Abstract
Canonical Wnt signaling is initiated by binding of Wnt proteins to members of the Frizzled family and subsequent complex formation with lipoprotein receptor-related proteins 5/6 (LRP5/6). Here, we show that LRP6 is palmitoylated on a juxtamembranous cysteine and that palmitoylation is required for exit from the endoplasmic reticulum (ER). We propose that palmitoylation serves to tilt the long, 23-residue transmembrane domain of LRP6 with respect to the plane of membrane to prevent a hydrophobic mismatch and subsequent recognition by the ER quality control. In support of this model, a palmitoylation-deficient LRP6 mutant could be rescued from ER retention by deletion of two to four residues in the transmembrane domain. Importantly, we found that palmitoylation-deficient LRP6 was retained in the ER by a completely novel monoubiquitination-dependent ER retention mechanism. Mutation of a specific lysine indeed abolished ubiquitination of palmitoylation-deficient LRP6 and led to a rescue from ER retention. Finally, at the cell surface, we found that interplay between palmitoylation and ubiquitination was necessary for efficient Wnt signaling.
Keywords: quality control, transmembrane, domain, tilting
The signaling of Wnt plays a critical role in a variety of developmental and adult processes in all metazoan and in diseases such as cancer (1). Canonical Wnt signaling via β-catenin is transduced by two receptor families: the Frizzled proteins and lipoprotein receptor-related proteins (LRPs) 5 and 6. Wnt binds to Frizzled, which then associates with LRP5/6. The cytoplasmic domains of LRP5/6, upon receptor activation by Wnt proteins, recruit the cytosolic scaffold protein axin to the membrane, displacing it from the so-called destruction complex (containing axin, glycogen synthase kinase 3β, Wilms tumor suppressor, and β-catenin) involved in the degradation of β-catenin by the proteasome (for review see ref. 2). This rescue of β-catenin allows it to translocate into the nucleus and interact with T cell factor (TCF)/lymphoid enhancer factor to activate transcription (3). As a result, Wnt activates multiple intracellular cascades, leading to cell differentiation, proliferation, migration, and polarity.
LRP6 is a type I membrane protein with a large, 1,351-aa extracellular domain, containing multiple β-propeller and EGF domains, and a 220-aa-long cytoplasmic tail. As all type I transmembrane proteins, LRP6 is synthesized with an N-terminal signal sequence that targets it to the endoplasmic reticulum (ER). Folding of the LRP6 β-propeller domains in the lumen of the ER requires the help of an ER-resident chaperone called mesoderm development candidate 2 (Mesd) in mammals and Boca in Drosophila (4, 5). In the absence of Mesd, LRP6 is retained in the ER and thus fails to reach the cell surface.
Human LRP6 contains six cysteine residues in its cytoplasmic tail, which are potential sites for palmitoylation (6). This lipid can serve to tether soluble proteins to membranes but is also found attached to the cytoplasmic domains of membrane proteins often close to the transmembrane region (7). The functional consequences of palmitoylation for membrane proteins are not clear, although a role in directing segregation to (6) or away (8) from membrane microdomains (lipid rafts) is often invoked. Palmitoylation is mediated by acyl transferases, 23 of which were identified in the human genome (9).
Our aim was to investigate whether LRP6 is palmitoylated and, if so, to determine the importance of this posttranslational modification for the localization of LRP6 and its ability to mediate Wnt signaling.
Results
LRP6 Is Palmitoylated on a Juxtamembranous Cysteine.
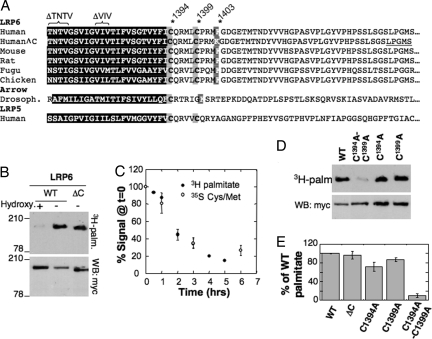
To determine whether LRP6 is palmitoylated, myc-LRP6 was immunoprecipitated from lysats of 3H-palmitic acid-labeled transiently transfected HeLa cells. A radiolabeled band with the same mobility, on SDS gels, as LRP6 was detected (Fig. 1B). When a similar analysis was performed upon expression of LRP6 with a deletion in the cytoplasmic tail (LRP6-ΔC) (10–12) (Fig. 1A), the radiolabeled band migrated at a lower molecular weight (Fig. 1B), indicating that LRP6 was labeled and not a coimmunoprecipitated protein. Labeling was lost by in vitro hydroxylamine treatment (Fig. 1B), indicating that the palmitate was attached to the protein via a thioester bond.
Fig. 1.
LRP6 is palmitoylated during its entire lifetime. (A) Alignment of the TMD (black box) and the 47 first amino acids of the cytoplasmic tails of LRP6 and LRP5 from diverse species. LRP6ΔC corresponds to LRP6 with a truncated cytoplasmic tail that ends by the sequence LPGMS as underlined. ΔTNTV and ΔVIV indicate deletion mutants analyzed in Fig. 3. (B) HeLa cells transfected with myc-LRP6 or myc-LRP6ΔC were labeled with 3H-palmitic acid before immunoprecipitation. Immunoprecipitates were split into two, run on 4–12% SDS gels, and analyzed either by autoradiography or Western blotting (anti-myc). Chemical removal of S-palmitoylation was performed by treating cell extracts for 1 h at room temperature with 1M hydroxylamine hydrochloride, pH 7.2. (C) HeLa cells transfected with myc-LRP6 WT and Mesd were pulsed 30 min either with 3H-palmitic acid or 35S cysteine/methionine and chased for different times at 37°C in complete medium. Myc-LRP6 was immunoprecipitated and subjected to quantitative autoradiography. Results were normalized to the values at time 0. Error bars correspond to SD (n = 3). (D) HeLa cells transiently transfected with myc-LRP6 WT, C1394A, C1399A mutant or double mutant were incubated with 3H-palmitic acid before immunoprecipitation against Myc. Immunoprecipitates were split into two, run on 4–12% gels, and analyzed either by radiography or Western blotting (anti-myc). (E) Radioactive gels from D were quantified by using the Typhoon Imager. Values were first normalized with respect to the expression levels, and then mutants were normalized with respect to WT. Error bars correspond to SD (n = 4).
Because palmitoylation is a reversible modification, we measured the half-life of the palmitate moiety and compared it with that of the protein. Pulse–chase experiments were performed using either 3H-palmitate or [35S]cysteine/methionine. As shown in Fig. 1C, kinetics of degradation of the palmitate moiety and the protein itself were very similar (≈2 h), indicating that LRP6 remained modified during its entire lifetime and thus did not undergo cycles of palmitoylation–depalmitoylation as observed for some proteins (13, 14).
Human LRP6 has two cytoplasmic cysteine residues close to the transmembrane domain, which are conserved in LRP6 from different species and in LRP5 (Fig. 1A). The only exception is the Drosophila homologue Arrow, which has only the most juxtamembranous cysteine (Fig. 1A). We mutated the two cysteines, Cys-1394 and Cys-1399 in human, to alanine. Whereas mutation of a single cysteine had a minor (30%) or no effect, mutation of both led to a complete inhibition of palmitoylation (Fig. 1 D and E). Mutations of palmitoylation sites often lead to the aberrant palmitoylation of neighboring cysteines (8, 15). Therefore, we can only conclude that human LRP6 is palmitoylated on Cys-1394 and/or Cys-1399. In principle, one or more of the three more distal cysteines in the cytoplasmic tail of LRP6 could also be palmitoylated, albeit in a manner that requires previous modification of the proximal cysteines as has been observed for a regulator of G protein signaling (16). This possibility is unlikely because LRP6ΔC, which has only the two juxtamembranous cysteines and lacks the three others (Fig. 1A), shows the same palmitoylation signal as the full-length protein (Fig. 1E).
LRP6 Palmitoylation Is Required for Wnt Signaling.
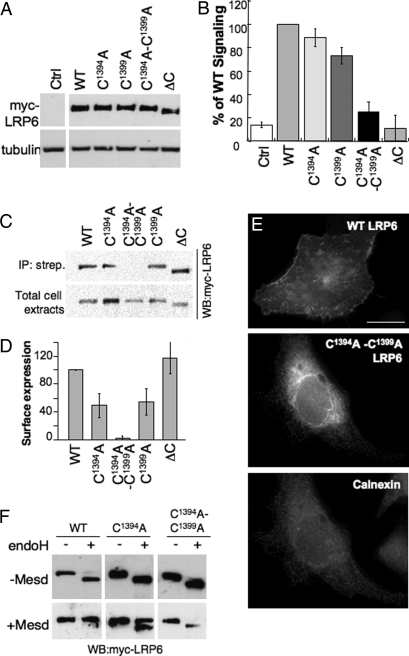
To determine whether palmitoylation of LRP6 is important for Wnt signaling, we made use of a well established reporter assay in HEK293 cells expressing the luciferase reporter construct TOPFLASH, carrying TCF-binding sites, which is directly activated by the TCF/β-catenin complex (17, 18). To ensure proper folding and cell surface delivery of LRP6 (Fig. 2), cells were cotransfected with the chaperone Mesd. Transfection of WT LRP6 induced the TOPFLASH reporter, as did the two single-cysteine mutants (Fig. 2B). In contrast, induction of the reporter was drastically lower in cells expressing palmitoylation-deficient LRP6 (Fig. 2B) whether the analysis was performed after 16, 24, or 48 h of LRP6 transfection [supporting information (SI) Fig. S1]. LRP6ΔC, which has a truncated cytoplasmic tail and is defective in Wnt signaling, was used as a negative control (10–12). The lower reporter induction was not caused by differences in expression levels, which were similar for all proteins (Fig. 2A).
Fig. 2.
Palmitoylation of LRP6 is required for ER exit. (A and B) HEK293 cells were transfected with WT and mutants LRP6 and with Mesd. (A) The expression levels of WT and mutant LRP6 were determined by Western blotting against myc. (B) Wnt signaling was probed with the TOPFLASH reporter assay. The results are shown as mean normalized to WT. Error bars represent the SD (n = 4 experiments performed in triplicate). (C and D) HeLa cells transfected with plasmids encoding WT or mutant myc-tagged LRP6 and Mesd were submitted to surface biotinylation. (C) The levels of LRP6 in streptavidin immunoprecipitate (Upper) and in total cell lysate (Lower) were determined by quantitative Western blotting against myc. The amount of ΔV at the cell surface varied somewhat between experiments as indicated by the error bars and the absence of signal of the chosen blot. (D) Results were first normalized to the expression level and the mutants were normalized to WT. Error bars correspond to SD (n = 4). (E) HeLa cells expressing WT or mutant myc-LRP6 were analyzed by immunofluorescence against myc and calnexin. (Scale bar: 10 μM.) (F) HeLa cells were transfected 24 h with plasmids encoding WT or mutant myc-tagged LRP6 with and without Mesd. Forty micrograms of cell extracts was treated or not with endo H and analyzed by SDS/PAGE and Western blotting against myc.
Palmitoylation-Deficient LRP6 Is Retained in the ER.
To understand why palmitoylation-deficient LRP6 was impaired in Wnt signaling, we first analyzed whether it was properly targeted to the plasma membrane by using a surface biotinylation assay. Whereas biotinylation could readily be observed for WT LRP6 and the single-cysteine mutants, no signal was detected for the double-cysteine mutant (Fig. 2 C and D), showing that palmitoylation of LRP6 is required for proper plasma membrane targeting. When analyzing the localization of palmitoylation-deficient LRP6 by immunofluorescence, we found staining of the nuclear membrane and a reticulate pattern throughout the cytoplasm, colocalizing with the ER marker calnexin (Fig. 2E Middle and Bottom), in contrast to staining of WT LRP6, which showed plasma membrane localization (Fig. 2E Top). To confirm the ER localization of the mutant, we analyzed whether the N-linked sugars on LRP6 (19) had undergone trimming by Golgi enzymes by monitoring the sensitivity to endoglycosidase H (endo H). In the absence of Mesd, WT LRP6 was endo H-sensitive, confirming previous findings that Mesd is required for proper folding of the protein and exit from the ER (Fig. 2F) (4, 10). In the presence of Mesd, WT LRP6 was, however, fully endo H-resistant (Fig. 2F). In contrast, the double-cysteine mutant remained endo H-sensitive in the presence of Mesd (Fig. 2F), confirming its retention in the early secretory pathway.
Palmitoylation and the Effective Length of the Transmembrane Domain.
The above results show that LRP6 is palmitoylated and that a palmitoylation-deficient mutant localizes to the ER, suggesting that palmitoylation occurs in the ER and is required for ER exit. This hypothesis was confirmed by the finding that inhibition of ER to Golgi transport by brefeldin A had no effect on the incorporation of 3H-palmitate into LRP6 (Fig. S2).
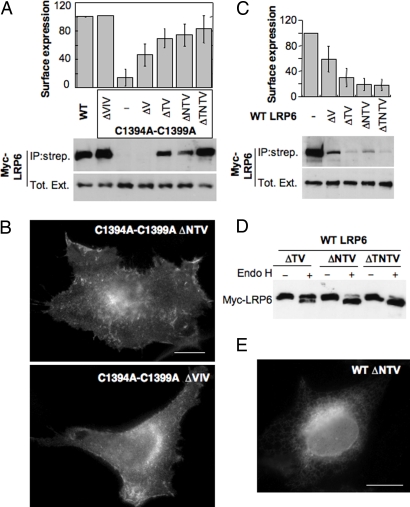
Palmitoylation of LRP6 occurs on juxtamembranous cysteines and therefore one could envision that this lipid modification affects the conformation of the transmembrane domain (TMD). More specifically, palmitoylation of a cysteine in very close proximity of the TMD could, because of steric hindrance upon insertion of the lipid into the membrane, induce the TMD to tilt. The TMD of LRP6 is predicted to be 23 aa long, which is two residues longer (≈3 Å in a helix configuration) than the vast majority of type I and type II transmembrane proteins (Fig. S3). Moreover, the ER membrane has been reported to be thinner by some 5 Å than the plasma membrane because of the lower cholesterol content and/or the protein composition (20, 21). If the TMD of LRP6 was oriented perpendicular to the plane of the ER membrane, the length of TMD of LRP6 would exceed the thickness of the membrane, thus leading to a so-called hydrophobic mismatch (22). This hydrophobic mismatch would presumably be detected by the ER quality control and lead to ER retention. We propose that palmitoylation of juxtamembranous cysteines leads to tilting of the LRP6 transmembrane helix, thus reducing the effective length of the helix and allowing it to evolve normally in the ER and exit the compartment. A prediction from this model is that if the TMD was shorter, there would be no requirement for tilting, and thus for palmitoylation, to exit the ER. To test this possibility, we generated, in the background of the palmitoylation-deficient LRP6, mutants truncated by one to four residues in the TMD (Fig. 1A): ΔTNTV from the N terminus of the helix, ΔNTV, ΔTV, and ΔV. To test whether the position of the deletion was important, we also generated a ΔVIV, in which three residues were removed from the middle of the TMD. To monitor ER exit, we directly performed surface biotinylation. Remarkably, deletion of two residues from the TMD of palmitoylation-deficient LRP6 was sufficient to rescue the protein from ER retention, leading to plasma membrane delivery (Fig. 3A). Confirming these findings, we found that these TMD truncation mutants acquired endo H resistance (data not shown). Surface localization was confirmed by immunofluorescence as shown in Fig. 3B for the ΔNTV and ΔVIV mutants.
Fig. 3.
Effect of TMD truncation on plasma membrane targeting of LRP6. (A) (Upper) HeLa cells were transfected 24 h with plasmids encoding Mesd and WT or palmitoylation-deficient myc-LRP6 mutants with deletions of one to four residues in their TMD (Fig. 1A): ΔTNTV, ΔNTV, ΔTV, ΔV, ΔVIV in the middle of the TMD. (Lower) The levels of LRP6 in the streptavidin immunoprecipitate and the total cell lysate were determined by quantitative anti-myc Western blotting. Error bars correspond to SD (n = 6 except for ΔTNTV, n = 4). Results were first normalized to the expression level and then to the surface expression of WT. (B) Localization of LRP6 mutants, coexpressed with Mesd, was analyzed by immunofluorescence. (Scale bar: 10 μm.) (C) (Upper) HeLa cells were transfected 24 h with plasmids encoding Mesd and LRP6 WT with deletions of zero to four residues in their TMD: ΔTNTV, ΔNTV, ΔTV, ΔV. (Lower) The levels of LRP6 in the streptavidin immunoprecipitate and the total cell lysate were determined by quantitative anti-myc Western blotting. Error bars correspond to SD (n = 3). (D) HeLa cells were transfected 24 h with the same plasmids as in C with and without Mesd. Forty micrograms of cell extracts were untreated or not with endo H and analyzed by SDS/PAGE and Western blotting against myc. (E) Localization of WT LRP6 ΔNTV, coexpressed with Mesd, was analyzed by immunofluorescence. (Scale bar: 10 μm.)
The above results support the proposed model that palmitoylation serves to tilt the long TMD of LRP6. A second prediction of the model is that truncation of the TMD in the WT palmitoylated background should lead to a too short effective transmembrane region, thus leading to a reverse hydrophobic mismatch, where the membrane would be thicker than the effective length of the TMD, which would presumably again be recognized by an ER quality-control mechanism. We therefore generated TMD truncation mutants in the WT background. As shown in Fig. 3C, truncations of the TMD of WT LRP6 prevented proper plasma membrane targeting. The truncation mutants showed endo H sensitivity (Fig. 3D) and localized to the ER as shown in Fig. 3E for the WT LRP6 ΔNTV mutant.
Palmitoylation Affects the Incorporation of LRP6 into COPII-Coated Vesicles.
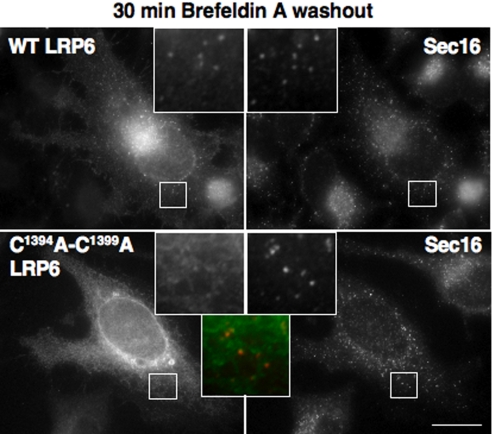
We next sought to understand why palmitoylation-deficient LRP6 localized the ER and investigated whether corporation into COPII-coated vesicles, which is required for ER exit (23), is impaired for the palmitoylation-deficient mutant. This possibility is supported by previous findings that the length of the TMD plays a role in the recruitment of type I membrane proteins into COPII vesicles (24, 25), the optimal length being 21 residues, whereas shorter, i.e., 18 residues, or longer TMDs lead to ER retention (25). To monitor ER exit, we first treated cells with brefeldin A for 1 h to block ER to Golgi transport and accumulate newly synthesized WT LRP6 in the ER [because the bulk of LRP6 was endo H-resistant (Fig. 2F) before brefeldin treatment]. Brefeldin A was subsequently washed out during 30 min, and LRP6 localization was performed. Sec16, an organizer of ER exit site (26), was used as a marker. Despite the low amount of WT LRP6 initially present in the ER, the protein rapidly accumulated in sec16-positive structures (Fig. 4). In contrast, despite the ER localization of the bulk of palmitoylation-deficient LRP6, sec16-positive structures only weakly stained for LRP6 (Fig. 4), suggesting that incorporation into ER exit sites was, not blocked, but strongly inhibited.
Fig. 4.
ER exit of WT and mutant LRP6. HeLa cells were transfected 24 h with plasmids encoding WT or mutant myc-LRP6 and cotransfected with Mesd. Cells were treated with brefeldin A for 1 h and then incubated in a brefeldin-free medium for 30 min at 37°C before fixation. Cells were analyzed by immunofluorescence against myc for LRP6 (green in the merged insets) and human sec16 (red in the merged insets). (Scale bar: 10 μM.)
Palmitoylation Deficiency Leads to Monoubiquitination of LRP6.
It has been observed that palmitoylation-deficient mutants of the yeast SNARE Tlg1 (27) and the anthrax toxin receptor TEM8 (8) undergo ubiquitination, which targets the proteins to degradation in the vacuole in yeast and in lysosomes in mammalian cells. This increase in ubiquitination was proposed to be caused by either an increased accessibility of the lysines in proximity of the palmitoylation site by E3 ubiquitin ligase (27) or a redistribution of the protein to membrane domains containing the E3 ligase (8). We here wondered whether in the absence of palmitoylation, LRP6 would also undergo ubiquitination. As shown in Fig. 5A, palmitoylation-deficient LRP6 was ubiquitinated, whereas the WT protein was not, whether Mesd had been coexpressed or not, i.e., whether LRP6 was misfolded in the ER or properly targeted to the plasma membrane. A single ubiquitinated band was observed, suggesting that LRP6 C1394A-C1399A underwent monoubiquitination (or multiubiquitination) rather than polyubiquitination, the latter being a signal for degradation (28). Consistent with this observation, palmitoylation-deficient LRP6 did not undergo premature degradation and in fact had a half-life very similar to that of the WT protein (Fig. S4). Degradation of the WT and palmitoylation-deficient LRP6 did, however, occur by different pathways because degradation of WT LRP6 was inhibited by leupeptin, an inhibitor of lysosomes enzymes, whereas that of palmitoylation-deficient LRP6 was not (Fig. S4).
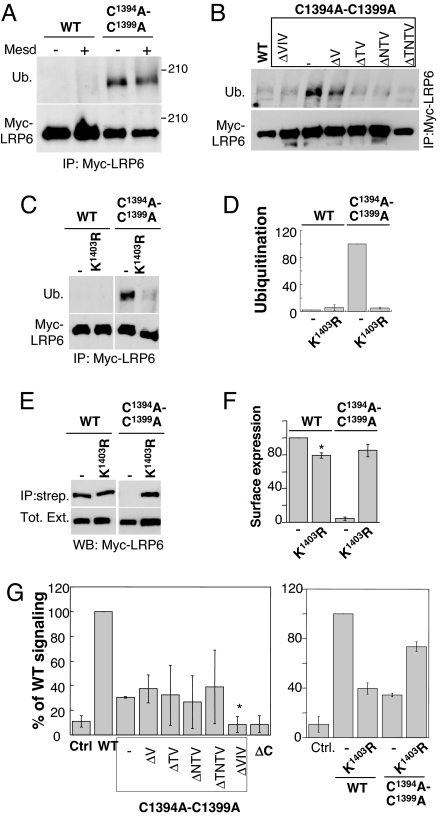
Fig. 5.
ER retention of palmitoylation-deficient LRP6 is ubiquitin-mediated. (A–C) HeLa cells were transfected 24 h with plasmids encoding WT or mutant myc-LRP6 and cotransfected or not with Mesd. Immunoprecipitates against myc-LRP6 were analyzed by SDS/PAGE and Western blotting against ubiquitin and myc. (D) The levels of WT and mutant LRP6 in the streptavidin immunoprecipitate and the total cell lysat from C were quantified with a Typhoon imager and normalized to that of the C1394A-C1399A-LRP6 mutant. Error bars correspond to SD (n = 3). (E and F) HeLa cells were transfected 24 h with plasmids encoding Mesd and WT or myc-LRP6 mutants. The levels of LRP6 in the streptavidin immunoprecipitate and the total cell lysate were determined by quantitative anti-myc Western blotting. Error bars correspond to SD (n = 4). Results were first normalized to the expression level and then to the surface expression of WT. (G) WT or mutant myc-tagged LRP6 cotransfected with Mesd into HEK293 cells were tested by using the TOPFLASH reporter assay. The results are shown as mean normalized to WT and standard deviations of the mean of three independent experiments done in triplicate. In Left none of the TMD truncation mutants were significantly different from the full-length C1394A-C1399A mutant except the ΔVIV mutant, which was significantly more deficient in signaling (*, P < 0.005).
Because ubiquitination did not seem to target palmitoylation-deficient LRP6 to degradation, we evaluated the possible existence of an ubiquitin-dependent ER retention mechanism. Consistent with such a mechanism, ubiquitination was no longer observed for palmitoylation-deficient LRP6 with deletions of two or more residues from the TMD (Fig. 5B), which as shown in Fig. 3A, exit the ER. We next searched for the site of ubiquitination. We (8) and others (27) have observed that absence of palmitoylation could in certain proteins lead to ubiquitination of a nearby lysine. We therefore mutated the first lysine after the palmitoylation sites, Lys-1403, to arginine. Remarkably, this mutation not only led to a >95% inhibition of ubiquitination (Fig. 5 C and D) but to a full recovery of plasma membrane targeting (Fig. 5 E and F). Altogether, these observations show the existence of a novel ER retention/quality-control system that is controlled by the monoubiquitination of the cytoplasmic domain(s) of transmembrane proteins that have a folding/assembly defect or have not yet reached their native conformation.
Wnt Signaling.
Having rescued cell surface localization of palmitoylation-deficient LRP6 either by truncation of the TMD or by the K1403R mutation, we measured their ability to mediate Wnt signaling by using the TOPFLASH reporter assay. Despite their surface localization, the TMD truncation mutants were inefficient in Wnt signaling (Fig. 5G Left), indicating that palmitoylation and/or the length of TMD domain are important for proper signaling. In contrast, the K1403R mutation in the background of the double C1394A-C1399A mutation led to an almost full recovery of the Wnt signaling ability (Fig. 5G Right). This observation suggests that palmitoylation is not per se required for efficient Wnt signaling. Interestingly however, the K1403R mutation in the WT background, which is the proper control for the C1394A-C1399A- K1403R mutant, led to a drastic inhibition of Wnt signaling (Fig. 5G Right) despite a proper surface localization (Fig. 5 E and F). The low signaling ability of a K1403R mutant suggests a requirement for ubiquitination in Wnt signaling. Taken together, these observations suggest that interplay, the details of which remain to be established, between palmitoylation and ubiquitination is required for efficient Wnt signaling.
Discussion
We found that the Wnt signaling coreceptor LRP6 is palmitoylated shortly after synthesis and remains palmitoylated during its entire lifetime. The cellular site for palmitoylation differs for each membrane protein: the ER for the influenza virus hemaglutinin, the Cis-Golgi for the vesicular stomatitis G protein, the Golgi for a subunit of the GABA receptor, or the plasma membrane and endosomes for the mannose-6-phosphate receptor (6, 7). We found that palmitoylation of LRP6 occurs in the ER and is required for exit from this compartment. A similar requirement has been reported for the polytopic membrane protein yeast chitin synthase Chs3 (29).
Our study indicates that palmitoylation of LRP6 serves multiple roles. First, it ensures ER exit, under conditions of proper folding of the ectodomain, i.e., in the presence of Mesd. Our data support the hypothesis that palmitoylation induces a tilt of the TMD with respect to the plane of the membrane to reduce the effective length of the TMD, allowing the protein to evolve in the thin lipid membrane of the ER. Second, once the protein has reached the plasma membrane, palmitoylation renders LRP6 dependent on Lys-1403, likely through its ubiquitination, to mediate Wnt signaling. Future studies will be required to elucidate the underlying mechanisms. An interesting possibility is, however, that palmitoylation of LRP6 orchestrates the optimal interaction with Casein kinase γ, a palmitoylated kinase that is required for signal transduction by LRP6 (30).
The last finding described here is the existence of a ubiquitin-dependent quality-control system in the ER. It is well known that exit of proteins from the ER requires proper folding and assembly (31). Assistance in folding can involve both luminal ER chaperones and folding enzymes and the cytosolic chaperone machineries Hsc-Hsp70 and Hsp-90. Quality control is best understood for glycosylated proteins that go through the calnexin/calreticulin cycle in a glucosidation-dependent manner (31). This glucose-dependent folding and quality-control system operates in the lumen of the ER. We now propose an ER-retention mechanism that operates on the cytosolic side of the ER that would control for correct conformations of the TMDs and cytosolic domains. One could envision that proteins, such as LRP6 that awaits palmitoylation, undergo cycles of ubiquitination–deubiquitination until the proper conformation is reached, allowing subsequent ER exit. Identifying the molecular players in this novel ER quality-control pathway is clearly of major interest.
Experimental Procedures
Reagents.
Anti-myc mAbs were obtained from Covance; anti-human transferrin receptor was from Zymed; anti-tubulin and streptavidin-Sepharose-conjugated beads were from Sigma–Aldrich; anti-caveolin 1 polyclonal, anti-ubiquitin (P4D1) antibodies, and anti-myc-Sepharose-conjugated beads were from Santa Cruz Biotechnology; HRP secondary antibodies were from Pierce; FITC-conjugated secondary antibodies were from Molecular Probes; and monoclonal anti-sec16 (KIAA0310) was from Bethyl Laboratories. Monoclonal anti-erlin-1 and polyclonal anti-calnexin antibodies were gifts from S. Robbins (University of Calgary, Calgary, Canada) and A. Helenius (ETH Zurich, Zurich), respectively. Endo H was from New England Biolabs and used as recommended. Plasmids encoding human myc-LRP6 were provided by G. Davidson (DKFZ, Heidelberg, Germany), and hMesd was provided by B. Holdener (State University of New York, Stony Brook, NY). Mutations were generated with the Quikchange mutagenesis kit (Stratagene).
Transfections.
Plasmids were transfected into HeLa cells for 24 h (2 μg myc-LRP6 WT or mutants, with 0.5 μg Mesd or empty vector, cDNA/9.6 cm2 per plate) using Fugene (Roche Diagnostics). For the dual luciferase assay, plasmids and reagents were from Promega. HEK293 cells were transfected with 0.1 μg TOP-luciferase, 50 ng TK-Renilla, and 0.5 μg myc-LRP6 WT or mutants, with 0.25 μg Mesd cDNA/9.6 cm2 per plate. After 24 h, cells were lysed, and luciferase activity was determined following the manufacturer's instructions (Promega).
Biochemical Assays.
For cell surface protein biotinylation, HeLa cells were transfected 24 h, treated 30 min with 0.2 mg/ml sulfo-NHS-SS-biotin (Pierce) at 4°C, quenched with 100 mM NH4Cl, and lysed in IP buffer as described below. The lysat was immunoprecipitated with streptavidin-coated Sepharose beads. For immunoprecipitations of LRP6, cells were lysed 30 min at 4°C in IP buffer (0.5% Nonidet P-40, 500 mM Tris·HCl (pH 7.4), 20 mM EDTA, 10 mM NaF, 2 mM benzamidine, and a mixture of protease inhibitors) and centrifuged 3 min at 2,000 × g, and supernatants were incubated 16 h at 4°C with anti-myc-coupled Sepharose beads. Autoradiography and Western blotting were quantified by using the Typhoon Imager (Image QuantTool; GE Healthcare).
Detection of palmitoylation and metabolic labeling was performed as described (8) and quantified with the Typhoon Imager.
Immunofluorescence Microscopy.
LRP6-expressing HeLa cells were fixed with 3% paraformaldehyde, permeabilized with 0.1% Triton X-100, and labeled with anti-myc mAbs followed by Alexa 488-conjugated goat anti-mouse IgG. For colocalization with calnexin, cells were fixed with methanol. For staining of ER exit sites with anti-sec16 antibodies, the fixation protocol described by Hammond et al. (32) was used. Images were acquired with a ×63 lens on an Axiovert 200 microscope equipped with a VisioCam camera (Zeiss).
Acknowledgments.
We thank G. Davidson for providing LRP6 cDNA, B. Holdener for Mesd cDNA, R. Watanabe (University of Geneva, Geneva, Switzerland) for anti-sec16 antibodies and advice for staining ER exit sites, A. Helenius for anti-calnexin, and members of F.G.v.d.G.'s laboratory for fruitful discussions and critical reading of the manuscript. This work was supported by a grant from the Swiss National Science Foundation. F.G.v.d.G. is an International Fellow of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. N.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710389105/DCSupplemental.
References
- 1.Cadigan KM, Nusse R. Wnt signaling: A common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 2.Kikuchi A, Yamamoto H, Kishida S. Multiplicity of the interactions of Wnt proteins and their receptors. Cell Signal. 2007;19:659–671. doi: 10.1016/j.cellsig.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 4.Koduri V, Blacklow SC. Requirement for natively unstructured regions of mesoderm development candidate 2 in promoting low-density lipoprotein receptor-related protein 6 maturation. Biochemistry. 2007;46:6570–6577. doi: 10.1021/bi700049g. [DOI] [PubMed] [Google Scholar]
- 5.Hsieh JC, et al. Mesd encodes an LRP5/6 chaperone essential for specification of mouse embryonic polarity. Cell. 2003;112:355–367. doi: 10.1016/s0092-8674(03)00045-x. [DOI] [PubMed] [Google Scholar]
- 6.Linder ME, Deschenes RJ. Palmitoylation: Policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 7.Bijlmakers MJ, Marsh M. The on-off story of protein palmitoylation. Trends Cell Biol. 2003;13:32–42. doi: 10.1016/s0962-8924(02)00008-9. [DOI] [PubMed] [Google Scholar]
- 8.Abrami L, Leppla SH, van der Goot FG. Receptor palmitoylation and ubiquitination regulate anthrax toxin endocytosis. J Cell Biol. 2006;172:309–320. doi: 10.1083/jcb.200507067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell DA, Vasudevan A, Linder ME, Deschenes RJ. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J Lipid Res. 2006;47:1118–1127. doi: 10.1194/jlr.R600007-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Lu W, He X, Bu G. Modulation of LRP6-mediated Wnt signaling by molecular chaperone Mesd. FEBS Lett. 2006;580:5423–5428. doi: 10.1016/j.febslet.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 11.Tamai K, et al. A mechanism for Wnt coreceptor activation. Mol Cell. 2004;13:149–156. doi: 10.1016/s1097-2765(03)00484-2. [DOI] [PubMed] [Google Scholar]
- 12.Tamai K, et al. LDL-receptor-related proteins in Wnt signal transduction. Nature. 2000;407:530–535. doi: 10.1038/35035117. [DOI] [PubMed] [Google Scholar]
- 13.Stockli J, Rohrer J. The palmitoyltransferase of the cation-dependent mannose 6-phosphate receptor cycles between the plasma membrane and endosomes. Mol Biol Cell. 2004;15:2617–2626. doi: 10.1091/mbc.E03-11-0808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rocks O, et al. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 2005;307:1746–1752. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- 15.Schweizer A, Kornfeld S, Rohrer J. Cysteine34 of the cytoplasmic tail of the cation-dependent mannose 6-phosphate receptor is reversibly palmitoylated and required for normal trafficking and lysosomal enzyme sorting. J Cell Biol. 1996;132:577–584. doi: 10.1083/jcb.132.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osterhout JL, et al. Palmitoylation regulates regulator of G proteinsignaling (RGS) 16 function. II. Palmitoylation of a cysteine residue in the RGS box is critical for RGS16 GTPase accelerating activity and regulation of Gi-coupled signaling. J Biol Chem. 2003;278:19309–19316. doi: 10.1074/jbc.M210124200. [DOI] [PubMed] [Google Scholar]
- 17.Korinek V, et al. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 18.Mao B, et al. Kremen proteins are Dickkopf receptors that regulate Wnt/β-catenin signalling. Nature. 2002;417:664–667. doi: 10.1038/nature756. [DOI] [PubMed] [Google Scholar]
- 19.Khan Z, Vijayakumar S, Villanueva de la Torre T, Rotolo S, Bafico A. Analysis of endogenous LRP6 function reveals a novel feedback mechanism by which Wnt negatively regulates its receptor. Mol Cell Biol. 2007;27:7291–7301. doi: 10.1128/MCB.00773-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitra K, Ubarretxena-Belandia I, Taguchi T, Warren G, Engelman DM. Modulation of the bilayer thickness of exocytic pathway membranes by membrane proteins rather than cholesterol. Proc Natl Acad Sci USA. 2004;101:4083–4088. doi: 10.1073/pnas.0307332101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bretscher MS, Munro S. Cholesterol and the Golgi apparatus. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- 22.Nyholm TK, Ozdirekcan S, Killian JA. How protein transmembrane segments sense the lipid environment. Biochemistry. 2007;46:1457–1465. doi: 10.1021/bi061941c. [DOI] [PubMed] [Google Scholar]
- 23.Watanabe R, Riezman H. Differential ER exit in yeast and mammalian cells. Curr Opin Cell Biol. 2004;16:350–355. doi: 10.1016/j.ceb.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 24.Letourneur F, Cosson P. Targeting to the endoplasmic reticulum in yeast cells by determinants present in transmembrane domains. J Biol Chem. 1998;273:33273–33278. doi: 10.1074/jbc.273.50.33273. [DOI] [PubMed] [Google Scholar]
- 25.Nufer O, Kappeler F, Guldbrandsen S, Hauri HP. ER export of ERGIC-53 is controlled by cooperation of targeting determinants in all three of its domains. J Cell Sci. 2003;116:4429–4440. doi: 10.1242/jcs.00759. [DOI] [PubMed] [Google Scholar]
- 26.Watson P, Townley AK, Koka P, Palmer KJ, Stephens DJ. Sec16 defines endoplasmic reticulum exit sites and is required for secretory cargo export in mammalian cells. Traffic. 2006;7:1678–1687. doi: 10.1111/j.1600-0854.2006.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valdez-Taubas J, Pelham H. Swf1-dependent palmitoylation of the SNARE Tlg1 prevents its ubiquitination and degradation. EMBO J. 2005;24:2524–2532. doi: 10.1038/sj.emboj.7600724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haglund K, Di Fiore PP, Dikic I. Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem Sci. 2003;28:598–603. doi: 10.1016/j.tibs.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Lam KK, et al. Palmitoylation by the DHHC protein Pfa4 regulates the ER exit of Chs3. J Cell Biol. 2006;174:19–25. doi: 10.1083/jcb.200602049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davidson G, et al. Casein kinase 1 γ couples Wnt receptor activation to cytoplasmic signal transduction. Nature. 2005;438:867–872. doi: 10.1038/nature04170. [DOI] [PubMed] [Google Scholar]
- 31.Hebert DN, Molinari M. In and out of the ER: Protein folding, quality control, degradation, and related human diseases. Physiol Rev. 2007;87:1377–1408. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- 32.Hammond AT, Glick BS. Dynamics of transitional endoplasmic reticulum sites in vertebrate cells. Mol Biol Cell. 2000;11:3013–3030. doi: 10.1091/mbc.11.9.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]