Abstract
Here, a scalable, accurate, reliable, and robust protein functional site comparison algorithm is presented. The key components of the algorithm consist of a reduced representation of the protein structure and a sequence order-independent profile–profile alignment (SOIPPA). We show that SOIPPA is able to detect distant evolutionary relationships in cases where both a global sequence and structure relationship remains obscure. Results suggest evolutionary relationships across several previously evolutionary distinct protein structure superfamilies. SOIPPA, along with an increased coverage of protein fold space afforded by the structural genomics initiative, can be used to further test the notion that fold space is continuous rather than discrete.
Keywords: functional site, structure
The evolutionary relationship between protein sequences, protein structures, and their associated function(s) remains a central topic of molecular biology and one resulting in the development of many computational methods (1–3). A central question is: What were the early protein folds and how did these folds change over long evolutionary time scales (4–7)? Comparative genomics studies and structural and phylogenetic analyses (8–10) have established that a subset of proteins, dominated by the structure classification of proteins (SCOP) (11) α/β class, were likely present in the last universal common ancestor (12, 13). Concurrently, growing evidence suggests that recurring substructures, that is, 3D fragments of noncontiguous sequence shared between different folds, may be clues that protein fold space is more continuous than discreet (14, 15). The sequence/structure similarity of such substructures correlates well with the similarity of function found between the different folds containing these substructures (16). The notion that protein fold space is a continuum is further supported by recent studies that show that protein domains can adopt different topologies through combination, swapping, deletion (4, 17, 18), and cyclic permutation (19, 20) of subdomains. Likewise, new folds can emerge from accretion (21) or embellishment (22) of substructures around a core of conserved secondary structures. Given these findings concerning the dynamic nature of protein structure and the possible continuous nature of protein fold space, it is important to distinguish proteins that share a common ancestor (divergent evolution) from those that have adopted common structural constraints (convergent evolution).
Typically, evolutionary relationships between protein sequence, structure, and function are deduced from the respective comparisons among known genes and their products. These comparisons are made at various levels, from genome sequences to protein domains and motifs to biochemical pathways. Such comparisons may miss important relationships because sequence relationships may be too weak to detect, and/or fail to identify complex evolutionary events such as domain swapping and cyclic permutation. Likewise, differences in global protein structure may disguise a true evolutionary relationship that exists between substructures. One approach, which involves the comparative analysis of substructures, including functional sites between proteins (1, 23–26), has been successful in detecting evolutionary relationships between different fold superfamilies and has been applied mostly to enzyme families. One study of 31 diverse enzyme superfamilies revealed that functional diversity during evolution is achieved by local sequence variation and domain shuffling (24). Such functional diversity can also be observed within a single SCOP superfamily. For example, within the protein kinase-like superfamily, it has been suggested that atypical kinases diverged early in evolution from protein kinases (26). In doing so, the overall catalytic mechanism is retained through a high level of conservation associated with the ATP binding cassette, thus preserving phosphorylation, yet the substrate binding motif exhibits significant diversity. In the case of mechanistically diverse enzymes, whose members catalyze different overall reactions but share a partial reaction, it has been found that these enzymes use a similar active site to generate a common intermediate, then direct the intermediate to different products in different active sites (25). Beyond these case studies, the global evolutionary relationship of functional sites across fold space has not been systematically studied and remains elusive. Global functional site comparison has been thwarted by the lack of efficient and accurate computational tools to undertake such a large scale comparison and a lack of rigorous statistics to test their similarity. The work described herein is a step toward accurate and efficient functional site comparison and analysis and is subsequently applied to seek out new evolutionary relationships.
Although the concept of functional site matching is not new, and a variety of approaches have been attempted (27–47), it has not proven an easy task to design and implement a practical software solution with performance that is close to that of routine sequence comparison. These site comparison algorithms usually consist of three interrelated components; the representation of the functional site, an algorithm to superimpose two sites and a method to score their similarity. The functional site is usually represented either by a coordinate set with certain physicochemical or evolutionary properties, or by 3D shape descriptors that define pockets within the protein (44). The coordinate set can consist of atoms (28), chemical groups (34) or surface points (33, 41). The optimum superimposition between two sites is achieved with geometric hashing (33, 42), graph theory (28, 30, 40), or other ad hoc algorithms (39, 43). Finally, the similarity between two sites is measured by using geometric criteria, such as RMSD (31), spherical harmonic expansion (44), residue conservation (29, 38), or physicochemical property similarity (28, 30, 34). With their inherent advantages and limitations, these algorithms have achieved considerable success. However, to perform protein functional site comparison on a large scale requires a scalable approach that remains accurate, reliable, and robust. Few, if any, of the current algorithms meet all these requirements, and hence a new algorithm is presented. We use this algorithm here to confirm or propose hitherto unseen evolutionary relationships and note that a larger scale study is underway. The algorithm proposed is different from previous algorithms in several respects. First, our algorithm searches for similar functional sites by scanning whole proteins in the spirit of local sequence alignments, i.e., it is not necessary to predefine functional sites during the search. Second, most of the available algorithms score similarities between whole functional sites. However, the complete ATP and NAD binding sites may look different overall, but they may share a similar subpocket for binding chemically and conformationally similar fragments, adenine in this case. This subsite similarity would be missed by most current algorithms. Our algorithm is designed to detect similar subsites across all of fold space. Third, we proposed a sequence order-independent profile–profile alignment (SOIPPA) algorithm, which is more general and sensitive than others (29, 38, 48), because the sequence order of amino acid residues in the functional site is not necessarily conserved. Fourth, with the aim of applying our approach to homology models and low resolution structures, the algorithm was purposely designed and tested with a Cα only representation of the protein structure. Finally, the statistical significance of the similarity is evaluated by using a nonparametric statistical method based on a fold distribution model. The algorithm is evaluated on a benchmark set and a control group that includes 247 and 101 nonredundant protein chains of diverse folds with and without adenine binding pockets, respectively. Our results show that SOIPPA is suitable for application on a large-scale. For example, in studies reported elsewhere, we have successfully applied SOIPPA to identify off-targets for pharmaceuticals for which the primary drug-receptor complex is present in the Protein Data Bank (PDB). Searching by using the primary site information, we have identified off-targets for selective estrogen receptor modulators (49) and have been able to repurpose an existing drug for treatment of drug resistant tuberculosis (S. Kinnings, L.X., and P.E.B., unpublished data). In both cases the primary and secondary targets are in different gene families. Here, we focus on a few examples that support the notion that fold and functional space may be continuous rather than discrete.
Results
Comparison of Functional Site Superposition Algorithms.
Two datasets were used in the comparison study. The 247-benchmark consisted of 247 nonredundant protein chains known to bind an adenine containing ligand. The 101-control consisted of 101 nonredundant protein chains believed not to bind an adenine containing moiety. From the 247-benchmark, 30,381 benchmark pairs were generated, and, from the 101-control, 24,947 control pairs were generated in an all-by-all pairwise comparison (see Methods). All benchmark and control pairs were used to evaluate the performance of the sequence order-independent profile–profile alignment (SOIPPA) algorithm [see Methods and supporting information (SI) Text]. We further implemented several existing functional site comparison algorithms that employ maximum cliques (28, 30, 40) and similarity measures, using substitution matrices (29, 38), physiochemical properties, and amino acid grouping (40, 43). Using the benchmark and control data, their performance was compared with SOIPPA with respect to alignment quality and sensitivity/specificity during site searches. We also include results from PSI-BLAST (50) and CE (51) in our comparisons to relate functional site similarity to more traditional sequence and structural similarity, respectively.
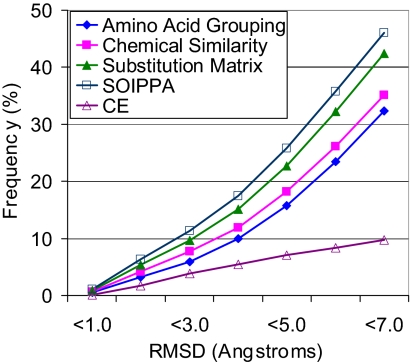
To measure the alignment quality, we use the root mean square deviation (RMSD) between ligands bound to the aligned functional sites. In our 247-benchmark, all proteins bind to ligands containing a chemically identical and conformationally rigid adenine plus a chemical and/or conformational variable component. Although the complete functional sites may be different as a result of binding to ligands with different compositions or conformations, they may share similar subsites that bind to similar ligand fragments. Our purpose is to detect such similar subsites. More specifically, the RMSD between common molecular moieties from diverse ligands is used to measure the alignment quality of the functional site comparison algorithm. SOIPPA provides the best performance when compared with other methods. As shown in Fig. 1, using SOIPPA, 6.5% and 25.9% of pairs are aligned with RMSD values of <2.0 and <5.0 Å, respectively. Lists of these pairs are given in Dataset S1 and Dataset S2. Using a BLOSUM45 substitution matrix (52), the percentage of aligned pairs drops to 5.3% and 22.3%, respectively. Using a chemical similarity-based method (53), only 4.2% and 18.1% of pairs are aligned with RMSD values of <2.0 and <5.0 Å, respectively. Using a maximum size clique method based on amino acid grouping (40), only 3.1% and 15.7% pairs are aligned with RMSD values of <2.0 and <5.0 Å, respectively. Moreover, the frequency of aligned ligands when performing global structure similarity, using CE, is significantly lower than that achieved with SOIPPA. Specifically, for pairs of proteins where the RMSD values of aligned ligands are <2.0 and <5.0 Å, the percentage of dissimilar structures (defined by a CE Z score <3.5) are ≈40% and ≈60%, respectively (Fig. S1). These results indicate that the functional site is more structurally conserved than the global protein structure. Although similar functional sites may arise from convergent evolution (69), the high occurrence among dissimilar structures raises the possibility of very significant divergent evolution. It is noteworthy that SOIPPA uses the whole-protein structure when performing the ligand site search and comparison and does not require that the ligand or functional site be known a priori. SOIPPA detects local similarity between two proteins by aligning two global structures in a manner that is independent of sequence order, important because sequence order is not always conserved (4) (Fig. S2, Fig. S3, Fig. S4, and Fig. S5). Beyond identifying similar subsites that bind to common molecular fragments with rigid conformations from different ligands, SOIPPA also detects similar subsites that bind flexible molecular fragments that adopt similar conformations (Fig. S3 and Fig. S5).
Fig. 1.
RMSD distribution of the aligned common fragments of ligands from the 247-benchmark, using maximum size clique with amino acid grouping (Amino Acid Grouping), maximum weight common subgraph with chemical similarity (Chemical Similarity), substitution matrix, SOIPPA, and CE.
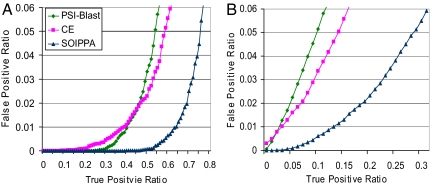
To evaluate the performance of SOIPPA in detecting evolutionary relationships, we first see whether the algorithm can identify known sequence and structural homologous within the same SCOP superfamily. Among the 247-benchmark pairs, <5% of them (1,230 pairs) are from the same SCOP superfamily. If only these 1,230 pairs are considered as true positives, Fig. 2a illustrates the performance of PSI-BLAST (50), CE (51), and SOIPPA in detecting remote homologous that belong to the same SCOP superfamily. For a false-positive ratio of 0.05, the coverage of PSI-BLAST, CE, and SOIPPA is 0.55, 0.60, and 0.75, respectively. If SCOP superfamilies are taken as the gold standard for defining remote evolutionary relationships, these results illustrate the well known fact that structure is more conserved than sequence. However, global structure comparison falls significantly short of SOIPPA, which takes both evolutionary profiles and structural constraints within the functional site into account. Consequently, it is more sensitive in detecting remote evolutional relationships than either PSI-BLAST or CE. The question then becomes, can SOIPPA detect functional similarities missed by SCOP, that is, relationships across superfamilies? In the 247-benchmark, there are 15,058 pairs aligned from different known SCOP superfamilies, and ≈30% of them are identified by SOIPPA with a false-positive ratio of 0.05 (Fig. 2b). Fig. 2b also shows that the sequence and structural similarity of these cross-superfamily pairs is not significant.
Fig. 2.
False-positive ratio vs. true-positive ratio for PSI-BLAST, CE, and SOIPPA. The aligned 247-benchmark pair is defined as a true positive if the two proteins are from the same (A) and different (B) SCOP superfamilies.
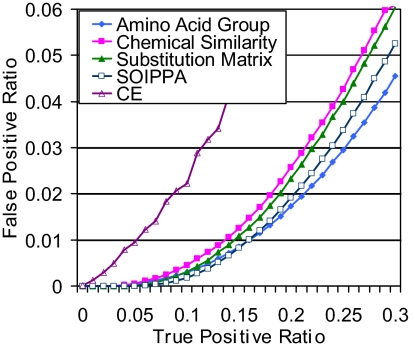
To illustrate that the sensitivity and specificity of functional site comparison, using SOIPPA, is improved over other methods, we define all of the 30,381 247-benchmark pairs to be true positives (Fig. 3). The false-positive rate for SOIPPA is ≈30% lower than that for a weighted method using an amino acid substitution matrix or chemical similarity at a 15% true-positive ratio. In the low false-positive region, the weighted methods (SOIPPA, substitution matrix, and chemical similarity) perform better than nonweighted amino acid grouping methods. However, if high false positives are tolerated, amino acid grouping performs best. As a baseline, the performance of CE is also included in the comparison. Clearly, a global structural comparison algorithm, such as CE, cannot detect the similarity between proteins when the overall structures have changed. For the 5,917 structurally related pairs (≈20% of 247-benchmark pairs) that have the same CATH topology, the coverage by CE is lower than SOIPPA at a false-positive ratio <0.05 (Fig. S6a). Moreover, the frequency of well aligned ligands from global alignment by CE is also lower than from SOIPPA (Fig. S7). For those cross-CATH topology pairs, it is not surprising that the coverage by CE is reduced to <10%. However, the coverage of SOIPPA is only slightly lower than that of the same CATH topology pairs (Fig. S6b). These results suggest that SOIPPA is not sensitive to overall structural changes, implying that local functional sites are more conserved than global sequence or structure. This then leads us to the question: Can evolutionary relationships be detected that are not defined by SCOP?
Fig. 3.
False-positive vs. true-positive ratios, using all 247-benchmark pairs, using maximum size clique with amino acid grouping (Amino Acid Group); maximum weight common subgraph with chemical similarity (Chemical Similarity); and substitution matrix, SOIPPA, and CE.
Evolutionary Linkage Across Fold and Functional Space.
Using SOIPPA to scan a nonredundant set of protein structures from the complete PDB (7,644 chains including both apo and holo structures) against several functional sites, we have detected statistically significant similarities between fold families whose global structures are very different yet whose local functional sites appear invariant over evolutionary time scales. Due to lack of structural and/or functional annotations on the whole set of structures, the true and false positives cannot be defined conventionally. However, the false-positive rate of the predication can be approximated by the P value from the fold distribution score (see SI Text).
One example is the relationship identified between the protein kinase-like, the SAICAR-synthesase-like, and the ATP grasp superfamilies after scanning the 7,644 proteins against ATP (PDB ID code 1ODB, SAICAR synthase-like) and ADP (PDB ID code 2HGS, ATP-grasp) binding sites. These three superfamilies show significant binding site similarities with each other (Fig. S8 and Fig. S9). Their evolutionary relationship has been proposed by others through manual analysis of structure (26, 54), even though their global structural similarities cannot be detected by structural comparison methods. For example, the CE similarity Z score and RMSD are 2.3 and 5.17 Å between PDB structure 1WBP (protein kinase-like) and 2HGS (ATP-grasp), respectively. Given that such remote evolutionary relationships can be established automatically by SOIPPA with high statistical significance we have a tool with the potential to discover unseen evolutionary relationships throughout protein fold space. We explore this notion further with some specific findings.
By searching against the 7,644 proteins, using the ATP binding site of PEP carboxykinase (PCK) (PDB ID code 1AYL), besides two top ranked PCK structures, a protein annotated as a P loop nucleotide triphosphate hydrolase (NTH) is ranked as the second most significant cross-superfamily hit (PDB ID code 1LS1; P = 8.4 × 10−5). Although there is no structural annotation for the most significant cross-superfamily hit (PDB ID code 1YRB; P = 8.4 × 10−5), its structure is similar to PDB 1LS1 (CE z score, 4.9; RMSD, 2.99 Å). Other high ranked hits with P < 2.0 × 10−3 are also similar to the P loop NTH fold (Table S1 and Table S2). The functional site alignment revealed by SOIPPA further suggests that these two superfamilies are evolutionarily related even though there is no observable global structure similarity. Besides the conserved Walker motif A (55) (GXXXXGKT/S) that is the common site for binding nucleotides in many proteins, several residues important for ATP binding and catalytic activity are also conserved. The N-terminal histidine residue (His-232) in PCK contacts the γ-phosphate of ATP and contributes to either activation of the substrate or stabilization of the transition state (56, 57). H232 of PCK can be superimposed onto H248 of NTPase, although the sequence order is switched to the C terminus. Likewise, residues D268 and D269 in PCK play roles in stabilizing a Mg2+–Mn2+ bimetal cluster at the active site (56, 58). An evolutionary linkage tree and multiple functional site alignment (Fig. S4 and Fig. S10) indicate that these two residues are strictly conserved in several other enzymes containing a P loop NTH fold. It is noteworthy that the overall conformation of the ATP molecule bound to these two classes of proteins is different, with an almost 180° flip in the adenine fragment. However, the conformations of the triphosphate tails are quite similar (Fig. S5). Our predication based on only a subsite similarity is consistent with previous speculation based on detailed structure analysis (59). However, both SCOP (11) and CATH (60) classify them differently missing an apparent evolutionary relationship.
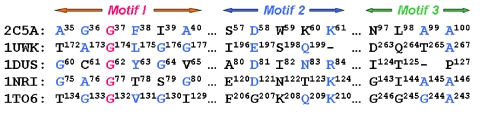
The NAD-binding Rossmann fold is one of the most common protein folds and observed in a large number of enzyme families (61) and believed to have evolved early (8). Evidence suggests that significant structural changes were accumulated from the original Rossmann fold (11, 61). By scanning structures against a NAD-binding site (PDB ID code 2C5A), SOIPPA detected similar functional sites from different SCOP superfamilies that have not been observed before. Except for the nicotinamide adenine dinucleotide (FAD) binding site from the FAD/NAD binding fold (P = 5.8 × 10−5), all other hits with a P < 2.0 × 10−3 contain the Rossmann fold, but with versatile domain insertions and/or sequence permutations (Table 1). Although their cofactor binding sites may bind different ligands, they share conserved sequence motifs (Fig. 4). Structural superimposition of the ligands indicates that the common adenine moiety from these cofactors is well aligned (Fig. S11, Fig. S12, and Fig. S13). Given that these proteins all adopt the same Rossmann fold topology, divergent evolution would seem the most likely scenario (62). Among the hits (Table 1), structures with SIS and glycerate kinase I domains do not have cocrystallized ligands. Their aligned residues are both located in deep pockets, indicating that they are potential ligand binding sites (Fig. S14). Moreover, their functions suggest that they can bind to nucleotides. For example, glycerate kinase catalyses the phosphorylation of (R)-glycerate to 3-phospho-(R)-glycerate in the presence of ATP (63).
Table 1.
List of proteins from different superfamilies shown by SOIPPA to have statistically significant similarity to the SCOP NAD(P)-binding Rossmann fold
| SCOP superfamily | PDB ID | Ligand in PDB | SOIPPA P value | CE Z score (RMSD) | Sequence identity, % |
|---|---|---|---|---|---|
| Urocanase | 1UWK | NAD | 1 × 10−5 | 4.2 (3.68) | 7.4 |
| SAM-dependent methyltransferase | 1DUS | SAM | 2.7 × 10−5 | 4.6 (3.14) | 9.7 |
| Sugar isomerase (SIS) domain | 1NRI | — | 1.1 × 10−4 | 3.7 (4.64) | 11.7 |
| Glycerate kinase I | 1TO6 | — | 1.5 × 10−3 | 3.1 (3.90) | 2.2 |
Fig. 4.
Multiple functional site alignment of a Rossmann NAD-binding protein (PDB ID code 2C5A) with four other proteins having the Rossmann topology but different SCOP superfamilies. The three conserved motifs are marked motif 1–3. The most conserved residues are labeled as red; and partially conserved residues are labeled as blue. Their corresponding SCOP superfamilies are listed in Table 1. The multiple functional site alignment is generated from pairwise SOIPPA alignments.
Discussion
Functional Sites as a Linkage to Trace Distant Evolutionary Relationships Across Fold Space.
It has been found that convergent evolution to achieve similar enzyme active sites is a common event (64). However, divergent evolution retaining only a ligand binding site cannot be discounted in some case given that the percentage of similar ligand binding motifs is high among dissimilar structures. Unlike enzyme active sites that typically only involve three or less spatially distinct and noncontiguous residues, many of the similar ligand binding motifs found here contain a number of conserved consecutive residues. We hypothesize that it is more efficient for nature to reuse such local features to bind the same or similar molecular moiety, but to modify other structural components to achieve different functions than to reinvent the whole structure from scratch. This is particularly true for adenine-binding folds studied here. They are found throughout fold space and have been predicated among the most ancient folds by several studies (12, 13, 65–68). It is possible that a small molecule containing an adenine fragment was the first ligand recognized by a protein (69).
Gross topology of protein can change dramatically in cases already attributed to divergent evolution, using mechanisms of structure drift, segment swapping, the insertion of additional structures, and fusion or permutation by duplication events (4, 70, 71).
Such events imply that the sequence order of conserved motifs may change. Thus, it is difficult to detect evolutionary relationships between different fold and functional families with overall sequence or structural similarity, although several studies have revealed that evolutionary relationships can be established within superfolds, using profile-profile comparison (72), or between different folds with similar short sequence motifs (73). Here, we assume that the functional site carries the evolutionary fingerprint, because it is one of the most important parts of the structure that must be conserved. Indeed, our results suggest that functional site residues and their spatial arrangements are conserved even when the global structure has changed. As such, the functional site can be used as a common linkage from which to potentially trace the evolutionary transition of the protein topology and discover underlying genetic events (15) and physical rules (17, 74) that govern structural changes.
A number of in silico and in vitro evolutionary studies have suggested that the sequence or profile similarity between proteins is a strong indication of divergent evolution (5, 6, 72, 73, 75–78). Moreover, it is necessary to establish the structural relationships based on a rigorous statistical test (79). By combining sensitive profile–profile alignment, structural constraints independent on the sequence order, and a rigorous statistics test, it is possible for us to propose divergent evolutionary relationships across fold space. Our results are consistent with existing observations, namely that convergent evolution of domain architectures is rare (62). However, to date, only a small percentage of such divergent structural relationships have been identified across fold space. Beyond new methods, structural genomics, because it strives to fill in gaps in the coverage of protein fold space (80), will also provide new clues as to the evolution and continuity of this space.
Conclusions
A new algorithm is introduced for functional site comparison that is based on a reduced protein structure representation and a sequence order independent profile-profile alignment. Using a well defined adenine-binding pocket from various folds as a benchmark, it has been shown that the proposed algorithm outperforms both nonweighted and sequence-weighted methods. Although there is still room for significant improvement in the performance of the current implementation, it provides a framework for developing a robust, reliable, accurate, and scalable functional site comparison algorithm. We show that the algorithm has the ability to detect new evolutionary relationships across existing current discreet descriptions of fold and functional space opening the door to tracing fold changes during evolution.
Methods
Benchmark and Control Data.
From the Research Collaboratory for Structural Bioinformatics Protein Data Bank (81), a set of 247 protein monomer chains, which are bound to the following ligands, ATP, ADP, NAD, FAD, S-adenosylmethionine (SAM), and SAH, were selected to use as a benchmark (247-benchmark). All of these ligands include adenine as a common molecular fragment. Structures with multiple heteromeric chains were not considered. The sequence identity between any pair of chains was <30%. These chains cover 106 SCOP superfamilies and 152 enzyme classifications (EC). In addition, 81 and 70 chains were yet to be given SCOP and EC assignments. As a control, a set of proteins not bound to a ligand containing ribose, adenine, flavin, and nicotinamide were extracted from the PDB. Subsequently, redundant chains were removed by using a 30% sequence identity cutoff against each other and with the 247-benchmark. The final control contained 101 protein chains (101-control).
The chains from the 247-benchmark were aligned against each other in a pairwise fashion to generate 247 × (247 − 1)/2 = 30,381 benchmark pairs. Among them, ≈5% of the pairs shared the same SCOP superfamily. The 247 × 101 = 24,847 control pairs were obtained by aligning the 247-benchmark against the 101 chains in the 101-control set.
Sequence Order Independent Profile–Profile Alignment of Functional Sites.
Protein structures are represented by Delaunay tessellation of Cα atoms and characterized with geometric potentials as described fully in ref. 82. Each Cα atom is assigned a probability distribution and position specific score matrix of 20 aa. The regular tessellation of the protein structure can be considered a graph representation in 3D space. The protein is scanned and aligned to the functional site by finding the maximum-weight common subgraph between two encoded protein graphs (83). Full details of the algorithm are provided in the SI Text.
Performance Evaluation.
The alignment quality between the functional sites of two protein structures is evaluated by the RMSD between the common molecular moieties associated with the ligands. In addition, search performance is evaluated by a true and false-positive rate defined as follows:
Acknowledgments.
We sincerely thank the anonymous reviewers and the editor for their constructive suggestions in improving the manuscript. This work was supported by National Institutes of Health Grants GM63208 and GM078596 and the Research Collaboratory for Structural Bioinformatics Protein Data Bank. The RCSB Protein Data Bank is supported by grants from National Science Foundation, the National Institute of General Medical Sciences, the Office of Science, the Department of Energy, the National Library of Medicine, the National Cancer Institute, the National Center for Research Resources, the National Institute of Biomedical Imaging and Bioengineering, and the National Institute of Neurological Disorders and Stroke.
Footnotes
Conflict of interest statement: This work is part of a provisional patent application filed by the University of California San Diego (University of California San Diego Reference No. SD2008-001-1).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704422105/DCSupplemental.
References
- 1.Orengo CA, Thornton JM. Protein families and their evolution—a structural perspective. Annu Rev Biochem. 2005;74:867–900. doi: 10.1146/annurev.biochem.74.082803.133029. [DOI] [PubMed] [Google Scholar]
- 2.Whisstock JC, Lesk AM. Prediction of protein function from protein sequence and structure. Q Rev Biophys. 2003;36:307–340. doi: 10.1017/s0033583503003901. [DOI] [PubMed] [Google Scholar]
- 3.Dobson PD, Cai YD, Stapley BJ, Doig AJ. Prediction of protein function in the absence of significant sequence similarity. Curr Med Chem. 2004;11:2135–2142. doi: 10.2174/0929867043364702. [DOI] [PubMed] [Google Scholar]
- 4.Andreeva A, Murzin AG. Evolution of protein fold in the presence of functional constraints. Curr Opin Struct Biol. 2006;16:399–408. doi: 10.1016/j.sbi.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Murzin AG. How far divergent evolution goes in protein. Curr Opin Struct Biol. 1998;8:380–387. doi: 10.1016/s0959-440x(98)80073-0. [DOI] [PubMed] [Google Scholar]
- 6.Grishin NV. Fold change in evolution of protein structures. J Struct Biol. 2001;134:167–185. doi: 10.1006/jsbi.2001.4335. [DOI] [PubMed] [Google Scholar]
- 7.Taylor WR. Evolutionary transitions in protein fold space. Curr Opin Struct Biol. 2007;17:354–361. doi: 10.1016/j.sbi.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Yang S, Doolittle RF, Bourne PE. Phylogeny determined through protein domain content. Proc Natl Acad Sci USA. 2005;102:373–378. doi: 10.1073/pnas.0408810102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mushegian AR, Koonin EV. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc Natl Acad Sci USA. 1996;93:10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carbone A. Computational prediction of genomic functional cores specific to different microbes. J Mol Evol. 2006;63:733–746. doi: 10.1007/s00239-005-0250-9. [DOI] [PubMed] [Google Scholar]
- 11.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 12.Choi I-G, Kim S-H. Evolution of protein structural classes and protein sequence families. Proc Natl Acad Sci USA. 2006;103:14056–14061. doi: 10.1073/pnas.0606239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winstanley HF, Abeln S, Deane CM. How old is your fold. Bioinformatics. 2005;21(Suppl 1):i449–i485. doi: 10.1093/bioinformatics/bti1008. [DOI] [PubMed] [Google Scholar]
- 14.Shindyalov IN, Bourne PE. An alternative view of protein fold space. Proteins. 2000;38:247–260. [PubMed] [Google Scholar]
- 15.Kolodny R, Petrey D, Honig B. Protein structure comparison: implications for the nature of “fold space,” and structure and function prediction. Curr Opin Struct Biol. 2006;16:393–398. doi: 10.1016/j.sbi.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Friedberg I, Godzik A. Connecting the protein structure universe by using sparse recurring fragments. Structure (London) 2005;13:1213–1224. doi: 10.1016/j.str.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Fong JH, Geer LY, Panchenko AR, Bryant SH. Modeling the evolution of protein domain architectures using maximum parsimony. J Mol Biol. 2007;366:307–315. doi: 10.1016/j.jmb.2006.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bashton M, Chothia C. The generation of new protein functions by the combination of domains. Structure (London) 2007;15:85–99. doi: 10.1016/j.str.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 19.Weiner J, Thomos G, Bornberg-Bauer E. Rapid motif-based prediction of circular permutations in multi-domain proteins. Bioinformatics. 2005;21:932–937. doi: 10.1093/bioinformatics/bti085. [DOI] [PubMed] [Google Scholar]
- 20.Vesterstrom J, Taylor WR. Flexible secondary structure based protein structure comparison applied to the detection of circular permutations. J Comput Biol. 2006;13:43–62. doi: 10.1089/cmb.2006.13.43. [DOI] [PubMed] [Google Scholar]
- 21.Pan JL, Bardwell JCA. The origami of thioredoxin-like folds. Protein Sci. 2006;15:2217–2227. doi: 10.1110/ps.062268106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reeves GA, Dallman TJ, Redfern OC, Akpor A, Orengo CA. Structural diversity of domain superfamilies in the CATH database. J Mol Biol. 2006;360:725–741. doi: 10.1016/j.jmb.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 23.Andreeva A, Prlic A, Hubbard TJP, Murzin AG. SISYPHUS-structural alignments for proteins with non-trivial relationships. Nucleic Acids Res. 2006;35:D253–D259. doi: 10.1093/nar/gkl746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Todd AE, Orengo CA, Thornton JM. Evolution of function in protein superfamilies, from a structural perspective. J Mol Biol. 2001;307:1113–1143. doi: 10.1006/jmbi.2001.4513. [DOI] [PubMed] [Google Scholar]
- 25.Gerlt JA, Babbitt PC. Divergent evolution of enzymatic function: Mechanistically diverse superfamilies and functionally distinct superfamilies. Annu Rev Biochem. 2001;70:209–246. doi: 10.1146/annurev.biochem.70.1.209. [DOI] [PubMed] [Google Scholar]
- 26.Scheeff ED, Bourne PE. Structural evolution of the protein kinase-like superfamily. PLoS Comput Biol. 2005;1:e49. doi: 10.1371/journal.pcbi.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cammer SA, et al. Structure-based active site profiles for genome analysis and functional family subclassification. J Mol Biol. 2003;334:387–401. doi: 10.1016/j.jmb.2003.09.062. [DOI] [PubMed] [Google Scholar]
- 28.Schmitt S, Kuhn D, Klebe G. A new method to detect related function among proteins independent of sequence and fold homology. J Mol Biol. 2003;323:387–406. doi: 10.1016/s0022-2836(02)00811-2. [DOI] [PubMed] [Google Scholar]
- 29.Binkowski TA, Adamian L, Liang J. Inferreing functional relationships of proteins from local sequence and spatial surface patterns. J Mol Biol. 2003;332:505–526. doi: 10.1016/s0022-2836(03)00882-9. [DOI] [PubMed] [Google Scholar]
- 30.Kinoshita K, Nakamura H. Identification of protein biochemical functions by similarity search using the molecular surface database eF-site. Protein Sci. 2003;12:1589–1595. doi: 10.1110/ps.0368703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stark A, Sunyaev S, Russell RB. A model for statistical significance of local similarities in structure. J Mol Biol. 2003;326:1307–1316. doi: 10.1016/s0022-2836(03)00045-7. [DOI] [PubMed] [Google Scholar]
- 32.Meng EC, Polacco BJ, Babbitt PC. Superfamily active site templates. Proteins. 2004;55:962–976. doi: 10.1002/prot.20099. [DOI] [PubMed] [Google Scholar]
- 33.Shulman-Peleg A, Nussinov R, Wolfson HJ. Recognition of functional sites in protein structures. J Mol Biol. 2004;339:607–633. doi: 10.1016/j.jmb.2004.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jambon M, Imberty A, Deleage G, Geourjon C. A new bioinformatics approach to detect common 3D sites in protein structures. Proteins. 2003;52:137–145. doi: 10.1002/prot.10339. [DOI] [PubMed] [Google Scholar]
- 35.Ivanisenko VA, Pintus SS, Grigorovich DA, Kolchanov NA. PDBSiteScan: A program for searching for active, binding and posttranslational modification sites in the 3D structures. Nucleic Acids Res. 2004;32:W549–W554. doi: 10.1093/nar/gkh439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barker JA, Thornton JM. An algorithm for constraint-based structural template matching: Application to 3D templates with statistical analysis. Bioinformatics. 2003;19:1644–1649. doi: 10.1093/bioinformatics/btg226. [DOI] [PubMed] [Google Scholar]
- 37.Torrance JW, Bartlett GJ, Porter CT, Thornton JM. Using a library of structural templates to recognize catalytic sites and explore their evolution in homologous families. J Mol Biol. 2005;347:565–581. doi: 10.1016/j.jmb.2005.01.044. [DOI] [PubMed] [Google Scholar]
- 38.Laskowski RA, Watson JD, Thornton JM. Protein function prediction using local 3D templates. J Mol Biol. 2005;351:614–626. doi: 10.1016/j.jmb.2005.05.067. [DOI] [PubMed] [Google Scholar]
- 39.Chen BY, et al. Algorithms for structural comparison and statistical analysis of 3D protein motifs. Pac Symp Biocomput. 2005:334–345. [PubMed] [Google Scholar]
- 40.Zhang Z, Grigorov MG. Similarity networks of protein binding sites. Protein Struct Funct Bioinform. 2006;62:470–478. doi: 10.1002/prot.20752. [DOI] [PubMed] [Google Scholar]
- 41.Kinoshita K, Furui J, Nakamura H. Identification of protein functions from a molecular surface database, eF-site. J Struc Func Genomics. 2001;2:9–22. doi: 10.1023/a:1011318527094. [DOI] [PubMed] [Google Scholar]
- 42.Brakoulias A, Jackson RM. Towards a structural classification of phosphate binding sites in protein-nucleotide complexes: An automated all-against-all structural comparison using geometric matching. Proteins. 2004;56:250–260. doi: 10.1002/prot.20123. [DOI] [PubMed] [Google Scholar]
- 43.Stark A, Russell RB. Annotation in three dimensions. PINTS: Patterns in non-homologous tertiary structures. Nucleic Acids Res. 2003;31:3341–3344. doi: 10.1093/nar/gkg506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morris RJ, Najmanovich RJ, Kahraman A, Thornton JM. Real spherical harmonic expansion coefficients as 3D shape descriptors for protein binding pocket and ligand comparisons. Bioinformatics. 2005;21:2347–2355. doi: 10.1093/bioinformatics/bti337. [DOI] [PubMed] [Google Scholar]
- 45.Siggers TW, Silkov A, Honig B. Structural alignment of protein-DNA interfaces: Insights into the determinants of binding specificity. J Mol Biol. 2005;345:1027–1045. doi: 10.1016/j.jmb.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 46.Campbell SJ, Gold ND, Jackson RM, Westhead DR. Ligand binding: Funcitonal site location, similarity and docking. Curr Opin Struct Biol. 2003;13:389–395. doi: 10.1016/s0959-440x(03)00075-7. [DOI] [PubMed] [Google Scholar]
- 47.Pickering SJ, Bulipitt AJ, Efford N, Gold ND, Westhead DR. AI-based alogrithms for protein surface comparison. Comput Chem. 2001;26:79–84. doi: 10.1016/s0097-8485(01)00102-4. [DOI] [PubMed] [Google Scholar]
- 48.Pazos F, Sternberg MJE. Automated prediction of protein function and detection of functional sites from structure. Proc Natl Acad Sci USA. 2004;101:14754–14759. doi: 10.1073/pnas.0404569101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie L, Wang J, Bourne PE. In silico elucidation of the molecular mechanism defining adverse effect of selective estrogen receptor modulators. PLoS Comp Biol. 2007;3:e217. doi: 10.1371/journal.pcbi.0030217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension. Protein Engng. 1998;9:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 52.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McLachlan AD. Repeating sequences and gene duplication in proteins. J Mol Biol. 1972;64:417–437. doi: 10.1016/0022-2836(72)90508-6. [DOI] [PubMed] [Google Scholar]
- 54.Grishin NV. Phosphatidylinositol phosphate kinase: A link between protein kinase and glutathione synthase folds. J Mol Biol. 1999;291:239–247. doi: 10.1006/jmbi.1999.2973. [DOI] [PubMed] [Google Scholar]
- 55.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tari LW, Matte A, Goldie H, Delbaere LT. Mg2+-Mn2+ clusters in enzyme-catalyzed phosphoryl-transfer reactions. Nat Struct Biol. 1997;4:990–994. doi: 10.1038/nsb1297-990. [DOI] [PubMed] [Google Scholar]
- 57.Tari LW, Matte A, Pugazhenthi U, Goldie H, Delbaere LT. Snapshot of an enzyme reaction intermediate in the structure of the ATP-Mg2+-oxalate ternary complex of Escherichia coli PEP carboxykinase. Nat Struct Biol. 1996;3:355–363. doi: 10.1038/nsb0496-355. [DOI] [PubMed] [Google Scholar]
- 58.Matte A, Tari LW, Goldie H, Delbaere LT. Structure and mechanism of phosphoenolpyruvate carboxykinase. J Biol Chem. 1997;272:8105–8108. doi: 10.1074/jbc.272.13.8105. [DOI] [PubMed] [Google Scholar]
- 59.Matte A, Goldie H, Sweet RM, Delbaere LT. Crystal structure of Escherichia coli phosphoenolpyruvate carboxykinase: A new structural family with the P-loop nucleoside triphosphate hydrolase fold. J Mol Biol. 1996;256:126–143. doi: 10.1006/jmbi.1996.0072. [DOI] [PubMed] [Google Scholar]
- 60.Orengo CA, et al. CATH—a hierarchical classification of protein domain structures. Structure (London) 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 61.Lesk AM. NAD-binding domains of dehydrogenases. Curr Opin Struct Biol. 1995;5:775–783. doi: 10.1016/0959-440x(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 62.Gough J. Convergent evolution of domain architectures (is rare) Bioinformatics. 2005;21:1464–1471. doi: 10.1093/bioinformatics/bti204. [DOI] [PubMed] [Google Scholar]
- 63.Cusa E, Obradors N, Baldoma L, Badia J, Aguilar J. Genetic analysis of a chromosomal region containing genes required for assimilation of allantoin nitrogen and linked glyoxylate metabolism in Escherichia coli. J Bacteriol. 1999;181:7479–7484. doi: 10.1128/jb.181.24.7479-7484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gherardini PF, Wass MN, Helmer-Citterich M, Sternberg MJE. Convergent evolution of enzyme active sites is not a rare phenomenon. J Mol Biol. 2007;372:817–845. doi: 10.1016/j.jmb.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 65.Caetano-Anolles G, Caetano-Anolles D. An evolutionary structured universe of protein architecture. Genome Res. 2003;13:1563–1571. doi: 10.1101/gr.1161903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Caetano-Anolles G, Caetano-Anolles D. Universal sharing patterns in proteomes and evolution of protein fold architecture and life. J Mol Evol. 2005;60:484–498. doi: 10.1007/s00239-004-0221-6. [DOI] [PubMed] [Google Scholar]
- 67.Wang M, Boca SM, Kalelkar R, Mittenthal JE, Caetano-Anolles G. Phylogenomic reconstruction of the protein world based on a genomic census of protein fold architecture. Complexity. 2006;12:27–40. [Google Scholar]
- 68.Ji H-F, Zhang H-Y. Protein architecture chronology deduced from structures of amino acid synthases. J Biomol Struct Dyn. 2007;24:321–323. doi: 10.1080/07391102.2007.10507122. [DOI] [PubMed] [Google Scholar]
- 69.Ji H-F, et al. Distribution patterns of small-molecule ligands in the protein universe and implications for origin of life and drug discovery. Genome Biol. 2007;8:R176. doi: 10.1186/gb-2007-8-8-r176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krishna SS, Grishin NV. Structural drift: A possible path to protein fold change. Bioinformatics. 2005;21:1308–1310. doi: 10.1093/bioinformatics/bti227. [DOI] [PubMed] [Google Scholar]
- 71.Peisajovich SG, Rockah L, Tawfik DS. Evolution of new protein topologies through multistep gene rearrangements. Nat Genet. 2006;38:168–174. doi: 10.1038/ng1717. [DOI] [PubMed] [Google Scholar]
- 72.Theobald DL, Wuttke DS. Divergent evolution within protein superfolds inferred from profile-based phylogenetics. J Mol Biol. 2005;354:722–737. doi: 10.1016/j.jmb.2005.08.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kunin V, Chan B, Sitbon E, Lithwick G, Pietrokovski S. Consistency analysis of similarity between multiple alignments: Prediction of protein function and fold structure from analysis of local sequence motifs. J Mol Biol. 2001;307:939–949. doi: 10.1006/jmbi.2001.4466. [DOI] [PubMed] [Google Scholar]
- 74.Deeds EJ, Shakhnovich EI. A structure-centric view of protein evolution, design and adaptation. Adv Enzymol Relat Areas Mol Biol. 2007;75:133–191. doi: 10.1002/9780471224464.ch2. [DOI] [PubMed] [Google Scholar]
- 75.Doolittle RF. Convergent evolution—the need to be explicit. Trends Biochem Sci. 1994;19:15–18. doi: 10.1016/0968-0004(94)90167-8. [DOI] [PubMed] [Google Scholar]
- 76.Krishna SS, Grishin NV. Structurally analogous proteins do exists. Structure (London) 2004;12:1125–1127. doi: 10.1016/j.str.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 77.Lo Surdo P, Walsh MA, Sollazzo M. A novel ADP- and zinc-binding fold from function-directed in vitro evolution. Nat Struct Mol Biol. 2004;11:382–383. doi: 10.1038/nsmb745. [DOI] [PubMed] [Google Scholar]
- 78.Keefe AD, Szostak JW. Functional proteins from a random-sequence library. Nature. 2001;410:715–718. doi: 10.1038/35070613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Taylor WR. Decoy models for protein structure score normalisation. J Mol Biol. 2006;357:676–699. doi: 10.1016/j.jmb.2005.12.084. [DOI] [PubMed] [Google Scholar]
- 80.Todd AE, Marsden RL, Thornton JM, Orengo CA. Progress of structural genomics initiatives: an analysis of solved target structures. J Mol Biol. 2005;348:1235–1260. doi: 10.1016/j.jmb.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 81.Deshpande N, et al. The RCSB Protein Data Bank: A redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res. 2005;33:D233–D237. doi: 10.1093/nar/gki057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xie L, Bourne PE. A robust and efficient algorithm for the shape description of protein structures and its application in predicting ligand binding sites. BMC Bioinformatics. 2007;8:S9. doi: 10.1186/1471-2105-8-S4-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ostergard PRJ. A new algorithm for the maximum-weight clique problem. Nordic J Computing. 2001;8:424–436. [Google Scholar]