Abstract
Imatinib inhibits Bcr-Abl, the oncogenic tyrosine kinase that causes chronic myeloid leukemia. The second-line inhibitors nilotinib and dasatinib are effective in patients with imatinib resistance resulting from Bcr-Abl kinase domain mutations. Bcr-AblT315I, however, is resistant to all Abl kinase inhibitors in clinical use and is emerging as the most frequent cause of salvage therapy failure. SGX393 is a potent inhibitor of native and T315I-mutant Bcr-Abl kinase that blocks the growth of leukemia cell lines and primary hematopoietic cells expressing Bcr-AblT315I, with minimal toxicity against Bcr-Abl-negative cell lines or normal bone marrow. A screen for Bcr-Abl mutants emerging in the presence of SGX393 revealed concentration-dependent reduction in the number and range of mutations. Combining SGX393 with nilotinib or dasatinib preempted emergence of resistant subclones, including Bcr-AblT315I. These findings suggest that combination of a T315I inhibitor with the current clinically used inhibitors may be useful for reduction of Bcr-Abl mutants in Philadelphia chromosome-positive leukemia.
Keywords: gatekeeper mutation, imatinib resistance, kinase inhibitor
Imatinib (Gleevec) inhibits Bcr-Abl, the deregulated tyrosine kinase that causes chronic myeloid leukemia (CML) (1) and is the first-line therapy for the disease. Responses in CML patients who begin imatinib treatment in chronic phase are durable (2), but relapse is common in accelerated and blastic phase (3–5). Even in patients reaching a complete cytogenetic response, residual disease remains detectable by sensitive PCR techniques, suggesting a continued threat of relapse (6, 7). Relapses can often be traced to emergence of CML subclones expressing Bcr-Abl with mutations in the kinase domain that confer varying degrees of resistance to imatinib (8, 9).
The imatinib family member nilotinib (AMN107; Tasigna) and the Src/Abl inhibitor dasatinib (BMS-354825; Sprycel) are Abl kinase inhibitors with enhanced potency against native Bcr-Abl compared with imatinib (≈30-fold and ≈325-fold, respectively), and these inhibitors are also effective against most imatinib-resistant Bcr-Abl mutants except Bcr-AblT315I (10–12).
We recovered only the Bcr-AblT315I mutant in vitro in the simultaneous presence of nilotinib and dasatinib (13). Emerging clinical data confirms that patients harboring Bcr-AblT315I are resistant to nilotinib (14) and dasatinib (15), and Bcr-AblT315I is frequently detected in patients with resistance to these inhibitors (14–16). Thus, an inhibitor of Bcr-AblT315I will be essential to circumvent resistance to Abl kinase inhibitor therapy for CML. Here, we report that the Bcr-AblT315I inhibitor SGX393 suppresses outgrowth of all Bcr-Abl escape mutants when combined with nilotinib or dasatinib.
Results
Catalytic Activity of AblT315I Is Inhibited by SGX393.
Application of x-ray crystallographic lead discovery and structure-guided optimization identified 1 [supporting information (SI) Fig. 6A] as an inhibitor of Abl and AblT315I, binding an active conformation of the Abl kinase domain (SI Fig. 6). Further development yielded SGX393, an inhibitor of native Bcr-Abl and most imatinib-resistant mutants, including Bcr-AblT315I, whose structure will be reported elsewhere. Recently, the crystal structure of AblT315I in complex with a pyrrolopyridine inhibitor related to 1 was reported (17).
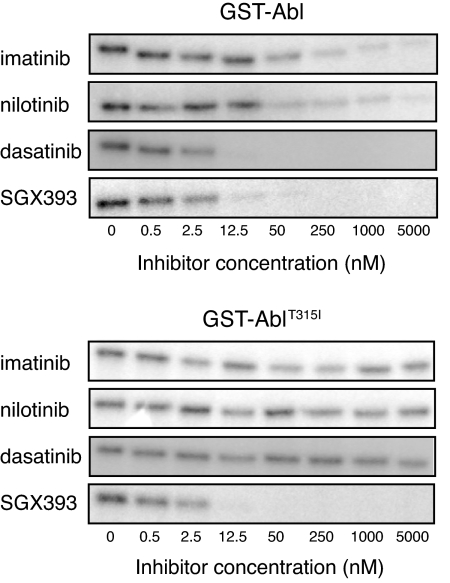
To establish that SGX393 directly targets Abl and AblT315I, we compared SGX393 to imatinib, nilotinib, and dasatinib in biochemical assays (10, 18, 19). SGX393 was comparable in potency to dasatinib in assays of Abl autophosphorylation and Abl-catalyzed peptide substrate phosphorylation (SI Fig. 7). In the case of AblT315I, only SGX393 was effective (Fig. 1).
Fig. 1.
Inhibition of GST-Abl and GST-AblT315I enzymatic activity by SGX393. Inhibition of in vitro autophosphorylation of purified GST-Abl and GST-AblT315I by imatinib, nilotinib, dasatinib, and SGX393. After incubation of purified, tyrosine-dephosphorylated enzyme with the indicated inhibitors in the presence of [γ-32P]-ATP and separation by gel electrophoresis, signal intensity was measured by autoradiography.
Among 177 kinases tested against SGX393, 4 exhibited an IC50 within 100-fold that of AblT315I (IC50 < 1 nM): CSFR1 (64 nM), FLT3 (15 nM), LCK (85 nM), and TRKC (39 nM). KIT, PDGFR, and most SRC family kinases were not sensitive to SGX393; the PDGFRαT674I gatekeeper mutant was inhibited (IC50 = 465 nM).
SGX393 Inhibits Growth of Ba/F3 Cells Expressing Native or Mutant Bcr-Abl, Including Bcr-AblT315I.
Proliferation assays performed with Ba/F3 cells revealed that SGX393 inhibited growth of cells expressing native Bcr-Abl (IC50, 12 nM) or Bcr-AblT315I (IC50, 7.3 nM) with similar potency (Table 1 and SI Fig. 8A). Sensitivity of Bcr-Abl mutants to SGX393 partitioned into three categories: IC50 < 25 nM (M244V, T315A, T315I, M351T, F359V, V379I, L387M, H396P, and H396R), IC50 < 300 nM (Q252H, Y253H, E255K, and F311L), and IC50 > 500 nM (G250E, Y253F, E255V, and F317L). SGX393 also inhibited growth of K562 and BV173 CML cell lines but not Bcr-Abl-negative leukemia cell lines. Growth of parental Ba/F3 cells was not affected by SGX393 at <2 μM (Table 1).
Table 1.
IC50 and IC90 values for SGX393 in cellular proliferation assays
| Cell Lines | SGX393 |
||
|---|---|---|---|
| IC50, nM | Fold IC50 | IC90, nM | |
| Ba/F3 cells | |||
| Native Bcr-Abl | 12 | 1.0 | 84 |
| M244V | 21 | 1.8 | 47 |
| G250E | 619 | 54 | 3,609 |
| Q252H | 241 | 21 | 894 |
| Y253F | 696 | 60 | 3,548 |
| Y253H | 176 | 15 | 440 |
| E255K | 207 | 18 | 585 |
| E255V | 2,210 | 191 | 4,795 |
| F311L | 250 | 22 | 465 |
| T315A | 12 | 1.0 | 39 |
| T315I | 7.3 | 0.6 | 28 |
| F317L | 598 | 52 | 3,242 |
| M351T | 15 | 1.3 | 117 |
| F359V | 10 | 0.9 | 47 |
| V379I | 13 | 1.1 | 56 |
| L387M | 12 | 1.0 | 57 |
| H396P | 5.6 | 0.5 | 15 |
| H396R | 18 | 1.5 | 49 |
| Parental | 3,632 | 314 | >5,000 |
| CML leukemia cells | |||
| K562 | 13 | NA | 24 |
| BV173 | 16 | NA | 70 |
| Non-CML leukemia cells | |||
| CMK | 3,129 | NA | >5,000 |
| CTV | 2,088 | NA | 4,742 |
| HL60 | 3,621 | NA | >5,000 |
| Marimo | 3,986 | NA | >5,000 |
SGX393 Inhibits Colony Formation by Murine Bone Marrow Cells Expressing Bcr-AblT315I.
To test the efficacy of SGX393 in primary hematopoietic cells, murine bone marrow was infected with retrovirus expressing native or mutant Bcr-Abl. All of the drugs inhibited colony formation of cells infected with native Bcr-Abl retrovirus, but only SGX393 was effective on cells expressing Bcr-AblT315I. Minimal toxicity of SGX393 was observed in cytokine-stimulated controls (SI Fig. 9).
Bcr-AblT315I-Driven Tumor Growth Is Inhibited by SGX393 in a Xenograft Model.
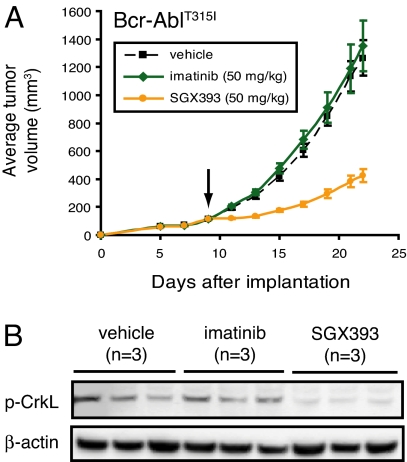
To evaluate SGX393 in vivo, nude mice were s.c. injected with Ba/F3 cells expressing Bcr-AblT315I. On day 9 (average tumor volume, 100 mm3), treatment was started with vehicle, imatinib, or SGX393 at 50 mg/kg, administered i.p. every 12 h for 12 days. SGX393 effectively inhibited tumor growth in mice compared with vehicle- or imatinib-treated mice (Fig. 2A). Phospho-CrkL-specific immunoblot analysis of tumor tissue collected 2 h after the final treatment confirmed inhibition of CrkL phosphorylation in mice receiving SGX393 but not imatinib or vehicle (Fig. 2B). Additionally, SGX393 and imatinib inhibited tumor growth in xenograft experiments that used K562 or Ba/F3 cells expressing native Bcr-Abl (SI Fig. 10). The relatively short half-life (2.9 h in BALB/c mice) of SGX393 in mice may explain the incomplete retardation of tumor growth.
Fig. 2.
Efficacy of SGX393 against Bcr-AblT315I in a murine xenograft model. (A) SGX393 mitigates increase in Bcr-AblT315I-driven tumor volume. Ba/F3 cells expressing Bcr-AblT315I were s.c. implanted in nude mice (2 × 106 cells per mouse). At a tumor volume of 100 mm3, treatment by i.p. injection was initiated with vehicle (black; 50% PEG400, 50% saline), imatinib (green), or SGX393 (orange) at 50 mg/kg every 2 days for 12–13 days. At study end, plasma levels of SGX393 were measured. (B) SGX393 inhibits phosphorylation of CrkL in a Bcr-AblT315I-driven tumor model. Two hours after administering vehicle or inhibitor for the final time in the study described in A, tumors from three mice per condition were collected and subjected to immunoblot analysis for CrkL phosphorylation status with an anti-phospho-CrkL antibody. β-actin was the loading control.
SGX393 Inhibits Bcr-AblT315I Kinase Activity in Primary CML and B-ALL Cells.
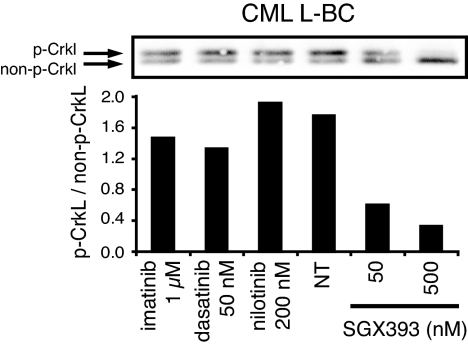
Peripheral blood mononuclear cells from a CML patient in lymphoid blast crisis harboring a T315I mutation were exposed to each of the inhibitors. Inhibition of Bcr-AblT315I kinase activity was assessed by immunoblot analysis with a total CrkL-specific antibody (Fig. 3). We observed a reduction in the level of phosphorylation of the Bcr-Abl substrate CrkL by SGX393, but not the other inhibitors. FACS analysis of intracellular phosphotyrosine levels yielded analogous results (SI Fig. 11A). Similar findings were observed for Bcr-AblT315I cells from a patient with Ph-positive B-ALL (SI Fig. 11B).
Fig. 3.
Inhibition of tyrosine phosphorylation by SGX393 in primary Bcr-AblT315I cells. Bcr-AblT315I CML lymphoid blast crisis (CML L-BC) mononuclear cells incubated with SGX393, imatinib, dasatinib, or nilotinib overnight were subjected to immunoblot analysis with a total CrkL-specific antibody (Upper), and the ratio of phosphorylated CrkL to nonphosphorylated CrkL was determined by densitometry (Lower).
To assess toxicity against normal hematopoietic progenitors, bone marrow mononuclear cells from three healthy individuals were cultured with SGX393 (SI Fig. 12). We observed minimal toxicity at concentrations of SGX393 up to 2 μM. At 5 μM SGX393, granulocyte/macrophage (CFU-GM) and erythroid (BFU-E) colony formation was reduced to 60–70% of controls.
The Resistance Profile of SGX393 Does Not Include Novel Sites.
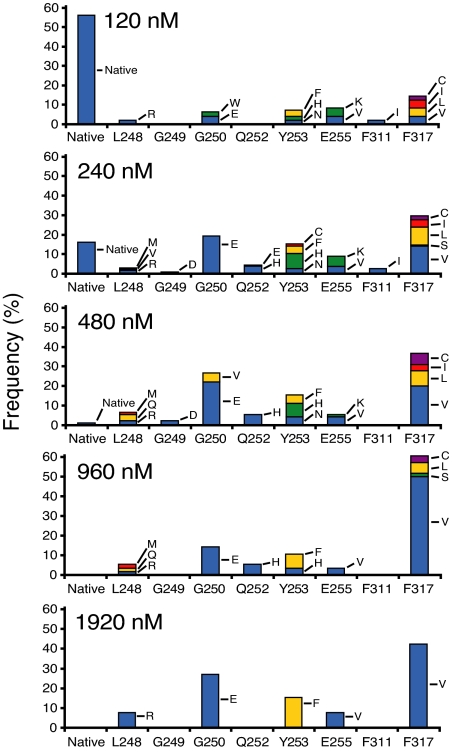
To establish a resistance profile for SGX393, we used our previously validated Ba/F3 cell line-based mutagenesis screen (13). Of note, not every possible mutation is necessarily represented in each experiment (13). We observed a concentration-dependent reduction of surviving subclones, ranging from 100% (120 nM SGX393) to 9% (1,920 nM SGX393) of wells exhibiting outgrowth (SI Table 2). Subclones expressing native Bcr-Abl were recovered rarely at SGX393 concentrations >240 nM (SI Table 2). Sequencing was confined to the kinase domain of Bcr-Abl.
At 240 nM SGX393, exchanges at 10 residues known to confer resistance to imatinib accounted for 98.5% of recovered mutated subclones (Fig. 4 and SI Table 2). At 480 nM SGX393, mutations were limited to seven positions, with exchanges at G250 and F317 accounting for 64.1% of recovered mutated subclones. Mutations recovered at 1,920 nM SGX393 were confined to L248R, G250E, Y253F, E255V, and F317V, with G250E and F317V accounting for 69.2% of the total.
Fig. 4.
Distribution of Bcr-Abl kinase domain mutations in subclones resistant to SGX393. ENU-treated Ba/F3 cells expressing native Bcr-Abl were cultured with graded concentrations of SGX393. Each bar represents the relative percentage of the indicated mutant among recovered subclones. Because the percentage of surviving resistant subclones and the concentration of SGX393 are inversely related, a different number of sequenced subclones is represented in the graph for each concentration of SGX393 (SI Table 2). The greatest number of resistant subclones was analyzed at 240 nM SGX393 [resolution of detection: one occurrence per 204 kinase domain-mutant-positive wells (0.5%)]. The detection limits for the remaining SGX393 concentrations are: 120 nM (2.1%; 48 subclones sequenced), 480 nM (1.1%; 90 subclones sequenced), 960 nM (1.8%; 56 subclones sequenced), and 1,920 nM (3.8%; 26 subclones sequenced). In addition, the following mutations accounted for 2.8% of recovered subclones at 240 nM SGX393 only: N322K, E355G, Y417S (1 of 243, 0.4% each), and E258K (4 of 243, 1.6%).
Because the L248R mutation persisted throughout the SGX393 screen but was not represented in the Ba/F3 cell panel, we tested GST-AblL248R in a peptide substrate-based tyrosine phosphorylation assay (11). The IC50 values were: >5,000 nM (imatinib), 2,328 nM (SGX393), 239 nM (nilotinib), and 0.8 nM (dasatinib) (data not shown).
No mutations involving residue T315 or unique exchanges that confer high level resistance to SGX393 were observed among the nearly 1,000 resistant subclones analyzed in this study. Rather, most of the recovered mutations had been documented in connection with imatinib resistance (7, 20–24).
Because T315I inhibitors, such as SGX393, may be used as second-line therapy for CML patients with the T315I mutation, we established the resistance profile when starting from Ba/F3 cells expressing Bcr-AblT315I. The distribution of mutations was similar to the native Bcr-Abl experiment, except for a striking decline in mutations at residue F317 (SI Table 3). This may indicate that two closely spaced mutations within the ATP binding site adversely affect kinase activity to an extent incompatible with survival of these subclones (25). Further studies will be necessary to explain this difference.
Exposure of Ba/F3 cells expressing native Bcr-Abl to a range of combinations of SGX393 and imatinib, dasatinib, or nilotinib revealed additive to synergistic cytotoxicity (SI Fig. 13) consistent with findings for imatinib combined with dasatinib (26) or nilotinib (10, 27).
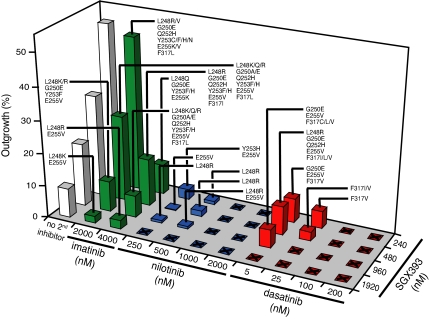
SGX393 in Combination with Nilotinib or Dasatinib Completely Suppresses Outgrowth of Resistant Clones.
To assess whether combinations of Abl kinase inhibitors offer an advantage over SGX393 alone, we carried out mutagenesis screens with combinations of SGX393 and imatinib, nilotinib, or dasatinib. Profiling of SGX393 with imatinib revealed a concentration-dependent reduction in both the percentage of wells with outgrowth and range of mutations (Fig. 5 and SI Table 4). However, outgrowth was not completely suppressed even at the highest tested combination (4,000 nM imatinib and 1,920 nM SGX393) with L248R, Y253H, and E255V persisting (Fig. 5).
Fig. 5.
Coadministration of SGX393 with a second Abl kinase inhibitor reduces the frequency of resistant subclones and the spectrum of Bcr-Abl kinase domain mutations. ENU-treated Ba/F3 cells expressing native Bcr-Abl were cultured with graded concentrations of SGX393 alone (white) and in dual combination with imatinib (green), nilotinib (blue), or dasatinib (red). Bars represent the percentage of wells from which drug-resistant subclones were recovered. A total of 288 wells were analyzed in all combination experiments except those involving 240 nM SGX393, for which 96 wells were examined. Labels indicate specific Bcr-Abl mutations identified for recovered subclones. Unlabeled squares with an X indicate inhibitor combinations for which no resistant subclones were recovered. See SI Tables 4–6 for further details pertaining to the Abl inhibitor combination experiments.
Consistent with the activity of nilotinib against most imatinib-resistant Bcr-Abl mutants and the potency of SGX393 against the T315I mutant, outgrowth at intermediate concentrations of these inhibitors was confined to three mutations: L248R, Y253H, and E255V. In the presence of 1,000 nM nilotinib and 240 nM SGX393, outgrowth of resistant subclones was completely suppressed. In total, 10 of 16 tested SGX393-nilotinib combinations abrogated outgrowth of resistant subclones (Fig. 5 and SI Table 5).
The combination of SGX393 with dasatinib was also tested for its potential to suppress outgrowth of resistant clones. At intermediate concentrations of SGX393 and dasatinib, resistance was confined to mutations of F317 (Fig. 5 and SI Table 6). However, no outgrowth was observed in the presence of 25 nM dasatinib and 960 nM SGX393. In total, 11/16 tested SGX393-dasatinib combinations were sufficient to completely suppress outgrowth of resistant subclones.
Discussion
Imatinib has changed the landscape of CML therapy, inducing complete cytogenetic responses in 87% of patients diagnosed with chronic phase disease (2). Resistance in early chronic phase is uncommon, although relapses do occur, particularly in patients with high-risk Sokal scores (2). In contrast, acquired resistance is frequent in patients who start imatinib in accelerated or blastic phase (3–5, 28). Relapse is frequently due to Bcr-Abl kinase domain mutations that impair imatinib binding (21, 22, 29). Nilotinib and dasatinib have largely addressed this issue, maintaining activity against most imatinib-resistant mutants except Bcr-AblT315I (14, 15, 30). The frequency of T315I in patients with resistance to nilotinib or dasatinib suggests that this mutant may emerge as a common mechanism of failure to second-line Abl kinase inhibitor therapy (14–16, 31, 32), consistent with predictions based on in vitro profiling and mutagenesis screens (11–13, 31, 33–36). Thus, the full potential of Abl kinase inhibitor therapy in patients with CML, particularly those with advanced disease, will depend on effectively targeting the Bcr-AblT315I mutant.
SGX393 activity in cell proliferation assays extended to a wide range of mutations, including T315A, which has been recovered from several patients resistant to dasatinib (16, 31). The key exceptions involve F311, F317, and certain P-loop mutations, especially E255V (IC50, 2,210 nM). However, E255V is inhibited to some extent by nilotinib (IC50, 430 nM) and dasatinib (IC50, 11 nM), providing a rationale for combined Abl inhibitor therapy (SI Fig. 3B) (26, 37, 38). Steady-state trough plasma levels in patients are 3,700 nM for nilotinib and 150 nM for dasatinib (14, 15).
In mutagenesis assays, SGX393 controlled the outgrowth of native Bcr-Abl, Bcr-AblT315I, and most mutants associated with imatinib resistance (Fig. 4 and SI Table 2). However, similar to results obtained in cell proliferation assays, mutations in and around the P-loop and at positions F311 and F317 remain vulnerabilities. Because dual combinations of currently available Abl inhibitors selected for T315I in vitro (13), we reasoned that combinations that include SGX393 might eliminate the outgrowth of resistant subclones. Indeed, when SGX393 was included with nilotinib or dasatinib, outgrowth of resistant subclones was reduced to zero (Fig. 5). Although further pharmacokinetic analysis of SGX393 and related compounds will be necessary, it is remarkable that even the lowest dose of SGX393 tested in combination with clinically relevant concentrations of nilotinib or dasatinib completely suppressed the emergence of resistant clones.
The T315I mutation is emerging as a common mechanism of failure to second-line Abl kinase inhibitors (16, 30, 31, 39, 40). Thus, even in these advanced cases, Bcr-Abl remains the critical therapeutic target. At this point, reports of successful salvage therapy for CML patients who acquire the T315I mutation are limited to small clinical trials and case reports (41, 42). Because SGX393 is active against native Bcr-Abl and the Bcr-AblT315I mutant, monotherapy with an inhibitor of this type might be sufficient to induce responses in patients harboring only Bcr-AblT3151. However, multiple mutation types are often detectable in patients with imatinib failure (22, 31) and patients subsequently failing dasatinib or nilotinib may harbor mutant clones other than T315I (30, 40, 41). Compound mutations, defined as more than one codon change in the same BCR-ABL mRNA, have also been detected in so far rare cases (31). Thus, the full clinical potential of T315I inhibitors may be realized in combinations with nilotinib or dasatinib. Of note, combination treatment is not expected to inhibit Bcr-Abl-independent resistance or to target CML stem cells. In light of evidence that nilotinib and dasatinib impinge on several nonkinase targets (43), there is also potential for side effects in patients because of off-target effects. The combination treatment suggested here may be useful to reduce the incidence of Bcr-Abl mutants in patients with Ph-positive leukemia and possibly eliminate Bcr-Abl-dependent resistance.
Materials and Methods
Inhibitors.
Inhibitor stock solutions (10.0 mM) were stored at −20°C, and diluted just before use. SGX393 may be requested from SGX Pharmaceuticals.
Kinase Autophosphorylation Assays with GST-Abl Kinase Domains.
Kinase assays, using tyrosine-dephosphorylated GST-Abl and mutant GST-Abl fusion proteins (c-Abl amino acids 220–498), were performed as described in ref. 44.
In Vitro Peptide Substrate Phosphorylation Assays with GST-Abl Kinase Domains.
The effects of SGX393 on the catalytic activity of tyrosine dephosphorylated GST-Abl kinase were assessed by using a synthetic peptide substrate as described in refs. 44 and 45. We also profiled the inhibitors imatinib, nilotinib, and dasatinib in all experiments for comparison.
Kinase Selectivity Profile of SGX393.
SGX393 (1,000 nM) was profiled against the T315I mutant of Abl and 177 off-target kinases, using the fluorescence resonance energy transfer-based SelectScreen assay (Invitrogen).
Cell Lines.
Ba/F3 transfectants expressing native or mutant Bcr-Abl were maintained in RPMI medium 1640 supplemented with 10% FCS, 1 unit/ml penicillin G, and 1 mg/ml streptomycin (complete media) at 37°C and 5% CO2. The Ba/F3 cell line expressing Bcr-AblT315A was a gift of N. Shah (University of California San Francisco, CA). Parental Ba/F3 cells were supplemented with IL-3, provided by WEHI-conditioned media.
Cell Proliferation Assays.
Ba/F3 cell lines were distributed in 96-well plates (4 × 103 cells per well) and incubated with graded concentrations of SGX393 for 72 h. Proliferation was measured by using a methanethiosulfonate-based viability assay (CellTiter96 aqueous one solution reagent; Promega). IC50 and IC90 values are reported as the mean of three independent experiments performed in quadruplicate. Combination index studies with Ba/F3 cells expressing native Bcr-Abl were done as described in ref. 26.
Colony-Forming Assays with Murine Bone Marrow Cells.
Murine bone marrow cells were prestimulated with IL-3 (3 ng/ml), IL-6 (5 ng/ml), and SCF (50 ng/ml) for 16 h. Cells were transduced twice with 1 × 106 plaque-forming units of murine stem cell retrovirus expressing native Bcr-Abl or mutant Bcr-Abl (Y253H, E255K, T315I, M351T, or H396P) and plated in Methocult without cytokines in the absence or presence of imatinib (500 or 5,000 nM), SGX393 (1–1,000 nM), nilotinib (1–1,000 nM), or dasatinib (0.1–100 nM). Colonies were quantified 7 days after plating. Control experiments were done in the presence of IL-3 (3 ng/ml), IL-6 (5 ng/ml), and SCF (50 ng/ml).
Bcr-AblT315I Murine Xenograft Model.
Nude mice (12 per condition) were s.c. implanted with K562 or Ba/F3 cells expressing native Bcr-Abl or Bcr-AblT315I (2 × 106 cells per mouse). At a tumor volume of 100 mm3, treatment by i.p. injection was initiated with vehicle (50% PEG400, 50% saline), imatinib, or SGX393 at 50 mg/kg every 2 days for 12–13 days. At study end, plasma levels of SGX393 were measured, and CrkL phosphorylation in tumor tissue was determined by immunoblot analysis with a phospho-CrkL-specific antibody. Inhibitor concentrations in plasma from three mice per group were determined by an internal standard LC/MS/MS method. Tissues for immunoblot analysis were harvested 2 h after the final dosing.
Hematopoietic Colony Forming Assays of Normal Bone Marrow.
Bone marrow mononuclear cells (AllCells) from three normal individuals were cultured with SGX393. Cells (5 × 104) were plated in methylcellulose containing 50 ng/ml SCF, 10 ng/ml GM-CSF, and 10 ng/ml IL-3 (Methocult GF H4534; Stem Cell Technologies) to assess CFU-GM colony formation or media also containing 3 units/ml erythropoietin (Methocult GF H4434; Stem Cell Technologies) to assess BFU-E colony formation. Colonies were scored at 14 days.
Ex Vivo Exposure of Bcr-AblT315I Patient Samples to SGX393.
After obtaining informed consent, peripheral blood mononuclear cells from a patient with CML L-BC and a patient with B-ALL, each with a Bcr-AblT315I mutation, were isolated by Ficoll centrifugation. RT-PCR and sequence analysis confirmed that each sample predominantly contained the Bcr-AblT315I mutant. Mononuclear cells (5 × 106 per well) were cultured overnight alone or with imatinib (1 μM), dasatinib (50 nM), nilotinib (200 nM), or SGX393 (50 nM, 500 nM). Cells lysates were subjected to immunoblot analysis with total CrkL-specific antibody C-20 (Santa Cruz Biotechnology). Phosphorylated and nonphosphorylated CrkL, distinguished based on band migration, were quantified by densitometry on a Lumi-Imager (Roche). In parallel experiments, mononuclear cells (2 × 105) were cultured overnight alone or with imatinib (1,000 nM), dasatinib (50 nM), nilotinib (200 nM), or SGX393 (10, 20, 50, 100, and 500 nM). Cells were fixed and permeabilized as per manufacturer's instructions (Caltag), incubated with 4G10-FITC phosphotyrosine antibody (2 μg; BD Biosciences) for 1 h, washed twice with PBS supplemented with 1% BSA and 0.1% NaN3, and fixed in 1% formaldehyde. FITC signal intensity was analyzed on a FACSAria instrument (BD Biosciences), and results are reported as fold increase in mean fluorescence intensity relative to unstained controls.
Accelerated Mutagenesis Screen.
Ba/F3 cells expressing native Bcr-Abl were treated overnight with N-ethyl-N-nitrosourea (ENU; 50 μg/ml), pelleted, resuspended, and distributed into 96-well plates (1 × 105 cells per well) in complete media supplemented with SGX393. Wells were visually inspected for media color change and for cell growth every 2 days throughout the 28-day experiment. The contents of wells exhibiting outgrowth were transferred to wells containing 2-ml of complete media supplemented with SGX393 at the same concentration as in the initial well. If growth was observed in all wells of a given condition, 24 representative wells were expanded. At confluency, cells were collected by centrifugation, DNA was extracted by using a DNEasy Tissue kit (Qiagen), the Bcr-Abl kinase domain was amplified by using primers B2A (5′-TTCAGAAGCTTCTCCCTGACAT-3′) and ABL4065 (5′-TGAGTTCATAGACCTTCTCTGG-3′), PCR products were bidirectionally sequenced (Agencourt), and chromatograms were analyzed by using Mutation Surveyor software (SoftGenetics). The accelerated mutagenesis screen for SGX393 in combination with imatinib, nilotinib, or dasatinib was conducted in the same manner except that a second inhibitor was included.
Supplementary Material
Acknowledgments.
This work is dedicated to the memory of Prof. Richard E. Moore (1933–2007), a scientist and role model of the highest realm. Dr. Moore assisted with the manuscript through valuable discussions on structure-function relationships between Abl and small-molecule inhibitors. He also made suggestions on experimental design and critiqued the manuscript. We thank Dr. Brian Druker and his group for valuable discussions, Caitlyn Smith for technical assistance, and Jeff Blaney for crystallographic figures. This work was supported in part by National Heart, Lung, and Blood Institute Grant HL082978-01 (to M.W.D.) and the Leukemia and Lymphoma Society (M.W.D.).
Footnotes
Conflict of interest statement: The sponsor is a consultant for the biotech company whose compound is reported in this manuscript. The sponsor receives less than $10,000 per year in consulting fees. S.B., K.H., K.A.J., C.T., H.A.L., R.D.R., and S.K.B. are employees of SGX Pharmaceuticals, Inc.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800587105/DC1.
References
- 1.Druker BJ, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 2.Druker BJ, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 3.Sawyers CL, et al. Imatinib induces hematologic and cytogenetic responses in patients with chronic myelogenous leukemia in myeloid blast crisis: Results of a phase II study. Blood. 2002;99:3530–3539. doi: 10.1182/blood.v99.10.3530. [DOI] [PubMed] [Google Scholar]
- 4.Talpaz M, et al. Imatinib induces durable hematologic and cytogenetic responses in patients with accelerated phase chronic myeloid leukemia: Results of a phase 2 study. Blood. 2002;99:1928–1937. doi: 10.1182/blood.v99.6.1928. [DOI] [PubMed] [Google Scholar]
- 5.Ottmann OG, et al. A phase 2 study of imatinib in patients with relapsed or refractory Philadelphia chromosome-positive acute lymphoid leukemias. Blood. 2002;100:1965–1971. doi: 10.1182/blood-2001-12-0181. [DOI] [PubMed] [Google Scholar]
- 6.Goldman J. Monitoring minimal residual disease in BCR-ABL-positive chronic myeloid leukemia in the imatinib era. Curr Opin Hematol. 2005;12:33–39. doi: 10.1097/01.moh.0000148551.93303.9e. [DOI] [PubMed] [Google Scholar]
- 7.Hughes T, et al. Monitoring CML patients responding to treatment with tyrosine kinase inhibitors: Review and recommendations for harmonizing current methodology for detecting BCR-ABL transcripts and kinase domain mutations and for expressing results. Blood. 2006;108:28–37. doi: 10.1182/blood-2006-01-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corbin AS, La Rosee P, Stoffregen EP, Druker BJ, Deininger MW. Several Bcr-Abl kinase domain mutants associated with imatinib mesylate resistance remain sensitive to imatinib. Blood. 2003;101:4611–4614. doi: 10.1182/blood-2002-12-3659. [DOI] [PubMed] [Google Scholar]
- 9.Nicolini FE, et al. Mutation status and clinical outcome of 89 imatinib mesylate-resistant chronic myelogenous leukemia patients: A retrospective analysis from the French intergroup of CML (Fi(phi)-LMC GROUP) Leukemia. 2006;20:1061–1066. doi: 10.1038/sj.leu.2404236. [DOI] [PubMed] [Google Scholar]
- 10.Weisberg E, et al. Characterization of AMN107, a selective inhibitor of wild-type and mutant Bcr-Abl. Cancer Cell. 2005;7:129–141. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 11.O'Hare T, et al. In vitro activity of Bcr-Abl inhibitors AMN107 and BMS-354825 against clinically relevant imatinib-resistant Abl kinase domain mutants. Cancer Res. 2005;65:4500–4505. doi: 10.1158/0008-5472.CAN-05-0259. [DOI] [PubMed] [Google Scholar]
- 12.Shah NP, et al. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 13.Bradeen HA, et al. Comparison of imatinib, dasatinib (BMS-354825), and nilotinib (AMN107) in an N-ethyl-N-nitrosourea (ENU)-based mutagenesis screen: High efficacy of drug combinations. Blood. 2006;108:2332–2338. doi: 10.1182/blood-2006-02-004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kantarjian H, et al. Nilotinib in imatinib-resistant CML and Philadelphia chromosome-positive ALL. N Engl J Med. 2006;354:2542–2551. doi: 10.1056/NEJMoa055104. [DOI] [PubMed] [Google Scholar]
- 15.Talpaz M, et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N Engl J Med. 2006;354:2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 16.Soverini S, et al. Resistance to dasatinib in Philadelphia-positive leukemia patients and the presence or the selection of mutations at residues 315 and 317 in the BCR-ABL kinase domain. Haematologica. 2007;92:401–404. doi: 10.3324/haematol.10822. [DOI] [PubMed] [Google Scholar]
- 17.Zhou T, et al. Crystal structure of the T315I mutant of AbI kinase. Chem Biol Drug Des. 2007;70:171–181. doi: 10.1111/j.1747-0285.2007.00556.x. [DOI] [PubMed] [Google Scholar]
- 18.Nagar B, et al. Crystal structures of the kinase domain of c-Abl in complex with the small molecule inhibitors PD173955 and imatinib (STI-571) Cancer Res. 2002;62:4236–4243. [PubMed] [Google Scholar]
- 19.Tokarski JS, et al. The structure of dasatinib (BMS-354825) bound to activated ABL kinase domain elucidates its inhibitory activity against imatinib-resistant ABL mutants. Cancer Res. 2006;66:5790–5797. doi: 10.1158/0008-5472.CAN-05-4187. [DOI] [PubMed] [Google Scholar]
- 20.Branford S, et al. Detection of BCR-ABL mutations in patients with CML treated with imatinib is virtually always accompanied by clinical resistance, and mutations in the ATP phosphate-binding loop (P-loop) are associated with a poor prognosis. Blood. 2003;102:276–283. doi: 10.1182/blood-2002-09-2896. [DOI] [PubMed] [Google Scholar]
- 21.Gorre ME, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 22.Shah NP, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 23.von Bubnoff N, Schneller F, Peschel C, Duyster J. BCR-ABL gene mutations in relation to clinical resistance of Philadelphia-chromosome-positive leukaemia to STI571: A prospective study. Lancet. 2002;359:487–491. doi: 10.1016/S0140-6736(02)07679-1. [DOI] [PubMed] [Google Scholar]
- 24.Al-Ali HK, et al. High incidence of BCR-ABL kinase domain mutations and absence of mutations of the PDGFR and KIT activation loops in CML patients with secondary resistance to imatinib. Hematol J. 2004;5:55–60. doi: 10.1038/sj.thj.6200319. [DOI] [PubMed] [Google Scholar]
- 25.Griswold IJ, et al. Kinase domain mutants of Bcr-Abl exhibit altered transformation potency, kinase activity, and substrate utilization, irrespective of sensitivity to imatinib. Mol Cell Biol. 2006;26:6082–6093. doi: 10.1128/MCB.02202-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Hare T, et al. Combined Abl inhibitor therapy for minimizing drug resistance in chronic myeloid leukemia: Src/Abl inhibitors are compatible with imatinib. Clin Cancer Res. 2005;11:6987–6993. doi: 10.1158/1078-0432.CCR-05-0622. [DOI] [PubMed] [Google Scholar]
- 27.Weisberg E, et al. Beneficial effects of combining nilotinib and imatinib in preclinical models of BCR-ABL+ leukemias. Blood. 2007;109:2112–2120. doi: 10.1182/blood-2006-06-026377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Druker BJ, et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001;344:1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- 29.Soverini S, et al. ABL mutations in late chronic phase chronic myeloid leukemia patients with up-front cytogenetic resistance to imatinib are associated with a greater likelihood of progression to blast crisis and shorter survival: A study by the GIMEMA Working Party on Chronic Myeloid Leukemia. J Clin Oncol. 2005;23:4100–4109. doi: 10.1200/JCO.2005.05.531. [DOI] [PubMed] [Google Scholar]
- 30.Kantarjian HM, et al. Nilotinib (formerly AMN107), a highly selective BCR-ABL tyrosine kinase inhibitor, is effective in patients with Philadelphia chromosome-positive chronic myelogenous leukemia in chronic phase following imatinib resistance and intolerance. Blood. 2007;110:3540–3546. doi: 10.1182/blood-2007-03-080689. [DOI] [PubMed] [Google Scholar]
- 31.Shah NP, et al. Sequential ABL kinase inhibitor therapy selects for compound drug-resistant BCR-ABL mutations with altered oncogenic potency. J Clin Invest. 2007;117:2562–2569. doi: 10.1172/JCI30890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sorel N, et al. Sequential emergence of ABL-kinase mutations with loss of unmutated BCR-ABL allele during targeted therapies of CML. Blood. 2006;108:1782–1783. doi: 10.1182/blood-2006-03-011668. [DOI] [PubMed] [Google Scholar]
- 33.von Bubnoff N, et al. A cell-based screen for resistance of Bcr-Abl-positive leukemia identifies the mutation pattern for PD166326, an alternative Abl kinase inhibitor. Blood. 2005;105:1652–1659. doi: 10.1182/blood-2004-06-2445. [DOI] [PubMed] [Google Scholar]
- 34.Azam M, Latek RR, Daley GQ. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell. 2003;112:831–843. doi: 10.1016/s0092-8674(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 35.Burgess MR, Skaggs BJ, Shah NP, Lee FY, Sawyers CL. Comparative analysis of two clinically active BCR-ABL kinase inhibitors reveals the role of conformation-specific binding in resistance. Proc Natl Acad Sci USA. 2005;102:3395–3400. doi: 10.1073/pnas.0409770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ray A, Cowan-Jacob SW, Manley PW, Mestan J, Griffin JD. Identification of BCR-ABL point mutations conferring resistance to the Abl kinase inhibitor AMN107 (nilotinib) by a random mutagenesis study. Blood. 2007;109:5011–5015. doi: 10.1182/blood-2006-01-015347. [DOI] [PubMed] [Google Scholar]
- 37.O'Hare T, Corbin AS, Druker BJ. Targeted CML therapy: Controlling drug resistance, seeking cure. Curr Opin Genet Dev. 2006;16:92–99. doi: 10.1016/j.gde.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 38.Komarova NL, Wodarz D. Drug resistance in cancer: Principles of emergence and prevention. Proc Natl Acad Sci USA. 2005;102:9714–9719. doi: 10.1073/pnas.0501870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khorashad JS, et al. In vivo kinetics of kinase domain mutations in CML patients treated with dasatinib after failing imatinib. Blood. 2007;111:2378–2381. doi: 10.1182/blood-2007-06-096396. [DOI] [PubMed] [Google Scholar]
- 40.Cortes J, et al. Dynamics of BCR-ABL kinase domain mutations in chronic myeloid leukemia after sequential treatment with multiple tyrosine kinase inhibitors. Blood. 2007;110:4005–4011. doi: 10.1182/blood-2007-03-080838. [DOI] [PubMed] [Google Scholar]
- 41.Giles FJ, et al. MK-0457, a novel kinase inhibitor, is active in patients with chronic myeloid leukemia or acute lymphocytic leukemia with the T315I BCR-ABL mutation. Blood. 2007;109:500–502. doi: 10.1182/blood-2006-05-025049. [DOI] [PubMed] [Google Scholar]
- 42.de Lavallade H, et al. Interferon-alpha or homoharringtonine as salvage treatment for chronic myeloid leukemia patients who acquire the T315I BCR-ABL mutation. Blood. 2007;110:2779–2780. doi: 10.1182/blood-2007-06-094508. [DOI] [PubMed] [Google Scholar]
- 43.Rix U, et al. Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib, and dasatinib reveal novel kinase and nonkinase targets. Blood. 2007;110:4055–4063. doi: 10.1182/blood-2007-07-102061. [DOI] [PubMed] [Google Scholar]
- 44.O'Hare T, et al. Inhibition of wild-type and mutant Bcr-Abl by AP23464, a potent ATP-based oncogenic protein kinase inhibitor: Implications for CML. Blood. 2004;104:2532–2539. doi: 10.1182/blood-2004-05-1851. [DOI] [PubMed] [Google Scholar]
- 45.Brasher BB, Van Etten RA. c-Abl has high intrinsic tyrosine kinase activity that is stimulated by mutation of the Src homology 3 domain and by autophosphorylation at two distinct regulatory tyrosines. J Biol Chem. 2000;275:35631–35637. doi: 10.1074/jbc.M005401200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.