Abstract
Human species C adenovirus serotype 5 (Ad5) is the most common viral vector used in clinical studies worldwide. Ad5 vectors infect liver cells in vivo with high efficiency via a poorly defined mechanism, which involves virus binding to vitamin K-dependent blood coagulation factors. Here, we report that the major Ad5 capsid protein, hexon, binds human coagulation factor X (FX) with an affinity of 229 pM. This affinity is 40-fold stronger than the reported affinity of Ad5 fiber for the cellular receptor coxsackievirus and adenovirus receptor, CAR. Cryoelectron microscopy and single-particle image reconstruction revealed that the FX attachment site is localized to the central depression at the top of the hexon trimer. Hexon-mutated virus bearing a large insertion in hexon showed markedly reduced FX binding in vitro and failed to deliver a transgene to hepatocytes in vivo. This study describes the mechanism of FX binding to Ad5 and demonstrates the critical role of hexon for virus infection of hepatocytes in vivo.
Keywords: gene transfer, virus targeting
Gene delivery systems based on human species C adenovirus serotype 5 (Ad5) are among the most frequently used in clinical studies, which aim to correct human genetic and acquired diseases, including cancer (1). The extreme propensity of the virus for hepatocyte infection after its intravascular delivery has made Ad5 the vector of choice for applications requiring high-level transgene expression in hepatocytes in vivo (2). However, the efficient interaction between Ad5 and liver cells, which sequester a large proportion of the delivered vector dose (3), represents a significant hindrance if gene delivery to extrahepatic cells and tissues is required. From in vitro analyses, it was found that Ad5 infection is initiated when the minor capsid protein, fiber, binds to the coxsackievirus and adenovirus receptor (CAR) on the cell surface (4, 5). Subsequent binding of the penton base protein to cellular integrins facilitates internalization of the attached particle into the cell (6). Although binding of both CAR and integrin is critical for cell infection in vitro, neither of these interactions is essential for Ad5 entry into hepatocytes in vivo (7–9).
We and others recently identified a pathway of hepatocyte infection by Ad5, which is mediated by the vitamin K-dependent blood coagulation factors, including factors VII, IX, X, and protein C (10, 11). When Ad5 particles are delivered intravenously, they interact with blood coagulation factors, which subsequently bind low-density lipoprotein receptor-related protein (LRP) and heparan sulfate proteoglycans (HSPGs) on hepatocytes to mediate virus entry. Although rather complex, this mechanism is very efficient and can be specifically blocked by lactoferrin, which competes for blood factor binding to LRP and HSPGs on hepatic cells (10). It is noteworthy that, although all of these blood factors support CAR-independent cell infection by Ad5 vectors to varying degrees, coagulation factor X (FX) appears the most efficient at mediating Ad5 entry into hepatic cells in vivo (11).
Based on numerous experiments that demonstrate reduced hepatocyte infection after i.v. delivery of fiber-modified vectors in mice, it was suggested that coagulation factors might bind to the Ad5 fiber to mediate hepatocyte infection. However, there is still no definitive evidence that a fiber–coagulation factor interaction is ultimately required to mediate CAR-independent Ad5 entry into hepatocytes in vivo.
In this study, we analyzed the interactions of human FX with human adenoviruses from species B–F. Using surface plasmon resonance (SPR), we found that FX binds with picomolar affinity to hexon, the major Ad capsid protein. Cryoelectron microscopy (cryoEM) and single-particle image reconstruction have localized the FX-binding area to the central depression at the top of each Ad5 hexon trimer. This cryoEM result, combined with sequence analysis of hexons that bind FX and those that do not, indicates there are likely two alternative sites for FX binding. One site is within hypervariable region 3 (HVR3), and the other is within hypervariable region 7 (HVR7). Both sites are predicted to form similar binding pockets within the central depression of the hexon trimer. An adenovirus mutant that binds FX in vitro with 10,000-fold reduced affinity compared with unmodified vector failed to deliver the red fluorescent protein (RFP) transgene in vivo. Thus, our study describes the mechanism of FX binding to Ad5 and demonstrates the critical role of hexon for virus infection of hepatocytes in vivo.
Results
Ad5 Hexon Binds FX with Picomolar Affinity.
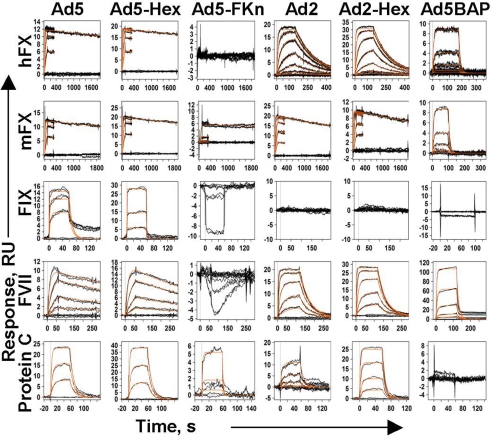
To quantitatively characterize binding between blood coagulation factors and the adenovirus capsid, we used SPR analysis (BIAcore). To avoid complications with interpretation of the results caused by the multivalent nature of the virus particle, Ad5 particles were immobilized on the sensor chip by cross-linking, and varying concentrations of blood factors were injected over sensor surfaces and binding responses were recorded and analyzed. Fig. 1(Ad5 column) shows the representative datasets obtained from the analysis of blood–factor interactions together with global fits to a 1:1 interaction model (orange lines). Among all blood factors tested [Fig. 1 and supporting information (SI) Fig. S1], human FX (hFX) and mouse FX (mFX) demonstrated the highest-affinity binding to Ad5. The association constant of the Ad5-hFX-binding reaction was determined to be 3.37 × 105 M−1·s−1, the dissociation constant 7.71 × 10−5·s−1 and KD equal to 228.7 pM. We detected no binding of FXI, FXII, and FX-desGla to Ad5 under the conditions used (Fig. S1). Ad5-hFX binding strongly depends on the presence of both Ca+2 and Mg+2 ions and could be completely eliminated in a buffer containing 3 mM EDTA (data not shown).
Fig. 1.
Kinetic response data for different blood coagulation factor binding to Ad5, Ad2, and adenovirus capsid proteins. Experimentally obtained data are shown by black lines. Global fits of these data to 1:1 single-site interaction model are shown in orange. The responses are shown in instrument response units (RU) vs. time in seconds (s). Columns represent six different ligands that were immobilized on the CM5 sensor chip. Ad5, adenovirus type 5; Ad5, Hex-Ad5 hexon; Ad5-FKn, Ad5 fiber knob domain; Ad2, adenovirus type 2; Ad2-Hex, Ad2 hexon; Ad5BAP, hexon-mutated Ad5-based vector. Rows represent eight different blood factors used as analytes in Biacore experiments. hFX, human factor X; mFX, mouse factor X; FIX, human factor IX; FVII, human factor VII; protein C, human protein C.
Because of the very high affinity of FX for Ad5 and its strong dependence on the presence of calcium and magnesium ions, we hypothesized that the FX-binding protein within the Ad5 capsid could be purified from virus-infected cells using affinity chromatography. Indeed, using the hFX affinity column, a 98% pure protein with an apparent molecular mass of 100 kDa (after separation by SDS/PAGE under denaturating conditions) was obtained from the lysate of cells infected with Ad5 (Fig. S2 and SI Text, Materials and Methods). The affinity-purified protein was digested with trypsin and analyzed by using data-dependent tandem mass spectrometry. Numerous peptides corresponding to the Ad5 hexon were readily identified by Mascot software. The trimeric state of the purified hexon was further confirmed by size-exclusion chromatography. The experimentally determined molecular mass of the protein was estimated to be 337 kDa, which corresponds well with the expected molecular mass of 324 kDa for the Ad5 hexon trimer (Fig. S2 A–C).
Using the SPR technique, we next analyzed the kinetic parameters of the blood factor interaction with purified Ad5 hexon and purified Ad5 fiber knob domain, expressed in Escherichia coli (Fig. S2 D–F and SI Text, Materials and Methods). The data obtained (Fig. 1, Ad5-Hex and Ad5-FKn columns, and Table S1) showed that, for all analyzed blood factors, interaction with purified Ad5 hexon trimer, but not the Ad5 fiber knob domain, closely follows the kinetics for the FX interaction with the whole virus (Fig. 1). In addition, we analyzed blood factor binding to another group C Ad serotype, Ad2, and commercially available purified Ad2 hexon protein. This analysis demonstrated that both human and mouse FX bind Ad2 particles and purified Ad2 hexon with identical kinetics and affinity (Fig. 1 and Table S1). Collectively, these data indicate that the Ad hexon possesses a high-affinity binding site for vitamin K-dependent blood factors. To further assess this possibility, we analyzed blood factor interaction with an Ad5-based vector, Ad5BAP, that possesses an insertion of a 71-aa biotin acceptor peptide (BAP) in the exposed hexon loop of the HVR5 (12). Our analysis showed that FX binding to Ad5BAP was reduced 10,000-fold compared with unmodified Ad5 vector, and the 71-aa insertion in HVR5 region of the hexon completely eliminated binding of FIX and Protein C (Fig. 1 and Table S1).
FX Interaction with Different Human Ad Serotypes and Reovirus.
We next analyzed the affinity of FX binding to various human Ad serotypes from species B–F and human reovirus T3D (13) by using the same SPR protocol. This analysis revealed that human Ad serotypes greatly vary in their ability to bind FX (Table 1). The highest-affinity FX binding was observed for species C Ad5, followed by species B Ad16, and then species C Ad2. Except for the observation that two species C serotypes (Ad2 and Ad5) exhibited high affinity for FX, no other correlation was noted between the Ad group and FX affinity among the 11 serotypes tested. Micromolar affinity binding was observed for species B (Ad3 and Ad21), E (Ad4), and F (Ad41) serotypes. No FX binding was observed for species B Ad35 or Ad50, species D Ad9 or Ad51 serotypes, or reovirus T3D. It is noteworthy that, in agreement with these data, vectors based on Ad5 and Ad2 serotypes were shown to infect hepatocytes with high efficiency (14, 15); however, Ad35-based vectors transduce hepatocytes poorly after i.v. delivery (16, 17).
Table 1.
Affinity of FX binding to different viruses
| Virus | Immobilized RU | Kd, nM |
|---|---|---|
| Adenovirus | ||
| Ad5 | 384 | 0.229 |
| Ad16 | 470 | 1.67 |
| Ad2 | 352 | 20.9 |
| Ad21 | 615 | 410 |
| Ad41 | 347 | 630 |
| Ad4 | 2,900 | 2,480 |
| Ad3 | 2,973 | 3,000 |
| Ad35 | 315 | No binding |
| Ad51 | 667 | No binding |
| Ad9 | 311 | No binding |
| Ad50 | 256 | No binding |
| Reovirus | ||
| T3D | 486 | No binding |
| Ad5-sCAR | 7.9 |
CryoEM Analysis of the Ad5-FX Complex.
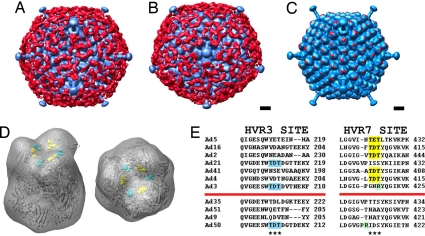
CryoEM and single-particle image reconstruction were performed to visualize the interaction of FX with Ad5. Comparison of cryoEM images of the Ad5-FX complex with those of Ad5 alone revealed that FX coats the virus with a layer of extra density. The FX coating effect was also clearly observed in a 3D reconstruction of the Ad5-FX complex (FX density is shown in red in Fig. 2 A and B). The reconstructed Ad5 virion appears to be covered with a ≈70-Å-thick mesh of FX density over the entire capsid, except within the vicinity of the pentons at the icosahedral fivefold symmetry axes.
Fig. 2.
CryoEM visualization of the Ad5-FX complex and FX-binding site on hexon. 3D single-particle reconstruction of the Ad5-FX complex. Ad capsid is shown in blue. FX density is shown in red. The view in A is along an icosahedral twofold axis. The view in B is along an icosahedral fivefold axis. (Scale bar, 100 Å.) (C) CryoEM structure of the Ad5-FX complex (blue) together with the strongest FX density (red). The FX density was generated by subtracting a cryoEM reconstruction of the Ad5 capsid from that of the Ad5-FX complex. The strongest FX density appears in the central depression of each hexon trimer. (Scale bar, 100 Å.) (D) Crystal structure of Ad5 hexon (PDB ID code 1P30) shown in a ribbon representation and as a density map filtered to 30-Å resolution (transparent gray). The hexon HVR7 residues 422–424 (TET) are shown in a space-filling representation in yellow. The position of HVR3 TDT residues found in Ad3, Ad21, and Ad50 hexons is shown by the corresponding Ad5 residues 212–214 in cyan. Both 45° tilt and top views are shown. (E) Amino acid sequence alignment of the hexon HVR3 and HVR7 regions for the 11 Ad serotypes tested for FX binding (Table 1). The positions of the two alternative sites proposed for FX binding are indicated by asterisks. The sequences TDT and TET that correlate with FX binding are highlighted in cyan within HVR3 and in yellow within HVR7. Nearby positively charged Arg residues that may ablate or reduce FX-binding affinity are highlighted in green. The red lines separate Ad serotypes that bind FX (above the line) from those that do not (below the line).
The Ad5-FX reconstruction compared with a previous reconstruction of the Ad5 capsid (18) revealed that the point of contact between FX and the Ad5 hexon is within the central depression of each hexon trimer (Fig. 2C). Fitting of the Ad5 hexon crystal structure [PDB ID code 1P30 (19)] into the cryoEM hexon density showed that the exposed hypervariable regions HVR3, HVR5, and HVR7 are in close proximity to FX density (Fig. 2D and Fig. S3). Amino acid sequence alignment of the hexons from the 11 Ad serotypes tested (Table 1) indicated that FX binding could be associated with the presence of amino acids TET or TDT within HVR7 (Fig. 2E). In addition, there are two serotypes (Ad3 and Ad21) that have the sequence TDT within HVR3. Both the HVR7 TET site in the Ad5 hexon and the HVR3 TDT site mapped onto the Ad5 hexon point into the central depression of hexon with a similar geometric arrangement (Fig. 2D). Although HVR5 is spatially adjacent to HVR7 and is also localized in the central depression of the hexon trimer (Fig. S3), its amino acid composition is highly diverse, and there is no apparent correlation between the presence of particular amino acids within HVR5 and the serotype affinity for FX.
FX-Binding-Ablated Virus Failed to Transduce Hepatocytes in Vivo.
Recent studies demonstrated that vitamin K-dependent blood coagulation factors mediate efficient hepatocyte transduction by Ad5-based vectors in vivo (10, 11). To analyze the role of the FX–hexon interaction in Ad hepatocyte transduction, we took advantage of a hexon-mutated vector, Ad5BAP (12). Analysis of blood factor binding to Ad5BAP by using SPR demonstrated a 4 order-of-magnitude loss in affinity of FX binding to Ad5BAP virions (Fig. 1 and Table S1). This suggested that this large insertion in HVR5 near the putative FX attachment site on hexon might sterically block FX binding or induce a conformational change within the FX attachment site that could inhibit binding. We first analyzed Ad5BAP infectivity in vitro. The virus particle-to-plaque forming unit ratio, determined on 293 cells, was 75 ± 23 and 79 ± 10 for control unmodified Ad5RFP vector and Ad5BAP, respectively. To further investigate whether a large insertion in the HVR5 area of hexon may adversely affect early steps of virus infection, we analyzed virus particle attachment by using quantitative Southern blot analysis (20) (Fig. S4A). Our analysis showed that both unmodified Ad5RFP vector and Ad5BAP attached to cells with similar efficacy. Next, we analyzed internalization rates for these vectors using a neutralizing antibody escape assay (21). The data obtained showed that internalization rates were identical for both Ad5RFP and Ad5BAP vectors (Fig. S4B). Confocal microscopy studies of intracellular virus trafficking revealed that both vectors rapidly migrated to the nuclear periphery. However, unlike the Ad5RFP vector, infection of cells with Ad5BAP induces significant rearrangement of Cathepsin D-positive late endosomal/lysosomal compartment and Ad5BAP virus particles colocalized with late endosomes/lysosomes (Fig. S4C).
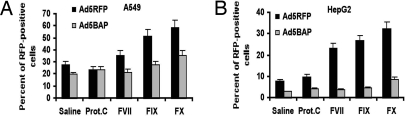
Analysis of virus infectivity on human lung epithelial (A549) or hepatoma (HepG2) cells revealed that, although no difference in infectivity between Ad5RFP and Ad5BAP was found in A549 cells, there was a marked reduction in infectivity of Ad5BAP, compared with Ad5RFP, in HepG2 cells (Fig. S4 D and E). When we analyzed virus infectivity on these cell lines in the presence of blood coagulation factors, we found that the infectivity of Ad5BAP was significantly lower compared with Ad5RFP when blood factors were added to the infection media (Fig. 3 A and B). This difference was most dramatic in hepatoma HepG2 cells, compared with A549 cells. Collectively, all these in vitro studies showed that virus attachment to cells and internalization into cells were identical for Ad5RFP and hexon-mutated Ad5BAP vectors. However, cell-type-specific factors may affect intracellular trafficking and infectivity of hexon-mutated vector. Importantly, addition of blood factors to infection media significantly increased infectivity of Ad5RFP vector and had no effect on infectivity of Ad5BAP in vitro.
Fig. 3.
Transduction of A549 and HepG2 cells by Ad5RFP and Ad5BAP vectors in the presence of human blood factors. Cells were infected with a multiplicity of infection of 25 pfu per cell for each indicated virus in the presence of 8 μg/ml of FIX, FX, or protein C or 0.5 μg/ml of FVII (physiological concentration of these factors in human plasma). Two hours after addition of virus to cells, cells were washed with new media, and RFP gene expression was analyzed 24 h later by flow cytometry. In control settings (Saline), viruses were added to cells in growth media only (n = 6).
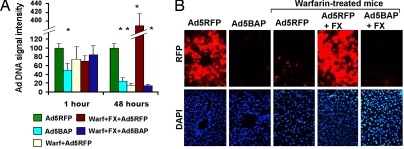
When equal doses of unmodified Ad5RFP and Ad5BAP vectors were injected i.v. into mice, the amounts of Ad5BAP vector trapped in the liver 1 h after virus administration were 50% lower than for Ad5RFP (Fig. 4A and Fig. S5A). Pretreatment of mice with warfarin, which blocks γ-carboxylation of all vitamin K-dependent zymogens and has been shown to reduce liver cell transduction by Ad5-based vectors (11), did not significantly reduce Ad vector trapping in the liver tissue at this time. Analysis of Ad5BAP DNA in the liver 48 h after virus injection revealed that these levels were 75% lower, compared with DNA levels of Ad5RFP. Treatment of mice with warfarin reduced levels of Ad5RFP DNA in the liver by 85%. Preinjection of warfarin-treated mice with FX before Ad5RFP injection resulted in a remarkable 24-fold increase in vector DNA levels in the liver at this time point (Fig. 4A). Administration of FX before Ad5BAP injection in warfarin-treated mice did not increase low levels of Ad5BAP DNA in the liver, confirming our findings on reduced affinity of blood factor binding to Ad5BAP (Fig. 1) and cell transduction in the presence of blood factors in vitro (Fig. 3).
Fig. 4.
In vivo analysis of hexon-mutated adenovirus vector. (A) Ad DNA deposition and persistence in the liver after i.v. injection of Ad5RFP or hexon-mutated Ad5BAP vectors. Amounts of vector genomic DNA in the liver were analyzed by Southern blot analysis and quantified after processing of band signal intensity by phosphorimager. The intensity of Ad5RFP bands at 1 and 48 h after virus injection was designated as 100. The graphs show the averaged data obtained from two independent experiments with four to six biological replicates per each group. *, P < 0.01. The representative image of Southern blot hybridization is shown in Fig. S6A. (B) Histological analysis of Ad-encoded RFP gene expression in mouse hepatocytes 48 h after i.v. injection of indicated vectors. Representative fields are shown. RFP expression is observed as red fluorescence on fixed liver sections. Corresponding fields in DAPI channel (nucleus-specific staining) are shown.
Evaluation of hepatocyte transduction by fluorescent microscopy 48 h after i.v. injection of Ad5RFP or Ad5BAP revealed (Fig. 4B) that RFP levels in hepatocytes mimicked vector genomic DNA levels, determined by Southern blot analysis.
The Ad5BAP vector failed to express RFP after intravascular delivery. Warfarin treatment of mice ablated RFP transgene expression in hepatocytes after injection of Ad5RFP vector, and this ablation could be completely reverted by restoration of physiological levels of FX in blood (Fig. 4B). Importantly, the Ad5BAP vector failed to transduce hepatocytes in warfarin-treated mice even in the presence of FX. Western blot analysis for RFP protein levels in hepatic tissue demonstrated high levels of RFP expression in livers of mice injected with Ad5RFP (Fig. S5 B and C). The levels of hepatic RFP expression were at the limit of detection by Western blot analysis in mice injected with Ad5BAP. The median of RFP band intensity was 68-fold stronger for Ad5RFP, compared with Ad5BAP.
Discussion
Virus binding to cellular receptors is a critical early step in initiation of infection. This interaction is under strong evolutionary pressure and is often mediated by specialized virus capsid proteins. For species C Ad serotype 5, CAR was shown to be the high-affinity attachment receptor at the cell surface (4, 5). The presence of CAR on epithelial cells determines the susceptibility of these cells to Ad5 infection both in vitro and in vivo (22, 23). Although the interactions of Ad5 with cells in vitro are known in great detail (24, 25), the pathways of cell-specific infection by Ad in vivo remain poorly understood. Specifically, after i.v. delivery, the vast majority of Ad particles are trapped in the liver (26–28). Unsuccessful attempts to prevent hepatocyte transduction by Ad5-based vectors through ablation of CAR and/or integrin binding implied that another mechanism must exist to allow Ad5 virus particles to enter hepatic cells in a CAR-independent manner (29, 30).
Recently, we and others identified a blood factor-dependent pathway that allows Ad5 to infect hepatocytes independently of virus binding to CAR (10, 11). These studies also demonstrated that FX is the most efficient at supporting Ad5 entry into hepatocytes. Despite this advance in our understanding of pathways of hepatocyte infection with Ad5 in vivo, the precise mechanism of blood coagulation factor interaction with Ad particles remained unclear.
In this study, we analyzed, in a systematic way, Ad capsid protein interaction with vitamin K-dependent coagulation factors and found that the Ad5 hexon possesses a high-affinity binding site for human FX. The determined picomolar affinity of FX binding to Ad5 implies that, when delivered via an intravascular route, Ad5 will form a very stable complex with FX and not other vitamin K-dependent zymogens, which bind Ad5 with lower affinity (Table S1) and are less abundant in plasma. CryoEM visualization of the Ad5-FX complex revealed a mesh of FX molecules covering almost the entire adenovirus capsid. Importantly, analysis of the contact point between FX and hexon showed that FX binds within the central depression at the top of each hexon trimer.
The hexon-mutated Ad5BAP vector failed to transduce hepatocyte when injected into wild-type mice or warfarin-treated mice with physiological levels of FX in circulation. Our in vitro data also showed that Ad5BAP infectivity cannot be increased by addition of blood factors into infection media (Fig. 3); however, unmodified Ad5RFP vector infectivity increases dramatically when FVII, FIX, or FX is added to the media during virus infection.
Hexon-mutated Ad5BAP virus could be efficiently propagated in 293 cells, and our in vitro analyses showed it is attached to and internalized into cells with high efficiency, which is equal to the unmodified Ad5RFP vector (Fig. S4). However, unlike Ad5RFP, Ad5BAP demonstrated colocalization with late endosomal/lysosomal cathepsin D-positive compartments and variable infectivity on human lung epithelial and hepatoma cells. These data indicate that such postinternalization steps of infection as release from the endosomes or viral DNA transport into the nucleus may be more significantly affected by cell-type-specific factors for hexon-modified Ads when compared with unmodified vectors.
Our earlier data suggested that FIX could efficiently support CAR-independent liver cell infection both in vitro and in vivo, and that the fiber knob domain is the likely moiety that binds FIX in the Ad capsid (10). Moreover, numerous studies of hepatocyte transduction by capsid-modified Ad vectors showed markedly reduced transgene expression, when vectors with short-shafted fibers were applied in vivo (29, 31, 32). Our SPR studies demonstrated that FIX binding to Ad5 follows a complex kinetic that cannot be described by a 1:1 single-site interacting model (Fig. 1). Moreover, the affinity of FIX binding to purified hexon is reduced, compared with its binding to complete Ad5 particles (Table S1). The observed kinetics of FIX binding to Ad5 particles fit well into a two-site interacting model (data not shown; O.K. and D.M.S., unpublished work). Although the precise localization of these sites remained to be determined, the hexon-mutated Ad5BAP vector failed to bind FIX in SPR assay and transduce cells in a FIX-dependent manner in vitro (Fig. 3). These data suggest that, similar to FX, for FIX, Ad5 hexon possesses the dominant binding site on the Ad capsid.
Our finding is critical for understanding the interactions of Ad with host cells in vivo and has important practical implications for development of safe and efficient cell- and tissue-type-specific adenovirus vectors for gene therapy applications.
Materials and Methods
Cells and Viruses.
The first-generation Ad5-based vectors, expressing the dsRED2 RFP, Ad5RFP, and Ad5BAP, were constructed and described in detail (12, 33). Ad5BAP is described as Ad-Hexon-BAP (33). Wild-type human Ad serotypes Ad2, Ad3, Ad4, Ad5, Ad9, Ad16, Ad21, Ad35, Ad41, Ad50, and Ad51 were obtained from American Type Culture Collection. Human reovirus T3D was kindly provided by Terence Dermody (Vanderbilt University, Nashville, TN). Viruses were banded in CsCl gradients, dialyzed, and stored in aliquots, as described (34). Adenovirus genome titers were determined by quantitative real-time PCR (for Ad5RFP and Ad5BAP) and OD260 measurement (for wild-type viruses).
Proteins.
All coagulation blood factors were purchased from Haematologic Technologies. All supplied blood factors were at ≥95% purity: human FX, HCX-0050, lot T1206; mouse FX, MCX-5050, lot R0903; human FIX, HCIX-0040, lot U1206; human FVII, HCVII-0030, lot U1206–0.02; human protein C, HCPC-0070, lot W0822; human FXI, HCXI-0150, lot U0607; human FXII, HCXII-0155, lot U1020; and human Gla-domainless FX, HCX-GD, lot U0629. Ad2 hexon (catalog no. R14800) was purchased from Meridian Life Sciences. Ad5 hexon and Ad5 fiber knob domain purification is described in detail in SI Text, Materials and Methods. BSA, catalog no. A0281-5G, was purchased from Sigma.
SPR Analyses.
All analyses were carried out on a Biacore 2000 machine. Research grade CM5 sensor chips, N-hydroxysuccinimide, N-ethyl-N′-(3-diethylaminopropyl)carbodiimide, ethanolaminehydrochloride, HBSP running buffer, and HBSEP regeneration buffer were purchased from the manufacturer (Biacore). All data were collected at 1 Hz by using two or three replicate injections for each concentration of analyte. Data processing and kinetic analysis were performed by using Scrubber software (version 2.0, BioLogic Software). Data processing included double referencing (35). Processed data were globally fit to a simple 1:1 interaction model.
Cryoelectron Microscopy.
An Ad5-FX complex was prepared by mixing 100 μl of the Ad5S vector (34), which has a short-shafted fiber (1 × 1013 virus particles per ml) with 1 μl of FX (2 mg/ml). Six-microliter aliquots of the Ad5-FX complex were applied to Quantifoil R2/4 holey carbon grids (Quantifoil Micro Tools) and a Vitrobot cryofixation device (FEI) was used for blotting and sample vitrification in liquid ethane. Data collection was performed on an FEI Tecnai 12 (120 kV, LaB6) transmission cryoelectron microscope equipped with a Gatan cryoholder and Gatan UltraScan 2kx2k charge-coupled device camera (Gatan). Twenty-six cryoelectron micrographs were collected at a nominal magnification of ×67,000, from which 40 particle images were selected by using the program VIRUS (36). Initial estimates for the defocus and astigmatism parameters were determined with CTFFIND3 (37). The FREALIGN package (38) was used for refinement of the orientational, defocus, and astigmatism parameters of each particle image and to calculate 3D reconstructions. The resolution of the icosahedral capsid is estimated to be 38 Å by the Fourier Shell Correlation 0.5 threshold with applied inner and outer spherical masks (radii = 300 and 463 Å). The resolution of the reconstruction including FX and fiber density (outer radius = 613 Å) is 40 Å.
Adenovirus Infection in Vivo.
All experimental procedures involving animals were conducted in accordance with the institutional guidelines set forth by the University of Washington. C57BL/6 mice (Charles River) were housed in specific pathogen-free facilities. For analysis of Ad-mediated gene transfer into liver cells, 1011 Ad particles in 200 μl of PBS were injected by tail-vein infusion. For in vivo transduction studies, mice were killed 1 or 48 h after virus infusion, and livers were processed for DNA, protein, and histological analyses. For analysis of Ad genome accumulation in the liver tissue 1 h after Ad vector administration, the blood was flushed from the liver by a cardiac saline perfusion, livers were harvested, and total DNAs were purified as described (39). Where applicable, mice were injected with warfarin and FX, as described in ref. 11.
Histological Analysis.
Liver samples from mice injected with Ad vectors were fixed, dehydrated in sucrose, and frozen in OCT compound. RFP gene expression in hepatic cells was analyzed on frozen sections using conventional UV fluorescent microscopy.
Acknowledgments.
We thank Daniel Stone for critical reading of this manuscript and Terence Dermody for providing reovirus T3D. This work was supported by funding from the National Institutes of Health (Grants AI062853, AI064882, and AI065429, to D.M.S.; Grant AI042929, to P.L.S.) and by grants to M.A.B. from the Muscular Dystrophy Association and the Propionic Acidemia Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711757105/DCSupplemental.
References
- 1. [Accessed March 20, 2008];J Gene Med. 2008 Gene Therapy Clinical Trials Worldwide, www.wiley.co.uk/genmed/clinical.
- 2.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 3.Wickham TJ. Ligand-directed targeting of genes to the site of disease. Nat Med. 2003;9:135–139. doi: 10.1038/nm0103-135. [DOI] [PubMed] [Google Scholar]
- 4.Bergelson JM, Cunningham JA, Droguett G, Kurt-Jones EA, Krithivas A, et al. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 5.Tomko RP, Xu R, Philipson L. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc Natl Acad Sci USA. 1997;94:3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wickham TJ, Mathias P, Cheresh DA, Nemerow GR. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 7.Alemany R, Curiel DT. CAR-binding ablation does not change biodistribution and toxicity of adenoviral vectors. Gene Ther. 2001;8:1347–1353. doi: 10.1038/sj.gt.3301515. [DOI] [PubMed] [Google Scholar]
- 8.Smith T, Idamakanti N, Kylefjord H, Rollence M, King L, et al. In vivo hepatic adenoviral gene delivery occurs independently of the coxsackievirus-adenovirus receptor. Mol Ther. 2002;5:770–779. doi: 10.1006/mthe.2002.0613. [DOI] [PubMed] [Google Scholar]
- 9.Smith TA, Idamakanti N, Marshall-Neff J, Rollence ML, Wright P, et al. Receptor interactions involved in adenoviral-mediated gene delivery after systemic administration in non-human primates. Hum Gene Ther. 2003;14:1595–1604. doi: 10.1089/104303403322542248. [DOI] [PubMed] [Google Scholar]
- 10.Shayakhmetov DM, Gaggar A, Ni SH, Li ZY, Lieber A. Adenovirus binding to blood factors results in liver cell infection and hepatotoxicity. J Virol. 2005;79:7478–7491. doi: 10.1128/JVI.79.12.7478-7491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parker AL, Waddington SN, Nicol CG, Shayakhmetov DM, Buckley SM, et al. Multiple vitamin K-dependent coagulation zymogens promote adenovirus-mediated gene delivery to hepatocytes. Blood. 2006;108:2554–2561. doi: 10.1182/blood-2006-04-008532. [DOI] [PubMed] [Google Scholar]
- 12.Campos SK, Parrott MB, Barry MA. Avidin-based targeting and purification of a protein IX-modified, metabolically biotinylated adenoviral vector. Mol Ther. 2004;9:942–954. doi: 10.1016/j.ymthe.2004.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stehle T, Dermody TS. Structural evidence for common functions and ancestry of the reovirus and adenovirus attachment proteins. Rev Med Virol. 2003;13:123–132. doi: 10.1002/rmv.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morral N, O'Neal W, Rice K, Leland M, Kaplan J, et al. Administration of helper-dependent adenoviral vectors and sequential delivery of different vector serotype for long-term liver-directed gene transfer in baboons. Proc Natl Acad Sci USA. 1999;96:12816–12821. doi: 10.1073/pnas.96.22.12816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parks RJ, Evelegh CM, Graham FL. Use of helper-dependent adenoviral vectors of alternative serotypes permits repeat vector administration. Gene Ther. 1999;6:1565–1573. doi: 10.1038/sj.gt.3300995. [DOI] [PubMed] [Google Scholar]
- 16.Seshidhar Reddy P, Ganesh S, Limbach MP, Brann T, Pinkstaff A, et al. Development of adenovirus serotype 35 as a gene transfer vector. Virology. 2003;311:384–393. doi: 10.1016/s0042-6822(03)00161-2. [DOI] [PubMed] [Google Scholar]
- 17.Sakurai F, Mizuguchi H, Yamaguchi T, Hayakawa T. Characterization of in vitro and in vivo gene transfer properties of adenovirus serotype 35 vector. Mol Ther. 2003;8:813–821. doi: 10.1016/s1525-0016(03)00243-0. [DOI] [PubMed] [Google Scholar]
- 18.Saban SD, Silvestry M, Nemerow GR, Stewart PL. Visualization of alpha-helices in a 6-angstrom resolution cryoelectron microscopy structure of adenovirus allows refinement of capsid protein assignments. J Virol. 2006;80:12049–12059. doi: 10.1128/JVI.01652-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rux JJ, Kuser PR, Burnett RM. Structural and phylogenetic analysis of adenovirus hexons by use of high-resolution X-ray crystallographic, molecular modeling, and sequence-based methods. J Virol. 2003;77:9553–9566. doi: 10.1128/JVI.77.17.9553-9566.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Paolo NC, Kalyuzhniy O, Shayakhmetov DM. Fiber shaft-chimeric adenovirus vectors lacking the KKTK-motif efficiently infect liver cells in vivo. J Virol. 2007;81:12249–12259. doi: 10.1128/JVI.01584-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shayakhmetov DM, Eberly AL, Li ZY, Lieber A. Deletion of penton RGD motifs affects the efficiency of both the internalization and the endosorne escape of viral particles containing adenovirus serotype 5 or 35 fiber knobs. J Virol. 2005;79:1053–1061. doi: 10.1128/JVI.79.2.1053-1061.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walters RW, Freimuth P, Moninger TO, Ganske I, Zabner J, et al. Adenovirus fiber disrupts CAR-mediated intercellular adhesion allowing virus escape. Cell. 2002;110:789–799. doi: 10.1016/s0092-8674(02)00912-1. [DOI] [PubMed] [Google Scholar]
- 23.Walters RW, van't Hof W, Yi SMP, Schroth MK, Zabner J, et al. Apical localization of the coxsackie-adenovirus receptor by glycosyl-phosphatidylinositol modification is sufficient for adenovirus-mediated gene transfer through the apical surface of human airway epithelia. J Virol. 2001;75:7703–7711. doi: 10.1128/JVI.75.16.7703-7711.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goosney DL, Nemerow GR. Adenovirus infection: Taking the back roads to viral entry. Curr Biol. 2003;13:R99–R100. doi: 10.1016/s0960-9822(03)00037-x. [DOI] [PubMed] [Google Scholar]
- 25.Meier O, Greber UF. Adenovirus endocytosis. J Gene Med. 2003;5:451–462. doi: 10.1002/jgm.409. [DOI] [PubMed] [Google Scholar]
- 26.Brunetti-Pierri N, Palmer DJ, Beaud, Carey KD, Finegold M, et al. Acute toxicity after high-dose systemic injection of helper-dependent adenoviral vectors into nonhuman primates. Hum Gene Ther. 2004;15:35–46. doi: 10.1089/10430340460732445. [DOI] [PubMed] [Google Scholar]
- 27.Hodges BL, Taylor KM, Chu QM, Scull SE, Serriello RG, et al. Local delivery of a viral vector mitigates neutralization by antiviral antibodies and results in efficient transduction of rabbit liver. Mol Ther. 2005;12:1043–1051. doi: 10.1016/j.ymthe.2005.06.475. [DOI] [PubMed] [Google Scholar]
- 28.Lievens J, Snoeys J, Vekemans K, Van Linthout S, de Zanger R, et al. The size of sinusoidal fenestrae is a critical determinant of hepatocyte transduction after adenoviral gene transfer. Gene Ther. 2004;11:1523–1531. doi: 10.1038/sj.gt.3302326. [DOI] [PubMed] [Google Scholar]
- 29.Nicol CG, Graham D, Miller WH, White SJ, Smith TAG, et al. Effect of adenovirus serotype 5 fiber and penton modifications on in vivo tropism in rats. Mol Ther. 2004;10:344–354. doi: 10.1016/j.ymthe.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 30.Akiyama M, Roelvink P, Einfeld D, Kovesdi I, King CR. Effect of Ablating CAR and integrin binding on the biodistribution and cellular localization of adenovirus vectors following intravenous or intraperitoneal delivery. Mol Ther. 2003;7:S173. [Google Scholar]
- 31.Ni SH, Bernt K, Gaggar A, Li ZY, Kiem HP, et al. Evaluation of biodistribution and safety of adenovirus vectors containing group B fibers after intravenous injection into baboons. Hum Gene Ther. 2005;16:664–677. doi: 10.1089/hum.2005.16.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schoggins JW, Gall JGD, Falck-Pedersen E. Subgroup B and F fiber chimeras eliminate normal adenovirus type 5 vector transduction in vitro and in vivo. J Virol. 2003;77:1039–1048. doi: 10.1128/JVI.77.2.1039-1048.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campos SK, Barry MA. Comparison of adenovirus fiber, protein IX, and hexon capsomeres as scaffolds for vector purification and cell targeting. Virology. 2006;349:453–462. doi: 10.1016/j.virol.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 34.Shayakhmetov DM, Lieber A. Dependence of adenovirus infectivity on length of the fiber shaft domain. J Virol. 2000;74:10274–10286. doi: 10.1128/jvi.74.22.10274-10286.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myszka DG. Improving biosensor analysis. J Mol Recognit. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 36.Adiga U, Baxter WT, Hall RJ, Rockel B, Rath BK, et al. Particle picking by segmentation: A comparative study with SPIDER-based manual particle picking. J Struct Biol. 2005;152:211–220. doi: 10.1016/j.jsb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 37.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 38.Grigorieff N. FREALIGN: High-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 39.Shayakhmetov DM, Li ZY, Ni S, Lieber A. Analysis of adenovirus sequestration in the liver, transduction of hepatic cells, and innate toxicity after injection of fiber-modified vectors. J Virol. 2004;78:5368–5381. doi: 10.1128/JVI.78.10.5368-5381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]