Abstract
Agriculture is a specialized form of symbiosis that is known to have evolved in only four animal groups: humans, bark beetles, termites, and ants. Here, we reconstruct the major evolutionary transitions that produced the five distinct agricultural systems of the fungus-growing ants, the most well studied of the nonhuman agriculturalists. We do so with reference to the first fossil-calibrated, multiple-gene, molecular phylogeny that incorporates the full range of taxonomic diversity within the fungus-growing ant tribe Attini. Our analyses indicate that the original form of ant agriculture, the cultivation of a diverse subset of fungal species in the tribe Leucocoprineae, evolved ≈50 million years ago in the Neotropics, coincident with the early Eocene climatic optimum. During the past 30 million years, three known ant agricultural systems, each involving a phylogenetically distinct set of derived fungal cultivars, have separately arisen from the original agricultural system. One of these derived systems subsequently gave rise to the fifth known system of agriculture, in which a single fungal species is cultivated by leaf-cutter ants. Leaf-cutter ants evolved remarkably recently (≈8–12 million years ago) to become the dominant herbivores of the New World tropics. Our analyses identify relict, extant attine ant species that occupy phylogenetic positions that are transitional between the agricultural systems. Intensive study of those species holds particular promise for clarifying the sequential accretion of ecological and behavioral characters that produced each of the major ant agricultural systems.
Keywords: Attini, divergence dating, Formicidae, phylogeny, symbiosis
Attine ants (subfamily Myrmicinae, tribe Attini) comprise a monophyletic group of >230 described species, exclusively New World and primarily Neotropical in distribution (1–4). All attine ants obligately depend on the cultivation of fungus gardens for food. So complete is this dependence that, upon leaving the maternal nest, a daughter queen must carry within her mouth a nucleus of fungus that serves as the starting culture for her new garden (5–7). Attine agriculture achieves its evolutionary apex in the leaf-cutting ants of the genera Acromyrmex and Atta, the dominant herbivores of the New World tropics (8, 9). Unlike more primitive attine ants that forage for and cultivate their fungus gardens on organic detritus, leaf-cutting ants have acquired the ability to cut and process fresh vegetation (leaves, flowers, and grasses) to serve as the nutritional substrate for their fungal cultivars. This key evolutionary innovation renders a mature Atta colony the ecological equivalent of a large mammalian herbivore in terms of collective biomass, lifespan, and quantity of plant material consumed (9).
Attine ant agriculture is the product of an ancient, quadripartite, symbiotic relationship between three mutualists and one parasite. The mutualists include the attine ants, their fungal cultivars (Leucocoprineae and Pterulaceae), and filamentous bacteria in the genus Pseudonocardia (Actinomycetes) that grow on the integuments of the ants. The parasite, a fungus in the genus Escovopsis (Ascomycetes) known only from attine fungus gardens, infects those gardens as a “crop disease” and is controlled, at least in part, by an antibiotic produced by the Pseudonocardia bacterial symbiont (4, 10, 11).
Based on nearly monolithic associations between broad phylogenetic groups of attine ants, cultivars, and Escovopsis parasites, attine agriculture has been divided into five biologically distinct agricultural systems, each representing a major transition in the evolution of ant agriculture. These systems are: (i) lower agriculture, practiced by species in the majority of attine genera (76 species), including those thought to retain more primitive features, which cultivate a wide range of fungal species in the tribe Leucocoprineae; (ii) coral fungus agriculture, practiced by species in the “pilosum group” (34 species), a subset of the attine genus Apterostigma, which cultivate a clade of fungi in the Pterulaceae; (iii) yeast agriculture, practiced by species in the “rimosus group” (18 species), a subset of the attine genus Cyphomyrmex, which cultivate a distinct clade of leucocoprineaceous fungi derived from the lower attine fungi; (iv) generalized higher agriculture, practiced by species in the three genera of non-leaf-cutting “higher attine” ants (63 species), which cultivate another distinct clade of leucocoprineaceous fungi separately derived from the lower attine fungi; and (v) leaf-cutter agriculture, a subdivision of higher attine agriculture practiced by species of ecologically dominant ants in the genera Atta and Acromyrmex (40 species), which cultivate a single highly derived species of higher attine fungus (4, 12–14).
In contrast to important advances in other areas of attine biology, including molecular phylogenies for the other three symbionts (10, 13–25), major features of fungus-growing ant phylogeny remain poorly understood (1, 26, 27). A well supported, resolved phylogeny of the attine ants is necessary for analyzing the coevolution of the ants and their three microbial symbionts as well as for understanding the historical sequence of evolutionary change that produced each of the five attine agricultural systems. To address this problem, we reconstructed the evolution of attine agriculture by inferring the first fossil-calibrated molecular phylogeny for the fungus-growing ants, based on data from four nuclear protein-coding genes and incorporating the full range of attine taxonomic diversity, particularly with regard to poorly understood, rarely collected, and potentially paraphyletic or polyphyletic taxa (1).
Results and Discussion
Origin of Ant Agriculture.
Based on the monophyly of the attine ants, on their exclusively New World distribution, and on their apparent center of diversity in the wet Neotropics, some researchers have speculated that ant agriculture arose a single time in the forests of South America after its isolation from Africa (1–3, 28–31). The results of our Bayesian codon-model and molecular-dating analyses (Fig. 1) provide strong corroboration for this view, indicating that ant agriculture had a single origin ≈50 million years ago and, because this date is far more recent than the last connection between South America and Africa ≈90 mya, indicating that ant agriculture originated on the South American continent. Significantly, the origin of fungus-growing coincides with the early Eocene climatic optimum (50–55 mya), a period of global warming in which an extraordinary diversity of plants with tropical affinities occurred at middle and high latitudes in South America (32). Unfortunately, our data are insufficient to identify the closest relative (i.e., sister group) of the Attini. Although in our phylogeny (Fig. 1) a clade consisting of Daceton and Orectognathus species is reconstructed as that sister group, this result is not significantly supported by any method of analysis, and we strongly caution against drawing any inferences based on it. Indeed, with few exceptions, the relationships of most nonattine myrmicines remain unresolved in this and in a previous study of ant relationships (33), indicating the critical need for additional data for resolving the profoundly important question of what group of ants is the closest non-fungus-growing relative of the Attini (1).
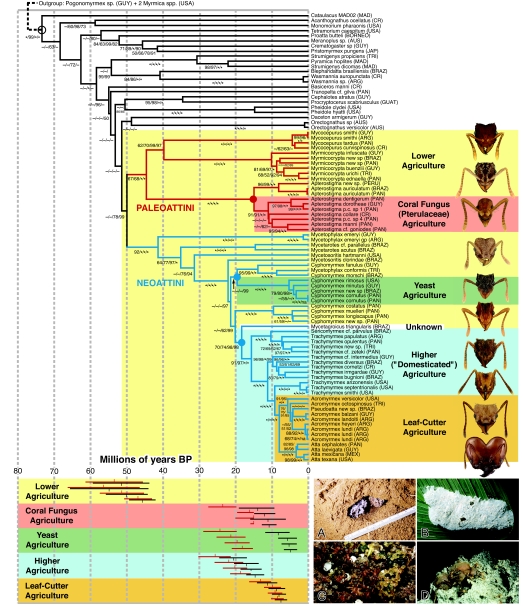
Fig. 1.
A time-calibrated phylogeny of the attine fungus-growing ants with age estimates for the origins of the five known ant agricultural systems. Agricultural systems, indicated by colored rectangles, are defined by phylogenetically distinct groups of associated fungal cultivars and were reconstructed under likelihood and parsimony methods with identical results. Tree topology is the maximum-likelihood reconstruction, identical with regard to attine phylogeny to the Bayesian codon-model result. Numbers on branches indicate support values from four analyses: parsimony bootstraps, ML bootstraps, Bayesian nucleotide-model posterior probabilities, and Bayesian codon-model posterior probabilities (“−,” < 50; “*,” 100). The three solid circles represent node assignments for Dominican amber fossil calibrations, and the open circle marks the root of the dating-analysis tree. Bars below the time scale summarize four separate relaxed-molecular-clock analyses dating the origin of the five agricultural systems. Black bars represent the most recent node containing all members of the system (“crown-group”) and red bars additionally include the branch leading to that node (“stem-group”). For each system, pairs of red and black bars from top to bottom correspond to (i) Bayesian uncorrelated lognormal, root age prior to 73.5 ± 4.5 mya; (ii) penalized likelihood, root age 81 mya; (iii) penalized likelihood, root age 73.5 mya; (iv) penalized likelihood, root age 66 mya. The tree shown here is the result of dating analysis (iii). Ant head photos (top to bottom): Mycocepurus tardus, Myrmicocrypta infuscata, Apterostigma collare, Mycetophylax emeryi, Cyphomyrmex rimosus, Cyphomyrmex longiscapus, Trachymyrmex opulentus, Trachymyrmex cornetzi, Acromyrmex octospinosus, Atta laevigata. Fungus gardens: (A) Lower attine agriculture. (B) Coral fungus agriculture. (C) Yeast agriculture. (D) Higher leaf-cutter agriculture. Country abbreviations: ARG, Argentina; AUS, Australia; BRAZ, Brazil; CR, Costa Rica; MAD, Madagascar; CR, Costa Rica; JAP, Japan; PAN, Panama; GUAT, Guatemala; GUY, Guayana; TRI, Trinidad; MEX, Mexico. Photo credits are given in Acknowledgments.
Lower Agriculture.
Our results (Fig. 1) indicate that the first fungus-growing ant practiced lower agriculture and that all extant members of a series of basally diverging lineages continue to practice this form of agriculture. This corroborates the hypothesis of some researchers that lower agriculture was the first attine agricultural system (31) but contradicts a long-standing hypothesis that yeast agriculture was the first system (9, 29, 30, 34) and a recently proposed hypothesis that coral-fungus agriculture was the first (35). Lower attine fungal cultivars all belong to a paraphyletic grade within the tribe Leucocoprineae (“parasol mushrooms”) and are, so far as is known, entirely capable of a feral, free-living existence outside of the attine symbiosis (17, 36). Current data indicate that a corresponding paraphyletic grade of Escovopsis (24, 37) infects lower attine fungal cultivars. It remains unknown whether Escovopsis infects cultivars while they are in the free-living phase.
Very early in their evolution, the Attini diverged into two lineages that would subsequently diversify into what Kusnezov (38) first recognized as the two major clades of attines, the “Paleoattini” and the “Neoattini” (Fig. 1). The three paleoattine genera are remarkably different from one another morphologically, a difference attributable to the span of time (≈40–45 mya) since they diverged from a common ancestor. Despite their morphological differences, these genera share a number of biologically important features (26, 38–40), the most striking of which is the consistent occurrence of a unique clear spot of unknown biological function on the wings of gynes (41). Early in the evolution of the Neoattini (50–30 mya) a temporal series of three successive divergences generated a grade of primitive lineages. These lineages are currently represented by, in order of oldest to youngest, the Mycetophylax emeryi species group, the genus Mycetarotes, and the species Mycetosoritis hartmanni (occurring in the southern U.S., with a sister species or conspecific in Central America) (42) (Fig. 1). Biological study of these extant, poorly known remnants of primitively diverged neoattine lineages may clarify the early evolution of ant agriculture.
Coral Fungus Agriculture.
During the 50-million-year evolution of the fungus-growing ants, there occurred only one known transition to a nonleucocoprineaceous fungal cultivar. Although the majority of paleoattine species, including one of the basally diverging clades within Apterostigma, practices lower attine (leucocoprineaceous) agriculture, all known species in the “pilosum group” clade of the genus Apterostigma cultivate a clade of coral fungi (Pterulaceae) closely related to the genera Pterula and Deflexula (21, 22). Our results clearly indicate that the earliest Apterostigma species cultivated leucoprineaceous fungi, but between 10 and 20 mya, an Apterostigma species acquired a radically different fungal cultivar in the Pterulaceae that all its descendant species continue to cultivate. Recent research indicates that coral fungus agriculture is infected by a specialized grade of Escovopsis that is derived from a lower attine Escovopsis species and, further, that this grade subsequently gave rise to a clade that infects higher agricultural cultivars (24). This pattern most likely indicates that, after the origin of coral fungus agriculture, a coral-fungus-infecting Escovopsis switched hosts and began infecting higher attine cultivars. The broad overlap in dates of origin of coral fungus and higher attine agriculture (Fig. 1) is consistent with this hypothesis.
Yeast Agriculture.
Another remarkable shift in cultivar type occurs in yeast-growing ants. Unlike typical attine mycelial gardens, yeast gardens consist of clusters of small, irregularly shaped nodules ≈0.5 mm in diameter (Fig. 1C) composed of fungal cultivars growing in a single-celled yeast phase rather than in the mycelial phase common to all other attine cultivars. Yeast agriculture is confined to the Cyphomyrmex “rimosus group,” which our results (Fig. 1) and prior work (1, 43, 44) indicate is monophyletic. The branch of the phylogeny subtending the C. rimosus group is remarkably long, indicating extensive evolutionary change and bracketing a broad potential time interval of 5–25 mya for the origin of yeast agriculture (Fig. 1). Significantly, this long branch in the ant phylogeny parallels a similarly long branch in the cultivar phylogeny (17) that subtends the attine yeast cultivars, members of a highly derived clade of leucocoprineaceous fungi that grow as yeast morphs when associated with attine ants. Like the lower attine cultivars from which they are derived, yeast cultivars are capable of a free-living, feral existence independent of the attine symbiosis (17) in which they grow on leaf litter in the mycelial phase typical for the rest of the tribe. Because yeast-phase growth is otherwise unknown in the order Agaricales, and because the attine yeast cultivars grow as yeasts only when associated with ants (or, depending on conditions, in artificial culture), yeast agriculture has been cited as a case of coadaptation and/or domestication (4). The parasite Escovopsis is unknown from yeast agriculture, suggesting that there may be some feature of the yeast morph that resists or prevents Escovopsis infection.
Higher Agriculture, Including Leaf-Cutter Agriculture.
The transition to higher agriculture and the subsequent origin of leaf cutting are arguably the two most ecologically significant events in the evolutionary history of the Attini. The cultivars of higher attine ants are descended from lower agricultural cultivars (4, 15) but are derived in two features that suggest a significant degree of “domestication,” i.e., modification for life with ants. First, higher attine fungi do not appear capable of a free-living existence separate from their ant hosts, and, second, only higher attine fungi produce “gongylidia,” nutritious swollen hyphal tips produced by the fungus and harvested by the ants for food.
Our analyses produced a series of unexpected results that hold the potential for reconstructing the origin and subsequent evolution of higher agriculture with a high degree of resolution. First, the Cyphomyrmex costatus species group is the sister group of the combined higher Attini and Mycetagroicus. The four described species in the C. costatus group have always been regarded as aberrant members of the genus (43–45), but a phylogenetic position entirely removed from Cyphomyrmex as the sister group to the higher attines is unexpected. Second, the most recently discovered attine genus, Mycetagroicus, is the sister group of the higher attines. Described in only 2001 (3), nothing is known of the biology of the three Mycetagroicus species, including the form of agriculture they practice. Given that both the C. costatus species group and Mycetagroicus belong to lineages that successively diverged during the transition from lower to higher agriculture, biological study of these groups promises to elucidate the sequence of evolutionary change that generated this transition. Third, ants formerly placed in two major groups of Trachymyrmex, including the T. opulentus and T. urichi groups (46, 47), form a well supported clade that includes the genus Sericomyrmex and that is the sister group to the remainder of the higher attines. Fourth, the Trachymyrmex septentrionalis species group, which includes T. diversus and allied species (48), is closely related to the leaf-cutting ants. In fact, a clade of North American species (including T. septentrionalis) is the sister group of the leaf-cutting ants. This surprising result suggests that renewed biological study of the T. septentrionalis group, broadly defined, is likely to yield new information about the transition from generalized higher agriculture to leaf-cutter agriculture, one of the most successful evolutionary transitions in the animal kingdom (8, 9). Importantly, members of this group (T. cornetzi and T. diversus) have been observed to cut leaves (1) (T.R.S., personal observation), and T. intermedius is morphologically one of the most “Acromyrmex-like” of all Trachymyrmex species. Finally, leaf-cutting ants are remarkably young, originating between 8 and 12 mya. Such a recent origin for this ecologically dominant group explains their conspicuous absence from Dominican amber (15–20 mya) and may help to explain why, so far as is known, most leaf-cutting ants cultivate the same cultivar species (12–14).
Concluding Remarks.
Agriculture is a specialized form of symbiosis that has evolved in only four known animal groups: humans, bark beetles, termites, and ants (11). Some researchers have hypothesized that similar evolutionary mechanisms may have driven the early evolution of agriculture in all of these groups (4, 49). Identifying those common mechanisms requires an understanding of the historical sequence of events that generated each system. Our results confirm that, like termites (50) but unlike humans (51, 52) and bark beetles (53), ants discovered agriculture a single time and discovered each of their derived agricultural systems a single time. We cannot know how many agricultural systems may have evolved during the 50-million-year-long evolutionary history of the Attini. Indeed, the attine ants are so poorly known (2) that it is possible that additional extant systems await discovery. Lineages that diverged at the critical evolutionary junctures that produced the five known attine agricultural systems are, fortunately, still represented by extant ant species that are available for biological study. Such study offers the most promising route for reconstructing the sequential accretion of ecological and behavioral characters that produced each ant agricultural system. Understanding the sequential evolution of the attine agricultural systems will, in turn, inform general hypotheses about the evolution of agricultural symbioses.
Methods
Data.
Our data, obtained by using standard PCR techniques, consist of 2,459 aligned nucleotide sites from the coding regions of four nuclear genes: elongation factor 1-α F1 (EF1αF1) (1,075 bp), elongation factor 1-α F2 (EF1αF2) (517 bp), wingless (409 bp), and long-wavelength rhodopsin (opsin) (458 bp). All data in this study represent protein-coding (exon) sequences; intervening introns in opsin and EF1αF1 were not used because they could not be aligned confidently. We sampled 65 attine taxa and 26 nonattine outgroups. All sequences generated are new to this study except for previously published fragments from 4 attine and 10 nonattine outgroup species (33). Primers used for PCR amplification and sequencing are found in supporting information (SI) Table S1. Of the total 2,459 included nucleotide positions from all genes, 952 were variable and 847 parsimony informative. Sequences are deposited in GenBank; taxa and accession numbers are listed in Table S2.
Phylogenetic Analyses.
Phylogenetic analyses used four methods: (i) parsimony, (ii) maximum likelihood, (iii) Bayesian nucleotide-model Markov Chain Monte Carlo (MCMC), and (iv) Bayesian codon-model MCMC.
Parsimony.
Maximum parsimony (MP) analyses were conducted in PAUP* v4.0b10 (54) using heuristic searches with tree bisection–reconnection (TBR) and 1,000 random-taxon-addition replicates. Nonparametric bootstrap analyses (55) used TBR branch-swapping and consisted of 1,000 pseudoreplicates, with 10 random-taxon-addition replicates per pseudoreplicate. Analyses identified 12 most-parsimonious trees (MPTs) of length = 4,383, CI = 0.270, RI = 0.704. Successive-approximations-weighting analyses identified a single tree, one of the MPTs.
Maximum Likelihood (ML).
The data and the MPT identified by successive-approximations weighting were evaluated under the Akaike information criterion (AIC) (56) as calculated in ModelTest v3.06 (57), identifying the GTR+I+Γ model of evolution. ML analyses consisted of four separate searches conducted in GARLI v0.951 (58) using the GTR+I+Γ model (with six Γ rate categories) and resulted in the topology presented in Fig. 1, with a log likelihood of −24,868.84927. A subsequent heuristic search in PAUP* using the most likely tree identified by the GARLI searches as the starting tree and employing TBR branch-swapping and the GTR+I+Γ model (with six Γ rate categories) resulted in exactly the same topology and likelihood score. Nonparametric bootstrap analyses consisted of 500 pseudoreplicates in GARLI under the same conditions as the ML search.
Bayesian MCMC.
Bayesian analyses were conducted in MrBayes v3.1.2 (59). Burn-in and run convergence were assessed by comparing the mean and variance of log likelihoods, both by eye and by using the program Tracer v1.3 (available at http://beast.bio.ed.ac.uk/Tracer) (60); by examination of the MrBayes “.stat” output file; and by examination of the split frequencies diagnostic. For the nucleotide-model analyses, sequence data were divided into eight character partitions, four partitions consisting of the combined first and second codon positions for each of the four genes and four partitions consisting of the third codon position for each of the four genes. Based on ModelTest results, the wingless third-position character partition was assigned the GTR+Γ model; opsin and EF1αF2 third positions were separately assigned the HKY+I+Γ model; and all other character partitions were separately assigned the GTR+I+Γ model. Nucleotide-model analyses consisted of two independent runs of 5 million generations, each distributed over eight chains (seven heated and one cold; temperature parameter 0.05) with trees sampled every 100 generations and with a burn-in of 4.2 million generations. Codon-model analyses used a 2,454-bp dataset, from which incomplete codon triplets were excluded, and 88 taxa, in which multiple exemplars representing two species (Cyphomyrmex cornutus and Acromyrmex lundi) were reduced to a single exemplar. Sequence data were divided into four character partitions, one for each gene. Each partition was separately assigned the codon model. Codon-model analyses consisted of two independent runs of 10 million generations, each distributed over eight chains (seven heated and one cold; temperature parameter 0.05) with trees sampled every 100 generations and with a burn-in of 9 million generations.
Phylogenetic Mapping of Agricultural Systems.
Terminal taxa were assigned states for a single six-state character representing the four attine agricultural systems and leaf-cutter agriculture (i.e., no agriculture, lower agriculture, yeast agriculture, higher agriculture, leaf-cutter agriculture, coral-fungus agriculture). Five species (Myrmicocrypta n. sp. Brazil, Mycetagroicus triangularis, Cyphomyrmex n. sp., Cyphomyrmex morschi, Trachymyrmex irmgardae, and Pseudoatta n. sp.) received “unknown” (i.e., “?”) state assignments, and Trachymyrmex papulatus received a “lower agriculture” state assignment based on a single garden collection from Argentina (a second colony from the same locality cultivated a typical higher attine garden). Character evolution was optimized onto the Bayesian codon-model consensus tree (with branch lengths) under both parsimony using MacClade (61) and maximum likelihood using the StochChar module provided in the Mesquite package (available at http://mesquiteproject.org) (62). Both methods produced the mappings shown in Fig. 1. Under parsimony, ancestral-state optimizations were unambiguous. Under the Markov k-state 1-parameter model (63), the likelihood that each agricultural system arose in the most recent common ancestor of the corresponding ant clade was, as a proportion of the total probability (= 1.0) distributed across the six character states, 0.9831 for lower agriculture, 0.9995 for yeast agriculture, 0.9905 for higher agriculture, 0.9924 for leaf-cutter agriculture, and 0.9998 for coral-fungus agriculture.
Divergence Dating.
We inferred divergence dates using both semiparametric and Bayesian relaxed clock methods. The first method used was the semiparametric penalized likelihood approach implemented in r8s v1.7 (64, 65). Branch lengths were first estimated on the ML topology using PAUP* under a GTR+I+Γ model. The Pogonomyrmex and two Myrmica species were used to root the tree during branch length estimation and were subsequently removed from all dating analyses. Thus, the root of the tree for all dating analyses represents the origin of the “core myrmicines,” a well supported clade established by previous work (33). Smoothing parameters were estimated by using the cross-validation feature in r8s. Confidence intervals were calculated by using 100 nonparametric bootstrap replicates of the dataset generated by Mesquite, followed by reestimation of branch lengths and divergence times for each replicate.
We calibrated three nodes with minimum-age constraints using attine Dominican amber fossils. These fossils are (i) Apterostigma electropilosum, a member of the A. pilosum group (40); (ii) Cyphomyrmex maya and Cyphomyrmex taino, both members of the C. rimosus group (66); and (iii) Trachymyrmex primaevus, a fossil of uncertain placement within the genus (67) (but see below). The fossils were used to calibrate stem-group nodes in the phylogeny (68). Because Dominican amber is dated between 15 and 20 mya (69), we calibrated these three nodes using a minimum age constraint of 15 mya. The r8s program requires that at least one node in the tree be either fixed or constrained with a maximum age. Using a maximum-age constraint for the root node proved unsatisfactory, because the program simply inferred the age of that node to be identical to the chosen maximum age, a common phenomenon in r8s that is underappreciated in many studies. We therefore conducted separate analyses in which the root node (i.e., “core myrmicines”) was fixed with ages representing the range of plausible dates for that node obtained from a separate study (33). The root ages were 81, 73.5, and 66 mya.
The second method used was the Bayesian relaxed clock uncorrelated lognormal approach implemented in BEAST v1.4.6 (70, 71) with the SRD06 two-partition codon-specific rates model of sequence evolution (72) and a Yule process for the tree prior. The root node was given a normal (mean = 73.5; SD = 4.5) age prior distribution. The stem-group nodes represented by the three attine fossils described above were given the following age prior distributions (all with zero offset lower bounds of 15 mya): Apterostigma pilosum-stem-group, lognormal (mean = 2.7; SD = 0.3); C. rimosus-stem-group, lognormal (mean = 2.2; SD = 0.5); Trachymyrmex stem-group, lognormal (mean = 1.5; SD = 0.5). MCMC searches were run for 10,000,000 generations, with the first 2,000,000 discarded as burn-in. The searches achieved adequate mixing as assessed by the high ESS values for all parameters, plateaus for divergence time estimates over generations after burn-in, and repeatability of results over multiple independent runs.
Based on direct examination of a fossil specimen of T. primaevus, we find the placement of this species within the genus uncertain. Because Mayhé-Nunes and Brandão (47, 48) suggest that T. primaevus belongs to the T. septentrionalis group, we additionally tested the effects of this placement on age estimates for the origins of higher agriculture and leaf-cutter agriculture. In analyses with the T. primaevus calibration assigned to the T. septentrionalis group (sensu lato) branch, we obtained ages 2–4 million years older for the origins of higher agriculture and leaf-cutter agriculture. With the T. primaevus calibration excluded entirely, age estimates are 0–2 million years older than those reported.
Numerical values of all divergence dates are listed in Table S3 and Table S4. For more information, see the SI Text.
Acknowledgments.
We gratefully acknowledge U. G. Mueller, S. A. Rehner, J. J. Boomsma, and C. R. Currie for attinological collegiality and collaboration; Nor F. Dahlan and E. Okonski for superior technical and organizational expertise; K. Pagenkopp and R. Adams for technical support; M. Braun, E. Zimmer, B. Danforth, L. Weigt, C. Huddleston, and J. Sakamoto for training and guidance; R. Wilson for help with the figures; and the following colleagues for specimens, field support, and other generous aid: D. Agosti (American Museum of Natural History), L. Alonso, M. Bacci, C. Bernard, C. Brandão, M. Cafaro, R. Camargo (University Estadual Paulista, Botucatu, Brazil), M. and M. Chan-A-Sue, I. Chapela (University of California, Berkeley, CA), P. and M. Charles-Dominique, S. Cover (Harvard University, Boston), F. Cuezzo, J. D'Arc, J. Delabie (CEPLAC, Ilhéus, Bahia, Brazil), P. de Silva, J. Diniz, M. Engel, F. Fernández, H. Fernández-Marin, B. Fisher (California Academy of Sciences, San Francisco) T. Friedlander, V. Funk, N. Gerardo, D. Grimaldi, A. Harada, S. Johnson, M. Kalamandeen, J. LaPolla, J. Lattke, A. Little (University of Wisconsin, Madison, WI), J. Longino, B. Lopes, A. Malsche, C. Marshall, A. Mayhe-Nunes, T. McGlynn (University of San Diego), A. Mikheyev, H. Morais, N. Pierce, N. Pitman, S. Price, J. Regier, F. Roces (University of Würzburg, Würzburg, Germany), J. Santisteban, R. Savolainen (University of Helsinki, Helsinki), S. Solomon (Smithsonian Institute, Washington, D.C.), J. Sosa-Calvo, A. Suarez, H. Vasconcelos, P. Villesen, P. Ward, J. Wetterer, R. Williams, E. O. Wilson, W. Wcislo, and A. Wild. This work was supported by National Science Foundation (NSF) Integrated Research Challenges in Environmental Biology (IRCEB) Grant EFB 0110073 (to U.G. Mueller, C. R. Currie, and T.R.S.) and by NSF Assembling the Tree of Life (AToL) Grant EF 0431330 (to P. S. Ward, B. L. Fisher, S.G.B., and T.R.S.). T.R.S. was additionally supported by awards from the National Geographic Society Committee for Research and Exploration, Smithsonian Scholarly Studies, and the Smithsonian Biodiversity of the Guianas Program. Fig. 1 ant head photographs by E. Okonski; Fig. 1 A and B by T.R.S.; Fig. 1C by A. Wild; Fig. 1D by J. Wetterer.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The DNA sequences reported in this paper have been deposited in the GenBank database (accession nos. EU204145–EU204615).
See Commentary on page 5287.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711024105/DCSupplemental.
References
- 1.Schultz TR, Meier R. A phylogenetic analysis of the fungus-growing ants (Hymenoptera: Formicidae: Attini) based on morphological characters of the larvae. Syst Entomol. 1995;20:337–370. [Google Scholar]
- 2.Mayhé-Nunes AJ, Jaffé K. On the biogeography of Attini (Hymenoptera: Formicidae) Ecotropicos. 1998;11:45–54. [Google Scholar]
- 3.Brandão CRF, Mayhé-Nunes AJ. A new fungus-growing ant genus, Mycetagroicus gen n, with the description of three new species and comments on the monophyly of the Attini (Hymenoptera: Formicidae) Sociobiology. 2001;38:639–665. [Google Scholar]
- 4.Schultz TR, Mueller UG, Currie CR, Rehner SA. Reciprocal illumination: A comparison of agriculture in humans and ants. In: Vega F, Blackwell M, editors. Ecological and Evolutionary Advances in Insect-Fungal Associations. New York: Oxford Univ Press; 2005. pp. 149–190. [Google Scholar]
- 5.Ihering RV. The founding of new colonies and fungus gardens in Atta sexdens (translated from German) Zool Anz. 1898;21:238–245. [Google Scholar]
- 6.Huber J. On colony founding in Atta sexdens (translated from German) Biol Centralbl. 1905;25:606–619. [Google Scholar]
- 7.Huber J. On colony founding in Atta sexdens (translated from German) Biol Centralbl. 1905;25:625–635. [Google Scholar]
- 8.Wheeler WM. The fungus-growing ants of North America. Bull Am Mus Nat Hist. 1907;23:669–807. [Google Scholar]
- 9.Hölldobler B, Wilson EO. The Ants. Cambridge, MA: Belknap Press; 1990. [Google Scholar]
- 10.Currie CR, Scott JA, Summerbell RC, Malloch D. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature. 1999;398:701–704. [Google Scholar]
- 11.Mueller UG, Gerardo NM, Aanen DK, Six DL, Schultz TR. The evolution of agriculture in insects. Ann Rev Ecol Evol Syst. 2005;36:563–595. [Google Scholar]
- 12.Silva-Pinhati ACO, et al. Low variation in ribosomal DNA and internal transcribed spacers of the symbiotic fungi of leaf-cutting ants (Attini: Formicidae) Braz J Med Biol Res. 2004;37:1463–1472. doi: 10.1590/s0100-879x2004001000004. [DOI] [PubMed] [Google Scholar]
- 13.Mikheyev AS, Mueller UG, Abbot P. Cryptic sex and many-to-one coevolution in the fungus-growing ant symbiosis. Proc Natl Acad Sci USA. 2006;103:10702–10706. doi: 10.1073/pnas.0601441103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikheyev AS, Mueller UG, Boomsma JJ. Population genetic signatures of diffuse coevolution between leaf-cutting ants and their cultivar fungi. Mol Ecol. 2007;16:209–216. doi: 10.1111/j.1365-294X.2006.03134.x. [DOI] [PubMed] [Google Scholar]
- 15.Chapela IH, Rehner SA, Schultz TR, Mueller UG. Evolutionary history of the symbiosis between fungus-growing ants and their fungi. Science. 1994;266:1691–1694. doi: 10.1126/science.266.5191.1691. [DOI] [PubMed] [Google Scholar]
- 16.Hinkle G, Wetterer JK, Schultz TR, Sogin ML. Phylogeny of the attine ant fungi based on analysis of small subunit ribosomal RNA gene sequences. Science. 1994;266:1695–1697. doi: 10.1126/science.7992052. [DOI] [PubMed] [Google Scholar]
- 17.Mueller UG, Rehner SA, Schultz TR. The evolution of agriculture in ants. Science. 1998;281:2034–2038. doi: 10.1126/science.281.5385.2034. [DOI] [PubMed] [Google Scholar]
- 18.Currie CR, Mueller UG, Malloch D. The agricultural pathology of ant fungus gardens. Proc Natl Acad Sci USA. 1999;96:7998–8002. doi: 10.1073/pnas.96.14.7998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green AM, Mueller UG, Adams RMM. Extensive exchange of fungal cultivars between sympatric species of fungus-growing ants. Mol Ecol. 2002;11:191–195. doi: 10.1046/j.1365-294x.2002.01433.x. [DOI] [PubMed] [Google Scholar]
- 20.Currie CR, et al. Ancient tripartite coevolution in the attine ant–microbe symbiosis. Science. 2003;299:386–388. doi: 10.1126/science.1078155. [DOI] [PubMed] [Google Scholar]
- 21.Villesen P, Mueller UG, Schultz TR, Adams RMM, Bouck MC. Evolution of ant-cultivar specialization and cultivar switching in Apterostigma fungus-growing ants. Evolution (Lawrence, Kans) 2004;58:2252–2265. doi: 10.1111/j.0014-3820.2004.tb01601.x. [DOI] [PubMed] [Google Scholar]
- 22.Munkascsi AB, et al. Convergent coevolution in the domestication of coral mushrooms by fungus-growing ants. Proc R Soc London Ser B. 2004;271:1777–1782. doi: 10.1098/rspb.2004.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cafaro MJ, Currie CR. Phylogenetic analysis of mutualistic filamentous bacteria associated with fungus-growing ants. Can J Microbiol. 2005;51:441–446. doi: 10.1139/w05-023. [DOI] [PubMed] [Google Scholar]
- 24.Gerardo NM, Mueller UG, Currie CR. Complex host–pathogen coevolution in the Apterostigma fungus-growing ant–microbe symbiosis. BMC Evol Biol. 2006;6:88. doi: 10.1186/1471-2148-6-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taerum SJ, Cafaro MJ, Little AEF, Schultz TR, Currie CR. Low host–pathogen specificity in the leaf-cutting ant–microbe symbiosis. Proc R Soc London Ser B. 2007;274:1971–1978. doi: 10.1098/rspb.2007.0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayhé-Nunes AJ. Caracas, Venezuela: Universidad Simón Bolivar; 1995. PhD thesis. [Google Scholar]
- 27.Wetterer JK, Schultz TR, Meier R. Phylogeny of fungus-growing ants (tribe Attini) based on mtDNA sequence and morphology. Mol Phylogenet Evol. 1998;9:42–47. doi: 10.1006/mpev.1997.0466. [DOI] [PubMed] [Google Scholar]
- 28.Wheeler WM. Social Life Among the Insects. New York: Harcourt Brace; 1923. [Google Scholar]
- 29.Wilson EO. The Insect Societies. Cambridge, MA: Belknap; 1971. [Google Scholar]
- 30.Weber NA. Gardening Ants: The Attines. Philadelphia: Am Philos Soc; 1972. [Google Scholar]
- 31.Mueller UG, Schultz TR, Currie CR, Adams RMM, Malloch D. The origin of the attine ant-fungus mutualism. Q Rev Biol. 2001;76:169–197. doi: 10.1086/393867. [DOI] [PubMed] [Google Scholar]
- 32.Wilf P, et al. High plant diversity in Eocene South America: Evidence from Patagonia. Science. 2003;300:122–125. doi: 10.1126/science.1080475. [DOI] [PubMed] [Google Scholar]
- 33.Brady SG, Schultz TR, Fisher BL, Ward PS. Evaluating alternative hypotheses for the early evolution and diversification of ants. Proc Natl Acad Sci USA. 2006;103:18172–18177. doi: 10.1073/pnas.0605858103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weber NA. Fungus ants. In: Hermann HR, editor. Social Insects. Vol 4. New York: Academic; 1982. pp. 255–363. [Google Scholar]
- 35.Sánchez-Peña SR. New view on origin of attine ant-fungus mutualism: Exploitation of a prexisting insect–fungus symbiosis (Hymenoptera: Formicidae) Ann Entomol Soc Am. 2005;98:151–164. [Google Scholar]
- 36.Vo TL, Mikheyev AS, Mueller UG. Free-living fungal symbionts (Lepiotaceae) of fungus-growing ants (Attini: Formicidae) Mycologia. 2008 doi: 10.3852/07-055. in press. [DOI] [PubMed] [Google Scholar]
- 37.Currie CR, Bot ANM, Boomsma JJ. Experimental evidence of a tripartite mutualism: Bacteria protect ant fungus gardens from specialized parasites. Oikos. 2003;101:91–102. [Google Scholar]
- 38.Kusnezov N. Zoogeography of ants in South America (translated from Spanish) Acta Zool Lilloana. 1963;19:25–186. [Google Scholar]
- 39.Bolton B. Synopsis and classification of Formicidae. Mem Am Entomol Inst. 2003;71:1–370. [Google Scholar]
- 40.Schultz TR. The fungus-growing ant genus Apterostigma in Dominican amber. Mem Am Entomol Inst. 2007;80:425–436. [Google Scholar]
- 41.Emery C. Studies of the Myrmicinae V. The genera of the Attini; descriptions of new forms of Mycocepurus and Myrmicocrypta (translated from French) Ann Soc Entomol Belg. 1913;57:250–262. [Google Scholar]
- 42.McKay WP. Two new species of ants in the tribe Attini of Costa Rica and Mexico: Mycetosoritis vinsoni y Mycocepurus curvispinosus (Hymenoptera: Formicidae) (translated from Spanish) Rev Biol Trop. 1998;46:421–426. [Google Scholar]
- 43.Kempf WW. (“1965”) Revision of the Neotropical ants of the genus Cyphomyrmex Mayr. Part II. Group of rimosus (Spinola) (Hym: Formicidae) Stud Entomol. 1966;8:161–200. [Google Scholar]
- 44.Snelling R, Longino JT. Revisionary notes on the fungus-growing ants of the genus Cyphomyrmex, rimosus group (Hymenoptera: Formicidae: Attini) In: Quintero D, Aiello A, editors. Insects of Panama and Mesoamerica. New York: Oxford Univ Press; 1992. pp. 479–494. [Google Scholar]
- 45.Schultz TR, et al. Cryptic speciation in the fungus-growing ants Cyphomyrmex longiscapus Weber and Cyphomyrmex muelleri Schultz and Solomon, new species (Formicidae: Attini) Insectes Soc. 2002;49:331–343. [Google Scholar]
- 46.Mayhé-Nunes AJ, Brandão CRF. Revisionary studies on the attine ant genus Trachymyrmex Forel. Part 1: Definition of the genus and the Opulentus group (Hymenoptera: Formicidae) Sociobiology. 2002;40:667–698. [Google Scholar]
- 47.Mayhé-Nunes AJ, Brandão CRF. Revisonary studies on the attine ant genus Trachymyrmex Forel. Part 3: The Jamaicensis group (Hymenoptera: Formicidae) Zootaxa. 2007;1444:1–21. [Google Scholar]
- 48.Brandão CRF, Mayhé-Nunes AJ. A phylogenetic hypothesis for the Trachymyrmex species groups, and the transition from fungus-growing to leaf-cutting in the Attini. Mem Am Entomol Inst. 2007;80:72–88. [Google Scholar]
- 49.Rindos D. The Origins of Agriculture: An Evolutionary Perspective. New York: Academic; 1984. [Google Scholar]
- 50.Aanen DK, et al. The evolution of fungus-growing termites and their mutualistic fungal symbionts. Proc Natl Acad Sci USA. 2002;99:14887–14892. doi: 10.1073/pnas.222313099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diamond J. Guns, Germs, and Steel: The Fates of Human Societies. New York: Norton; 1997. [Google Scholar]
- 52.Smith BD. The Emergence of Agriculture. New York: Sci Am Library; 1998. [Google Scholar]
- 53.Farrell BD, et al. The evolution of agriculture in beetles (Curculionidae: Scolytinae and Platypodinae) Evolution (Lawrence, Kans) 2001;55:2011–2027. doi: 10.1111/j.0014-3820.2001.tb01318.x. [DOI] [PubMed] [Google Scholar]
- 54.Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) v. 4.0b10. Sunderland, MA: Sinauer; 2002. [Google Scholar]
- 55.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution (Lawrence, Kans) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 56.Posada D, Crandall KA. Selecting the best-fit model of nucleotide substitution. Syst Biol. 2001;50:580–601. [PubMed] [Google Scholar]
- 57.Posada D, Crandall KA. Modeltest: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 58.Zwickl DJ. Austin, TX: University of Texas; 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Ph.D. dissertation. [Google Scholar]
- 59.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 60.Rambaut A, Drummond AJ. Tracer. Edinburgh, UK: University of Edinburgh; 2007. Version 1.4. [Google Scholar]
- 61.Maddison DR, Maddison WP. MacClade v4.0. Sunderland, MA: Sinauer; 2000. [Google Scholar]
- 62.Maddison WP, Maddison DR. Mesquite: A modular system for evolutionary analysis. Tucson, AZ: University of Arizona; 2006. Version 1.1. [Google Scholar]
- 63.Lewis PO. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst Biol. 2001;50:913–925. doi: 10.1080/106351501753462876. [DOI] [PubMed] [Google Scholar]
- 64.Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: A penalized likelihood approach. Mol Biol Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- 65.Sanderson MJ. r8s: Inferring absolute rates of evolution and divergence time estimates by fossil calibrations and fossil-based model selection. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- 66.de Andrade ML. First descriptions of two new amber species of Cyphomyrmex from Mexico and the Dominican Republic (Hymenoptera: Formicidae) Beitr Entomol. 2003;53:131–139. [Google Scholar]
- 67.Baroni Urbani C. First description of fossil gardening ants (Amber Collection Stuttgart and Natural History Museum Basel; Hymenoptera: Formicidae. I: Attini) Stuttg Beitr Naturkd Ser B (Geol Paläontol) 1980;54:1–13. [Google Scholar]
- 68.Magallón SA, Sanderson MJ. Absolute diversification rates in angiosperm clades. Evolution (Lawrence, Kans) 2001;55:1762–1780. doi: 10.1111/j.0014-3820.2001.tb00826.x. [DOI] [PubMed] [Google Scholar]
- 69.Iturralde-Vinent MA, MacPhee RDE. Age and paleogeographical origin of Dominican amber. Science. 1996;273:1850–1852. [Google Scholar]
- 70.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Drummond AJ, Rambaut BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shapiro B, Rambaut A, Drummond AJ. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol. 2006;23:7–9. doi: 10.1093/molbev/msj021. [DOI] [PubMed] [Google Scholar]