostic tests read with and without clinical information: a systematic review. Meta-analysis of prostate-specific antigen and digital rectal examination as screening tests for prostate carcinoma. Prediction of rodent carcinogenicity utilizing a battery of in vitro and in vivo genotoxicity tests. JAMA 292: 1602–1609 [DOI] [PubMed] [Google Scholar]
- Matzuk MM, Lu N, Vogel H, Sellheyer K, Roop DR, Bradley A (1995) Multiple defects and perinatal death in mice deficient in follistatin. Nature 374: 360–363 [DOI] [PubMed] [Google Scholar]
- Michiels S, Koscielny S, Hill C (2005) Prediction of cancer outcome with microarrays: a multiple random validation strategy. Lancet 365: 488–492 [DOI] [PubMed] [Google Scholar]
- Mistry K, Cable G (2003) Meta-analysis of prostate-specific antigen and digital rectal examination as screening tests for prostate carcinoma. J Am Board Fam Pract 16: 95–101 [DOI] [PubMed] [Google Scholar]
- Natsoulis G, El Ghaoui L, Lanckriet GR, Tolley AM, Leroy F, Dunlea S, Eynon BP, Pearson CI, Tugendreich S, Jarnagin K (2005) Classification of a large microarray data set: algorithm comparison and analysis of drug signatures. Genome Res 15: 724–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie A, McMillian M, Brandon Parker J, Leone A, Bryant S, Yieh L, Bittner A, Nelson J, Carmen A, Wan J (2006) Predictive toxicogenomics approaches reveal underlying molecular mechanisms of nongenotoxic carcinogenicity. Mol Carcinog 45: 914–933 [DOI] [PubMed] [Google Scholar]
- Peters JM, Cattley RC, Gonzalez FJ (1997) Role of PPAR alpha in the mechanism of action of the nongenotoxic carcinogen and peroxisome proliferator Wy-14,643. Carcinogenesis 18: 2029–2033 [DOI] [PubMed] [Google Scholar]
- Quackenbush J (2001) Computational analysis of microarray data. Nat Rev Genet 2: 418–427 [DOI] [PubMed] [Google Scholar]
- Ramm GA, Carr SC, Bridle KR, Li L, Britton RS, Crawford DH, Vogler CA, Bacon BR, Tracy TF (2000) Morphology of liver repair following cholestatic liver injury: resolution of ductal hyperplasia, matrix deposition and regression of myofibroblasts. Liver 20: 387–396 [DOI] [PubMed] [Google Scholar]
- Simon R, Radmacher MD, Dobbin K, McShane LM (2003) Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. J Natl Cancer Inst 95: 14–18 [DOI] [PubMed] [Google Scholar]
- Slatter J, Templeton I, Castle J, Kulkarni A, Rushmore T, Richards K, He Y, Dai X, Cheng O, Caguyong M (2006) Compendium of gene expression profiles comprising a baseline model of the human liver drug metabolism transcriptome. Xenobiotica 36: 938–962 [DOI] [PubMed] [Google Scholar]
- Takahara Y, Takahashi M, Wagatsuma H, Yokoya F, Zhang QW, Yamaguchi M, Aburatani H, Kawada N (2006) Gene expression profiles of hepatic cell-type specific marker genes in progression of liver fibrosis. World J Gastroenterol 12: 6473–6499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsunomiya T, Okamoto M, Hashimoto M, Yoshinaga K, Shiraishi T, Tanaka F, Mimori K, Inoue H, Watanabe G, Barnard GF, Mori M (2004) A gene-expression signature can quantify the degree of hepatic fibrosis in the rat. J Hepatol 41: 399–406 [DOI] [PubMed] [Google Scholar]
- Varma S, Simon R (2006) Bias in error estimation when using cross-validation for model selection. BMC Bioinformatics 7: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring J, Ulrich R (2000) The impact of genomics-based technologies on drug safety evaluation. Annu Rev Pharmacol Toxicol 40: 335–352 [DOI] [PubMed] [Google Scholar]
- Zhu HJ, Burgess AW (2001) Regulation of transforming growth factor-beta signaling. Mol Cell Biol Res Commun 4: 321–330 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Material 1
Supplementary Material 2