Abstract
We have constructed a collection of single-gene deletion mutants for all dispensable genes of the soil bacterium Acinetobacter baylyi ADP1. A total of 2594 deletion mutants were obtained, whereas 499 (16%) were not, and are therefore candidate essential genes for life on minimal medium. This essentiality data set is 88% consistent with the Escherichia coli data set inferred from the Keio mutant collection profiled for growth on minimal medium, while 80% of the orthologous genes described as essential in Pseudomonas aeruginosa are also essential in ADP1. Several strategies were undertaken to investigate ADP1 metabolism by (1) searching for discrepancies between our essentiality data and current metabolic knowledge, (2) comparing this essentiality data set to those from other organisms, (3) systematic phenotyping of the mutant collection on a variety of carbon sources (quinate, 2-3 butanediol, glucose, etc.). This collection provides a new resource for the study of gene function by forward and reverse genetic approaches and constitutes a robust experimental data source for systems biology approaches.
Keywords: Acinetobacter baylyi ADP1, deletion mutants, essential genes, growth phenotype, metabolism, systems biology
Introduction
The genome sequences of approximately 500 bacteria have already been released in public databases, providing a mine of information that contributes to the understanding of prokaryotic physiology. However, the function of a large fraction of these bacterial genes is still unknown. Experimental and in silico analyses have elucidated the role of numerous genes although about 13.9% of Escherichia coli genes have no assigned function and an additional 32% have only a predicted function (Riley et al, 2006). It is thus likely that a number of biological processes remain to be understood, and that the identification of gene function represents the next big challenge for the forthcoming years. Furthermore, many processes are not ubiquitous and, although important, are only shared by a subset of bacterial species (or strains) that are not well-studied models. Understanding and modelling the complexity of a living organism require global elucidation of gene function as well as the identification of the essential genes. For many years, the analysis of mutant phenotypes has been a powerful tool for understanding gene function, but until recently, the production of mutants has been performed on a gene-to-gene basis. Recently, the availability of complete genome sequences has permitted the construction of large-scale collections of mutants. Two different methods have been used to construct mutant libraries on a genomic scale. The most frequently used method has been a random approach by whole-genome transposon mutagenesis such as for Haemophilus influenzae (Akerley et al, 2002), Pseudomonas aeruginosa PA14 (Liberati et al, 2006) and PA01 (Jacobs et al, 2003), E. coli (Gerdes et al, 2003) and Corynebacterium glutamicum (Suzuki et al, 2006). An alternative strategy has been represented by directed approaches such as the genome-wide gene-by-gene transposon mutagenesis (Kang et al, 2004) or the complete gene deletion method used for Saccharomyces cerevisiae (Giaever et al, 2002), Bacillus subtilis (Kobayashi et al, 2003) and E. coli (Baba et al, 2006). As the random approach is both cost and time effective, it has progressively become the most popular one, although it suffers from three main drawbacks: (1) due to the heterogeneous distribution of transposon insertions along the chromosome, the production of mutants for each nonessential gene turns out to be difficult, (2) the insertion of a transposon within a gene is not strictly correlated with gene inactivation, (3) the growth of each mutant occurs in a mixed population, which leads to the counter-selection of those with decreased fitness. The more laborious and expensive directed approaches do not suffer from these limitations and appear to be the least error-prone (Gerdes et al, 2006).
Despite numerous studies on genes and the enzymatic activities of their corresponding proteins, there are still gaps in some metabolic pathways, and a large number of genes for which function has yet to be assigned. Recently, a complete collection of single-gene knockout mutants of E. coli has been reported by Keio University (Baba et al, 2006). This resource constitutes a powerful tool for the elucidation of parts of metabolism that remain to be characterized. However, as E. coli lives in a very specific environment (e.g., the human gut), it is important to extend the analysis of prokaryotic metabolism to other organisms living in different environments.
Species of genus Acinetobacter (a γ-proteobacterium) commonly found in aquatic and soil environments are capable of utilizing a very diverse range of compounds, such as phytochemicals (e.g., aromatic compounds), as sources of carbon/energy. Furthermore, their robust metabolism is responsible for a high capacity for adaptation. For this reason, they have aroused a growing interest because of the possible biotechnological and environmental applications. Some strains of this genus are indeed known to be involved in the biodegradation of a number of different pollutants (biphenyl, chlorinated biphenyl, benzoate, crude oil, and so on) and in the removal of phosphate or heavy metals. They are also well represented in the production of a number of extra- and intracellular economical products (lipases, proteases, cyanophycine, bioemulsifiers, and so on) and several kinds of biopolymers (Abd El-Haleem, 2003).
Acinetobacter baylyi ADP1 (ADP1) has been considered particularly interesting because of its remarkable competence for natural transformation. This highly transformable strain was obtained by mutagenesis of a strain that had been isolated from soil. ADP1 was sequenced and annotated at Genoscope with special focus on reconstruction of metabolism (Barbe et al, 2004). Annotation data are available through the Magnifying Genome platform (MaGe; http://www.genoscope.fr/agc/mage/; Vallenet et al, 2006). The expert annotation is an ongoing process, and updated releases of the annotation are regularly deposited in public databases. ADP1 has a relatively small genome (3.6 Mb) in which genes encoding most catabolic functions are clustered in several genetic islands (Young et al, 2005). Moreover, it is an ideal organism for high-throughput genetic analysis (Metzgar et al, 2004), as it shows a high competence for natural transformation (Palmen and Hellingwerf, 1997) and can be easily manipulated by homology-directed recombination with linear fragments (de Vries and Wackernagel, 2002). In nature, new genes of functional interest can be recovered in ADP1 using DNA from strains isolated from soil or other natural environments. ADP1, a strict aerobe, shares about one-third of its genes with E. coli, thus providing a convenient and complementary model for studying metabolism for two major reasons: (1) it does not show metabolic ambiguity (aerobic/anaerobic), which sometimes leads to difficult interpretations, and (2) it has been known to present important metabolic differences with E. coli due to the specificity of their respective ecological niches (Juni, 1978; Barbe et al, 2004; Young et al, 2005). Moreover, the presence of a smaller number of genes in ADP1 genome compared with other bacteria (around 3300 genes versus ∼4000 genes for E. coli and ∼5600 genes for P. aeruginosa) suggests an equally smaller number of redundant genes, which would increase the detection of single-gene deletion phenotypes.
Here, we report the construction of a nearly complete collection of ADP1 mutants by single-gene deletion on minimal media. Of the 3197 annotated protein-encoding genes in ADP1, essential/dispensable status was assigned for 3093 genes (∼97% of the total). We have started to explore ADP1 metabolism by analyzing these mutant data with respect to our current knowledge of its metabolism, based on the genome annotation, and by comparison with essentiality data from other organisms, namely E. coli and a phylogenetically closer bacterium, P. aeruginosa. Genomic comparative analyses with diverse genomes have been used as a complementary approach that should facilitate the prediction of the physiological function of genes located in synteny in other bacterial genomes. Furthermore, a number of mutants correspond to genes involved in catabolic processes. Indeed, when the whole mutant collection is grown on a defined metabolite as carbon or nitrogen source, the genes involved in its assimilation (enzymes, regulators, transporters, etc.) will then appear as essential genes. To characterize ADP1 catabolic pathways, we have thus initiated a systematic phenotyping of the mutant collection grown on diverse carbon sources.
Results and discussion
Construction of the ADP1 mutant collection
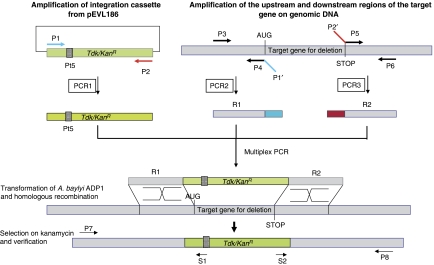
The approach used for gene deletion in ADP1 has already been described (Metzgar et al, 2004) and is based on ADP1 natural competence for transformation combined with homology-directed recombination with linear fragments. The gene deletion strategy, summarized in Figure 1, comprises the following steps: (1) a cassette carrying both positive (KanR) and negative (tdk gene used to generate unmarked deletions and to construct double mutants) selection markers was amplified by PCR amplification; (2) two approximately 300 bp regions (R1 and R2) located upstream and downstream from the targeted gene were also amplified using primers that included 5′-extension complementary to the primers used for the cassette amplification; (3) these three PCR products were then combined and re-amplified to generate a linear fragment containing the cassette flanked by the two specific regions; (4) the resulting product was used directly to transform ADP1 cells selected on minimal medium agar plates supplemented with kanamycin.
Figure 1.
Method of construction of the single-gene deletion mutants by creation of a spliced PCR integration cassette. P1–P6 are used for integration cassette construction and P7, P8, S1 and S2 for verifications. The kanR integration cassette is obtained by PCR amplification using P1 and P2 primers on pEVL186 DNA template. The flanking regions, specific for the target gene, are amplified on wild-type DNA template by P3/P4 primers (R1 region) and P5/P6 primers (R2 region). Designations followed by a prime (′) represent reverse complement sequences. The primers P7 and P8 are used for external PCR verification of the correct replacement of the targeted gene by the integrative cassette. The primers S1 and S2 located within the kanR cassette are used to sequence junctions of the cassette on the P7/P8 PCR product.
Selection of primers
As a general rule, the primers named P4 and P5 were designed to delete the whole gene from the start to the stop codons (Figure 1). For overlapping genes or genes separated by a short intergenic region, these primers were designed to preserve the expression of the two flanking genes (see Materials and methods). According to the genome annotation, 21% of ADP1 genes are overlapping and 70% of these overlaps span less than nine nucleotides. Moreover, 36% of the genes are separated by intergenic regions shorter than 20 nucleotides.
Choice of the selective medium for mutant library construction
Four different media supplemented with kanamycin (LB, 2YT, YT, minimal medium supplemented with succinate) were tested for deletion on 96 genes (one 96-well plate) and the number of mutants obtained was scored for each condition. Surprisingly, a higher rate of mutant recovery was obtained on minimal medium, which may suggest that the soil bacterium ADP1 is adapted to a lifestyle in which carbon sources can be rare and scarce. This could influence its competence and ability to recover from the transformation process. To optimize our mutation screen, we have therefore chosen to realize the mutant library on the minimal medium supplemented with succinate, an efficient carbon source for ADP1, which is directly assimilated through central metabolism. A number of mutants that could not be obtained on minimal medium (e.g., genes involved in the biosynthesis of amino acids, cofactors, and so on), will be recovered on the same medium with the ad hoc supplementation.
Construction and validation of deletion mutants
For each gene deletion experiment, we picked two KanR colonies and re-isolated each of them twice. Two correct clones of each mutant corresponding to nonessential genes (dispensable genes) were obtained in the first set of experiments in about 90% of the cases. A second set of mutations were produced in all cases for which mutants were not obtained, the amplification of at least one PCR product failed or the second clone was missing. Finally, the mutants that could not be obtained in the high-throughput process were reattempted individually (up to five times for genes with unknown function).
Mutants were checked for correct structure and size by three PCR amplifications combining locus-specific, kanamycin (P1/P6 and P3/P2) and external primers (P7/P8) (Figure 1). A clone was validated if all three PCR products had the correct size. However, in some cases, a clone was validated with only two correct PCR products if one of the PCR products generated by either locus-specific or kanamycin primers was missing. The analysis of the PCR products generated with primers P7/P8 resulted in the detection of 196 mutants, which showed two amplification products, one corresponding to the genomic region with the intact target gene and the other to the genomic region harboring the correct replacement of the targeted gene by the cassette. These mutants are called ‘Double Band' mutants (DB mutants). Using a Comparative Genomic Hybridization microarray, we have shown that 9 out of 10 DB mutants tested present a duplication ranging from 300 kb to more than 1 Mb (Gyapay et al, in preparation). This result could be explained by the high plasticity of ADP1 genome as shown previously by Reams and Neidle (2004) with the construction of mutants carrying large tandem head-to-tail repeats (less than 260 kb). For the construction of the Keio collection (Baba et al, 2006), mutants carrying both a bona fide deletion and an intact copy of the deleted gene localized elsewhere in the genome were suggested to be the cause of the rare recovery of mutants for essential genes (frequency estimated to less than 1%). As the frequency was significantly higher in our case (∼7.5% of the mutants), we assessed the presence/absence of the targeted gene using internal primers on a subset of 360 mutants including (1) 220 mutants with clones appearing after more than 3 days of growth, (2) 120 mutants corresponding to genes with an ortholog described as essential in the analysis of other mutant collections, i.e. in E. coli (Gerdes et al, 2003; Baba et al, 2006), P. aeruginosa (Jacobs et al, 2003; Liberati et al, 2006) or B. subtilis (Kobayashi et al, 2003) and (3) 20 mutants flanked by essential genes in ADP1. We confirmed the deletion of the targeted gene in 350 mutants, whereas it was still present in 10 mutants. As these 10 genes have a size similar to the integration cassette (∼1.7 kb), the two P7/P8 PCR products are thus undistinguishable by electrophoresis. These 10 real DB mutants thus raise to 206, the total number of DB mutants.
To double-check the integration sites, the vast majority of PCR products obtained with primers P7/P8 were sequenced using internal primers, S1 and S2, located within the cassette (Figure 1). We detected an incorrect structure for a small gene, which showed the integration of a cassette lacking the R2 flanking region beside the targeted gene.
Preliminary analysis of the collection
From the 3310 coding sequences (CDS) of ADP1, 113 genes previously annotated as insertion sequences (IS), gene remnants and doubtful CDSs were discarded from the study, while the remaining 3197 were targeted for deletion. Primers could not be designed using the standard automated procedure for 30 genes and will later be developed manually. The gene deletion procedure was thus carried out on a set of 3167 genes.
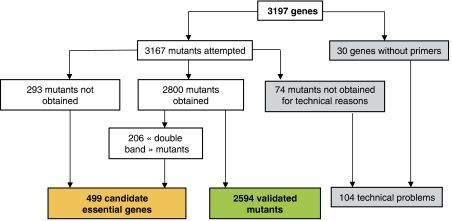
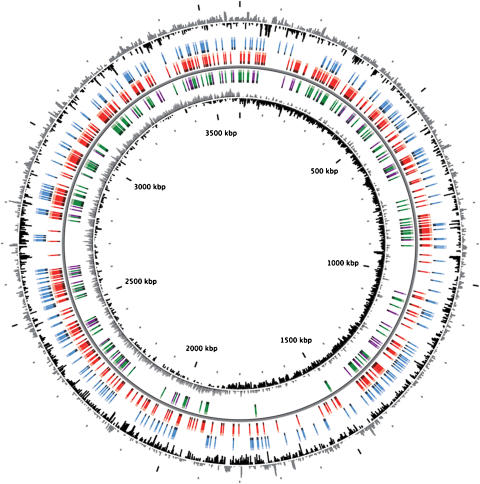
We generated a total of 2594 mutants with the correct structure (Figure 2) and obtained two independent clones for 92% of them. About 85% of the mutants were obtained after one day of incubation and 92% of them grew in less than 3 days. Mutants could not be obtained or were produced with an incorrect structure (e.g., DB mutants) for 499 genes (15.6% of the targeted genes). These genes thus represent the set of candidate genes essential for growth on a minimal medium supplemented with succinate as the sole carbon source (Figure 3; Supplementary Table I). These essential genes are distributed all along the chromosome except in four regions: the two archipelago catabolic islands, the prophagic region and a siderophore region. In ADP1, CDSs show an asymmetry in their distribution along the chromosome between the leading and the lagging strands (60 versus 40%). The distribution of essential genes is not biased between the two strands (15 versus 13% of the genes). The mutant collection is currently being extended to mutants of biosynthetic genes using media supplemented by the appropriate component (arginine, histidine, leucine, methionine, tryptophan), which has already resulted in the production of 30 auxotrophic mutants. The 74 mutants that could not be generated due to various technical problems will be produced manually.
Figure 2.
Flow diagram of the ADP1 mutant collection.
Figure 3.
Chromosomal representation of the ADP1 mutant collection. Circles display (from the outside): (1) GC percent deviation (GC window—mean GC) in a 1000-bp window. (2) CDSs in blue are classified into three biosynthesis TIGR role categories: (i) amino acid, (ii) cofactors, prosthetic groups, and carriers, and (iii) purines, pyrimidines, nucleosides, and nucleotides. (3) CDSs in red corresponds to ADP1 essential genes. (4) ADP1 CDSs in purple are sharing orthologs with E. coli K12 genes, described as essential (Keio collection, in green) or conditionally essential (mutants growing very poorly on glucose minimal media (OD<0.1)). (5) GC skew (G+C/G−C) in a 1000-bp window.
As mentioned above, the pool of 499 essential genes includes 206 DB mutants (41% of essential genes). A large proportion of the genes resulting in DB mutants present evidence of essentiality. Indeed, 121 have orthologs in E. coli and P. aeruginosa, and 87% of them correspond to candidate essential genes in at least one of these two organisms (Supplementary Table II; Supplementary Annex 1). To investigate further the essentiality of these genes, we have started to generate the mutants corresponding to the genes involved in amino-acid biosynthesis by complementing our basic medium with the suitable amino acid. Mutants were obtained with a correct structure, and their amino-acid auxotrophy was confirmed for all the genes tested so far (metE, metR, metW, argB, argC, argF, arG, argJ, leuB, leuC, leuD, trpF).
Essentiality studies: a comparison between ADP1 and other prokaryotes
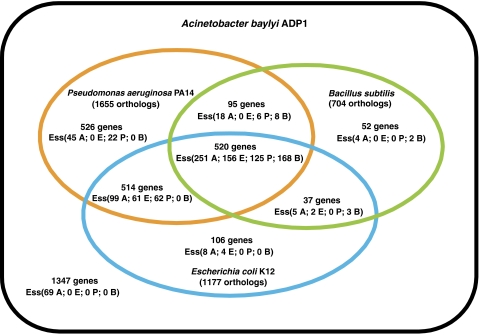
A comparative analysis of essential genes across different organisms should reveal differences between metabolic networks and, therefore, define the set of essential genes shared by bacteria living in diverse environments. It should be noted that interpretations of differences between pools of essential genes have to be performed cautiously, as these data are crucially dependent on the technique and the medium used for generating the mutant collection (Gerdes et al, 2006). Only restricted sets of gene essentiality data are available to date. A general analysis on orthologous gene essentiality data between ADP1 and other bacteria (E. coli, P. aeruginosa and B. subtilis) is summarized in Figure 4. Among the 499 candidate essential genes, 350 are shared either by the three other organisms, or by E. coli and P. aeruginosa. Moreover, 69 ADP1 genes with no obvious orthologs in these organisms happen to be essential and encode proteins of unknown or putative function (82%).
Figure 4.
Analysis of orthologous gene essentiality data between ADP1 and three other bacteria (E. coli, P. aeruginosa and B. subtilis). The number of orthologous genes for each comparison and the number of genes that are essential in the four organisms, are reported as Ess(A: ADP1, E: E. coli, P: P. aeruginosa, B: B. subtilis).
Functional categories
Of the 499 candidate essential genes, 397 (79.6%) correspond to known genes, 56 (11.2%) to genes with a putative function and 46 (9.2%) to genes of unknown function (Table I). These data are consistent with those of previous reports in which the majority of essential genes are known genes (Baba et al, 2006; Liberati et al, 2006). However, the number of essential genes is far higher in our analysis than those estimated in previous studies of other bacterial mutant collections such as E. coli (303 mutants or 7% of the collection; Baba et al, 2006), P. aeruginosa (335 mutants or 6% of the collection; (Liberati et al, 2006)) and B. subtilis (271 mutants or 6.6% of the collection; Kobayashi et al, 2003). All these collections were constructed on rich media, and their sets of essential genes are thus restricted to genes required for viability under favorable conditions, whereas in the present study, the genes involved in the biosynthesis of essential compounds, which are necessary for growth on a minimal medium, should be added to the aforementioned number of essential genes (Supplementary Table III).
Table 1.
Summary of the A. baylyi ADP1 knockout collection
| Number of genes | Number of attempted mutants | Number of mutants | Number of essential genes | |
|---|---|---|---|---|
| Protein with known function | 1160 | 1150 | 731 | 397 |
| Protein with putative function | 928 | 920 | 835 | 56 |
| Conserved hypothetical proteinsa | 889 | 881 | 825 | 36 |
| Hypothetical proteinsb | 220 | 216 | 203 | 10 |
| All categories | 3197 | 3167 (97%) | 2594 (82%) | 499 (16%) |
aHomologs of previously reported genes of unknown function.
bNo homology to any previously reported sequences.
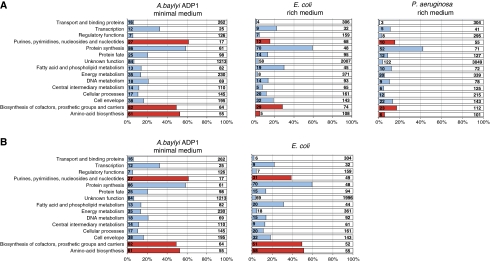
ADP1 CDS were classified into different functional roles according to the TIGR classification. The distribution of the essential genes of ADP1, E. coli (Baba et al, 2006) and P. aeruginosa (Liberati et al, 2006) into functional categories is shown in Figure 5A. For three functional categories (biosynthesis of amino acids, cofactors/prosthetic groups and purines/pyrimidines/nucleosides/nucleotides), the percentage of essential genes is significantly higher in ADP1 than in the other species, which could reflect the different media used in the construction of the libraries (minimal versus rich medium). We, therefore, compared our data to an extended set of essential genes that included conditional essential genes (or CEG; mutants growing very poorly (OD<0.1) on minimal medium supplemented with glucose), which was available for E. coli in the Keio collection (EEGS for ‘Extended Essential Gene Set'; see Figure 5B). For two of the three categories of essential genes that appeared significantly lower in E. coli, the comparison with the EEGS resulted in a ratio and a number of essential genes that were very similar to the ones observed in ADP1. For the last category (synthesis of purines, pyrimidines, nucleosides and nucleotides), although the number of essential genes is similar, the percentage difference is only reduced, mainly because a higher number of genes were assigned to this category in E. coli (80 versus 44 in ADP1). For example, the salvage pathway of nucleosides and nucleotides, which does not exist in ADP1, involves at least 20 genes in E. coli. The other functional categories are not notably influenced by the medium used for the mutant construction.
Figure 5.
Comparison of ADP1, E. coli and P. aeruginosa essentiality data according to TIGR role classification on (A) the medium used to obtain the mutants, (B) on minimal medium. White and blue areas correspond to nonessential and essential genes, respectively. The red area concerns the main categories notably impacted by the medium used for the mutant construction.
Comparison with Pseudomonas aeruginosa essential genes
P. aeruginosa is phylogenetically the closest organism to ADP1 for which collections of transposon mutants are available (Jacobs et al, 2003; Liberati et al, 2006). These mutant collections were obtained on rich medium (LB) and a set of 335 essential genes was defined as genes with no transposon integration in either library. P. aeruginosa and ADP1 share 1655 orthologous genes, including 209 that are described as essential in P. aeruginosa, and have essentiality data in ADP1. A large majority of these genes (80%) are also essential in ADP1. In contrast, 246 essential genes in ADP1 are described as dispensable in P. aeruginosa (Supplementary Table IV). Among these 246 genes, about 100 genes are associated with biosynthesis function (e.g., amino acids, cofactors, etc.) and are expected to be essential only on minimal media. Surprisingly, an unexpectedly high number of discrepancies (33 cases) were found for genes involved in protein synthesis. Most of these genes encode tRNA synthetases or ribosomal proteins that are essential for life as previously described in a number of organisms. They could thus represent the background of false dispensable genes detected with a random transposon mutagenesis approach.
Another example of the background caused by the use of a transposon library is described below and illustrates how a similar gene essentiality status between two mutant collections may conceal different features. In riboflavin biosynthesis, ribA and ribB encode a GTP cyclohydrolase and a riboflavin synthase, respectively. Both these genes and a bifunctional ribB/A gene are found in ADP1; ribB is dispensable as expected, but surprisingly ribA appears to be essential, suggesting that RibB/A does not complement the RibA function. In P. aeruginosa, where only ribA and ribB/A are present, ribA is essential, but despite the absence of ribB, ribB/A is unexpectedly reported as dispensable. A close examination of the insertion sites within this gene shows that all the insertions are located in the ribA region of ribB/A, whereas no insertion is found in the ribB region, revealing its essentiality. Taken together, these observations clearly show the essentiality of both RibA and RibB functions in these two bacteria and cast a doubt on the commonly annotated function of the ribA-like region of ribB/A. In agreement with our hypothesis, the genes ribB/A from both Helicobacter pylori strain P1 (Fassbinder et al, 2000) and Streptomyces davawensis (Grill et al, 2007) fail to complement an E. coli mutant lacking RibA function. This example illustrates the advantage of the gene deletion method for a better determination of multifunctional protein essentiality.
Comparison with Escherichia coli essential genes
The Keio collection of E. coli is the only library for which a detailed comparison of the essential gene data sets could be performed with accuracy, as both sets are based on single-gene knockout mutant collections and were obtained or profiled for growth on minimal media. A status has been assigned in our mutant collection for 1144 genes out of the 1177 orthologous genes between E. coli and ADP1. We compared our set of essential genes with the EEGS of the Keio collection, and the results are consistent in 88% of the cases (i.e., genes found to be essential or dispensable in both sets; see Table II). In comparison, Baba et al. (2006) have reported that 67% of their predicted essential genes overlap the 620 essential genes estimated by Gerdes et al. (2003) in their E. coli transposition mutant collection. This result highlights the importance of the techniques and the conditions used for the construction of the library in the prediction of essential genes.
Table 2.
Orthologous genes of A. baylyi ADP1 and Escherichia coli and their essentiality in E. coli
|
E. coli |
||
|---|---|---|
| Acinetobacter baylyi ADP1 | Dispensable genes | Essential genes (E+CEG) |
| Dispensable genes (781 genes)a | 745 | 36 (24 E+12 CEG) |
| Essential genes (363 genes)a | 98 | 265 (189 E+76 CEG) |
| All categories | 843 | 301 (213 E+88 CEG) |
E, essential gene from the Keio collection data corresponding to mutants not obtained on rich medium; CEG, conditional essential gene from the Keio collection data corresponding to mutants growing very poorly on glucose (OD<0.1); discrepancies between ADP1 and E.coli are written in bold characters.
aFor each category, the number of ADP1 genes that has an ortholog in E. coli is reported in brackets. This number corresponds to the sum of the number of dispensable and essential genes in E. coli.
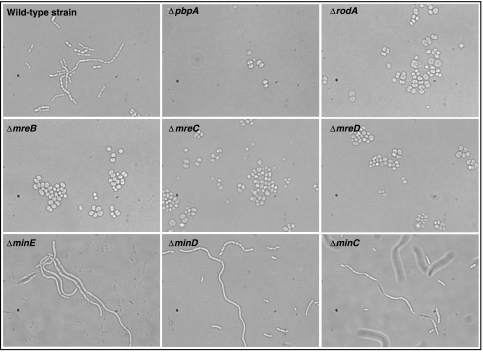
The essentiality status is different for 134 genes between the Keio and our collection (Table II; Supplementary Table V). Of these, 98 genes found to be essential in ADP1 are dispensable in E. coli, and 36 are dispensable in ADP1, but essential in E. coli. Although we cannot exclude that a few of these cases could be due to undetected technical problems that occurred during the deletion process or polar effect, these differences cannot be explained by major factors such as isofunctional genes. Therefore, these discrepancies mainly reflect the following points. (1) Lifestyle differences (strict aerobe/facultative aerobe) as shown for instance by the dispensability of five ATP synthase subunits in E. coli. (2) Various carbon sources (glucose versus succinate) used to obtain/grow the mutants as illustrated by the essentiality of both succinate dehydrogenase subunits and the succinate transporter in ADP1 or by that of phosphoenolpyruvate carboxylase (ppc) in E. coli clones grown on glucose (this gene is dispensable when the mutant is grown in the presence of an intermediate of the tricarboxylic acid cycle). (3) The presence of isofunctional genes such as the genes involved in the gluconeogenesis and the pentose phosphate pathway (gpmI, fbp, rpiA) in E. coli, and map, folD, icd, ribB, lip in ADP1. (4) The redundancy of metabolic pathways. This latter point is illustrated for example by the biosynthesis of iron-sulfur clusters (ISC). E. coli contains at least two well-described biosynthetic systems, ISC (Iron Sulfur Cluster) and SUF (sulfur assimilation) (Barras et al, 2005). Both isc and suf operons encode proteins with similar biochemical activities in vitro (Fontecave et al, 2005), and each pathway is dispensable in E. coli, as confirmed by the data of the Keio collection. In ADP1, similar to Pseudomonas species, only the isc operon is found in the genome. Moreover, the essentiality of the iscSUA-hscBA-fdx genes in these two species reinforces the hypothesis that only one pathway is active or even exists. (5) Variations in metabolic pathways, such as the linear/cyclic arginine biosynthetic pathway, are illustrated by the dispensability of argE and argA, which raises questions about their role in ADP1. (6) Physiological differences. For instance, a number of genes responsible for the formation of the rod-shaped cell and polar differentiation (mreBCD, minCDE, pbpA/mrdA and rodA/mrdB) are considered as essential in the Keio collection or the PEC database (except for minCDE; www.shigen.nig.ac.jp/ecoli/pecv3/). In E. coli, the cells depleted of mreB, mreC or mreD become spherical, enlarge and finally undergo lysis (Kruse et al, 2005). We have observed the same phenomenon in ADP1, in which the deletion of these genes also produces a dramatic change in cell morphology from the normal rod shape to a spherical form, but no lysis was observed after a 24h-culture (Figure 6). A spherical form was also observed after the deletion of pbpA and rodA genes in ADP1. Moreover, the deletion of minCDE involved in Z-ring formation (Zhou and Lutkenhaus, 2005) leads to the formation of giant long cells in ADP1 (Figure 6). In some other cases, the dispensability of genes in ADP1 will require further investigation, such as genes for the carbon storage regulator (csrA), ribonuclease III (rnc) and rmpA (the 50S ribosomal subunit protein L27), which are all described as essential in E. coli.
Figure 6.
Comparison of mutants corresponding to deletion of genes responsible for the formation of the rod shape or polar differentiation and wild-type strain phenotypes observed by microscopy (× 1000). The mreBCD, pbpA and rodA genes are involved in cell-shape determination in rod-shaped bacteria, whereas the minCDE genes are involved in cellular division. The cells were grown overnight in liquid media (minimal medium complemented with succinate as carbon source).
Exploration of gene function and pathways through the mutant status
Acinetobacter genome annotation is a prerequisite for the expert reconstruction of metabolic pathways, which can provide us with a global view of metabolism. The comparison of the current knowledge of metabolism with our experimental data set raises questions about the validity of annotation-based knowledge. However, we found a number of exceptions that allowed us to formulate new hypotheses, including the two examples described below. To help us in this comparison, an ADP1 Genome Browser was implemented to present data of our mutant collection in their genomic context (KEGG and BioCyc metabolic pathways, SEED subsystems, operon predictions, orthologous genes and essentiality data from several organisms, and phenotyping data from high-throughput experiments), which is available (http://www.genoscope.cns.fr/cgi-bin/ggb/thesaurus/gbrowse/thesaurus).
The first step in Coenzyme-A biosynthesis involves the formation of β-alanine from aspartate. The panD gene encodes the 1-aspartate decarboxylase activity, and is essential for growth on minimal medium in a large number of prokaryotes including E. coli. In ADP1 a panD mutant was obtained, suggesting either the presence of another gene that could perform this activity or an alternative pathway leading to β-alanine biosynthesis. Similarly, the dispensability of erythonate-4-phosphate dehydrogenase (pdxB) suggests the presence of an isofunctional gene or an alternative pathway resulting in the formation of the pyridoxal 5-phosphate cofactor.
In de novo biosynthesis of pyrimidine ribonucleotides, two dihydroorotases, ACIAD1150 (pyrC) and ACIAD1419, which share no homology, were identified. We could not delete pyrC, indicating that this gene cannot be complemented by the other dihydroorotase in vivo. In Pseudomonas, the presence of two functional dihydroorotases PyrC and PyrC2, the orthologs of ACIAD1150 and ACIAD1419, respectively, was experimentally demonstrated. It was shown that pyrC is constitutively expressed, whereas pyrC2 is expressed only on a pyrC mutant background due to its possible induction by carbamoyl L-aspartate accumulation (Brichta et al, 2004). In ADP1, we hypothesize that pyrC essentiality could be explained by a delay in pyrC2 induction, which would then be incompatible with the survival of the cell depleted in dihydroorotase activity during the construction of our mutant library.
Methionine biosynthesis
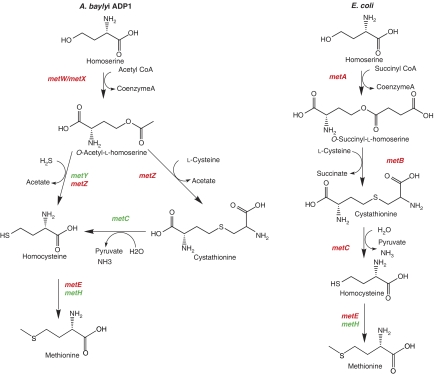
Prokaryotic organisms synthesize methionine through different biosynthetic pathways from homoserine (Figure 7). The first step corresponds to the acylation of homoserine using succinate or acetate to generate O-succinyl-L-homoserine mediated by metA in E. coli or O-acetyl-L-homoserine mediated by the concerted action of both metX and metW as in Pseudomonas syringae (Andersen et al, 1998) and Pseudomonas putida (Alaminos and Ramos, 2001). Little is known about metW, whose function remains unclear (Picardeau et al, 2003). In ADP1, we have shown that metX and metW mutants are auxotrophic for methionine (Table III) as in P. syringae. In addition, E. coli metA complements both mutants, confirming that metX and metW are involved in homoserine acylation. Surprisingly, we have shown that the overexpression of metX can complement not only the deletion of metX but also the deletion of metW or metX/metW, whereas metW can complement only the metW mutant (Table IV). These results strongly suggest that the constitutive expression of metX can compensate for the absence of metW and that MetW is essential for acetylation of homoserine in vivo through its direct or indirect action on MetX.
Figure 7.
Methionine biosynthesis. The first steps of methionine biosynthesis in ADP1 and E. coli. Essential genes are shown in red, whereas nonessential genes are in green.
Table 3.
Media used for the deletion of genes involved in methionine biosynthesis
| Mutants | Media |
|---|---|
| ΔmetX | Succinate+methionine |
| ΔmetW | Succinate+methionine |
| ΔmetX/metW | Succinate+methionine |
| ΔmetY | Succinate |
| ΔmetZ | Succinate+methionine |
| ΔmetC | Succinate |
| ΔmetR | Succinate+methionine |
| ΔACIAD3524 | Succinate+methionine |
| ΔmetE | Succinate+coenzyme-B12 |
| ΔmetH | Succinate |
Table 4.
Functional complementation experiments of A. baylyi ADP1 mutants involved in the acetylation of L-homoserine
| Genes used for complementation | ΔmetX | ΔmetW | ΔmetX/metW |
|---|---|---|---|
| pEVL174a | − | − | − |
| pEVL174/metXW | + | + | + |
| pEVL174/metAb | + | + | + |
| pEVL174/metX | + | + | + |
| pEVL174/metW | − | + | − |
aE. coli-A. baylyi ADP1 shuttle expression vector.
bmetA gene of E. coli.
The second step of methionine biosynthesis generates homocysteine by two major pathways in microorganisms: (1) the transsulfuration pathway through cystathionine (CTT) by the CTT γ-synthase (metB in Enterobacteria or metZ in Pseudomonales) and the CTT β-lyase (metC); (2) the sulfhydrylation pathway leading to the formation of homocysteine in one step by metY and/or metZ. Only a few sequenced bacteria have both pathways (Alaminos and Ramos, 2001; Hwang et al, 2002; Picardeau et al, 2003; Kobayashi et al, 2003), and, according to its genome annotation, ADP1 seems to have both. As expected, the deletion of metY or metC is not lethal but intriguingly, the metZ mutant is a methionine auxotroph, suggesting that MetY does not catalyze the direct sulfhydrylation in our conditions. Further analyses have been initiated to define whether MetZ catalyzes direct sulfhydrylation or this route is absent in ADP1.
The final step in the methionine pathway is performed by the methionine synthase encoded by B12 dependent metH or B12 independent metE. As expected, the metH mutant is prototrophic, whereas the metE mutant, obtained in presence of coenzyme-B12, is auxotrophic for methionine. In Enterobacteria, metE is usually flanked by the transcriptional regulator metR, whereas in ADP1, metR is located at another position in the genome, whereas a gene of unknown function (ACIAD3524) is found next to metE. We have shown that the ACIAD3524 mutant is auxotrophic for methionine, whereas prototrophy is restored by complementation with ACIAD3524. The colocalization of metE and ACIAD3524 is conserved in many proteobacteria, such as Pseudomonas (but not P. aeruginosa), Xanthomonas, Agrobacterium, Gluconobacter, Shewanella and Vibrio genera. These findings thus suggest a role for this gene in methionine biosynthesis or its regulation.
Ubiquinone biosynthesis
A second example of pathway exploration focuses on the ubiquinone biosynthesis through chorismate (Supplementary Figure 1), which has previously been described in E. coli (Meganathan, 2001) and seems to be widely shared by γ-proteobacteria. The gene encoding UbiC appears to be dispensable in both ADP1 and E. coli, which indicates the existence of at least one other enzyme to perform the chorismate pyruvate lyase reaction. Moreover, the presence of an essential gene of unknown function (ACIAD0383), which is localized between ubiB and ubiE in ADP1 (yigp in E. coli), suggests its potential implication in ubiquinone biosynthesis. In addition, the absence of 3-octaprenyl-4-hydroxybenzoate decarboxylase is particularly intriguing. In general, this activity is fulfilled by two isofunctional genes, ubiX and ubiD. Although these two genes are not colocalized, both of them are always present in a given genome (see ubiquinone subsystem on SEED platform; http://seed-viewer.theseed.org/). Nevertheless, some bacteria like Gluconobacter oxydans and Xanthomonas species do not have these genes, reinforcing the possibility of the presence of at least one other decarboxylase fulfilling the same function.
Analysis of gene function and pathways using growth phenotypes on several carbon sources
The nearly complete collection of mutants was analyzed for growth on different carbon sources. Out of the 220 carbon sources tested by Phenotype MicroArrays (PM1&2 plates of Biolog Inc-CA-, Bochner et al, 2001) and in-house growth experiments, 50 led to significant growth and were used to define our preliminary set of carbon sources. The growth profiling of approximately 2450 mutants was performed on both solid and liquid media (Supplementary Table VI).
Evaluation of this approach
To evaluate the specificity and sensitivity of the growth phenotyping approach for detecting genes involved in catabolic pathways, we tested two carbon sources assimilated through known pathways in ADP1. One of the best characterized catabolic pathways is the dissimilation of quinate, which occurs through protocatechuate, a central metabolite for a number of significant phytochemical degradation pathways (quinate, benzoate, vanillate, shikimate, and so on), through the β-ketoadipate pathway (Stanier and Ornston, 1973; Neidle et al, 1991). Quinate dissimilation is performed by 13 genes corresponding to 10 enzymatic activities and two transporters (Supplementary Figure 2). The results we obtained are consistent with the current knowledge on ADP1 quinate assimilation (Supplementary Annex 2). However, some mutants (e.g. mutants of NADH dehydrogenase subunits, panD) show a growth defect that is not explained by current knowledge of quinate metabolism and would thus require more investigation.
The glucose dissimilation has been described biochemically by Juni in 1978, and genes have been associated with these known enzymatic activities during the genome annotation process (Barbe et al, 2004). This pathway involves two transporters (glucose and gluconate) and five genes corresponding to five enzymatic activities (Supplementary Figure 3). Our phenotypic screening for growth on glucose confirms its dissimilation by periplasmic oxidation, leading to the formation of gluconate, which is further assimilated through the final steps of the Entner–Doudoroff pathway and confirms that this is the only route for glucose dissimilation (Supplementary Annex 2). Moreover, four mutants of genes of unknown function that are not able to grow on glucose require more investigation.
Catabolism of 2,3-butanediol
Using the growth phenotyping strategy, we have carefully explored the degradation of 2,3-butanediol, which is historically important for ADP1, as this strain was initially selected for its ability to use this compound as a carbon source. However, the catabolic pathway has not been entirely understood (for a review, see Xiao and Xu, 2007). Two different routes for 2,3-butanediol dissimilation have been described to date: (1) the 2,3-butanediol cycle involving 2,3-butanediol dehydrogenase and acetoin dehydrogenase (Juni and Heym, 1956), which generates acetate that is further oxidized and (2) the degradation of 2,3-butanediol via the acetoin dehydrogenase complex most likely generating acetyl-CoA and acetaldehyde (Xiao and Xu, 2007). To obtain a clearer picture of this pathway, all mutants were profiled for growth on minimal medium supplemented with 2,3-butanediol or acetate. A total of 21 mutants were reproducibly unable to grow on 2,3-butanediol, including 8 mutants unable to grow on acetate (Table V). These results clearly confirm that the degradation of 2,3-butanediol is dependent on acetate utilisation, as genes of the glyoxilic shunt (isocitrate lyase/ACIAD1084 and malate synthase/ACIAD2335) are essential for growth on both 2,3-butanediol and acetate. Moreover, a putative transcriptional regulator (ACIAD1082) is found to be essential for growth on the two carbon sources, suggesting its potential implication in the regulation of the glyoxilic shunt. Our findings also show the essentiality of genes clustered on the genome such as the putative transcriptional regulator of acetoin metabolism (ACIAD1014), which has no homology with acoR and three of the four subunits of the acetoin dehydrogenase complex (acoABC/ACIAD1017-1019). The dispensability of the acoL gene (ACIAD1020) encoding a dihydrolipoamide dehydrogenase could be explained by the presence of Lpd, an isofunctional enzyme involved in the pyruvate/oxoglutarate dehydrogenase complex. The deletion of the putative butanediol dehydrogenase (ACIAD1021) results in severe growth impairment of the mutant, indicating that another dehydrogenase may perform this function, albeit weakly. The data clearly show that the putative acetaldehyde dehydrogenase (ACIAD2018) is also involved in 2,3-butanediol degradation. Altogether these results strongly suggest that 2,3-butanediol is degraded into two C2 compounds, similar to in Bacillus subtilis (Lopez et al, 1975), Pelobacter carbinolicus (Oppermann et al, 1991) and Clostridium magnum (Kruger et al, 1994). Moreover, the acetoin dehydrogenase (budC/ACIAD1022) is present in ADP1 and its deletion does not impair growth on 2,3-butanediol nor on acetate, reinforcing the probable absence of the 2,3-butanediol cycle in ADP1. Furthermore, we have detected several other genes encoding proteins with putative functions that are essential for growth on 2,3-butanediol, but not on acetate. The implication of these genes in this specific catabolic pathway needs to be further analyzed to complete our understanding of 2,3-butanediol degradation.
Table 5.
Summary of 2,3 butanediol and acetate growth analysis based on high throughput data and confirmed by individual analysis
| Name | Gene product | 2,3 butanediol | Acetate |
|---|---|---|---|
| ACIAD0772 | Putative ATP-binding protein | − | + |
| ACIAD0808 | Putative protease | − | + |
| ACIAD1014 | Putative transcriptional regulator | − | + |
| ACIAD1017 | Acetoin:2,6-dichlorophenolindophenol oxidoreductase α subunit (acoA) | − | + |
| ACIAD1018 | Acetoin:2,6-dichlorophenolindophenol oxidoreductase β subunit (acoB) | − | + |
| ACIAD1019 | Dihydrolipoamide acetyltransferase (acoC) | − | + |
| ACIAD1021 | Putative (R,R)-butanediol dehydrogenase | Growth defect | + |
| ACIAD1052 | Putative membrane-bound protein in GNT I transport system | − | + |
| ACIAD1550 | Putative oxidoreductase, short-chain dehydrogenase/reductase family | − | + |
| ACIAD2018 | Aldehyde dehydrogenase (ald1) | − | + |
| ACIAD2084 | Putative transcriptional regulator | − | + |
| ACIAD2346 | Integration host factor (himA) | − | + |
| ACIAD3071 | Cysteine synthase B (cysM) | − | + |
| ACIAD3549 | γ-Glutamate-cysteine ligase (gshA) | − | + |
| ACIAD0273 | DnaK suppressor protein (dksA) | Growth defect | − |
| ACIAD0922 | Putative D-amino acid oxidase | − | − |
| ACIAD1082 | Putative transcriptional regulator | − | − |
| ACIAD1084 | Isocitrate lyase (aceA) | − | − |
| ACIAD1135 | Putative NADH pyrophosphatase (NUDIX hydrolase family) | − | − |
| ACIAD1951 | Putative thioesterase | − | − |
| ACIAD2236 | Putative bifunctional protein (dGTP-pyrophosphohydrolase; thiamine phosphate synthase) | − | − |
| ACIAD2335 | Malate synthase G (glcB) | − | − |
| ACIAD3469 | Putative two-component response regulator | − | − |
Growth phenotype on solid media
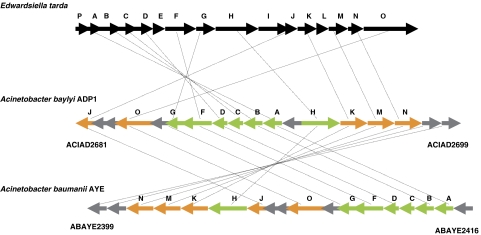
The analysis of our mutant collection for growth phenotypes on agar plates supplemented with various carbon sources has allowed the identification of mutants that generate colonies with a size over twice the average. This phenotype could suggest a membrane or cell adhesion disorder. The distribution of these genes, gathered in two main clusters in the genome, is shown in Supplementary Figure 4. One cluster is located between ACIAD0068 and ACIAD0102 and is composed of genes involved in exopolysaccharide biosynthesis (panel A). When grown in liquid medium, these mutants usually aggregate leading to an apparent null OD. The other cluster, ACIAD2681–2699 is characterized by a large number of genes of unknown function (panel B). Moreover, a closer study of this cluster has revealed that 12 of these genes present a genomic organization similar to the evp operon of Edwardsiella tarda (Figure 8), which encodes a type VI protein secretion system (Zheng and Leung, 2007). Furthermore, these ADP1 genes are also found in synteny in Acinetobacter baumanii AYE (Vallenet et al, 2008), Ralstonia solanaecerum, Dechloromonas aromatica and in many Burkholderia species showing homologies up to 50%. Mutants of four additional genes (ACIAD0852, ACIAD0854, ACIAD1205 and ACIAD2603) encoding proteins related to bacterioferritin have also produced larger colonies on solid media.
Figure 8.
Acinetobacter genomic organization of the evp homolgous gene cluster involved in a type VI protein secretion system in E. tarda. Genes in green are those homologous to E. tarda genes; genes in orange are those with a conserved domain (COG or PUF domain) with E. tarda. Genes in gray are genes with no known function annotated as conserved hypothetical proteins in Acinetobacter species.
Conclusion
We took advantage of the natural competence of ADP1 combined with homology-directed recombination with linear DNA to construct a complete collection of gene-by-gene deletion mutants selected on minimal medium. A total of 2594 genes covering 81% of the predicted genes of ADP1 were successfully disrupted, and 499 genes, including 47 genes of unknown function, were found to be essential for prototrophic life. The library is currently being completed for auxotrophic mutants, such as those of genes involved in amino-acid biosynthetic pathways. A simple examination of the essential/dispensable genes has highlighted several discrepancies between these experimental data, and the current knowledge of metabolic pathways, which has led to the identification of interesting new features and differences with known pathways in other bacteria. It should be noted that most of the discrepancies were automatically detected by using the computational model of the ADP1 metabolic network (Durot et al, in preparation), which confirmed the efficiency and importance of such in silico models to identify quickly the differences between our annotation-based knowledge of metabolism and a set of experimental data derived from a mutant library.
This collection also provides a powerful tool for the exploration of gene function, in particular, catabolic pathways through the analysis of growth phenotypes on defined compounds that can be assimilated. A number of essential genes for some catabolic pathways have already been identified while other pathways remain to be investigated using different carbon or nitrogen sources. Furthermore, the clustering of similar growth phenotypic profiles in the different mutants should allow the identification of a potential role for genes with unknown function by comparison with well-known genes (Ito et al, 2005; Dudley et al, 2005).
This new collection of mutants is also likely to be a valuable resource for other bacteria, as ADP1 shares a large number of genes with other bacteria. Genetic tools developed for ADP1, such as multiple gene deletions and the existence of E. coli/ADP1 shuttle vectors (Metzgar et al, 2004), should allow heterologous functional complementation. ADP1 could thus be an alternative and complementary model for the study of gene function of other bacteria, especially those that are not easily amenable to genetic manipulations.
In conclusion, this collection is not only important for the identification of gene function, alternative pathways and regulatory networks, but also for the genome-scale reconstruction of ADP1 metabolism based on high throughput phenotyping and essentiality data (Oh et al, 2007; Durot et al, in preparation). Moreover, to precisely analyze the effect of genetic perturbations at a metabolite level, we have initiated a large-scale and systematic study of our mutant collection using mass spectrometry aiming at collecting information on the metabolome of several hundred mutants. The comparison of these metabolic profiles will provide information on the effect of a well-defined mutation on quantitative/qualitative variation of metabolites and will therefore be of great importance for collecting information on genes of unknown function, as well as providing a dynamic and global view of ADP1 metabolism.
Materials and methods
Bacteria, media, chemicals and reagents
Cells are routinely grown on MA (Medium for Acinetobacter) minimal medium (31 mM Na2HPO4, 25 mM KH2PO4, 18 mM NH4Cl, 41 μM nitrilotriacetic acid, 2 mM MgSO4, 0.45 mM CaCl2, 3 μM FeCl3, 1 μM MnCl2, 1 μM ZnCl2, 0.3 μM (CrCl3, H3BO3, CoCl2, CuCl2, NiCl2, Na2MoO2, Na2SeO3)) supplemented with 25 mM of carbon source and used with (MASK) or without (MAS) kanamycin at 30 μg/ml. The carbon sources and other chemicals were from Sigma-Aldrich. In special cases (e.g., deletion of genes involved in amino acid biosynthesis), the MAS/MASK was complemented with 1 mM of the appropriate amino acid compound.
PCR primers
The choice of the primers P3–P8 (Figure 1) was based on the Primer3 program (Rozen and Skaletsky, 2000). All primers were chosen to be as close as possible to 20 bp with a melting temperature of 55°C to maintain consistent PCR conditions between all constructions. When two neighboring genes were in the same orientation, primers P4 and P5 used for the deletion of the target gene were designed to preserve at least 25 nucleotides in the 5′ region of the flanking gene to keep the ribosome-binding site intact. The preserved region was extended to 75 nucleotides when genes were located on opposite strands, both to preserve the ribosome-binding site and to include the promoter region. These rules were relaxed to up to 100 nucleotides inside the target gene in very AT-rich regions (AT% higher than 70%). The primers P4 and P5 were extended by the complementary sequences of the integration cassette amplification primers as already described (Metzgar et al, 2004). Primers P3 and P6 for the amplification of the flanking regions were chosen to obtain regions of about 300±50 bp. The ‘external primers' P7 and P8, used for the verification of the correct locus integration, were chosen to be 800±200 pb upstream and downstream of the target gene, respectively. The primers used for each mutant construction are available through the Artemis interface via the MaGe platform.
Generation of the integration cassette
The strategy followed to obtain gene deletions is depicted in Figure 1. Splicing PCR was performed as described previously (Murphy et al, 2000; Metzgar et al, 2004) to generate recombinant DNA fragments consisting of the tdk/kan cassette flanked by 250–350 pb chromosomal regions upstream and downstream of the gene to be deleted. PCR to amplify the tdk/kan markers for positive and negative (tdk gene used to generate unmarked deletions and construct double mutants) selection was performed on plasmid pEVL386. We have constructed our plasmids from the pUC18-derived E. coli/ADP1 shuttle vector pKT40 (Metzgar et al, 2004). The plasmid pEVL174 was constructed from pKT40 by insertion of the tdk gene expression cassette, containing the E. coli tdk gene under the control of the bacteriophage T5 promoter into EcoRI/SacI sites. The plasmid pEVL386 is a derivative of plasmid pEVL174 containing the ahp gene for kanamycin resistance inserted into XbaI/PstI restriction sites. PCR to amplify the upstream and downstream flanking regions and the recombinant PCR were carried out following published protocols (Metzgar et al, 2004). PCRs are performed in 96-well microplates using 1.5 μl of an overnight culture of ADP1. At each step, the 96 PCR products were run on a 1% agarose electrophoresis gel using 0.5 × TBE buffer and analyzed manually for length.
Transformation and mutant selection
A 5 μl volume of an overnight culture of the ADP1 was added to 500 μl of MAS in 96-well microtiter plates. After 2 h of culture, 100 μl of the recombinant PCR product were added to the culture. The microtiter plates were incubated with shaking at 30°C for 24 h and spread onto MASK agar plates. The number of clones and the time of clone appearance were stored in the database. For each mutant, two colonies were isolated twice before further verifications. Three PCRs were carried out to test the correct structure of each mutant. Two of them use a combination of locus-specific and kanamycin primers (P3/P2 and P6/P1) and the last uses a couple of external primers (P7/P8). PCR reactions were carried out as described previously, except that in this case 1 μl of an overnight mutant culture was used. The P7/P8 PCR products were sequenced with primers S1 (5′-CTCCTTCATTACAGAAACGGC) and S2 (5′-CTTACCCGCATTCATTGCGG) specific for the integration cassette (see Figure 1), and their integration site was verified. Mutants were stored individually and in microplates at −80°C in MASK supplemented with 10% glycerol.
Auxotrophy and complementation assays
About 30 mutants of genes involved in amino acid biosynthesis not obtained on MASK medium during the high-throughput process were obtained in presence of 1 μM of the appropriate amino acid (arginine, methionine, histidine, leucine, tryptophan) and their auxotrophy was confirmed on MASK. To perform the complementation tests, the coding sequences of E. coli metA and metX, metW, metXW, metY and of ACIAD3524 from ADP1 were cloned into the PacI/NotI sites of plasmid pEVL174 (primers are shown in Supplementary Table VII). The correct sequence of each plasmid construction was confirmed before transformation.
Growth tests
For growth tests, an overnight preculture was inoculated with 5 μl of glycerol stock solution. Liquid growth studies were carried out in duplicate and solid growth in triplicate, starting with independent precultures. Mutants were tested for growth in 200 μl of MA with 25 mM of a carbon source and directly inoculated with the preculture using 96 inoculation pins (Genetix) and incubated at 30°C for 24 h with shaking. The plates are closed by an air pore pad (Qiagen) for correct aeration. The 30°C room is equipped with Kühner shakers and allows incubation of 240 plates simultaneously (four carbon/nitrogen sources in duplicate experiments for instance). The absorbance at 600 nm of the cultures was measured with a plate reader (Spectramax 384plus, Molecular Devices). Normalization of OD data was performed for each plate. Discrepancies between replications were noted and these data were excluded from the analyses. The mutants that are auxotrophic for acetate or/and 2,3-butanediol found in the high-throughput analyses were confirmed individually. Solid growth phenotypes were scored by replica printing the entire collection onto agar containing MA with 25 mM of carbon sources. Twelve 96-well overnight culture plates could be replicated per agar plate (20 × 20 cm) by using a colony-picking robot (Qbot from Genetix) with a 96-pin head. The replicas were incubated for 24 or 36 h at 30°C, photographed and analyzed automatically using the picking program of the robot. Normalization of colony diameter was performed using 12 wild-type colonies spotted on each plate.
Distribution of the ADP1 mutant collection
Individual mutants have already been distributed and used for several studies such as DNA damage (Chakravorty et al, 2008) and analysis of metabolism (Aghaie et al, in preparation). The mutant distribution is being handled through a Genoscope web page including primers data used for deletions (http://www.genoscope.cns.fr/spip/Strain-request-for-mutants-of,749.html).
Supplementary Material
Supplementary Information
Supplementary Table 1
Supplementary Table 2
Supplementary Table 4
Supplementary Table 5
Supplementary Table 6
Acknowledgments
We thank the Genoscope bioinformatic team for support throughout the project. We are grateful to AGC team for their dynamic and reactive support in the development of MaGe platform. We especially thank the Genoscope services team for the daily support and to Jean-Louis Petit, Céline Orvain-Durand and Laurie Bertrand for their help. We thank Valérie Barbe, Nuria Fonknechten and Alain Perret for critical reading and Susan Cure for correcting the manuscript. We are deeply indebted to Jamile Hazan for improving the manuscript. This project was financially supported by Consortium National de Recherche en Génomique.
References
- Abd El-Haleem D (2003) Acinetobacter: Environmental and Biotechnological Applications. Af J Biotechnol 2: 71–74 [Google Scholar]
- Akerley BJ, Rubin EJ, Novick VL, Amaya K, Judson N, Mekalanos JJ (2002) A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc Natl Acad Sci USA 99: 966–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaminos M, Ramos JL (2001) The methionine biosynthetic pathway from homoserine in Pseudomonas putida involves the metW, metX, metZ, metH and metE gene products. Arch Microbiol 176: 151–154 [DOI] [PubMed] [Google Scholar]
- Andersen GL, Beattie GA, Lindow SE (1998) Molecular characterization and sequence of a methionine biosynthetic locus from Pseudomonas syringae. J Bacteriol 180: 4497–4507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006.0008 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbe V, Vallenet D, Fonknechten N, Kreimeyer A, Oztas S, Labarre L, Cruveiller S, Robert C, Duprat S, Wincker P, Ornston LN, Weissenbach J, Marliere P, Cohen GN, Medigue C (2004) Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res 32: 5766–5779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barras F, Loiseau L, Py B (2005) How Escherichia coli and Saccharomyces cerevisiae build Fe/S proteins. Adv Microb Physiol 50: 41–101 [DOI] [PubMed] [Google Scholar]
- Bochner BR, Gadzinski P, Panomitros E (2001) Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res 11: 1246–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brichta DM, Azad KN, Ralli P, O'Donovan GA (2004) Pseudomonas aeruginosa dihydroorotases: a tale of three pyrCs. Arch Microbiol 182: 7–17 [DOI] [PubMed] [Google Scholar]
- Chakravorty A, Klovstad M, Peterson G, Lindeman R, Gregg-Jolly LA (2008) Sensitivity of Acinetobacter baylyi mpl mutant to DNA damage. Appl Environ Microbiol 74: 1273–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J, Wackernagel W (2002) Integration of foreign DNA during natural transformation of Acinetobacter sp. by homology-facilitated illegitimate recombination. Proc Natl Acad Sci USA 99: 2094–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley AM, Janse DM, Tanay A, Shamir R, Church GM (2005) A global view of pleiotropy and phenotypically derived gene function in yeast. Mol Syst Biol 1: 2005.0001. 10.1038/msb4100004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassbinder F, Kist M, Bereswill S (2000) Structural and functional analysis of the riboflavin synthesis genes encoding GTP cyclohydrolase II (ribA), DHBP synthase (ribBA), riboflavin synthase (ribC), and riboflavin deaminase/reductase (ribD) from Helicobacter pylori strain P1. FEMS Microbiol Lett 191: 191–197 [DOI] [PubMed] [Google Scholar]
- Fontecave M, Choudens SO, Py B, Barras F (2005) Mechanisms of iron-sulfur cluster assembly: the SUF machinery. J Biol Inorg Chem 10: 713–721 [DOI] [PubMed] [Google Scholar]
- Gerdes S, Edwards R, Kubal M, Fonstein M, Stevens R, Osterman A (2006) Essential genes on metabolic maps. Curr Opin Biotechnol 17: 448–456 [DOI] [PubMed] [Google Scholar]
- Gerdes SY, Scholle MD, Campbell JW, Balazsi G, Ravasz E, Daugherty MD, Somera AL, Kyrpides NC, Anderson I, Gelfand MS., Bhattacharya A, Kapatral V, D′Souza M, Baev MV, Grechkin Y, Mseeh F, Fonstein MY, Overbeek R, Barabasi AL, Oltvai ZN, Osterman AL (2003) Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol 185: 5673–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G et al. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Grill S, Yamaguchi H, Wagner H, Zwahlen L, Kusch U, Mack M (2007) Identification and characterization of two Streptomyces davawensis riboflavin biosynthesis gene clusters. Arch Microbiol 188: 377–387, advance online publication 1 June 2007; doi: advance online publication 8 July 2004; doi: 10.1007/s00203-007-0258-1 [DOI] [PubMed] [Google Scholar]
- Hwang BJ, Yeom HJ, Kim Y, Lee HS (2002) Corynebacterium glutamicum utilizes both transsulfuration and direct sulfhydrylation pathways for methionine biosynthesis. J Bacteriol 184: 1277–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Baba T, Mori H, Mori H (2005) Functional analysis of 1440 Escherichia coli genes using the combination of knock-out library and phenotype microarrays. Metab Eng 7: 318–327 [DOI] [PubMed] [Google Scholar]
- Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C (2003) Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci USA 100: 14339–14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juni E (1978) Genetics and physiology of Acinetobacter. Annu Rev Microbiol 32: 349–371 [DOI] [PubMed] [Google Scholar]
- Juni E, Heym GA (1956) A cyclic pathway for the bacterial dissimilation of 2,3-butanediol, acetylmethylcarbinol, and diacetyl. I. General aspects of the 2,3-butanediol cycle. J Bacteriol 71: 425–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Durfee T, Glasner JD, Qiu Y, Frisch D, Winterberg KM, Blattner FR (2004) Systematic mutagenesis of the Escherichia coli genome. J Bacteriol 186: 4921–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Kondo M, Fukuda H, Nishimura M, Ohta H (2003) Essential Bacillus subtilis genes. Proc Natl Acad Sci USA 100: 4678–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger N, Oppermann FB, Lorenzl H, Steinbuchel A (1994) Biochemical and molecular characterization of the Clostridium magnum acetoin dehydrogenase enzyme system. J Bacteriol 176: 3614–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T, Bork-Jensen J, Gerdes K (2005) The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol Microbiol 55: 78–89 [DOI] [PubMed] [Google Scholar]
- Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM (2006) An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci USA 103: 2833–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez JM, Thoms B, Rehbein H (1975) Acetoin degradation in Bacillus subtilis by direct oxidative cleavage. Eur J Biochem 57: 425–430 [DOI] [PubMed] [Google Scholar]
- Meganathan R (2001) Ubiquinone biosynthesis in microorganisms. FEMS Microbiol Lett 203: 131–139 [DOI] [PubMed] [Google Scholar]
- Metzgar D, Bacher JM, Pezo V, Reader J, Doring V, Schimmel P, Marliere P, de Crecy-Lagard V (2004) Acinetobacter sp. ADP1: an ideal model organism for genetic analysis and genome engineering. Nucleic Acids Res 32: 5780–5790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KC, Campellone KG, Poteete AR (2000) PCR-mediated gene replacement in Escherichia coli. Gene 246: 321–330 [DOI] [PubMed] [Google Scholar]
- Neidle EL, Hartnett C, Ornston LN, Bairoch A, Rekik M, Harayama S (1991) Nucleotide sequences of the Acinetobacter calcoaceticus benABC genes for benzoate 1,2-dioxygenase reveal evolutionary relationships among multicomponent oxygenases. J Bacteriol 173: 5385–5395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh YK, Palsson BO, Park SM, Schilling CH, Mahadevan R (2007) Genome-scale reconstruction of metabolic network in Bacillus subtilis based on high-throughput phenotyping and gene essentiality data. J Biol Chem 282: 28791–28799 [DOI] [PubMed] [Google Scholar]
- Oppermann FB, Schmidt B, Steinbuchel A (1991) Purification and characterization of acetoin:2,6-dichlorophenolindophenol oxidoreductase, dihydrolipoamide dehydrogenase, and dihydrolipoamide acetyltransferase of the Pelobacter carbinolicus acetoin dehydrogenase enzyme system. J Bacteriol 173: 757–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmen R, Hellingwerf KJ (1997) Uptake and processing of DNA by Acinetobacter calcoaceticus—a review. Gene 192: 179–190 [DOI] [PubMed] [Google Scholar]
- Picardeau M, Bauby H, Saint Girons I (2003) Genetic evidence for the existence of two pathways for the biosynthesis of methionine in the Leptospira spp. FEMS Microbiol Lett 225: 257–262 [DOI] [PubMed] [Google Scholar]
- Reams AB, Neidle EL (2004) Selection for gene clustering by tandem duplication. Annu Rev Microbiol 58: 119–142 [DOI] [PubMed] [Google Scholar]
- Riley M, Abe T, Arnaud MB, Berlyn MK, Blattner FR, Chaudhuri RR, Glasner JD, Horiuchi T, Keseler IM, Kosuge T, Mori H, Perna NT, Plunkett G III, Rudd KE, Serres MH, Thomas GH, Thomson NR, Wishart D, Wanner BL (2006) Escherichia coli K-12: a cooperatively developed annotation snapshot—2005. Nucleic Acids Res 34: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Srinivasa Rao PS, Yamada Y, Leung KY (2003) A major catalase (KatB) that is required for resistance to H2O2 and phagocyte-mediated killing in Edwardsiella tarda. Microbiology 149: 2635–2644 [DOI] [PubMed] [Google Scholar]
- Stanier RY, Ornston LN (1973) The beta-ketoadipate pathway. Adv Microb Physiol 9: 89–151 [PubMed] [Google Scholar]
- Suzuki N, Okai N, Nonaka H, Tsuge Y, Inui M, Yukawa H (2006) High-throughput transposon mutagenesis of Corynebacterium glutamicum and construction of a single-gene disruptant mutant library. Appl Environ Microbiol 72: 3750–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallenet D, Labarre L, Rouy Z, Barbe V, Bocs S, Cruveiller S, Lajus A, Pascal G, Scarpelli C, Medigue C (2006) MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res 34: 53–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallenet D, Nordmann P, Barbe V, Poirel L, Mangenot S, Bataille E, Dossat C, Gas S, Kreimeyer A, Lenoble P, Oztas S, Poulain J, Segurens B, Robert C, Abergel C, Claverie JM, Raoult D, Medigue C, Weissenbach J, Cruveiller S (2008) Comparative analysis of Acinetobacters: three genomes for threee lifestyles. Plos ONE (in press) [DOI] [PMC free article] [PubMed]
- Xiao Z, Xu P (2007) Acetoin metabolism in bacteria. Crit Rev Microbiol 33: 127–140 [DOI] [PubMed] [Google Scholar]
- Young DM, Parke D, Ornston LN (2005) Opportunities for genetic investigation afforded by Acinetobacter baylyi, a nutritionally versatile bacterial species that is highly competent for natural transformation. Annu Rev Microbiol 59: 519–551 [DOI] [PubMed] [Google Scholar]
- Zheng J, Leung KY (2007) Dissection of a type VI secretion system in Edwardsiella tarda. Mol Microbiol 66: 1192–1206 [DOI] [PubMed] [Google Scholar]
- Zhou H, Lutkenhaus J (2005) MinC mutants deficient in MinD- and DicB-mediated cell division inhibition due to loss of interaction with MinD, DicB, or a septal component. J Bacteriol 187: 2846–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Table 1
Supplementary Table 2
Supplementary Table 4
Supplementary Table 5
Supplementary Table 6