Abstract
Epstein–Barr virus (EBV), like other DNA tumor viruses, induces an S-phase in the natural host cell, the human B lymphocyte. This is linked with blast transformation. It is believed that the EBV-encoded nuclear antigen 6 (EBNA-6) is involved in the regulation of cell cycle entry. However, the possible mechanism of this regulation is not approached. In our current study, we found that EBNA-6 binds to a MRPS18-2 protein, and targets it to the nucleus. We found that MRPS18-2 binds to both hypo- and hyperphosphorylated forms of Rb protein specifically. This binding targets the small pocket of pRb, which is a site of interaction with E2F1. The MRPS18-2 competes with the binding of E2F1 to pRb, thereby raising the level of free E2F1. Our experimental data suggest that EBNA-6 may play a major role in the entry of EBV infected B cells into the S phase by binding to and raising the level of nuclear MRPS18-2, protein. This would inhibit pRb binding to E2F1 competitively and lift the block preventing S-phase entry.
Keywords: cell transformation, cell cycle, surface plasmon resonance, S-phase entry
Epstein-Barr virus (EBV) is one of the most highly transforming viruses known (1). EBV-immortalized lymphoblastoid cell lines (LCLs) express a virally encoded growth transformation program, characterized by the expression of six nuclear proteins (EBNAs) and three membrane proteins (LMPs), of which five, EBNA-2, -3, -5, -6, and LMP1, are essential for transformation. EBNA-6 (EBNA-3C, according to the alternative nomenclature) belongs to the EBNA-3 protein family. EBNA-3 (EBNA-3A), EBNA-4 (EBNA-3B), and EBNA-6 are encoded by tandemly arranged genes and have a similar genomic organization (for review, see refs. 2 and 3). They contain short intron sequences that may be retained in the mRNA. The level of the three proteins is differently regulated by cellular factors (4).
Among the EBNA-3 family proteins, EBNA-6 has been most extensively studied. It was found to bind to histone deacetylase (HDAC)1 (5), HDAC2 (6), acetyltransferase p300, and prothymosin alpha (7). EBNA-6 was also shown to interact with the DEAD-box protein DP103 (8) that has helicase activity (9). This suggests that EBNA-6 may play a role in chromatin remodeling.
EBNA-6 also associates with RBP-Jκ, suggesting it may be involved in transcriptional regulation (10, 11). EBNA-6 expressed exclusively in immunoblasts of B cell origin binds RBP-2N, the major isoform of RBP-Jκ in B lymphocytes (12). In view of the fact that EBNA-6 also associates with corepressors CtBP (13), mSin3A, and NcoR (6), it is likely that EBNA-6 can either activate or repress transcription. EBNA-6 and its homologs encoded by the baboon and rhesus relatives of EBV (BaLCV and RhLCV) were shown to bind RBP- Jκ and Spi1 (14). When bound to RBP-Jκ, the EBNA-6 homologs of both viruses could repress EBNA-2-mediated transcription in vitro. When bound to Spi1, they activated EBNA-2-mediated transcription.
It was also shown that EBNA-6 can activate transcription from the LMP1 promoter in vitro in the presence of EBNA-2 (15–17) or in its absence (18).
EBNA-6 is also believed to play a role in cell cycle control. Reportedly, it binds to pRb protein in vitro (19–21). EBNA-6 transfection was found to decrease the level of phosphorylated pRb and inhibit p27 protein accumulation in primary rat fibroblasts (20).
The decrease of the p27 level was later related to the finding that EBNA-6 binds cyclin A (22, 23) and the E3 ubiquitin ligase SCF SKP1-CUL1-F-box complex (24). It was proposed that the SCF complex is involved in the EBNA-6 promotional degradation of pRb (21) and p27 as well (24).
In LCL, however, EBNA-6 is not responsible for the degradation of pRb (25): the level of total pRb and p27 did not change upon inactivation of EBNA-6 in an LCL that carried a conditionally active (hydroxytamoxifen-dependent) EBNA-6. Eventually, the portion of lymphoblastoid cells in S and G2/M phases was reduced within 1 week after the inactivation of EBNA-6. In parallel, there was an increase of p16 expression and a decrease in pRb phosphorylation. Taken together, these findings indicate that EBNA-6 interferes with some components of the pRb-E2F pathway.
Using the yeast two-hybrid system, we found that MRS18-2, a pRb-binding protein, associates with EBNA-6. EBNA-6 targets cellular MRS18-2 to the nucleus and promotes binding between pRb and MRS18-2. This disrupts the association of E2F1 with pRb, as indicated by the increased level of free E2F1 in the nucleus. It appears likely that the pRb control of the cell cycle is at least partially reduced by this interaction.
Results
EBNA-6 Binds to MRPS18-2 in the Yeast Two-Hybrid System.
Screening of a human lymphoblast cDNA library with EBNA-6 as a bait identified a clone, P33 (plate P, clone 33), that could grow on His-, Leu-, and Trp-deficient synthetic dextrose medium. This clone expressed β-galactosidase from a lacZ reporter, activated by protein–protein interaction. The strength of the binding was ≈12% of the strength of positive pRb-SV40LT control. The P33 clone contained a ≅900-bp-long insert (at its XhoI site).
The interaction between EBNA-6 and P33 was confirmed by reintroduction of the corresponding plasmids into the SFY526 yeast strain. Empty pGBT9/BR vector and EBNA-1, ΔEBNA-3, EBNA-5, and ΔEBNA-4 constructs in the GAL4-binding domain yeast vector were used as negative controls. None showed any significant interaction, according to the β-galactosidase test.
Sequencing of the insert showed that the EBNA-6 interactive clone was identical to part of human mitochondrial ribosomal protein MRPS18-2 or MRPS18B (GenBank accession nos. NM_014046 and protein, NP_054765). This protein is encoded by a cellular gene, located at 6p21.3. S18-2 cDNA was cloned during an analysis of 300 previously undefined genes with ORFs expressed in CD34+ hematopoietic stem/progenitor cells by Zhang et al. (26). It was shown later that this gene encodes one of the three MRPS18 family proteins localized on the surface of the small subunit (28S) of the mammalian mitochondrial ribosome (27, 28). Importantly, the human genome encodes three S18-2 genes, in contrast to one in bacteria. S18-2 homologs are found in mammals, Drosophila melanogaster, and Caenorhabditis elegans.
The 5′ end of the MRPS18-2 gene, encoding the first 104 residues, and the 3′ end were missing from the cDNA library insert.
We have used a heart c-DNA library to obtain the full-length MRPS18-2 gene. The full-length cDNA, encoding a 258-aa-long protein, was cloned into the green fluorescence protein vector pEGFP-C1 (GFP), the pCMV-Tag 3A c-myc-tagged vector (MT), and the GST bacterial expression vector [GST2TK (GST)].
EBNA-6 Binds MRPS18-2 in Vitro.
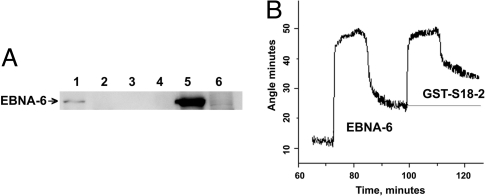
To verify the binding between EBNA-6 and MRPS18-2 (abbreviated as S18-2), we used lysates of freshly EBV-infected B cells (72 h after infection), EBV+ latency I MUTUI cells, and GST-S18-2 beads. GST-S18-2, but not GST, could precipitate EBNA-6 from the EBV-infected cell lysate. A Western blot is shown in Fig. 1A.
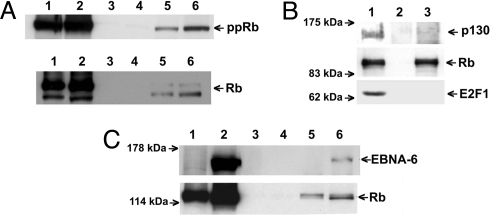
Fig. 1.
EBNA-6 binds S18-2 in vitro. (A) GST-S18-2 precipitates EBNA-6 from freshly EBV-infected B cells in the GST pulldown assay. The membrane was probed with anti-EBNA-6 antibody. Lane 1: GST-S18-2 precipitated EBNA-6 from freshly EBV-infected B cells (72 h, 5 × 106 cells); lane 2: GST-S18-2 precipitation from MUTUI cell lysates; lane 3: GST precipitation from freshly infected B cells (as in lane 1); lane 4: GST precipitation from MUTUI cell lysate; lane 5: lysate of 0.5 × 106 freshly infected B cells; lane 6: lysate of 0.5 × 106 MUTUI cells. (B) SPR responses at the stage of protein interaction. The stepwise changes are due to solution replacement and correspond to changes in refractive index. EBNA-6 from LCL cell lysate binds to anti-EBNA-6 on the surface (first curve). GST-S18-2 binds to surface EBNA-6 (second curve). The binding of protein A to the chip surface and the anti-EBNA-6 antibody to protein A is not shown.
The GST pulldown data have been verified by surface plasmon resonance (SPR) technique to monitor the protein–protein interaction. A protein-capturing layer was created by using anti-EBNA-6 mouse monoclonal antibody, coupled to protein A. EBNA-6 was captured from the LCL cell lysate by the surface anti-EBNA-6 antibody. No SPR signal was detected with the MUTUI cell lysate (data not shown). Surface EBNA-6, bound to antibody, gave a signal with GST-S18-2 (Fig. 1B).
EBNA-6 Raises the Level of S18-2 in the Nucleus and Shows Extensive Colocalization.
We have shown that EBNA-6 and S18-2 can bind each other in yeast cells and in vitro, and so the next step was to study whether these two proteins can form complexes in mammalian cells.
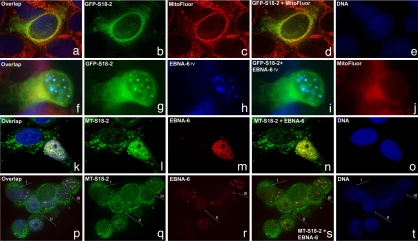
First of all, we wanted to study the cellular distribution of the S18-2 protein. To do so, MCF7 cells were transfected with the GFP-S18-2 plasmid. A major portion of the GFP-S18-2 protein was expressed in the cytoplasm(the cell with the green signal in Fig. 2 a, b, and d). MitoFluor staining visualized the mitochondria (the red signal in Fig. 2 a, c, and d). The signals of the GFP-S18-2 protein and of mitochondria were quite similar (Fig. 2 a and d).
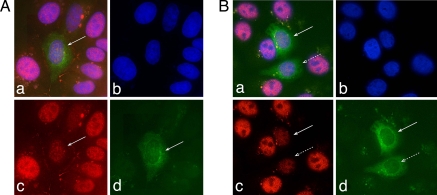
Fig. 2.
EBNA-6 targets S18-2 to the nucleus of the transfected cells. (a–e) MCF7 cells transfected with GFP-S18-2 (green signal). Mitochondria are stained with MitoFluor in red (see a, c, and d) and DNA in blue (a and e). Notice that GFP-S18-2 (green signal in a, b, and d) shows predominantly cytoplasmic distribution. (f–j) MCF7 cells that expressed GFP-S18-2 for 24 h were infected with recombinant vaccinia virus that expressed EBNA-6 (stained in blue; see f, h, and i). Mitochondria are stained in red (see f and j). Notice the translocation of GFP-S18-2 (green signal in f, g, and i) to the nucleus and colocalization with EBNA-6 (see overlapping blue and green signals in layers f and i). MitoFluor red was used to visualize mitochondria. They remain in cytoplasm (see red signal in f and j). (k–o) MCF7 cells that expressed MT-S18-2 constitutively (green signal) were transfected with pBabe-EBNA-6 (red signal). Cells after 24 h were stained with anti-EBNA-6 mouse monoclonal antibody, followed with horse anti-rabbit TR conjugated antibody. After blocking with mouse serum, we stained MT-S18-2 with FITC-conjugated anti-c-myc monoclonal antibody. Predominantly cytoplasmic MT-S18-2 (green signal in k, l, and n) translocated to the nucleus in the presence of EBNA-6. Moreover, the green S18-2 signal followed the red EBNA-6 pattern in the EBNA-6-expressing cell (k and n). (p–t) MCF7 cells that expressed MT-S18-2 constitutively were infected with EBNA-6-expressing recombinant vaccinia virus. Images were captured by using a confocal microscope. EBNA-6 was stained with anti-EBNA-6 mouse monoclonal antibody and TR-conjugated secondary horse anti-mouse antibody (red signal in p, r, and s). After blocking with total mouse serum, MT-S18-2 was stained with anti-c-myc mouse FITC-conjugated antibody (green signal in p, q, and s). Notice the predominantly cytoplasmic expression of MT-S18-2 in cells where EBNA-6 was absent (type I cell). Observe the change in S18-2 distribution (types II and III cells) when EBNA-6 was expressed. Moreover, EBNA-6 and S18-2 showed a high degree of colocalization (types II and III cells, see p and s).
To monitor the fate of the GFP-S18-2 protein in the presence of EBNA-6, the GFP-S18-2-transfected cells were infected 24 h after transfection with recombinant vaccinia virus that expressed EBNA-6 protein. After 24 h, the cells were stained with anti-EBNA-6 mouse monoclonal antibody, followed by AMKA-streptavidin on a rabbit anti-mouse biotinylated antibody. We also used MitoFluor red to visualize mitochondria. EBNA-6 (EBNA-6 rv in Fig. 2 f–j) formed nuclear inclusions when expressed from recombinant vaccinia virus (blue signal in Fig. 2 f, h, and i). A large portion of GFP-S18-2 (green signal in Fig. 2 f, g, and i) was expressed in the nucleus, where it colocalized with EBNA-6 (Fig. 2 f and i). Mitochondria (red signal on Fig. 2j) remained in the cytoplasm.
To obtain cells that expressed S18-2 at the lower level, compared with the GFP construct, we established a MCF7 subline that expressed S18-2 constitutively, using a MT-S18-2 plasmid for transfection. In these cells, MT-S18-2 was expressed mainly in the cytoplasm, but a small portion of protein was observed in the nucleus as well.
To follow the S18-2 protein expression pattern in the presence of EBNA-6, we transfected this subline with pBabe-EBNA-6. After 24 h, cells were stained with anti-EBNA-6 mouse monoclonal antibody, followed by horse anti-rabbit Texas red-conjugated antibody. After blocking with mouse serum, we stained MT-S18-2 with FITC-conjugated anti-c-myc monoclonal antibody. Predominantly cytoplasmic MT-S18-2 (green signal in Fig. 2 k, l, and n) translocated to the nucleus in the presence of EBNA-6. It is seen in Fig. 2 k and n that the green S18-2 signal followed the red EBNA-6 pattern in the EBNA-6-expressing cell.
To test whether a degree of S18-2 translocation could be linked to the EBNA-6 expression level, we infected the MCF7 cells that expressed MT-S18-2 constitutively with EBNA-6-expressing vaccinia recombinant virus (Fig. 2 p–t). Cells were stained after 24 h, as described earlier.
In Fig. 2 p–t, we draw attention to the three types of cells: (I) cells that do not express EBNA-6, (II) cells that express homogeneous EBNA-6, and (III) cells in which EBNA-6 formed nuclear inclusions (see the red signal for EBNA-6 in Fig. 2 p, r, and s). As seen in Fig. 2, MT-S18-2 was mainly cytoplasmic in cells of type I, where EBNA-6 was not expressed (Fig. 2 p, q, and s) When EBNA-6 was expressed at high levels in the nucleus, forming nuclear inclusions, MT-S18-2 relocated to the nucleus and colocalized to a high degree with EBNA-6 (cells of type III) (Fig. 2 p and s). In cells of type II, nuclear translocation of MT-S18-2 was also observed. Moreover, the red signal of EBNA-6 and green signal of MT-S18-2 showed a high degree of colocalization in the nucleus.
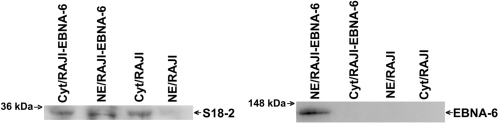
To monitor changes in S18-2 distribution in the presence of endogenous EBNA-6, we compared RAJI cells that carry an EBNA-6-defective EBV strain with a RAJI-EBNA-6 transfectant that expresses EBNA-6 constitutively from a CMV promoter. We prepared nuclear extracts and cytoplasmic fractions from RAJI and RAJI-EBNA-6 cells. Using Western blot and anti-S18-2 rabbit serum, we detected an increased amount of endogenous S18-2 in the nuclei of RAJI-EBNA-6 cells compared with RAJI cells (Fig. 3Left).
Fig. 3.
Increased level of nuclear S18-2 in the EBNA-6-expressing cells; Western blot of the nuclear extracts and cytoplasmic fractions of RAJI and RAJI-EBNA-6 cells. (Left) S18-2 expression in RAJI and RAJI-EBNA-6 cells. Ten micrograms of total protein in nuclear extracts and cytoplasmic fractions was loaded on the gel. The membrane was probed with rabbit serum (76-2) raised against the S18-2 peptide. Notice the increase of nuclear S18-2 signal in RAJI-EBNA-6 cells. (Right) EBNA-6 expression. The mouse monoclonal ani-EBNA-6 antibody was used to monitor EBNA-6 in the nuclear extract and the cytoplasmic fraction of RAJI-EBNA-6 and RAJI cells.
From the experiments, we can conclude that expression of EBNA-6 led to targeting of S18-2 to the nucleus, where these two proteins showed a high degree of colocalization.
EBNA-6 Forms Complexes with S18-2 in Vivo.
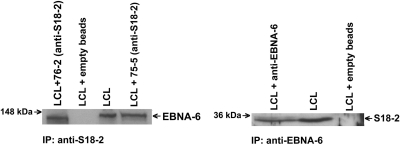
To show whether EBNA-6 and S18-2 could form protein complexes in an LCL, we performed immunoprecipitations, using LCL cell lysates, mouse monoclonal anti-EBNA-6 antibody, and polyclonal anti-S18-2 sera.
As shown in Fig. 4, EBNA-6 was detected in immunocomplexes with anti-S18-2 antibody (Fig. 4 Left) in lymphoblastoid cells. S18-2 was also detected in protein complexes that contained the mouse anti-EBNA-6 antibody (Fig. 4 Right).
Fig. 4.
EBNA-6 and S18-2 are components of the one protein complex in vivo. For immunoprecipitations, 5 μg of antibodies was coupled to the CN-Br Sepharose, and such supports were used to capture the corresponding proteins from the Nonidet P-40 LCL cell lysates. After extensive washing, the protein complexes were eluted by heat and separated on the polyacrylamide gel. Beads not coupled to antibodies were used as a negative control. Ten micrograms of LCL lysate was used as a control for protein expression. (Left) EBNA-6 from LCL was precipitated by anti-S18-2 antibody (with two clones, 75-5 and 76-2). The EBNA-6 antibody was used to probe the membrane. (Right) Precipitation of S18-2 from LCL by the anti-EBNA-6 mouse antibody. Clone 76-2 was used as a probe for Western blot analysis. Notice that beads not coupled to antibodies did not give any signal.
MRS18-2 Is Associated with Hypo- and Hyperphosphorylated pRb.
We have shown (29) that S18-2 can bind to pRb, using a modified SPR method. Here, using a GST-pulldown assay, we observed that GST-S18-2, but not GST, could precipitate pRb and ppRb from the MUTUI and MUTUIII cell lysates (Fig. 5A, lanes 5 and 6). This indicates that the S18-2 and pRb proteins can associate, regardless of whether EBNA-6 is present. Importantly, GST-pRb beads precipitated the endogenous S18-2 and E2F1 proteins, as we have shown. S18-2–pRb binding was specific, because GST-S18-2 beads could not precipitate p130, another pocket protein, from MUTUIII cell lysate, as also shown earlier (29). Interestingly, pRb bound to GST-S18-2 beads was not able to bind E2F1 (Fig. 5B). We have mapped the interaction site between S18-2 and pRb to a small pocket (Fig. 5A, box spacer; Fig. 5B, box, residues 380–785) of pRb, as we reported (29). It is known that the same portion of pRb is the site of E2F1 binding.
Fig. 5.
S18-2 binds EBNA-b and pRb in vitro specifically. (A) GST-S18-2 precipitates pRb regardless of EBNA-6 presence in GST pulldown assay. The membranes were probed with anti-pRb (Upper) and anti-ppRb (Lower) antibodies. Lane 1: lysate of 0.4 × 106 MUTUI cells; lane 2: lysate of 0.4 × 106 MUTUIII cells; lane 3: GST precipitation from MUTUI cell lysate (10 × 106 cells); lane 4: GST precipitation from MUTUIII cell lysate (10 × 106 cells); lane 5: GST-S18-2 precipitated pRb and ppRb from the MUTUI cell lysates (10 × 106 cells); lane 6: GST-S18-2 precipitated pRb and ppRb from MUTUIII cell lysates (10 × 106 cells). (B) GST-S18-2 precipitates pRb specifically in GST pulldown assay. The membranes were probed with anti-p130, anti-pRb, and anti-E2F1 antibodies. Lane 1: MUTUIII cell lysate; lane 2: GST beads; lane 3: GST-S18-2 precipitated pRb, but not p130 neither E2F1. (C) GST-S18-2 precipitates EBNA-6 and pRb simultaneously in GST pulldown assay. Membranes were probed with anti-EBNA-6 and pRb antibody. Lanes 1, 3, and 5: RAJI cell lysate, lanes 2, 4, and 6: RAJI-EBNA-6 cell lysate. Lanes 1 and 2: cell lysates (5% of input); lanes 3 and 4: GST beads; lanes 5 and 6: GST-S18-2 beads.
EBNA-6 May Form Ternary Complexes with MRS18-2 and pRb.
As we described earlier, we have detected both EBNA-6 and pRb on the surface of the GST-S18-2 beads (Figs. 1 and 5). These two proteins were pulled down also simultaneously by GST-S18-2 beads (Fig. 5C). Importantly, we could not detect any direct EBNA-6 and pRb binding in any of the methods used, including SPR and the yeast two-hybrid system. We propose that ternary complexes, containing EBNA-6-S18-2 and pRb, could be formed with S18-2 serving as a bridge between EBNA-6 and pRb.
In conclusion, EBNA-6 appears to raise the level of S18-2 in the nucleus and enhances S18-2–pRb association.
Expression of S18-2 Is Accompanied by Decreased Levels of pRb and ppRb.
We wanted to study the fate of pRb when S18-2 was overexpressed (endogenous S18-2 was quite low). To do so, we transfected MCF7 cells with GFP-S18-2 and stained them with anti-pRb and -ppRb antibodies. Both the ppRb (Fig. 6 A a and c) and total pRb (Fig. 6B a and c) signals decreased when the cells expressed GFP-S18-2 (Fig. 6 A a and d and B a and d; cells indicated with arrows). We could not detect any change of the p53 signal (data not shown). A similar decrease of the pRb expression was observed in MT-S18-2-transfected cells (data not shown).
Fig. 6.
S18-2 expression decreases pRb levels. (A) MCF7 cells, transfected with GFP-S18-2 (green). Phosphorylated pRb (ppRb) is stained in red, and DNA is stained in blue. Notice the red signal decrease in the GFP-S18-2-expressing cells (a and c). The arrow indicates a transfected cell. (B) MCF7 cells, transfected with GFP-S18-2 (green). The total pRb protein is stained in red (see the red signal in a and c), and DNA is in blue. Notice that the pRb signal is essentially decreased in GFP-S18-2-expressing cells (red signal in a and c). The arrows indicate the transfected cells.
EBNA-6 Raises the Level of Free E2F1.
To assess the consequence of EBNA-6–S18-2–pRb binding in relation to E2F1, we compared RAJI and RAJI-EBNA-6 transfectant. RAJI-EBNA-6 expressed higher levels of pRb and E2F1 than the EBNA-6-negative RAJI parental cell line (Fig. 7A).
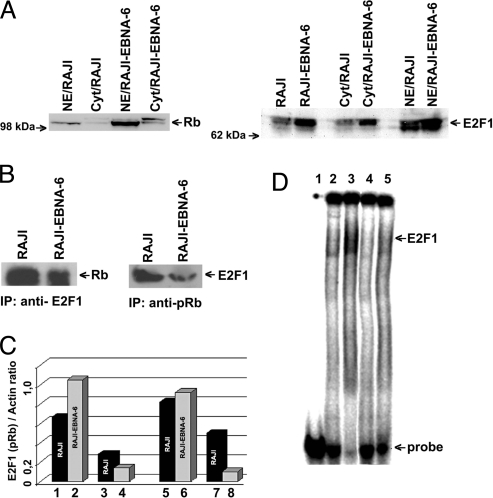
Fig. 7.
S18-2 expression decreases pRb levels. (A) Western blot showing protein levels in the nucleus and cytoplasm of the RAJI-EBNA-6 cells compared with RAJI. Notice the increase in pRb and E2F1 levels when EBNA-6 is expressed. (B) Immunoprecipitations from RAJI and RAJI-EBNA-6 cell lysates. (Left) Precipitations with anti-E2F1 antibody. Notice the decrease in the amount of pRb, bound to E2F1. (Right) Precipitations with anti-pRb antibody. The amount of pRb in complex with E2F1 protein is lower in RAJI-EBNA-6 cells. (C) Levels of total E2F1 and pRb proteins and in complex with each other. Lanes 1 and 2: amount of E2F1 in the RAJI and RAJI-EBNA-6 cell lysate (as a ratio to actin); lanes 3 and 4: amount of E2F1, precipitated with anti-pRb antibody from cell lysates; lanes 5 and 6: amount of total pRb in the cell lysates; lanes 7 and 8: 10-fold magnified amount of pRb, precipitated with anti-E2F1. (D) EMSA of DNA oligo, containing the two binding sites for E2F1. Lane 1: labeled DNA probe; lane 2: DNA probe with RAJI nuclear extract; lane 3: DNA probe with RAJI-EBNA-6 nuclear extract; lane 4: as lane 2 and anti-E2F1 mouse monoclonal antibody; lane 5: as lane 3 and anti-E2F1 antibody. Notice that almost all DNA is bound to E2F1 in RAJI-EBNA-6 cells compared with RAJI cells (lane 3 compared with lane 2). Observe that treatment with anti-E2F1 antibody leads to disappearance of DNA shift (lanes 4 and 5).
To monitor the levels of the E2F1–pRb protein complex, we performed immunoprecipitations from RAJI and RAJI-EBNA-6 cell lysates, using mouse monoclonal anti-pRb and anti-E2F1 antibodies. We used the same amount of cell lysates and antibodies in each experiment. The results of Western blot analysis after immunoprecipitation are shown in Fig. 7B. Less pRb was precipitated with anti-E2F1 from the RAJI-EBNA-6 cell lysate, and less E2F1 associated with pRb, compared with the RAJI cell lysate. We have plotted the ratios of actin/protein after densitometry (Fig. 7C).
Despite the increased levels of E2F1 and pRb in the RAJI-EBNA-6 cells, compared with RAJI, there was lower binding between E2F1 and pRb in EBNA-6-expressing cells. This suggests that more free E2F1 may be available for DNA binding in the presence of EBNA-6.
Using DNA oligos that contained two binding sites for E2F1, we have performed EMSA with RAJI and RAJI-EBNA-6 cell lysates. We have observed specific binding of E2F1 to DNA in both cases (Fig. 7D). The band shift suggested E2F1 binding to DNA. Monoclonal anti-E2F1 antibody abolished the band shift, indicating a specific reaction. We have introduced equal amounts of probe in each experiment (see details in Material and Methods), but nearly all DNA was shifted by E2F1 from the RAJI-EBNA-6 cell lysate, not from the RAJI cell lysate. The EMSA data allowed us to draw the same conclusion that more free E2F1 is available for DNA binding in RAJI-EBNA-6 cells compared with RAJI, as evident from immunoprecipitation.
Discussion
S18-2 Serves as a Bridge Between EBNA-6 and pRb.
It has been proposed that EBNA-6 is involved in the regulation of cell cycle entry (19). The possible mechanism of this regulation was not approached, however. Binding between pRb and EBNA-6 was demonstrated by GST–GST in vitro assay (19).
Using the SPR method that permits the detection of protein–protein interactions at the molecular level, we could not observe any direct binding between EBNA-6 and pRb. Interestingly, S18-2, found by the yeast two-hybrid system with EBNA-6 as bait, could also bind pRb. On the basis of GST-pulldowns from LCL lysates and SPR data, we proposed that a ternary complex of these three proteins could be formed, where S18-2 served as a bridge between EBNA-6 and pRb. This may explain the cohabitation of pRb and EBNA-6 in the GST-pulldown and immunoprecipitation experiments, as reported by Knight et al. (21). Immunoprecipitations and GST precipitations may be informative with regard to the presence of individual components in a protein complex, but they do not answer the question of direct molecular interactions.
EBNA-6 and pRb Degradation.
Based on the hypothesis that EBNA-6 may bind directly to pRb, it has been speculated that EBNA-6 may mediate pRb degradation by complexing with SCF ubiquitin ligase (21). The levels of pRb and ppRb decreased upon transient transfection of EBNA-6 into HEK, 293T, and BJAB cells. We have observed similar changes upon transient EBNA-6 transfection into MCF7 cells (data not shown). This was a short-lived effect, however, with pRb returning to its original level in a few days.
It is noteworthy that pRb and ppRb levels were higher in RAJI-EBNA-6 than in the parental RAJI cells (Fig. 7 A–C). Earlier, it was shown that pRb is phosphorylated and dephosphorylated normally in relation to the cell cycle, irrespective of the presence or absence of EBNA-6 in RAJI (16).
Taken together with the fact that a decrease of the EBNA-6 level did not lead to an increase of the pRb level in an LCL carrying a conditional EBNA-6 (25), it appears unlikely that EBNA-6 promotes the degradation of pRb in the LCLs.
EBNA-6 Promotes Binding of S18-2 to pRb, Freeing E2F1.
EBNA-6 inactivation in the same conditional LCL (25) has led to the decrease in the proportion of the S and G2/M fractions and of the ppRb, cyclin A, and cyclin D2/cdk4 levels, without changing the levels of pRb, cdk2, cdk4, and p27 (25). The E2F1 level was not monitored.
We have detected a higher E2F1 level in the EBNA-6-expressing, compared with the EBNA-6-deleted RAJI cells (Fig. 7A). Although the EBNA-6-expressing cells showed only a slight increase of pRb, the free pRb unbound E2F1, detected by immunoprecipitations, was significantly increased (Fig. 7 B and C). Moreover, the level of DNA-bound E2F1 was higher in the EBNA-6-expressing RAJI cells, according to EMSA (Fig. 7D).
We have also found that EBNA-6 binds to S18-2 and targets it to the nucleus (Figs. 2 and 3). S18-2 binds to both pRb and ppRb, regardless of the presence of EBNA-6 (Fig. 5A). The binding of S18-2 to pRb is specific, as shown by the GST-pulldown assay. It targets the small pocket of pRb (29), the site of the interaction with E2F1. This binding competes with the binding of E2F1 to pRb, thereby raising the level of free E2F1 (Figs. 5B and 7 B–D).
Our findings suggest that the ubiquitously expressed S18-2 may participate in regulating the level of free E2F1.
EBNA-6 May Inhibit pRb Control on S-Phase Entry.
According to our findings, the level of pRb and ppRb is significantly decreased in the cell nucleus upon transient expression of S18-2 as GFP- and c-myc-tag fusion protein (Fig. 6 A and B). We speculate that the decrease in pRb level observed after transient transfection by EBNA-6 was due to the enrichment of endogenous S18-2 in the cell nucleus, following its binding to EBNA-6. The mechanism of pRb level decrease should be further elucidated.
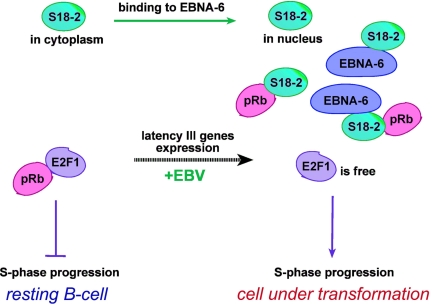
The translocation of S18-2 to the nucleus by EBNA-6 may facilitate the binding of S18-2 to pRb, as already discussed. As long as it is bound to S18-2, pRb may not influence the level of free E2F1. This may promote the entry of the EBV-infected B cell into the cell cycle (Fig. 8).
Fig. 8.
Functional consequences of the binding between S18-2 and EBNA-6: S18-2 bound to EBNA-6 translocates to the nucleus. This may facilitate the binding of S18-2 to pRb. As long as it is bound to S18-2, pRb may not influence the level of free E2F1. This may promote the entry of the EBV infected B cell into the cell cycle.
Herpes Viruses Induce S-Phase in the Infected Cells.
EBV, like other DNA tumor viruses (1), induces an S-phase in the natural host cell, the human B lymphocyte. This is linked with blast transformation. It is noteworthy, in this context, that EBNA-6 is expressed only in immunoblasts.
In the case of the small and medium-sized DNA tumor viruses (SV40, polyoma, transforming adenoviruses), the induction of an S-phase is linked to the inactivation of the Rb and the p53 pathway. In the much larger and more complex herpesviruses, this is not as straightforward. It may be relevant in this context that other herpesviruses can increase the level of free E2F1, although by different mechanisms. ORF72 (ORF75) of human herpesvirus 8 (Kaposi's sarcoma herpesvirus) encodes a cyclin, homologous to cyclin D2, that binds to cdk6 and more weakly to cdk4. This leads to the phosphorylation of pRb and release of E2F1 (30). The ORF eclf2 of herpes virus saimiri (HSV) encodes a D cyclin homolog, which associated with cdk6 and can phosphorylate pRb, thereby increasing the level of free E2F1 (31).
In conclusion, our findings suggest that EBNA-6 may play a major role in the entry of EBV-infected B cells into the S-phase by binding to and raising the level of nuclear S18-2, a pRb-binding protein. This would inhibit pRb binding to E2F1 competitively and lift the block preventing S-phase entry.
Materials and Methods
Plasmids.
EBNA-6 cDNA was subcloned into the GAL4-binding domain containing vector (BD) (Clontech) and into pBabe (puro) retroviral vector from pZip-EBNA-6 (kind gift of Elliott Kieff, Harvard Medical School, Boston). All of the plasmids used in our study are described in supporting information supporting information (SI) Materials and Methods. MSR18-2 cDNA (from a heart cDNA Marathon library, Clontech) was cloned into pEGFP-C1 (GFP, Clontech), the pCMV-Tag 3A c-myc-tagged vector (MT; Stratagene), and the GST bacterial expression vector [GST2TK (GST); Clontech). These constructs encoded the fusion protein that contained 266 aa (full length) of MRS18-2 protein.
Yeast Strains and cDNA Library Screening.
Detailed description is given in SI Materials and Methods.
GST Protein Production and GST Pulldown Assay.
Details of GST production and the GST pulldown assay are described in SI Materials and Methods.
SPR Spectrometer, Surface Modification, and Protein Immobilization.
We have used a scanning SPR spectrometer (BioHelper-01, Institute of Semiconductor Physics, National Academy of Sciences, Kiev) with open measurement architecture (for a description, see ref. 29). More details are presented in SI Materials and Methods.
Antibodies.
A list of the commercial antibodies used in our experiments is presented in SI Materials and Methods. Anti-MRPS18-2 was produced in rabbits to the peptides, corresponding to the residues 4-37 and 177-215 of the MRPS18-2 sequence (clones 75-5 and 76-2).
Cells, Cell Culture, Immunostaining, and Imaging.
A list of the cell lines, conditions of culturing, detailed transfection, staining and imaging protocols and necessary additional information is provided in SI Materials and Methods.
Immunoprecipitation.
The immunoprecipitation procedure is described in SI Materials and Methods.
EMSA.
A detailed protocol of EMSA experiments is given in SI Materials and Methods.
Acknowledgments.
This work was supported by the Swedish Cancer Society, a matching grant from the Concern Foundation (Los Angeles), the Cancer Research Institute (New York), the Swedish Institute, and the Swedish Foundation for Strategic Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801053105/DCSupplemental.
References
- 1.Klein G. Perspectives in studies of human tumor viruses. Front Biosci. 2002;7:d268–d274. doi: 10.2741/A726. [DOI] [PubMed] [Google Scholar]
- 2.Kieff E, Rikinson A. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 2511–2574. [Google Scholar]
- 3.Bornkamm GW, Hammerschmidt W. Molecular virology of Epstein–Barr virus. Philos Trans R Soc London Ser B. 2001;356:437–459. doi: 10.1098/rstb.2000.0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kienzle N, et al. Intron retention may regulate expression of Epstein–Barr virus nuclear antigen 3 family genes. J Virol. 1999;73:1195–1204. doi: 10.1128/jvi.73.2.1195-1204.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radkov SA, et al. Epstein–Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J Virol. 1999;73:5688–5697. doi: 10.1128/jvi.73.7.5688-5697.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knight JS, Lan K, Subramanian C, Robertson ES. Epstein–Barr virus nuclear antigen 3C recruits histone deacetylase activity and associates with the corepressors mSin3A and NCoR in human B-cell lines. J Virol. 2003;77:4261–4272. doi: 10.1128/JVI.77.7.4261-4272.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotter MA, II, Robertson ES. Modulation of histone acetyltransferase activity through interaction of Epstein–Barr nuclear antigen 3C with prothymosin alpha. Mol Cell Biol. 2000;20:5722–5735. doi: 10.1128/mcb.20.15.5722-5735.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grundhoff AT, et al. Characterization of DP103, a novel DEAD box protein that binds to the Epstein–Barr virus nuclear proteins EBNA2 and EBNA3C. J Biol Chem. 1999;274:19136–19144. doi: 10.1074/jbc.274.27.19136. [DOI] [PubMed] [Google Scholar]
- 9.Yan X, Mouillet JF, Ou Q, Sadovsky Y. A novel domain within the DEAD-box protein DP103 is essential for transcriptional repression and helicase activity. Mol Cell Biol. 2003;23:414–423. doi: 10.1128/MCB.23.1.414-423.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robertson ES, et al. Epstein–Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein J kappa. J Virol. 1995;69:3108–3116. doi: 10.1128/jvi.69.5.3108-3116.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robertson ES, Lin J, Kieff E. The amino-terminal domains of Epstein–Barr virus nuclear proteins 3A, 3B, and 3C interact with RBPJ(kappa) J Virol. 1996;70:3068–3074. doi: 10.1128/jvi.70.5.3068-3074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krauer KG, Kienzle N, Young DB, Sculley TB. Epstein–Barr nuclear antigen-3 and -4 interact with RBP-2N, a major isoform of RBP-J kappa in B lymphocytes. Virology. 1996;226:346–353. doi: 10.1006/viro.1996.0662. [DOI] [PubMed] [Google Scholar]
- 13.Touitou R, Hickabottom M, Parker G, Crook T, Allday MJ. Physical and functional interactions between the corepressor CtBP and the Epstein–Barr virus nuclear antigen EBNA3C. J Virol. 2001;75:7749–7755. doi: 10.1128/JVI.75.16.7749-7755.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao B, et al. Transcriptional regulatory properties of Epstein–Barr virus nuclear antigen 3C are conserved in simian lymphocryptoviruses. J Virol. 2003;77:5639–5648. doi: 10.1128/JVI.77.10.5639-5648.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allday MJ, Crawford DH, Thomas JA. Epstein–Barr virus (EBV) nuclear antigen 6 induces expression of the EBV latent membrane protein and an activated phenotype in Raji cells. J Gen Virol. 1993;74:361–369. doi: 10.1099/0022-1317-74-3-361. [DOI] [PubMed] [Google Scholar]
- 16.Allday MJ, Farrell PJ. Epstein–Barr virus nuclear antigen EBNA3C/6 expression maintains the level of latent membrane protein 1 in G1-arrested cells. J Virol. 1994;68:3491–3498. doi: 10.1128/jvi.68.6.3491-3498.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin J, Johannsen E, Robertson E, Kieff E. Epstein–Barr virus nuclear antigen 3C putative repression domain mediates coactivation of the LMP1 promoter with EBNA-2. J Virol. 2002;76:232–242. doi: 10.1128/JVI.76.1.232-242.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshall D, Sample C. Epstein–Barr virus nuclear antigen 3C is a transcriptional regulator. J Virol. 1995;69:3624–3630. doi: 10.1128/jvi.69.6.3624-3630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker GA, et al. Epstein–Barr virus nuclear antigen (EBNA)3C is an immortalizing oncoprotein with similar properties to adenovirus E1A and papillomavirus E7. Oncogene. 1996;13:2541–2549. [PubMed] [Google Scholar]
- 20.Parker GA, Touitou R, Allday MJ. Epstein–Barr virus EBNA3C can disrupt multiple cell cycle checkpoints and induce nuclear division divorced from cytokinesis. Oncogene. 2000;19:700–709. doi: 10.1038/sj.onc.1203327. [DOI] [PubMed] [Google Scholar]
- 21.Knight JS, Sharma N, Robertson ES. Epstein–Barr virus latent antigen 3C can mediate the degradation of the retinoblastoma protein through an SCF cellular ubiquitin ligase. Proc Natl Acad Sci USA. 2005;102:18562–18566. doi: 10.1073/pnas.0503886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knight JS, Robertson ES. Epstein–Barr virus nuclear antigen 3C regulates cyclin A/p27 complexes and enhances cyclin A-dependent kinase activity. J Virol. 2004;78:1981–1991. doi: 10.1128/JVI.78.4.1981-1991.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knight JS, Sharma N, Kalman DE, Robertson ES. A cyclin-binding motif within the amino-terminal homology domain of EBNA3C binds cyclin A and modulates cyclin A-dependent kinase activity in Epstein–Barr virus-infected cells. J Virol. 2004;78:12857–12867. doi: 10.1128/JVI.78.23.12857-12867.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knight JS, Sharma N, Robertson ES. SCFSkp2 complex targeted by Epstein–Barr virus essential nuclear antigen. Mol Cell Biol. 2005;25:1749–1763. doi: 10.1128/MCB.25.5.1749-1763.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maruo S, et al. Epstein–Barr virus nuclear protein EBNA3C is required for cell cycle progression and growth maintenance of lymphoblastoid cells. Proc Natl Acad Sci USA. 2006;103:19500–19505. doi: 10.1073/pnas.0604919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang QH, et al. Cloning and functional analysis of cDNAs with open reading frames for 300 previously undefined genes expressed in CD34+ hematopoietic stem/progenitor cells. Genome Res. 2000;10:1546–1560. doi: 10.1101/gr.140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki T, et al. Proteomic analysis of the mammalian mitochondrial ribosome. Identification of protein components in the 28 S small subunit. J Biol Chem. 2001;276:33181–33195. doi: 10.1074/jbc.M103236200. [DOI] [PubMed] [Google Scholar]
- 28.Cavdar Koc E, Burkhart W, Blackburn K, Moseley A, Spremulli LL. The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. J Biol Chem. 2001;276:19363–19374. doi: 10.1074/jbc.M100727200. [DOI] [PubMed] [Google Scholar]
- 29.Snopok B, Yurchenko M, Szekely L, Klein G, Kashuba E. SPR-based immunocapture approach to creating an interfacial sensing architecture: Mapping of the MRS18-2 binding site on retinoblastoma protein. Anal Bioanal Chem. 2006;386:2063–2073. doi: 10.1007/s00216-006-0867-6. [DOI] [PubMed] [Google Scholar]
- 30.Li M, et al. Kaposi's sarcoma-associated herpesvirus encodes a functional cyclin. J Virol. 1997;71:1984–1991. doi: 10.1128/jvi.71.3.1984-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jung JU, Stager M, Desrosiers RC. Virus-encoded cyclin. Mol Cell Biol. 1994;14:7235–7244. doi: 10.1128/mcb.14.11.7235. [DOI] [PMC free article] [PubMed] [Google Scholar]