Abstract
DNA, whether it is microbe-derived or host-derived, evokes immune responses when exposed to the cytosol of a cell. We previously reported that DNA-dependent activator of IFN regulatory factors (DAI), also referred to as DLM-1/ZBP1, functions as a DNA sensor that activates the innate immune system. In the present study, we examined the regulation of the complex DNA-sensing system by DAI and other molecules. We first show that DAI directly interacts with DNA in vitro and that it requires three DNA-binding domains for full activation in vivo. We also show that the artificially induced dimerization of DAI results in the DNA-independent activation of type I IFN genes, thereby providing a better understanding for the molecular basis of DAI activation. Furthermore, we provide evidence for the presence of additional DNA sensors, either positively or negatively regulating cytosolic DNA-mediated innate immune responses. These results in toto provide insights into the mechanism of DAI activation and reveal the complex regulatory mechanisms underlying DNA-mediated protective and pathologic immune responses.
Keywords: ADAR1, E3L, interferon, IFN regulatory factor
Non-self-nucleic acids from invading microbes or self-nucleic acids exposed in a cell by infection or incomplete clearance during cell damage commonly evoke immune responses (1–5). In addition to membrane-type Toll-like receptors (TLRs), such as TLR3, TLR7, and TLR9, that are activated by RNA or DNA (6, 7), there are cytosolic receptors that also evoke the nucleic acid-mediated activation of innate immune responses, the hallmark of which is the induction of type I IFN genes (8–11). Indeed, RIG-I and MDA5 molecules function as cytosolic RNA sensors and activators, whereas LGP2 acts as a negative regulator, probably by competing with RIG-I and MDA5 for RNA binding (8, 10, 12). Recent attention has focused on the regulation of DNA-sensing systems, because they also relate to protective and pathologic immune responses. In this context, we identified DAI [also referred to as DLM-1 and ZBP1 (13); we refer to it as DAI hereafter for convenience] as the first cytosolic DNA sensor and activator of innate immune responses activated by cytosolic DNAs (14).
We have shown that the artificial expression of otherwise IFN-inducible DAI selectively enhances the DNA-mediated induction of type I IFN genes and other genes involved in innate immunity and that the RNA interference of DAI mRNA in cells, on the other hand, strongly inhibits this gene induction program in mouse fibroblast L929 cells (14). In addition to two binding domains for left-handed Z-form DNA (Z-DNA), termed Zα and Zβ (15, 16), DAI also contains an additional candidate DNA-binding region/domain, termed the D3 region, that may also interact with DNA (Fig. 1a) (14). Furthermore, we adduced evidence that DAI recruits TBK1 serine/threonine kinase and IFN regulatory factor 3 (IRF3) transcription factor, both of which play critical roles in the induction of type I IFN genes and other genes (14). A DAI mutant lacking the DNA-binding region, interestingly, cannot spontaneously activate its downstream signaling pathway, supporting the notion that DNA is critical for not only initiating but also sustaining the active DAI signaling complex.
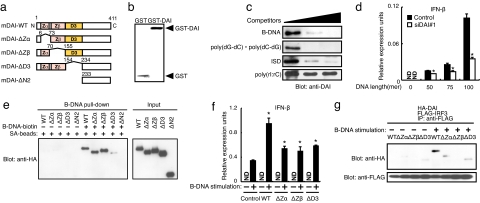
Fig. 1.
Recognition of DNAs by DAI protein. (a) Schematic illustration of DAI (DLM-1/ZBP1; DAI-WT) and its deletion mutants. Zα and Zβ DNA-binding domain (pink) and DNA-binding domain D3 (orange) are shown. DAI-ΔN2 is a deletion mutant of all three DNA-binding domains (14). (b) Coomassie brilliant blue (CBB) staining of GST or GST-DAI recombinant proteins. (c) Binding analysis of DAI protein with B-DNA. Recombinant DAI protein was incubated with biotin-conjugated B-DNA and with streptavidin (SA)-conjugated magnetic beads in the absence or presence of unconjugated B-DNA, poly(dG-dC)·poly(dC-dG), ISD, or poly(rI:rC) (0, 0.5, and 5.0 μg/ml; wedge above gels). DAI protein was analyzed by immunoblotting with anti-DAI antibody. (d) DNA length-dependent type I IFN production. L929 cells were transiently transfected with a plasmid vector for Renilla siRNA (Control) or for siRNA targeting DAI (siDAI#1). IFN-β mRNA induction was analyzed by qRT-PCR. Data are mean ± SD (n = 3). ND, not detected. *, P < 0.01 siDAI#1 versus control. (e) DNA pull-down analysis of DAI and DAI mutants. HEK293T cells were transiently transfected with plasmid for HA-tagged DAI-WT or deletion mutants as in a, and whole-cell lysate was incubated with biotin-conjugated B-DNA and SA-conjugated magnetic beads. (Left) Bound protein was analyzed by immunoblotting with anti-HA antibody. (Right) Input protein used for this assay is shown. (f) Type I IFN gene induction by DAI and DAI mutants. L929 cells that retrovirally expressed mock (Control), DAI-WT, or deletion mutants of DAI as in a were stimulated with 6 μg/ml B-DNA for 9 h. Induction of IFN-β mRNA was analyzed by qRT-PCR. Data are mean ± SD (n = 3). *, P < 0.01 DAI-WT or mutants versus control. (g) Interaction of DAI-WT and DAI mutants with IRF3. DAI-WT or deletion mutant of DAI as in a was transiently expressed in L929 cells with FLAG-tagged IRF3. Cells were stimulated with B-DNA for 2 h and analyzed by immunoprecipitation with anti-FLAG antibody, followed by immunoblotting with anti-HA (Upper) and anti-FLAG (Lower) antibodies.
Although these previous results indicate that DAI functions as a DNA sensor for the activation of innate immune responses, several interesting issues remain to be clarified. First, does DAI directly interact with DNA and, if so, how does such binding contribute to DAI activation? Indeed, it was not clarified in our previous study whether DAI alone is capable of binding DNA in vitro. Neither was it studied how DAI becomes an active form, although it was conjectured that the DNA-mediated molecular assembly of DAI may be critical to its activation. Second, is DAI the sole DNA sensor? Although the RNA interference of DAI in L929 cells resulted in a strong inhibition of the DNA-mediated induction of type I IFN genes and other genes, the induction was not completely abolished, suggesting the presence of a redundant cytosolic DNA-sensing molecule(s). Third, are there any negative regulators of DNA-mediated immune responses? In this context, it is interesting that host- and virus-derived proteins are found to also contain DNA-binding motifs similar to those of DAI, i.e., Zα and Zβ domains (17, 18). Indeed, host-derived adenosine deaminase acting on RNA 1 (ADAR1) contains domains homologous to both domains, and the vaccinia virus (VV)-derived E3L protein contains a Zα domain (17, 18). Thus, these molecules may participate in the cytosolic DNA-sensing system to regulate innate immune responses. In the present study, we addressed these issues and provide insights into the complex regulation of the DNA-sensing system in innate immunity.
Results
Interaction of Recombinant DAI with DNAs.
In our previous study, we adduced evidence that DAI interacts with DNA by fluorescence resonance energy transfer analysis and by a DNA pull-down assay. However, whether DAI directly interacts with DNA was not rigorously examined (14). To address this issue, we prepared recombinant DAI in Escherichia coli and subjected it to an in vitro pull-down assay. As shown in Fig. 1b, the GST-fused DAI was purified to almost homogeneity. The recombinant DAI was then precipitated (or pulled down) with biotin-labeled synthetic B-form DNA, poly(dA-dT)·poly(dT-dA) (hereafter referred to as B-DNA) (2), which shows a potent immunostimulatory activity (2, 14). This precipitation was inhibited by adding an excess amount of nonconjugated B-DNA but not poly(rI:rC) (Fig. 1c). Interestingly, the precipitation was also inhibited by addition of a DNA, termed IFN stimulatory DNA (ISD) (3) that activates IRF3 for type I IFN gene induction, as well as by a synthetic poly(dG-dC)·poly(dC-dG) that may take on a Z-form (19). These results indicate that DAI directly and specifically binds to DNA, irrespective of its sequence.
DNA Length-Dependent Activation of DAI.
To gain further insights into the mechanism of DAI activation by DNA, we next examined to what extent the DAI activation was affected by the length of DNA. The induction of IFN-β mRNA was monitored from L929 cells in whose cytosol B-DNAs of various lengths, as indicated in Fig. 1d, were delivered. As shown in Fig. 1d, longer DNA sequences were progressively more effective in inducing IFN-β mRNA than shorter sequences. Moreover, this effect was significantly suppressed upon the expression of an siRNA vector targeting DAI, termed siDAI#1 (14). It is worth mentioning that the B-DNA of 100-bp length was far weaker than “conventional” B-DNA, which has an average length of ≈500 to 1,000 bp and is used to activate cytosolic DNA-sensing mechanisms (2, 14). In a separate experiment, we observed that the IFN mRNA induction level achieved using 100-bp B-DNA is ≈100-fold lower compared with that achieved using conventional B-DNA (data not shown). These results, therefore, indicate that DNA-mediated activation of the IFN gene, as mediated by DAI and other proteins, is dependent on DNA length. This is interesting when compared with the RNA-sensing mechanism that is efficiently activated by relatively short pieces of RNA that contain a triphosphate moiety (9, 20), and it may provide clues to better understanding the DNA-sensing and activation mechanisms (see below).
Contribution of Three DNA-Interacting Regions for DAI Activation.
We previously reported that DAI contains, in addition to the Zα and Zβ domains, a third domain termed the D3 region (14). To examine the contribution of each region to DAI activation, we constructed expression vectors for DAI mutants lacking each one of these regions and then evaluated their biologic function (Fig. 1a). First, HEK293T cell lysates containing transiently expressed, HA-tagged WT or a DAI mutant protein were subjected to a pull-down assay using biotin-labeled B-DNA and streptavidin-conjugated magnetic beads. As shown in Fig. 1e, WT as well as the mutants lacking either the Zα or Zβ domain all precipitated with the conjugated B-DNA; however, the mutant lacking the D3 region failed to do so or did so only very poorly. Therefore, these results indicate the essential contribution of the D3 region to the interaction of DAI with DNA.
We next examined whether these mutants can activate the Ifnb gene in L929 cells by expressing the mutants, which was followed by B-DNA stimulation. As reported previously, the B-DNA-induced expression level of IFN-β mRNA was higher in cells expressing WT DAI than in control cells (14). Interestingly, however, such an enhancement of mRNA induction was markedly weak in cells expressing the mutants lacking either of the three DNA-binding regions, suggesting that these regions are all required for the full activation of DAI (Fig. 1f). We also examined whether these mutants can recruit IRF3, the essential transcription factor for DAI-mediated activation of the Ifnb gene, upon B-DNA stimulation in L929 cells (2, 14). As shown in Fig. 1g, although WT DAI can recruit IRF3, the mutants can do so very weakly or negligibly, an observation congruent with the above results indicates the requirement of all of the DNA-binding regions for efficient DAI activation.
Dimerization of DAI and Activation of Innate Immune Responses.
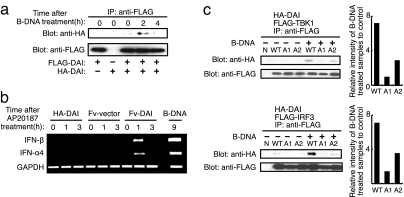
How is DAI activated upon DNA stimulation? Because we observed a DNA length-dependent activation of the Ifnb gene, we hypothesized that DNA may serve as a scaffold to mediate the formation of a tandem array of DAI molecules, which then recruit and activate downstream signaling molecules, such as TBK1 and IRF3 (2, 14). To test this possibility, we first examined whether DAI undergoes intermolecular association induced by B-DNA stimulation. Two cDNA expression vectors, each encoding either HA-tagged or FLAG-tagged DAI, were cotransfected into L929 cells and stimulated with B-DNA. Cell extracts were then prepared and subjected to immunoprecipitation using an anti-FLAG Ab followed by Western blotting using an anti-HA Ab. As shown in Fig. 2a, coimmunoprecipitation was observed between the two distinctly labeled DAI molecules, reaching a maximum 2 h after B-DNA stimulation. We next asked whether artificial dimerization of DAI results in its activation without DNA by constructing an expression vector for DAI cDNA encoding DAI fused to the Fv protein (supporting information (SI) Fig. S1). The Fv protein allows dimerization of the DAI molecule upon administration of a FK506 derivative, AP20187, to a cell (21). Interestingly, the induction of type I IFN mRNAs was observed 1 h after the administration of AP20187, indicating that artificially dimerized DAI can activate transcription of type I IFN genes in the absence of DNA, albeit to levels lower than those achieved by B-DNA stimulation (Fig. 2b). Thus, these results further demonstrate that DAI is indeed an activator of innate immune responses and support our hypothesis that DAI must undergo the DNA-mediated formation of a multimeric complex, to then evoke robust innate immune responses.
Fig. 2.
Dimerization/oligomerization of DAI activates immune response. (a) Dimer/oligomer formation of DAI by B-DNA stimulation. HA-tagged DAI (HA-DAI) and FLAG-tagged DAI (FLAG-DAI) were transiently coexpressed in L929 cells. The cells were stimulated with 6 μg/ml B-DNA for the indicated periods and analyzed by immunoprecipitation with anti-FLAG antibody, followed by immunoblotting with anti-HA (Upper) and anti-FLAG (Lower) antibodies. (b) Dimerization of DAI induces type I IFN production. HA-DAI or Fv (Fv-vector) or Fv-fused DAI (Fv-DAI) was transiently expressed in L929 cells. The cells were treated with AP20187 for the indicated periods, and the expressions of IFN-β (Top), IFN-α4 (Middle), and GAPDH (Bottom) mRNAs were assessed by RT-PCR. Results from B-DNA treatment on L929 cells are also shown. (c) DAI-WT, DAI-A1, or DAI-A2 was transiently expressed in L929 cells with either FLAG-TBK1 (Upper) or FLAG-IRF3 (Lower). These cells were stimulated with B-DNA and analyzed by immunoprecipitation with anti-FLAG antibody, followed by immunoblotting with anti-HA and anti-FLAG antibodies. The relative band intensities depicted in the graphs are of B-DNA-treated samples measured by a densitometer normalized to those of unstimulated sample.
Activation of DAI by Phosphorylation.
Because we have shown previously that DAI can recruit TBK1 upon DNA stimulation (14), we next envisaged that DAI may undergo phosphorylation and that this may be an event critical to the recruitment of these downstream signaling molecules: Indeed, the cotransfection of DAI cDNA with TBK1 cDNA resulted in a mobility shift of DAI that could be abolished upon treatment with calf intestinal alkaline phosphatase (Fig. S2), suggesting an extensive phosphorylation of DAI by TBK1. A query for predicted phosphorylation sites was performed on DAI's C-terminal amino acid sequence by using the DISPHOS program (22), and the Phospho.ELM database (23) revealed a number of potential serine/threonine phosphorylation sites in amino acids spanning from 338 to 366. In particular, serines 352 and 353 were found to be the particularly attractive phosphorylation sites for TBK1 (24).
To examine the importance of these phosphorylation sites, we next generated separate S→A substitution protein expression constructs (DAI-A1 for serine 352 and DAI-A2 for serine 353) (Fig. S3) and then tested their ability to recruit IRF3 or TBK1 by coprecipitation assay. Both DAI mutants still showed a mobility shift essentially similar to that observed in WT DAI (Fig. S2), indicating that DAI is likely phosphorylated by TBK1 at multiple sites. Interestingly, however, the DNA-dependent recruitment of DAI for TBK1 and IRF3 was impaired for these two mutants, particularly for DAI-A1 (Fig. 2c). Furthermore, the overexpression of these mutants in L929 cells revealed that DAI-A1 cannot induce IFN-β mRNA upon B-DNA stimulation and, although not as pronounced as in the case of DAI-A1, the activity of DAI-A2 is also impaired (Fig. S4). These results support the notion that DAI needs to be phosphorylated at these residues to efficiently recruit and activate TBK1 and IRF3 for IFN gene induction.
Evidence for Redundancy in Cytosolic DNA-Sensing System.
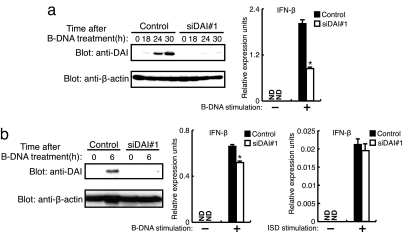
We next examined whether DAI is the sole DNA sensor. To monitor the expression level of the endogenous DAI protein, we raised polyclonal rabbit sera against mouse DAI (mDAI Ab). As shown in Fig. 3a, in mouse L929 cells expressing siDAI#1, DAI protein expression was strongly suppressed; concomitantly, IFN-β mRNA induction by B-DNA was also inhibited (≈60% inhibition), an observation consistent with that in a previous study (14). Interestingly, when similar experiments were performed using mouse embryonic fibroblasts (MEFs), a different picture emerged (Fig. 3b): The DAI protein was suppressed, even more strongly than that in L929 cells, owing to the expression of siDAI#1, but the IFN-β mRNA induction was inhibited only marginally (≈15% inhibition). Furthermore, in these MEFs a weak induction of IFN-β mRNA by ISD remained almost unaffected. These results clearly indicate the presence of an additional, redundant cytosolic DNA-sensing molecule(s) whose contribution to activating innate immune responses in relation to DAI may depend on the type of cell.
Fig. 3.
Redundancy in the cytosolic DNA-sensing system. (a) L929 cells were transiently transfected with a plasmid vector for Renilla siRNA (Control) or for siRNA-targeting DAI (siDAI#1), stimulated with 6 μg/ml B-DNA for the indicated periods. (Left) Immunoblots with anti-DAI (Upper) and anti-β-actin (Lower) are shown. (Right) The induction of IFN-β mRNA in siRNA-expressing L929 cells after 6 h of treatment of B-DNA was assessed by qRT-PCR. Data are mean ± SD (n = 3). *, P < 0.01 siDAI#1 versus control. (b) MEFs were transiently transfected with Renilla siRNA (Control) or siDAI#1 plasmid vector, stimulated with B-DNA. (Left) Immunoblots with anti-DAI (Upper) and anti-β-actin (Lower) are shown. (Center and Right) The induction of IFN-β mRNA in siRNA-expressing MEFs after B-DNA stimulation (Center) or 10 μg/ml of ISD stimulation (Right) was assessed by qRT-PCR. Data are mean ± SD. (n = 3). *, P < 0.001 siDAI#1 versus control.
Negative Regulation of DNA-Sensing System by Host and Viral Proteins.
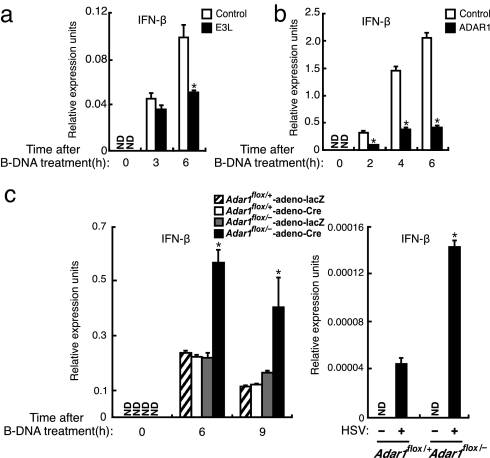
Pathogens often encode a molecule(s) with which they evade the host's immune responses, whereas the host would generally require the negative regulation of immune responses so as to avoid the harmful aspects of the responses, such as development of autoimmunity (25). There are proteins encoded by virus or host that possess DNA-binding domains similar to those found in DAI, namely, VV-encoded E3L (18) and ADAR1 (17) (Fig. S5). VV is a double-stranded DNA virus that replicates in the cytoplasm of infected cells, and E3L plays an essential role in the pathogenesis induced by VV by blocking the IFN response (18). We then examined whether E3L interferes with the B-DNA-mediated induction of IFN-β mRNA by overexpressing E3L in MEFs. As shown in Fig. 4a, E3L inhibited IFN-β mRNA induction, supporting the notion that E3L inhibits the cytosolic DNA-mediated activation of the immune system for the replication of the DNA virus.
Fig. 4.
Negative regulation of DNA-sensing system. (a and b) MEFs that retrovirally expressed mock (Control, white bars), E3L (a) (black bars), or ADAR1 (b) (black bars) were stimulated with 6 μg/ml B-DNA for the indicated periods. Induction of IFN-β mRNA was analyzed by qRT-PCR. *, P < 0.01; E3L versus control (a) or ADAR1 versus control (b). Data are mean ± SD (n = 3). (c) Adar1flox/+ and Adar1flox/− MEFs were stimulated with B-DNA (Left) for the indicated periods or infected with 1 moi of HSV-1 (Right) for 18 h. Induction of IFN-β mRNA was analyzed by qRT-PCR. *, P < 0.01 Adar1flox/−-adeno-Cre versus Adar1flox/+-adeno-Cre. Data are mean ± SD (n = 3).
We next examined the role of ADAR1 in the cytosolic DNA-sensing system, first by overexpressing the molecule in MEFs and then treating the cells with B-DNA. As shown in Fig. 4b, the ectopic ADAR1 expression strongly inhibited the B-DNA-mediated induction of IFN-β mRNA. To consolidate the role of ADAR1 further, we next examined the DNA-mediated induction of IFN-β mRNA in MEFs derived from mice carrying a conditional mutation of the Adar1 gene (17). Interestingly, when the Adar1 gene was ablated by expressing Cre recombinase, the induction level of IFN-β mRNA, either upon B-DNA stimulation or by HSV-1 infection, was markedly higher (≈3-fold) than that in the control cells (Fig. 4c). Additionally, the level of IFN-β mRNA induction in these cells was strongly suppressed by retroviral gene expression of ADAR1, further indicating the role of ADAR1 in the negative regulation of DNA-mediated immune responses (Fig. S6). It is worth mentioning that E3L and ADAR1 both bind to dsRNA as well, and they also interfere with the IFN gene induction by dsRNAs, such as poly(rI:rC) (data not shown).
Discussion
In the present study, we aimed at gaining insight into the cytosolic DNA-sensing system and its regulation. We first addressed the issue of whether DAI, a cytosolic DNA sensor for activating innate immunity, directly interacts with DNA. Our results obtained by using recombinant DAI protein indicate that DAI indeed interacts with various kinds of DNA but not RNA in vitro. Although not rigorously examined, our results suggest that DAI has no strict preference for the sequence and configuration of DNA, which is consistent with the fact that various DNAs, whether pathogen-derived or host-derived, can activate the innate immune system when exposed en masse to the cytosol (2–5). Given that there is little structural discrimination between microbial and host DNAs by DAI, we envisage that the DNA-sensing system may be regulated in the following manner so as to prevent the aberrant activation of the sensor with self-DNA: First, both the DNA sensor(s) and self-DNA should be compartmentalized separately to prevent their encounter with each other; second, aberrantly exposed self-DNA should be rapidly eliminated by degradation. The abrogation of these regulatory mechanisms may contribute to the development of excessive inflammatory responses and autoimmune abnormalities. It will, therefore, be interesting to examine the contribution of DAI in this context, for example, the artificial overexpression of DAI in the mouse.
The DNA length-dependent activation of innate immune responses suggests that the formation of a tandem array of DAI and other sensors is critical to the efficient activation of the innate immune system. In this regard, it is interesting that the D3 region but not the other two DNA-binding domains of DAI is essential for the interaction with DNA, whereas all of the three regions are required for the DNA-dependent recruitment of IRF3 and the subsequent induction of IFN-β mRNA (Fig. 1 e–g). Although further work is required to elucidate the detailed mechanism of DAI activation, we infer that the initial interaction of the D3 region of DAI results in a protein conformational change such that the other two DNA-binding domains become accessible; the interaction of the two domains with DNA is contingent on initial binding of the D3 region to DNA. DAI then forms a stable complex with DNA, a state that none of the regions can achieve alone, resulting in the activation of downstream signaling pathway(s) (Fig. S7).
The fact that the artificial dimerization of DAI results in the DNA-independent induction of IFN genes further underscores the contribution of DAI to the activation of innate immune responses. It is currently not fully clarified how DAI is activated by DNA, but we can propose the following events on the basis of our present results. In view of our results that the dimerization of DAI could activate IFN gene induction to relatively low levels compared with B-DNA stimulation (Fig. 2b), it is most likely that the oligomerization/multimerization of DAI is required for the full-blown activation of the innate immune responses. On the basis of these observations, we envisage the following scenario for the DNA-mediated activation of DAI: When DNA is exposed to the cytosol, DAI first interacts with the DNA through its D3 region; subsequently, a tighter DNA–DAI complex is formed via the Zα and Zβ domains of DAI. The formation of this complex results in the oligomerization/multimerization of DAI, which in turn recruits, only at low levels initially, TBK1 and IRF3. The phosphorylation of DAI by TBK1 at serine 352/353 (and possibly other serine/threonine residues) amplifies the recruitment of additional TBK1 and IRF3.
Although DAI may be the first identified DNA sensor for the activation of innate immune responses (14), it is clear from the present results that an additional cytosolic sensor exists. This reveals further the complex nature of the DNA-mediated activation of innate immunity. On the basis of our data (Fig. 3), it is tempting to speculate that DNA sensors differentially contribute to the initiating the immune response, depending on the type of cell. Further studies, including the generation of DAI-deficient mice and identification of another sensor(s), are required to clarify these issues.
The negative regulation of the immune system is critical to both pathogens and hosts for efficient propagation and maintenance of homeostasis, respectively. Our present study indicates that ADAR1 is a host protein that negatively regulates the immune response that is activated by cytosolic DNA. There are three separate ADAR gene family members, namely, ADAR1, ADAR2, and ADAR3 (26–28). Two isoforms of ADAR1, a full-length ADAR1p150 and a shorter N-terminus-truncated ADAR1p110, are known. One of the three promoters that drive the Adar1 gene is IFN-inducible, and the mRNA transcribed from this promoter directs the translation of ADAR1p150 containing two Z-DNA-binding domains (29). ADAR1p110 localizes in the nucleus, whereas ADAR1p150 is detected both in the nucleus and cytoplasm (29, 30). Thus, we infer that ADAR1 interferes with DNA sensors by competing for cytosolic DNAs and that this allows the down-regulation of innate immune responses which, if permitted to continue in excess, may be harmful to the host. In a similar vein, we also envisage that E3L of VV is critical to virus replication because of its suppressive activity on the induction of IFN responses by cytosolic DNA and RNA sensors, at least in part (18). We infer that the inhibition of IFN gene induction by ADAR1 or E3L is mediated by their Zα or Zβ domain. Indeed, we observed that the overexpression of ADAR1 or E3L in HEK293T cells resulted in the suppression of DAI's interaction with B-DNA in the same pull-down assay system as in Fig. 1e. Furthermore, the overexpression of ADAR1 or E3L in HEK293T cells resulted in the suppression of the interaction of DAI with B-DNA (Z.W., unpublished data). In view of the presence of DAI and other “activating” DNA sensors, it is possible that there are additional “inhibitory” DNA sensors.
Finally, how DAI and other DNA sensor(s) are involved in protective and pathologic immune responses awaits further investigation. In this context, the generation and analysis of DAI knockout mice may be useful in helping to clarify DAI's role during infections by various DNA-containing intracellular pathogens. However, as revealed in our present study, the DAI knockout mice may tell us only part of the story, due to the existence of redundant DNA-sensing mechanisms and until the additional cytosolic sensor(s) are identified and its function in relation to DAI determined.
Materials and Methods
Mice, Cells, and Reagents.
Conditional Adar1 knockout mice were used as described previously (17). MEFs, L929 cells, and HEK293T cells were maintained as previously described (14). Poly(dA-dT)·poly(dT-dA) (B-DNA), poly(dG-dC)·poly(dC-dG), and poly(rI:rC) were used (14). Each length of B-DNA and other oligonucleotides were purchased from Hokkaido System Science.
Plasmids and Gene Transfer.
DAI and DAI mutants expression vectors have been described previously (14). Mouse ADAR1 and VV E3L cDNAs were obtained by standard PCR method and cloned into pMSCVpac-FLAG vector. AxCANLacZ and AxCANCre adenovirus expression vectors were kindly provided by Izumu Saito (University of Tokyo). Retroviral gene transfer was carried out as described previously (14). Transient transfection was performed as described previously (4). Because transfection of plasmid DNAs could potentially evoke a type I IFN response, we stimulated the cells by B-DNA >24 h after plasmid DNA transfection, so as to avoid or minimize any effects due to the transfected plasmids.
Purification of Recombinant DAI Protein.
pGEX4T3-DAI expression vector was constructed by inserting DAI cDNA into the SalI and NotI sites of pGEX4T3 vector (GE Healthcare). The fusion protein was purified by using Glutation–Sepharose beads (GE Healthcare). DAI and GST protein were separated by treatment of Thrombin proteinase (Novagen).
Quantitative RT-PCR Analysis.
Analysis of gene expression by quantitative RT-PCR (qRT-PCR) was performed as described previously (14).
RNA Interference.
siRNA-mediated knockdown of DAI was performed as described previously (14).
Statistical Analysis.
Differences between control and experimental groups were evaluated using the Student t test.
Pull-Down, Coimmunoprecipitation, and Immunoblotting analysis.
Pull-down and coimmunoprecipitation assays were performed as described previously (14). Polyclonal antibody against DAI was produced by immunizing a rabbit with peptide identical to C-terminal residues (398–411) of mouse DAI (Sigma).
Homodimer Formation.
The induction of homodimer formation was performed by using an ARGENT regulated homodimerization Kit (ARIAD). AP20187, a synthetic dimerizer, can induce homodimer formation of FV (Phe36Val point mutant of FKBP) domain-containing fusion proteins. AP20187 was used at a concentration of 1 nM.
Phosphatase Treatment.
Cells were solubilized with lysis buffer. The lysate was subjected to treatment with 2 units of calf intestinal alkaline phosphatase (TaKaRa).
Phospholylation Site Prediction.
Potential phosphorylation sites were computationally predicted by using the DISPHOS program (http://core.ist.temple.edu/pred). Disorder profile was verified by DISOPRED2 disorder prediction server (http://bioinf.cs.ucl.ac.uk/disopred). Comparison between known TBK1 phosphorylation sites and predicted sites was done by using information from Phospho.ELM (http://phospho.elm.eu.org).
Adar1flox/− MEF Generation.
Adar1flox/+ and Adar1flox/− MEFs were infected with 12 moi cre recombinase-expressing retrovirus to obtain Adar1flox/− and control WT MEFs.
Viral Infection.
Cells were infected for 18 h with 1.0 moi HSV-1 (KOS strain; American Type Culture Collection no. VR-1493;) as described previously (14).
Acknowledgments.
We thank M. Shishido and R. Takeda for technical assistance and David Savitsky for kind advice and critical reading of the manuscript. This work was supported by Grant-in-Aid for Scientific Research on Priority Areas “Integrative Research Toward the Conquest of Cancer,” Grant-in-Aid for Scientific Research (A) and (C), and Global Center of Excellence Program “Integrative Life Science Based on the Study of Biosignaling Mechanisms” from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. Z.W. is a research fellow of the Japan Society for the Promotion of Science. M.K.C. is a research fellow of the Korea Science and Engineering Foundation.
Note Added in Proof.
K. Ishii et al. (31) have recently reported data on DAI-deficient mice that also indicate that there is a redundant cytosolic DNA-sensing molecule(s) that may compensate for the absence of DAI.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801295105/DCSupplemental.
References
- 1.Tokunaga T, Yamamoto T, Yamamoto S. How BCG led to the discovery of immunostimulatory DNA. Jpn J Infect Dis. 1999;52:1–11. [PubMed] [Google Scholar]
- 2.Ishii KJ, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 3.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida H, Okabe Y, Kawane K, Fukuyama H, Nagata S. Lethal anemia caused by interferon-β produced in mouse embryos carrying undigested DNA. Nat Immunol. 2005;6:49–56. doi: 10.1038/ni1146. [DOI] [PubMed] [Google Scholar]
- 5.Napirei M, et al. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nat Genet. 2000;25:177–181. doi: 10.1038/76032. [DOI] [PubMed] [Google Scholar]
- 6.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 7.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 8.Yoneyama M, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 9.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 10.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 11.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 12.Venkataraman T, et al. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol. 2007;178:6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- 13.Fu Y, et al. Cloning of DLM-1, a novel gene that is up-regulated in activated macrophages, using RNA differential display. Gene. 1999;240:157–163. doi: 10.1016/s0378-1119(99)00419-9. [DOI] [PubMed] [Google Scholar]
- 14.Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 15.Schwartz T, Behlke J, Lowenhaupt K, Heinemann U, Rich A. Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat Struct. Biol. 2001;8:761–765. doi: 10.1038/nsb0901-761. [DOI] [PubMed] [Google Scholar]
- 16.Ha SC, et al. Biochemical characterization and preliminary X-ray crystallographic study of the domains of human ZBP1 bound to left-handed Z-DNA. Biochim Biophys Acta. 2006;1764:320–323. doi: 10.1016/j.bbapap.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 17.Wang Q, et al. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- 18.Langland JO, et al. Suppression of proinflammatory signal transduction and gene expression by the dual nucleic acid binding domains of the vaccinia virus E3L proteins. J Virol. 2006;80:10083–10095. doi: 10.1128/JVI.00607-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rich A, Zhang S. Z-DNA: The long road to biological function. Nat Rev Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- 20.Malathi K, Dong B, Gale MJ, H, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2070;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amara JF, et al. A versatile synthetic dimerizer for the regulation of protein-protein interactions. Proc Natl Acad Sci USA. 1997;94:10618–10623. doi: 10.1073/pnas.94.20.10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iakoucheva LM, et al. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diella F, Gould CM, Chica C, Via A, Gibson TJ. Phospho.ELM: A database of phosphorylation sites. Nucleic Acids Res. 2008;36:D240–D244. doi: 10.1093/nar/gkm772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McWhirter SM, et al. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc Natl Acad Sci USA. 2004;101:233–238. doi: 10.1073/pnas.2237236100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson AJ, Locarnini SA. Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immune response. Immunol Cell Biol. 2007;85:435–445. doi: 10.1038/sj.icb.7100100. [DOI] [PubMed] [Google Scholar]
- 26.Melcher T, et al. A mammalian RNA editing enzyme. Nature. 1996;379:460–464. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- 27.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. (1994) Proc Natl Acad Sci USA. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen CX, et al. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 2000;6:755–767. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishikura K. Editor meets silencer: Crosstalk between RNA editing and RNA interference. Nat Rev Mol Cell Biol. 2006;7:919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patterson JB, Samuel CE. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: Evidence for two forms of the deaminase. Mol Cell Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishii KJ, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]