Abstract
The neural-related genes Sox2, Pax6, Otx2, and Rax have been associated with severe ocular malformations such as anophthalmia and microphthalmia, but it remains unclear as to how these genes are linked functionally. We analyzed the upstream signaling of Xenopus Rax (also known as Rx1) and identified the Otx2 and Sox2 proteins as direct upstream regulators of Rax. We revealed that endogenous Otx2 and Sox2 proteins bound to the conserved noncoding sequence (CNS1) located ≈2 kb upstream of the Rax promoter. This sequence is conserved among vertebrates and is required for potent transcriptional activity. Reporter assays showed that Otx2 and Sox2 synergistically activated transcription via CNS1. Furthermore, the Otx2 and Sox2 proteins physically interacted with each other, and this interaction was affected by the Sox2-missense mutations identified in these ocular disorders. These results demonstrate that the direct interaction and interdependence between the Otx2 and Sox2 proteins coordinate Rax expression in eye development, providing molecular linkages among the genes responsible for ocular malformation.
Keywords: anophthalmia, comparative genomics, microphthalmia, rx1, Xenopus
Severe forms of ocular malformation, such as anophthalmia (absence of the eye) and microphthalmia (very small eye), appear in the human population at a frequency of ≈1 per 5,000–10,000 persons (1). These malformations of the eye are caused by genetic and molecular disruption of the development of the anterior neuroectoderm and forebrain, which contribute to the nascent eye. The vertebrate eye develops from a part of the forebrain that is called the optic vesicle. Progress has been made in understanding the molecular and cellular mechanisms underlying the formation of the optic vesicle, revealing the involvement of several transcription factors in its development (2). Recent advances in human genetics have identified the causative genes for ocular malformations, which include Sox2, Pax6, Otx2, and Rax (3–7). These genes all encode transcription factors that are highly conserved among vertebrates.
Sox2 encodes a high-mobility group (HMG) domain-containing transcription factor, which is a member of the SOXB1 subfamily in the larger family of SOX proteins. Heterozygous mutations in Sox2 have been reported as the cause of 10–20% of cases of anophthalmia and severe microphthalmia (3, 8). The requirement for Sox2 during eye development has been confirmed by the generation of a gene-dosage allelic series of Sox2 mutations in the mouse (9). The Sox2 protein is thought to exert its function in cooperation with other transcription factors (10). During lens development, Sox2 interacts with Pax6 to bind cooperatively to DNA, thereby regulating δ-crystallin expression (11). Pax6, which is an essential eye regulator gene, was the first causative gene for anophthalmia to be identified (6, 12).
A candidate gene approach subsequently identified Otx2 as another causative gene for anophthalmia and microphthalmia (4). Otx2 encodes a bicoid-type homeodomain transcription factor and is a vertebrate homolog of otd, which was identified in the fruit fly as being required for the formation of the anterior neural structure (13, 14). The requirements of Otx2 for the formation of the brain and eye in vertebrates have been investigated by generating targeted mutant mice (15). Murine Otx2 is expressed in the visceral endoderm and anterior neuroectoderm, which eventually develop into the eye and brain. Otx2-null mice show a severe head defect, which is accompanied by abnormal development of the visceral endoderm that comes in contact with the neuroectoderm and directs its fate. A study using chimerae has demonstrated that Otx2 in the visceral endoderm is required for induction of the forebrain and midbrain and that Otx2 in the anterior neuroectoderm is required for its regional specification (16).
Rax, which is a paired-type homeobox gene, is another causative gene for anophthalmia (5, 17, 18). Rax-null mice lack eyes (18). The Rax protein directly or indirectly regulates the expression of downstream genes, including Xhmgb3, IRBP, arrestin, Xhairy2, Zic2, and XOptx2 (19–21). These genes are involved in the specification of the eye field and the proliferation of retinal progenitor cells. Furthermore, previous studies have shown that the 5′-upstream regions of Rax in Xenopus laevis and Xenopus tropicalis contain cis-regulatory elements that direct Rax expression in the developing eye (22, 23). However, the trans-acting factors that bind directly to the cis-regulatory elements to regulate Rax expression remain unknown.
Initially, we explored the upstream regulation of Rax expression in the African clawed frog (X. laevis). The present work provides evidence for two direct linkages between the causative genes for ocular malformation: (i) the transcription of Rax is regulated directly by the Otx2 and Sox2 proteins; and (ii) the Otx2 protein interacts directly with the Sox2 protein, and their interdependence coordinates transcriptional activation.
Results
Conserved Noncoding Sequence 1 (CNS1) Has cis-Regulatory Activity.
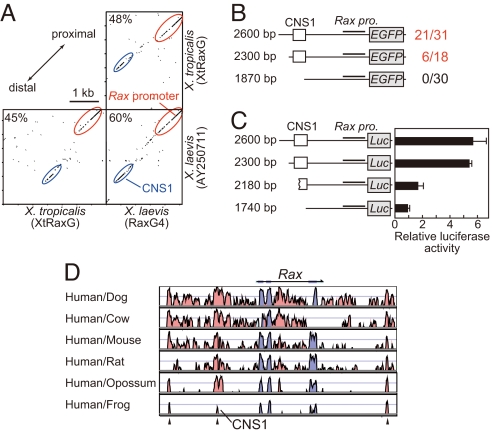
Initially, we compared three upstream sequences of frog Rax (X. laevis, AY250711 and RaxG4; X. tropicalis, XtRaxG) by pairwise alignment and dot-matrix analyses (Fig. 1A). Although the total sequence of ≈3 kb showed low similarity among the three clones, two restricted regions showed high sequence similarity. One of these regions contained a potential TATA motif for the Rax gene promoter, whereas the other, located 2 kb upstream of the promoter, neither contained a promoter nor represented a gene; we named this latter region CNS1. Although CNS1 is a noncoding region, it is specifically conserved among frog genomes, which suggests that it has cis-regulatory activity. Transgenic analyses demonstrated that regions upstream of X. laevis Rax drove EGFP expression in optic vesicles where endogenous Rax mRNA is expressed and that loss of CNS1 abolished this expression [Fig. 1B and supporting information (SI) Fig. S1A]. Similarly, the upstream sequences that contained CNS1 showed strong transcriptional activities in the luciferase assay, whereas upstream sequences that partially or completely lacked CNS1 showed markedly lower activities (Fig. 1C). These two reporter assays showed that CNS1 exerts a cis-regulatory activity in X. laevis embryos.
Fig. 1.
Conservation and cis-regulatory activity of the sequences upstream of Rax. (A) Comparison of three upstream sequences of frog Rax (X. laevis, AY250711 and RaxG4; X. tropicalis, XtRaxG). A proximal region that contains a minimal promoter and transcriptional start site (red), as well as a distal noncoding region (blue), are highly conserved among the three clones. The distal region is termed the conserved noncoding sequence 1 (CNS1). The percentages indicate sequence similarities. (B) Transgenesis in X. laevis. Two constructs contain CNS1 and up-regulate the EGFP reporter. The number of transgenic embryos that express EGFP in the optic vesicles of normally developing embryos obtained in this assay is indicated on the right. Black lines indicate the Rax promoter. (C) Luciferase reporter activities of sequences upstream of Rax in X. laevis embryos. (D) VISTA view of the occurrence of the conserved sequence domain in the genomic region that encompasses the Rax gene. Colored peaks (purple, coding; pink, noncoding) indicate regions of at least 100 bp and 60% similarity. There are three conserved noncoding regions (arrowheads) in a region of ≈20 kb, and the most proximal one is CNS1.
CNS1 Is Conserved Among Vertebrates and Contains both Otx- and Sox-Binding Sites.
Because CNS1 is conserved and has transcriptional activity in frogs and as Rax is a highly conserved transcriptional factor among vertebrates, we investigated whether CNS1 is conserved exclusively among frogs or more broadly among vertebrates by using the VISTA Browser (24). We plotted sequence similarities over a 20-kb genomic region that contained Rax, with the human genome as the base sequence (Fig. 1D). Thus, we identified three conserved noncoding sequences in the Rax regions of the human, dog, cow, mouse, rat, opossum, and frog genomes (Fig. 1D, arrowhead). The most-proximal sequence was CNS1, which is described above as a sequence that is conserved among frog genomes.
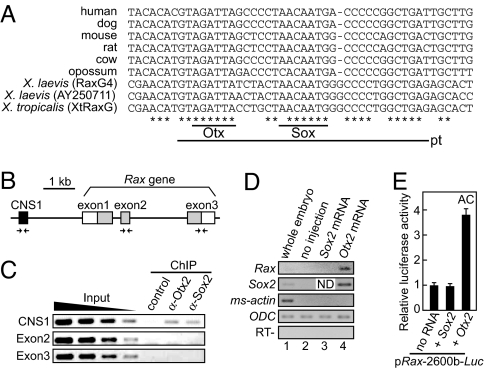
Phylogenetic footprinting using vertebrate CNS1 sequences identified conserved 35-nucleotide regions (pentatriacontamer, pt) in the CNS1 domain of Rax, including putative Otx- and Sox-binding sites (Fig. 2A). Given the causative role of Rax in anophthalmia, we hypothesized that the anophthalmia-associated proteins Otx2 and Sox2 would bind to these sites. In Xenopus at the late neurula stage, we observed that both genes were coexpressed with Rax in the optic vesicle (Fig. S1B). Electrophoretic mobility shift assays (EMSAs) confirmed that the Xenopus Otx2 and Sox2 proteins bound to these putative binding sites in vitro (Fig. S2 A–D). To investigate whether endogenous Otx2 and Sox2 proteins bind to Rax CNS1 in the optic vesicle, we performed chromatin immunoprecipitation (ChIP) assays (Fig. 2 B and C). Genomic fragments bound by the Otx2 or Sox2 protein were immunoprecipitated with specific antibodies and analyzed by PCR with primer sets designed to amplify the CNS1 region. As a result, Rax CNS1 was immunoprecipitated by the anti-Otx2 and anti-Sox2 antibodies but not by nonspecific IgG (normal rabbit IgG) or when using primers that amplify a part of Rax exon 2 or exon 3. We also confirmed the binding capabilities of overexpressed myc-Otx2 and myc-Sox2 proteins to Rax CNS1 using the ChIP assay with the anti-myc antibody (Fig. S2 E and F). Taken together, these experiments demonstrate specific binding of the Otx2 and Sox2 proteins to Rax CNS1.
Fig. 2.
Otx2 and Sox2 are upstream regulators of Rax. (A) Multiple DNA sequence alignment of vertebrate CNS1. CNS1 contains a specially conserved 35-nucleotide sequence (pentatriacontamer, pt) that contains consensus binding sites for Otx and Sox. (B) Genomic structure of the Xenopus Rax locus. Arrows indicate the primers used in the ChIP assays. CNS1, the coding region and untranslated region of Rax are indicated as black, gray, and white boxes, respectively. (C) The ChIP assay demonstrates that endogenous Otx2 and Sox2 proteins bind to CNS1 in vivo. (D) RT-PCR analysis showing that overexpression of Otx2, but not of Sox2, induces Rax in Xenopus animal cap cells. (E) Luciferase assays using Xenopus animal cap (AC) cells shows that overexpression of Otx2 induces transcriptional activation of pRax-2600b-Luc, whereas Sox2 overexpression does not. (D and E) Sox2 or Otx2 mRNA (100 pg) was injected.
Overexpression of Otx2, but Not of Sox2, Induces Rax Expression in Xenopus Animal Cap Cells.
Based on the results of the in vitro and in vivo DNA-binding assays, Otx2 and Sox2 were identified as candidates for upstream proteins that regulate the transcription of Rax. We then used RT-PCR to examine whether overexpression of Otx2 or Sox2 causes up-regulation of Rax in Xenopus animal cap cells. Animal cap cells, which are part of the undifferentiated ectodermal tissue of the Xenopus blastula, are competent to respond to an inductive signal or overexpressed genes. RT-PCR analysis revealed that overexpression of Otx2 increased the level of Rax mRNA in animal cap cells, whereas overexpression of Sox2 had no effect (Fig. 2D, topmost row). In addition, Sox2 expression was induced in animal cap cells by Otx2 overexpression (Fig. 2D, second row from top).
We also used the luciferase assay to examine the response patterns of the sequences upstream of Rax to the overexpression of Sox2 and Otx2 in animal cap cells. In this assay, we used the reporter construct pRax-2600b-Luc, which contains all of the sequences required to drive appropriate expression of a reporter gene in transgenic embryos (Fig. S1A). Overexpression of Otx2 increased luciferase activity 3.7-fold, whereas overexpression of Sox2 again had no effect (Fig. 2E). These results are in accordance with the RT-PCR results, in that they implicate Otx2 as a positive regulator of Rax. However, it should be noted that Otx2 overexpression was accompanied by increased expression of Sox2.
Otx2-Dependent Transactivation in Xenopus Animal Cap Cells Requires Both the Otx- and Sox-Binding Sites of CNS1.
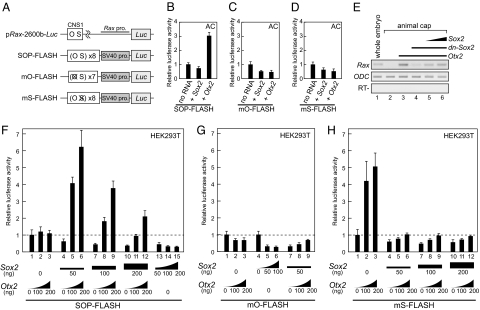
For further luciferase assays, we generated a SOP-FLASH vector that contained multiple Otx- and Sox-binding sites derived from CNS1 (Fig. 3A). Microinjection of Otx2 mRNA, but not Sox2 mRNA, increased by 3-fold the transcription from SOP-FLASH in animal cap cells (Fig. 3B). Therefore, the responses of SOP-FLASH and pRax-2600b-Luc to overexpression of Otx2 and Sox2 were similar (Fig. 2E). To examine whether the sequence of CNS1 is responsible for Otx2-dependent transactivation, we introduced point mutations into the Otx- and Sox-binding sites of SOP-FLASH to generate mO-FLASH and mS-FLASH, respectively (Fig. 3A). Overexpression of Otx2 did not cause an increase in transcription from mO-FLASH (Fig. 3C), which indicates that transactivation by Otx2 requires an Otx-binding site on CNS1. Surprisingly, Otx2 overexpression did not drive transcription from mS-FLASH (Fig. 3D). A competitive EMSA confirmed that a substitution at the Sox-binding site of CNS1 did not affect the binding of Otx2 to a neighboring Otx-binding site (Fig. S2 G and H). The observation that a Sox-binding site affects transactivation without exogenous Sox2 protein suggests that Otx2-dependent transactivation requires the binding of endogenous Sox2 protein to the Sox-binding site in animal cap cells.
Fig. 3.
Synergistic actions of Otx2 and Sox2 on transcription via CNS1. (A) Diagram of luciferase reporter constructs used in the present work. (B–D) Luciferase assays using Xenopus animal cap (AC) cells. Transcription from SOP-FLASH (B) is induced by the injection of Otx2 mRNA (100 pg) but not by the injection of Sox2 mRNA (100 pg). Overexpression of Otx2 does not induce transcription from either mO-FLASH (C) or mS-FLASH (D). (E) RT-PCR analysis shows that dominant-negative Sox2 (dn-Sox2) represses Otx2-induced Rax expression in animal cap cells. The amounts of mRNA injected were: Otx2, 100 pg; dn-Sox2, 1,000 pg; and Sox2, 10 or 30 pg. (F–H) Luciferase assays using HEK293T cells. (F) Synergistic effect of Otx2 and Sox2 on transcription from SOP-FLASH. (G) Transcription from mO-FLASH is not induced by the combination of Otx2 and Sox2 in HEK293T cells. (H) From mS-FLASH, transcription is induced by transfection with Otx2 alone in HEK293T cells. This increase is attenuated rather than enhanced by the addition of Sox2.
Both Otx2 and Sox2 Are Required for Transcriptional Activation Through CNS1 in Xenopus Animal Cap Cells and HEK293T Cells.
As mentioned above, Otx2 overexpression induced the up-regulation of Sox2 in animal cap cells (Fig. 2D). It is possible that the induced Sox2 protein collaborates with Otx2 protein in transactivation via CNS1. To test whether Sox2 is required for Rax up-regulation induced by Otx2, we performed loss-of-function experiments by using a dominant-negative construct of Sox2 (dn-Sox2), which lacks most of the HMG domain (25). RT-PCR analysis showed that Otx2-induced Rax expression in animal cap cells was repressed by coinjection of dn-Sox2 mRNA (Fig. 3E). This repression was rescued by the additions of wild-type Sox2 mRNA, which suggests that Sox2 is required for Otx2-induced Rax expression in vivo. To elucidate the underlying mechanisms in more detail, we used the HEK293T cell line, which is derived from a human embryonic kidney and does not express either Sox2 or Otx2 endogenously. In addition, Sox2 expression was not induced by overexpression of Otx2 in these cells (Fig. S3). Similar to the results obtained for the Xenopus animal cap cells, overexpression of Sox2 in HEK293T cells did not induce transcription from SOP-FLASH (Fig. 3F, lanes 1 and 13–15). However, overexpression of Otx2 alone did not increase transcription in these cells (Fig. 3F, lanes 1–3), in contrast to the earlier animal cap experiments, in which Otx2 overexpression induced transcription from SOP-FLASH (Fig. 3B). Surprisingly, cotransfection of Otx2 and Sox2 markedly increased transcription (Fig. 3F, lanes 4–6), and this synergistic transactivation was dose-dependent (Fig. 3F, lanes 4–12). Although an appropriate dose of Sox2 (50 ng) was required for synergistic transactivation with Otx2, an excess of Sox2 (100 or 200 ng) attenuated the increase in transcription. To confirm that the observed synergism between Otx2 and Sox2 requires the binding of these proteins to the CNS1 region, we performed additional luciferase assays by using Otx2 that was mutated at a critical lysine residue (K50) in the homeodomain as the expressed protein (26) and mO-FLASH or mS-FLASH as the reporter. K50-mutated Otx2 did not activate transcription in a cooperative manner with Sox2 (Fig. S4A), whereas mO-FLASH expression was not associated with synergistic transactivation under any condition tested (Fig. 3G). Experiments with mS-FLASH showed that Otx2 expression alone increased luciferase activity and that the addition of Sox2 induced no further increase; indeed, luciferase activity was strongly suppressed in the latter case (Fig. 3H). Possible explanations for the differences in responsiveness between SOP-FLASH and mS-FLASH are addressed in Discussion.
Physical Interactions Between the Otx2 and Sox2 Proteins in Vitro and in Vivo.
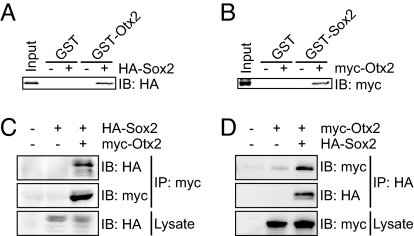
The luciferase assays showed that Otx2 and Sox2 activated transcription interdependently. In addition, the six-nucleotide gap between the Sox-binding and Otx-binding sites on CNS1 is very short and is conserved among vertebrates (i.e., a six-nucleotide gap rather than a specific six-nucleotide motif) (Fig. 2A). These observations prompted us to look for a physical interaction between the Otx2 and Sox2 proteins. GST pulldown assays with tagged proteins demonstrated that Otx2 and Sox2 physically bound to each other in vitro (Fig. 4 A and B). Pulldown assays using deletion mutants of these proteins demonstrated the DNA-binding domains of Otx2 and Sox2, the HMG domain of Sox2, and the homeodomain of Otx2 were essential for this binding (Fig. S5 A–D). These domains are all remarkably conserved among vertebrates (Fig. S5 J and K), suggesting similar conservation of the interaction between Otx2 and Sox2. Assays using additional deletion constructs further implicated helices 2 and 3 of the Sox2 HMG domain and the N- and C-flanking amino acids of the Otx2 homeodomain in the modulation of this interaction (Fig. S5 E–I). To examine these interactions in vivo, we performed coimmunoprecipitation assays using the two-step lysis method, which was developed for the detection of nuclear complexes (27). Proteins were collected from HEK293T cells that transiently expressed tagged Sox2 and/or Otx2. HA-Sox2 expressed alone was not immunoprecipitated by the anti-myc antibody. However, when HA-Sox2 and myc-Otx2 were coexpressed, they were coimmunoprecipitated by the anti-myc antibody (Fig. 4C). Reverse immunoprecipitation experiments confirmed this result (Fig. 4D). Immunocytochemistry showed that the two proteins colocalized to the nucleus (Fig. S5 L and M). Collectively, these results demonstrate that Otx2 and Sox2 interact directly with each other both in vitro and in vivo.
Fig. 4.
Physical interactions between Otx2 and Sox2 proteins in vitro and in vivo. (A) GST pulldown by immobilized GST-Otx2 of in vitro-translated HA-Sox2 protein. (B) GST pulldown by immobilized GST-Sox2 of in vitro-translated myc-Otx2 protein. (C and D) Coimmunoprecipitation assays demonstrating in vivo interactions between the Otx2 and Sox2 proteins. (C) HA-Sox2 is coimmunoprecipitated with myc-Otx2 by the anti-myc antibody (9E10). (D) Myc-Otx2 is coimmunoprecipitated with HA-Sox2 by the anti-HA antibody (Y-11). Full scans of the Western blotting data are presented in Fig. S7.
Missense Mutations Identified in the Sox2 HMG Domain Affect Sox2 Activity Associated with Otx2.
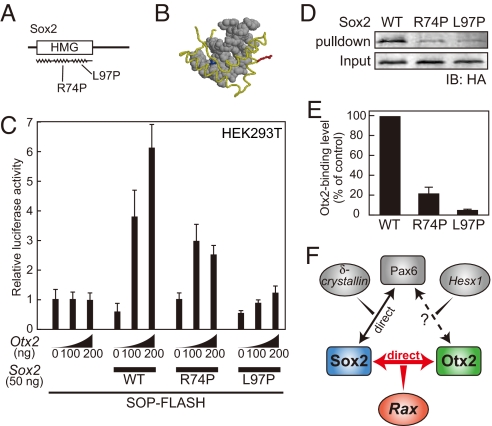
In severe ocular malformations, the majority of the mutations identified in the Sox2 locus are frameshift or nonsense mutations and are expected to produce a truncated Sox2 protein (7). However, missense mutations that lead to amino acid changes have been found in three cases (8, 28, 29). Intriguingly, two of these three missense mutations are predicted to alter the conserved residues in helices 2 and 3 of the Sox2 HMG domain (R74P and L97P, respectively), which in the pulldown assays of the present work were revealed to have a role in the interaction with the Otx2 protein (Fig. S5 E and F).
To characterize the mutated Sox2 proteins in the cases of missense mutations in Sox2, we introduced point mutations into the corresponding residues of the Xenopus Sox2 protein to generate R74P-Sox2 and L97P-Sox2 (Fig. 5A). The helices and tail of the HMG domain come in contact with the minor groove of the double-stranded DNA, and the determined 3D structure of the HMG domain shows that the side chains of the 74th arginine and the 97th lysine protrude opposite to the DNA and are not directly in contact with the DNA (Fig. 5B) (30). Initially, we performed luciferase assays to examine whether these mutated proteins could activate transcription in cooperation with Otx2 (Fig. 5C). Although Otx2 could induce transcription synergistically with wild-type Sox2, Otx2 could not increase luciferase activity in combination with L97P-Sox2. In addition, R74P-Sox2 drove transcription in combination with Otx2, albeit to a much lesser extent than the wild-type Sox2. We then tested the DNA-binding abilities of these mutated Sox2 proteins by EMSA (Fig. S2I). Wild-type Sox2 protein potently bound to the CNS1 fragment, whereas the R74P-Sox2 and L97P-Sox2 proteins did not bind to CNS1. Finally, we examined the binding activities of the mutated Sox2 proteins to Otx2 protein in a GST pulldown assay (Fig. 5 D and E). Whereas the wild-type Sox2 protein was efficiently pulled down by GST-Otx2, the R74P-Sox2 and L97P-Sox2 proteins were pulled down to markedly lesser extents. These results suggest that the missense mutations in the Sox2 gene are associated with reductions in the transactivational activity on Rax CNS1 and a loss of ability to bind to both DNA and the Otx2 protein.
Fig. 5.
Missense mutations in the Sox2 HMG domain affect the activities of Sox2 protein associated with Otx2. (A) Missense mutations identified in helices 2 and 3 of the human Sox2 HMG domain (R74P and L97P). (B) The 3D structure of the Sox2 HMG domain (yellow) binding to DNA (gray), based on a previous study (30). The side chains of the 74th arginine (blue) and 97th lysine (red) do not come in direct contact with the DNA. (C) Mutated Sox2 proteins do not induce transcription via CNS1 in cooperation with Otx2. (D) GST pulldown assay showing that the mutated Sox2 proteins lose Otx2-binding activity. HA-tagged Sox2 proteins were in vitro-synthesized and pulled down by GST-Otx2, followed by Western blotting with antibody against HA-tag. (E) Quantification of Otx2-binding levels by using the Odyssey infrared imaging system. Error bars indicate SD values. (F) Molecular relationships among ocular malformation-associated genes. The present work demonstrates that a direct interaction between the Otx2 and Sox2 proteins coordinately regulates Rax expression (colored). Upstream proteins and target genes are indicated as round rectangles and ovals, respectively. A full scan of the Western blotting data is shown in Fig. S7.
Discussion
The transcription factor-encoding genes Pax6, Sox2, and Otx2 have been identified as the causative genes for human ocular malformation. In the present work, we define the regulatory relationships between these genes (Fig. 5F). Previous studies have shown that Pax6 and Sox2 form a complex to coregulate the expression of δ-crystallin (11). In addition, the findings that the Otx2-binding sites are required for activation of the chick Hesx1 promoter and that overexpression of Pax6 in the chick or loss-of-function in mice causes repression or expansion of Hesx1 expression, respectively (31), indicate that the Pax6 and Otx2 proteins coregulate Hesx1 expression, although the nature of the interdependence and the form of the direct interaction between Pax6 and Otx2 remain unknown. The final combination of Otx2 and Sox2 was not addressed before the present work. We demonstrate that Otx2 and Sox2 interact directly with each other and synergistically activate Rax expression through CNS1. We propose that the genetic and molecular interactions among these three key transcription factors organize the developmental program of the vertebrate eye.
We have also discovered that Otx2 increases the level of Sox2 mRNA in Xenopus animal cap cells. A previous study revealed that multiple cis-regulatory elements spatiotemporally control Sox2 expression in neural development and are conserved among the chicken, mouse, and human (32). We found that the N-2 and N-3 elements contain Otx-binding sites (N-2, AGATTA; N-3, GGATTA) that are perfectly conserved among vertebrates, including the frog (data not shown). This observation raises the possibility that the Otx2 protein directly up-regulates Sox2 expression in Xenopus animal cap cells.
To understand the molecular mechanisms underlying transactivation via Rax CNS1 regulated by Otx2 and Sox2, we note the unexpected nature of the CNS1 response to Otx2 and Sox2. Our luciferase assays show that combined overexpression of Otx2 and Sox2 drives transactivation of SOP-FLASH (wild-type CNS1), whereas overexpression of Otx2 alone is sufficient for the transactivation of mS-FLASH (CNS1 mutated at the Sox-binding sites), as reported for other regulatory elements (33, 34). We attribute this difference to the existence of a Sox-binding site close to the Otx-binding site (Fig. S6). These results imply that a Sox-binding site possesses repressive activity. It is possible that, in the absence of Sox2, a repressive protein binds preferentially to the Sox-binding site of CNS1 and prevents Otx2 from binding to DNA or transactivating the basic transcription complex. In contrast, in the presence of Sox2, Sox2 may occupy the Sox2-binding sites in place of the repressive protein, thereby causing synergistic transactivation with Otx2. Further studies are needed to understand these intricate regulation mechanisms.
As reported, Sox2 protein has an inhibitory function (35); in mouse embryonic stem cells, elevation of Sox2 levels suppressed the expression levels of Sox2:Oct3/4 target genes. These investigators also showed that the suppression was mediated by the C-terminal region, not the DNA-binding domain, of the Sox2 protein. These findings are consistent with the results obtained in the present work. In the luciferase assay using HEK293T cells, whereas a low level of Sox2 expression (50 ng) caused synergistic activation of SOP-FLASH in a cooperative manner with Otx2, excess Sox2 (100 or 200 ng) inhibited the transcription induced by Otx2 and Sox2 (Fig. 3F). In addition, the transcription of mS-FLASH (mutated SOP-FLASH at Sox-binding sites), which was caused by Otx2 alone, was also suppressed by Sox2 (50, 100, or 200 ng; Fig. 3H). The fact that the mS-FLASH vector lacks Sox-binding sites suggests that this inhibitory function of Sox2 protein is not dependent on DNA binding but instead requires the C-terminal region of Sox2. Low levels of Sox2 expression (50 ng) sufficiently repressed Otx2-driven mS-FLASH activation, which suggests that transcription from SOP-FLASH is also inhibited by a low level of Sox2 expression (50 ng), whereas the interaction of Sox2 and Otx2 overrides this inhibition, inducing a high level of transactivation.
The stoichiometric association of Sox2 with Otx2 may explain why Rax is not expressed in the presumptive brain, in which Otx2 and Sox2 mRNA are coexpressed (Fig. S1B). It is possible that the ratio of Sox2 to Otx2 is in the range required for transcriptional activation of Rax in the optic vesicle, whereas this ratio is too high or too low in the presumptive brain, resulting in the loss of Rax expression. Of course, the involvement of other transcription factors in the regulation of Rax expression cannot be excluded. Furthermore, previous studies on Pax6–Sox2 cooperation have reported that differences in the sequences of cis-regulatory elements result in differences in the threshold protein levels required for the cooperative action to occur (36). The present work permits us to speculate that Otx2 and Sox2 coregulate the expression patterns of multiple target genes in various subdomains of the brain and eye and that the expression levels of the different target genes are controlled by the different stoichiometric ratios of the individual subdomains. Comprehensive identification of the target genes coregulated by Otx2 and Sox2 will allow us to compose a conceptual model for regionalization of the developing brain and eye.
We noted that a gap of six nucleotides between the Otx- and Sox-binding sites in CNS1 was conserved, even if the specific sequence was not conserved. Previous studies have reported that the distance between the Sox2- and Pax6-binding sites is conserved between two cis-regulatory elements and that insertions of a few base pairs between these sites ablate the cooperative action of Sox2 and Pax6 (11, 36), which raises the possibility that, in addition to the individual sequences of the binding sites, the gap between two binding sites contributes to the stoichiometric association and the interdependence of Otx2 and Sox2.
In conclusion, our analysis of the upstream region of the frog Rax gene reveals molecular linkages among three genes associted with human ocular malformation. The direct interaction and interdependence of the Otx2 and Sox2 proteins coordinate Rax expression through a conserved noncoding sequence. These findings of coordinated transcriptional regulation improve our understanding of eye development and ocular malformation in humans.
Materials and Methods
DNA Constructs.
SOP-FLASH, mO-FLASH, and mS-FLASH were generated by inserting the multiple repeats of pt, mO-pt, and mS-pt, respectively, into the pGL3-Promoter vector (Promega). Details of the DNA construction are presented in SI Materials and Methods.
Xenopus Embryos.
Manipulation of X. laevis embryos and explants, whole-mount in situ hybridization (WISH), and transgenesis were performed as described (37, 38). DNA and mRNA were microinjected into four animal hemispheres of eight-cell-stage embryos. The expression vectors used for mRNA synthesis were: pCS2-XOtx2, pCS2-XSox2, pCS2-myc-XOtx2, pCS2-myc-XSox2, pCS2-dn-XSox2, and pCS2-nls-βgal.
RT-PCR and ChIP.
For RT-PCR, total RNA was extracted from 15 animal cap explants or two whole embryos at the midneurula stage by using ISOGEN (Wako). ChIP assays were performed using anti-Otx2, anti-Sox2, or anti-myc antibodies. Details of the RT-PCR and ChIP assays are provided in SI Materials and Methods.
Luciferase Assay.
The pRL-TK or pGL4.74 (Promega) plasmid was used as an internal control. Xenopus embryos and explants injected with the reporter vectors (firefly, 30 pg; Renilla, 10 pg) and the mRNA of Otx2 and Sox2 (100 pg) were harvested from stage-13 embryos. Cultured cells were transfected with the reporter vectors (firefly, 100 ng; Renilla, 10 ng) and the expression vectors pCS2-myc-XOtx2, pCS2-HA-XSox2, pCS2-HA-XSox2-R74P, pCS2-HA-XSox2-L97P, and pCS2-nls-βgal and were harvested 48 h after transfection. Reporter activities were measured by using the dual-luciferase reporter assay system (Promega). Each assay was performed in duplicate, and all results are shown as mean ± SD for at least three independent assays.
GST Pulldown Assays, Coimmunoprecipitation Assays, and Western Blotting.
GST pulldown assays were performed according to standard procedures. Coimmunoprecipitation assays were performed according to the two-step lysis method (27). The details of these assays are provided in SI Materials and Methods.
Bioinformatics.
The EMBOSS software was used for total sequence analyses (39). For comparative genomic analyses, the VISTA Browser (http://genome.lbl.gov/vista) was used (24). The 3D model of the Sox2 HMG domain was created by using Rasmol based on Protein Data Bank (PDB) ID code 1GT0.
Acknowledgments.
We thank Drs. M. Pannese (Istituto Scientifico H San Raffaele, Milan, Italy), H. Clevers (Hubrecht Institute, Utrecht, The Netherlands), and K. Nitta (Institut de Biologie du Développement de Marseille Luminy, Marseille, France) for generous gifts of plasmids. This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to M.A. and T.M.) and by Wako Pure Chemical Industries, Ltd. H.D. is also supported by the Japan Society for the Promotion of Science for Young Scientists.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB365789).
This article contains supporting information online at www.pnas.org/cgi/content/full/0710954105/DCSupplemental.
References
- 1.Morrison D, et al. National study of microphthalmia, anophthalmia, and coloboma (MAC) in Scotland: Investigation of genetic aetiology. J Med Genet. 2002;39:16–22. doi: 10.1136/jmg.39.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adler R, Canto-Soler MV. Molecular mechanisms of optic vesicle development: Complexities, ambiguities and controversies. Dev Biol. 2007;305:1–13. doi: 10.1016/j.ydbio.2007.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fantes J, et al. Mutations in SOX2 cause anophthalmia. Nat Genet. 2003;33:461–463. doi: 10.1038/ng1120. [DOI] [PubMed] [Google Scholar]
- 4.Ragge NK, et al. Heterozygous mutations of OTX2 cause severe ocular malformations. Am J Hum Genet. 2005;76:1008–1022. doi: 10.1086/430721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voronina VA, et al. Mutations in the human RAX homeobox gene in a patient with anophthalmia and sclerocornea. Hum Mol Genet. 2004;13:315–322. doi: 10.1093/hmg/ddh025. [DOI] [PubMed] [Google Scholar]
- 6.Glaser T, et al. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7:463–471. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- 7.Hever AM, Williamson KA, van Heyningen V. Developmental malformations of the eye: The role of PAX6, SOX2, and OTX2. Clin Genet. 2006;69:459–470. doi: 10.1111/j.1399-0004.2006.00619.x. [DOI] [PubMed] [Google Scholar]
- 8.Ragge NK, et al. SOX2 anophthalmia syndrome. Am J Med Genet A. 2005;135:1–7. doi: 10.1002/ajmg.a.30642. discussion 8. [DOI] [PubMed] [Google Scholar]
- 9.Taranova OV, et al. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20:1187–1202. doi: 10.1101/gad.1407906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson M, Koopman P. Matching SOX: Partner proteins and cofactors of the SOX family of transcriptional regulators. Curr Opin Genet Dev. 2002;12:441–446. doi: 10.1016/s0959-437x(02)00323-4. [DOI] [PubMed] [Google Scholar]
- 11.Kamachi Y, Uchikawa M, Tanouchi A, Sekido R, Kondoh H. Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes Dev. 2001;15:1272–1286. doi: 10.1101/gad.887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- 13.Finkelstein R, Boncinelli E. From fly head to mammalian forebrain: The story of otd and Otx. Trends Genet. 1994;10:310–315. doi: 10.1016/0168-9525(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 14.Pannese M, et al. The Xenopus homolog of Otx2 is a maternal homeobox gene that demarcates and specifies anterior body regions. Development. 1995;121:707–720. doi: 10.1242/dev.121.3.707. [DOI] [PubMed] [Google Scholar]
- 15.Simeone A, Acampora D. The role of Otx2 in organizing the anterior patterning in mouse. Int J Dev Biol. 2001;45:337–345. [PubMed] [Google Scholar]
- 16.Rhinn M, et al. Sequential roles for Otx2 in visceral endoderm and neuroectoderm for forebrain and midbrain induction and specification. Development. 1998;125:845–856. doi: 10.1242/dev.125.5.845. [DOI] [PubMed] [Google Scholar]
- 17.Furukawa T, Kozak CA, Cepko CL. rax, a novel paired-type homeobox gene, shows expression in the anterior neural fold and developing retina. Proc Natl Acad Sci USA. 1997;94:3088–3093. doi: 10.1073/pnas.94.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathers PH, Grinberg A, Mahon KA, Jamrich M. The Rx homeobox gene is essential for vertebrate eye development. Nature. 1997;387:603–607. doi: 10.1038/42475. [DOI] [PubMed] [Google Scholar]
- 19.Terada K, Kitayama A, Kanamoto T, Ueno N, Furukawa T. Nucleosome regulator Xhmgb3 is required for cell proliferation of the eye and brain as a downstream target of Xenopus rax/Rx1. Dev Biol. 2006;291:398–412. doi: 10.1016/j.ydbio.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 20.Andreazzoli M, et al. Xrx1 controls proliferation and neurogenesis in Xenopus anterior neural plate. Development. 2003;130:5143–5154. doi: 10.1242/dev.00665. [DOI] [PubMed] [Google Scholar]
- 21.Kimura A, et al. Both PCE-1/RX and OTX/CRX interactions are necessary for photoreceptor-specific gene expression. J Biol Chem. 2000;275:1152–1160. doi: 10.1074/jbc.275.2.1152. [DOI] [PubMed] [Google Scholar]
- 22.Hirsch N, et al. Xenopus tropicalis transgenic lines and their use in the study of embryonic induction. Dev Dyn. 2002;225:522–535. doi: 10.1002/dvdy.10188. [DOI] [PubMed] [Google Scholar]
- 23.Zhang L, et al. Targeted expression of the dominant-negative FGFR4a in the eye using Xrx1A regulatory sequences interferes with normal retinal development. Development. 2003;130:4177–4186. doi: 10.1242/dev.00626. [DOI] [PubMed] [Google Scholar]
- 24.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kishi M, et al. Requirement of Sox2-mediated signaling for differentiation of early Xenopus neuroectoderm. Development. 2000;127:791–800. doi: 10.1242/dev.127.4.791. [DOI] [PubMed] [Google Scholar]
- 26.Gehring WJ, et al. Homeodomain-DNA recognition. Cell. 1994;78:211–223. doi: 10.1016/0092-8674(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 27.Klenova E, Chernukhin I, Inoue T, Shamsuddin S, Norton J. Immunoprecipitation techniques for the analysis of transcription factor complexes. Methods. 2002;26:254–259. doi: 10.1016/S1046-2023(02)00029-4. [DOI] [PubMed] [Google Scholar]
- 28.Faivre L, et al. Recurrence of SOX2 anophthalmia syndrome with gonosomal mosaicism in a phenotypically normal mother. Am J Med Genet A. 2006;140:636–639. doi: 10.1002/ajmg.a.31114. [DOI] [PubMed] [Google Scholar]
- 29.Williamson KA, et al. Mutations in SOX2 cause anophthalmia–esophageal–genital (AEG) syndrome. Hum Mol Genet. 2006;15:1413–1422. doi: 10.1093/hmg/ddl064. [DOI] [PubMed] [Google Scholar]
- 30.Remenyi A, et al. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 2003;17:2048–2059. doi: 10.1101/gad.269303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spieler D, et al. Involvement of Pax6 and Otx2 in the forebrain-specific regulation of the vertebrate homeobox gene ANF/Hesx1. Dev Biol. 2004;269:567–579. doi: 10.1016/j.ydbio.2004.01.044. [DOI] [PubMed] [Google Scholar]
- 32.Uchikawa M, Ishida Y, Takemoto T, Kamachi Y, Kondoh H. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev Cell. 2003;4:509–519. doi: 10.1016/s1534-5807(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 33.Mailhos C, et al. Drosophila Goosecoid requires a conserved heptapeptide for repression of paired-class homeoprotein activators. Development. 1998;125:937–947. doi: 10.1242/dev.125.5.937. [DOI] [PubMed] [Google Scholar]
- 34.Martinez-Morales JR, et al. OTX2 activates the molecular network underlying retina pigment epithelium differentiation. J Biol Chem. 2003;278:21721–21731. doi: 10.1074/jbc.M301708200. [DOI] [PubMed] [Google Scholar]
- 35.Boer B, et al. Elevating the levels of Sox2 in embryonal carcinoma cells and embryonic stem cells inhibits the expression of Sox2:Oct-3/4 target genes. Nucleic Acids Res. 2007;35:1773–1786. doi: 10.1093/nar/gkm059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inoue M, et al. PAX6 and SOX2-dependent regulation of the Sox2 enhancer N-3 involved in embryonic visual system development. Genes Cells. 2007;12:1049–1061. doi: 10.1111/j.1365-2443.2007.01114.x. [DOI] [PubMed] [Google Scholar]
- 37.Kroll KL, Amaya E. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development. 1996;122:3173–3183. doi: 10.1242/dev.122.10.3173. [DOI] [PubMed] [Google Scholar]
- 38.Michiue T, et al. XIdax, an inhibitor of the canonical Wnt pathway, is required for anterior neural structure formation in Xenopus. Dev Dyn. 2004;230:79–90. doi: 10.1002/dvdy.20037. [DOI] [PubMed] [Google Scholar]
- 39.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]