Abstract
To ensure efficient and timely replication of genomic DNA, organisms in all three kingdoms of life possess specialized translesion DNA synthesis (TLS) polymerases (Pols) that tolerate various types of DNA lesions. It has been proposed that an exchange between the replicative DNA Pol and the TLS Pol at the site of DNA damage enables lesion bypass to occur. However, to date the molecular mechanism underlying this process is not fully understood. In this study, we demonstrated in a reconstituted system that the exchange of Saccharomyces cerevisiae Polδ with Polη requires both the stalling of the holoenzyme and the monoubiquitination of proliferating cell nuclear antigen (PCNA). A moving Polδ holoenzyme is refractory to the incoming Polη. Furthermore, we showed that the Polη C-terminal PCNA-interacting protein motif is required for the exchange process. We also demonstrated that the second exchange step to bring back Polδ is prohibited when Lys-164 of PCNA is monoubiquitinated. Thus the removal of the ubiquitin moiety from PCNA is likely required for the reverse exchange step after the lesion bypass synthesis by Polη.
Keywords: translesion DNA synthesis, ubiquitin binding domain, holoenzyme stability
The eukaryotic replicative DNA polymerase δ (Polδ) forms a stable holoenzyme complex with proliferating cell nuclear antigen (PCNA). The holoenzyme is responsible for the highly accurate and processive DNA synthesis in eukaryotes (1). However, in the presence of DNA damage Polδ faces difficulties in synthesizing through the damaged base, which results in replication fork stalling and interruption in genomic DNA duplication. Prolonged stalling of the replication fork causes premature replication fork collapse and generates deleterious DNA damage in the form of dsDNA breaks that compromises genome stability (2, 3).
Both error-free and error-prone damage avoidance mechanisms have been discovered in eukaryotic cells. In the error-free branch a template switch to sister chromatid after replication fork regression is proposed to ensure accurate DNA synthesis through the lesion (4–6). In the error-prone branch a specialized translesion DNA synthesis (TLS) Pol is believed to release the replication fork blockage by carrying out TLS through the damaged site (7, 8). Although the essential role of the specialized Pols in TLS has been well documented, it is not clear how a specific Pol is selected and how an exchange between replicative and TLS Pols occurs. The answers to these questions are crucial for our understanding of TLS in view that it is essential to restrict the actions of TLS Pol only to the site of DNA damage to avoid further undesirable mutagenesis during genome replication.
The phenomenon of “polymerase exchange” was first uncovered in T4 bacteriophage DNA replication. Using a catalytically impaired T4 replicative Pol (gp43) trap, it was found that gp43 from solution undergoes active exchange with gp43 in the holoenzyme (9). A model was proposed to explain the Pol exchange process in T4 phage based on known x-ray crystal structures of both gp43 and gp45. In this model T4 clamp protein gp45 serves as a platform that interacts with both the resident and the incoming Pols. Because gp45 exists as homotrimer, a transient intermediate with two Pols tethered to the same clamp is possible with no major steric clashes given the flexibility in the gp43 C-terminal tail. Interestingly, DNA Pol exchange has also been shown in the bacteriophage T7 system through an interaction between Pol gp5 and helicase gp4 (10, 11).
A recent study in the Escherichia coli replication system directly demonstrated that the E. coli processivity factor β clamp is able to bind the replicative DNA Pol III and the TLS Pol IV simultaneously (12). Indiani et al. (12) also found that such an intermediate is essential for the exchange between Pol IV and Pol III on DNA and is instrumental for the TLS in E. coli.
In eukaryotes TLS is also indispensable for the fitness of the organism. Elegant genetic studies in Saccharomyces cerevisiae revealed the complex nature of the initiation and regulation of TLS inside the cell (13–15). Hoege et al. (13) found that in the yeast cell TLS function is directly linked to the covalent modification of PCNA by monoubiquitin. In response to DNA-damaging agents, PCNA is ubiquitinated at the conserved Lys-164 residue by Rad6, an E2 ubiquitin (Ub)-conjugating enzyme, and Rad18, a RING finger-containing E3 Ub ligase. It was later established that the monoubiquitination of PCNA by Rad6/Rad18 activates the TLS by Polη (14, 15). Recently, it was found that many Y-family TLS Pols contain an Ub-binding domain (UBD) (16–18), and from coimmunoprecipitation studies, it has been inferred that human Polη interacts with monoubiquitinated PCNA through both the UBD and the PCNA-interacting protein (PIP) motif (16, 18, 19). Despite the recent advance in our knowledge of TLS, the molecular basis of the regulation of TLS in eukaryotes is still not fully understood.
In this study, we demonstrated in a reconstituted system that Pol exchange transpires between the yeast replicative DNA Polδ and the TLS Polη. We also found that an efficient exchange of S. cerevisiae Polδ with Polη requires both the stalling of the holoenzyme and monoubiquitination of PCNA. By using Polη mutants that are defective in the PIP motif we showed that PIP is strictly required for Pol exchange. Furthermore, our results suggest that the monoubiquitin moiety needs to be removed from PCNA after the lesion-bypass synthesis to resume normal DNA synthesis by Polδ. Our results indicate a safety mechanism in regulating the TLS that is crucial to keep the mutagenic load low in eukaryotic cells.
Results
Native Monoubiquitinated PCNA and PCNA-Ub Fusion Protein.
The S. cerevisiae PCNA has been successfully monoubiquitinated at Lys-164 in the reconstituted system comprising Rad6/Rad18, Uba1, and Ub (20, 21). It was found that replication factor C (RFC) and DNA are required for the efficient monoubiquitination of PCNA. Previously, PCNA–Ub fusion proteins were constructed to mimic the native monoubiquitinated PCNA (16, 22). Bienko et al. (16) showed that an in-frame fusion of Ub to the C terminus of PCNA allowed the coprecipitation of human Polι and the chimeric PCNA–Ub from transfected cells. Parker et al. (22) fused Ub to either the N or C terminus of PCNA through a 6-aa linker and found the fusion proteins can partially sustain the DNA damage tolerance pathway in vivo in a yeast strain compromised in PCNA ubiquitination. Here, we have compared the effects of native monoubiquitinated PCNA and the PCNA–Ub fusion protein on Pol exchange.
In this study, the Lys-164 monoubiquitinated PCNA was prepared following the published protocol (20). The reaction solution contained Uba1, Rad6/Rad18, Ub, RFC, and DNA. The DNA was a primer-template DNA oligo with both ends blocked with streptavidin through interaction with the biotin moieties attached to DNA ends. The purification procedure after the ubiquitination reaction ensured that all PCNA trimers contained at least one Ub moiety at Lys-164 (20).
We also constructed a PCNA–Ub fusion protein by genetically fusing the Ub gene in-frame to the C terminus of PCNA. The recombinant protein was purified to homogeneity as described in Materials and Methods. To ensure the PCNA–Ub fusion functioned normally, we first determined whether the protein was loaded onto DNA by RFC by using an ATPase activity assay as described before (23). The results showed that at the same molar concentration both PCNA–Ub and PCNA stimulate the ATPase activity of RFC to a similar level in the presence of a forked DNA substrate, suggesting that fusion of the Ub moiety to the C terminus of PCNA does not affect the normal loading of PCNA onto DNA.
Next, we compared the processivity of the Polδ holoenzyme assembled with either unmodified PCNA or monoubiquitinated PCNA (native monoubiquitinated K164–Ub–PCNA or PCNA–Ub fusion). A singly primed single-stranded M13 DNA (9) was used to measure the processivity of the Polδ–PCNA holoenzyme. RFC was included in the reaction solution for loading PCNA onto the primer end and for the formation of Polδ–PCNA holoenzyme. Processive DNA synthesis was initiated by the addition of a full set of dNTPs. Processive DNA synthesis by Polδ holoenzyme was observed in all cases [supporting information (SI) Fig. S1]. The rate of processive DNA synthesis by the Polδ–PCNA holoenzyme was measured to be ≈110 bp per second on a primed single-stranded M13 DNA coated with E. coli ssDNA-binding protein (SSB). When PCNA was substituted with the PCNA–Ub fusion protein or K164–Ub–PCNA, the rate of processive synthesis was close to the value measured for the unmodified PCNA (see Fig. S1). Therefore, both forms of monoubiquitinated PCNA support normal processive DNA synthesis by Polδ.
The Polδ–PCNA Holoenzyme Is Resistant to a PolδAA Trap.
Previously, an active exchange process was observed for the T4 Pol holoenzyme (9). To test whether a similar Pol exchange occurs for yeast Polδ we constructed a Polδ trap (PolδAA) by mutating the two active-site aspartates of the catalytic subunit of Polδ to alanine (Pol3 D762A, D764A). Polδ is a heterotrimeric protein comprised of subunits Pol3, Pol31, and Pol32. The mutated Pol3 subunit is void of Pol activity (data not shown). However, a residual Pol activity (<5% of WT Polδ activity) was detected for the purified mutant Polδ complex, presumably because of the incorporation of the WT Pol3 subunit endogenous to the S. cerevisiae host cell (the Polδ mutant was overexpressed and purified from the yeast S. cerevisiae cell strain). Nonetheless, the residual Pol activity is unlikely to be problematic for the trapping experiment because the PolδAA mutant preparation contains >95% inactive species and should have a dominant negative effect in the assay.
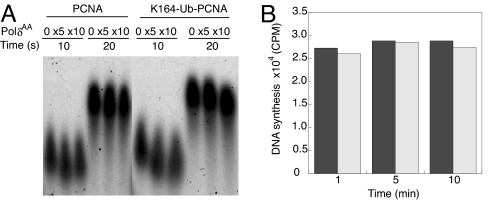
The processive DNA synthesis by Polδ–PCNA holoenzyme in the presence of a PolδAA trap was assayed by using the single-stranded M13 DNA substrate described above. Our results indicated that addition of PolδAA did not significantly affect the processive DNA synthesis by Polδ–PCNA holoenzyme (see Fig. 1A). At the highest PolδAA concentration (10-fold excess to WT Polδ) we still observed a tight band of DNA synthesis product with unchanged size and intensity (Fig. 1A). Conversely, preincubation with the PolδAA trap followed by addition of Polδ showed only background level of DNA synthesis as observed for the PolδAA trap alone (data not shown), indicating the PolδAA trap forms a stable holoenzyme complex with PCNA at the DNA end. These results clearly suggest that the integrity of the holoenzyme is not compromised in the presence of the PolδAA trap. In a parallel experiment using K164–Ub–PCNA, normal processive DNA synthesis was observed in the presence of ×5 and ×10 PolδAA trap. Furthermore, we incubated PolδAA with the assembled holoenzyme for an extended period (up to 10 min) and measured the DNA synthesis by Polδ. The holoenzyme assembled with PCNA or K164–Ub–PCNA showed similar levels of DNA synthesis by Polδ at each time point (Fig. 1B). These results suggest that the attachment of Ub moiety to PCNA does not compromise the stability of the Polδ holoenzyme under our assay condition. Collectively, the absence of trapping by PolδAA suggests that there is no measurable dissociation of Polδ from DNA during this time period.
Fig. 1.
Stability of the Polδ holoenzyme assembled with PCNA or K164–Ub–PCNA. (A) The processive DNA synthesis by Polδ holoenzyme assembled with PCNA or K164–Ub–PCNA in the presence of increasing PolδAA trap concentration is shown. The experiment was carried out under stalled condition (see Materials and Methods). (B) The DNA synthesis by Polδ holoenzyme assembled with PCNA (black bars) or K164–Ub–PCNA (gray bars) after 1, 5, and 10 min of incubation with ×10 PolδAA trap is shown.
Because both the Pol32 and Pol31 subunits of Polδ have been shown to interact with PCNA (24), a binary subcomplex of Pol δ consisting of subunits Pol31 and Pol32 was also tested as a trap in the same exchange experiment. We observed no changes in either the size or the amount of DNA product.
Polη Exchanges with Polδ More Efficiently with Monoubiquitinated PCNA Compared with Unmodified PCNA.
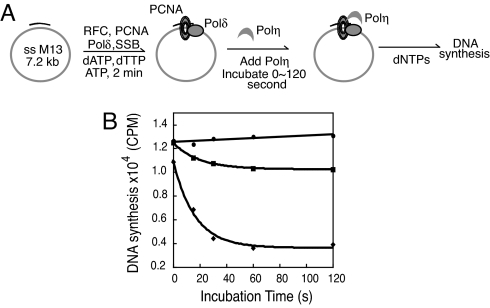
We examined the exchange between Polη and the holoenzyme Polδ under a stalled condition. To mimic this scenario in which the Polδ–PCNA holoenzyme is stalled by a DNA lesion, we assembled the holoenzyme in the presence of two deoxynucleotides (dATP and dTTP). After incorporation of the two deoxynucleotides the holoenzyme stalls because of nucleotide omission.
A control experiment was first carried out to determine the time required for completing the holoenzyme assembling process. By varying the incubation time for Polδ–PCNA holoenzyme formation from 0 to 4 min we found that the holoenzyme assembly was completed within 2 min under our condition. To ensure no unwanted dissociation of the Polδ holoenzyme complex occurred during the time frame of our experiments, the full set of dNTPs was added as a control at different times after the assembling of Polδ–PCNA holoenzyme and DNA synthesis was quantified. As shown in Fig. 2B the final DNA synthesis is constant for all incubation times used, thus suggesting that there is no significant dissociation of the assembled Polδ holoenzyme within the time frame of our experiment.
Fig. 2.
The exchange between Polη and Polδ under stalled condition. (A) Schematic illustration of the reaction sequence is shown. (B) The DNA synthesis by Polδ holoenzyme at different times of incubation with Polη (270 nM) is shown. The DNA synthesis in the presence of PCNA (■) or K164–Ub–PCNA (◆) are compared. In a control reaction (●) PCNA was used and buffer was added instead of Polη.
Next, we introduced Polη after holoenzyme assembly and incubated Polη with assembled Polδ–PCNA holoenzyme. The assay was carried out as depicted in Fig. 2A. Briefly the Polδ–PCNA holoenzyme is assembled on the primed single-stranded M13 DNA. After 2 min Polη at a concentration of 270 nM was added to the solution that contained 6 nM Polδ. The reaction solution was incubated for varying times up to 120 s before the addition of dNTPs to initiate the processive DNA synthesis by Polδ–PCNA holoenzyme for another 20 s. It should be noted that Polη is not processive in DNA synthesis even in the presence of loaded PCNA (25). Therefore, the DNA product synthesized by Polη was not detected by alkaline agarose gel electrophoresis because of its small size. Only the DNA synthesis product by Polδ–PCNA holoenzyme that survives the exchange by Polη was detected and quantified.
We did parallel experiments with both unmodified PCNA and K164–Ub–PCNA. As shown in Fig. 2B, only a small time-dependent decrease in DNA synthesis was observed when unmodified PCNA was used for assembling the Polδ holoenzyme (≈15% decrease in DNA synthesis). However, when K164–Ub–PCNA instead was used, a more pronounced decrease in DNA synthesis was observed. The time-dependent decrease of DNA synthesis was fit to a single-exponential equation to obtain an observed rate constant of 0.06 s−1 for K164–Ub–PCNA. The curve reaches a plateau at 1 min with a ≈60% decrease in DNA synthesis, which suggests that almost two-thirds of holoenzyme undergoes exchange with Polη. Although the amplitude of decrease in DNA synthesis is smaller for unmodified PCNA, the kinetics of the exchange process is similar with a measured rate constant of 0.05 s−1. From the fact that the presence of Polη, but not PolδAA (see Fig. 1), leads to exchange with the Polδ holoenzyme, we infer that this process is active (dependent on the identity of the Pol) rather than passive.
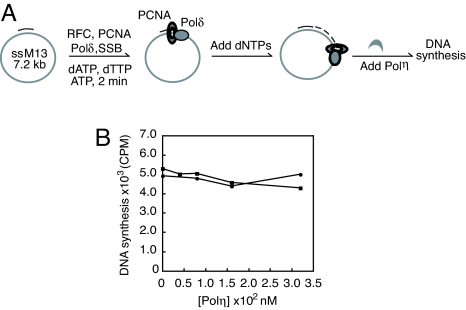
A Moving Polδ–PCNA Holoenzyme Is Resistant to Exchange with Polη.
After demonstrating that a stalled Polδ holoenzyme is prone to exchange with Polη, we investigated how a moving Polδ holoenzyme responds to the challenge by Polη. This was achieved by following the reaction sequence shown in Fig. 3A. The primed single-stranded M13 DNA substrate allows processive DNA synthesis by Polδ holoenzyme up to ≈1 min. In our experiment Polη was added while Polδ was undergoing processive DNA synthesis. In a marked difference from what was observed for a stalled holoenzyme, the presence of Polη at concentrations up to 320 nM did not significantly inhibit the DNA synthesis by Polδ holoenzyme (Fig. 3B). When K164–Ub–PCNA was used for assembling the holoenzyme only a slight decrease in DNA synthesis (<10%) was observed with increasing Polη concentration. A similar trend was observed when the unmodified PCNA was used for assembling the holoenzyme. This observation suggests that a moving Polδ–PCNA holoenzyme is refractory to the exchange with Polη, even with a Polη concentration 50-fold higher than Polδ.
Fig. 3.
The exchange between Polη and Polδ under moving condition. (A) Schematic illustration of the reaction sequence is shown. (B) The DNA synthesis by Polδ holoenzyme in the presence of increasing Polη concentration is shown. The DNA synthesis in the presence of PCNA (●) or K164–Ub–PCNA (■) are compared.
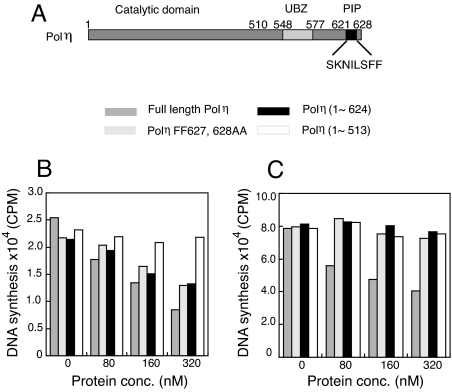
The Requirement of Polη PIP Motif for Pol Exchange.
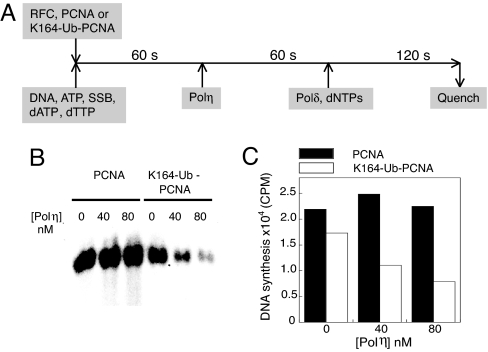
The yeast Polη contains a PIP motif at its C terminus encompassing residues 621–628, which is required for the interaction of Polη with PCNA (25). To probe the role of PIP motif in Pol exchange we used two Polη PIP mutants, Polη (1–624) and Polη FF627,628AA (see Fig. 4A for a schematic illustration of Polη domains). In Polη (1–624) the C-terminal eight amino acid residues of Polη are deleted, and in Polη FF627,628AA the two conserved phenylalanine residues in PIP motif are mutated to alanine. Both mutants were shown to possess identical Pol activity as the WT Polη (25). The exchange experiments were carried out under the stalled condition with increasing concentrations of full-length or mutant Polη. We first used the PCNA–Ub fusion protein for assembling the holoenzyme. The full-length Polη resulted in the largest decrease in DNA synthesis at each protein concentration tested (Fig. 4B). With Polη concentration increasing from 0 to 320 nM an exponential decrease in DNA synthesis (Fig. 4B) was observed. Varying NaCl concentration in the assay buffer from 25 to 100 mM had little effect on the extent of Pol exchange. At the highest Polη concentration a large decrease in DNA synthesis (66% when normalized to the DNA synthesis in the absence of Polη) was observed. When the PIP motif mutants, Polη (1–624) and Polη FF627,628AA, were used, a decrease in DNA synthesis was also observed, albeit to a less extent. The effects of the two PIP mutants were comparable. Approximately a 38% decrease in DNA synthesis was measured at the highest protein concentration of Polη (1–624) and Polη FF627,628AA that was used.
Fig. 4.
The exchange between Polδ with various forms of Polη under stalled condition. (A) Schematic illustration of PIP motif and UBZ domain in Polη sequence is shown. (B) The quantification of DNA product by Polδ holoenzyme assembled with PCNA–Ub in the presence of different forms of Polη as indicated is shown. (C) The quantification of DNA product by Polδ holoenzyme assembled with K164–Ub–PCNA is shown.
In yeast Polη the UBD is an Ub binding zinc finger (UBZ) between residues 548 and 577 (22, 26). We used a truncated Polη (residues 1–513) missing both the PIP motif and UBZ domain to test the effect of the loss of both Polη PIP motif and UBZ domain on Pol exchange. Varying the Polη (1–513) concentration from 0 to 320 nM resulted in no significant decrease in DNA synthesis (see Fig. 4B). Because Polη (1–513) retains normal deoxynucleotidyl transfer activity (27, 28) and thus normal DNA binding ability, the lack of impact of Polη (1–513) on the processive DNA synthesis of Polδ holoenzyme suggests that with the PCNA–Ub fusion protein, both PIP and UBZ domains contribute to Pol exchange.
We also tested the effect of the same Polη mutants on the Polδ holoenzyme assembled with K164–Ub–PCNA. Similar to the PCNA-Ub fusion, with K164–Ub–PCNA, the inhibitory effect of full-length Polη on the DNA synthesis by Polδ holoenzyme was most prominent, although the amplitude of decrease in Polδ synthesis was smaller compared with the PCNA–Ub fusion (≈50% decrease) (Fig. 4C). However, unlike the PCNA–Ub fusion, with K164–Ub–PCNA we observed very little inhibitory effect on Polδ synthesis on the addition of either Polη PIP mutant (see Fig. 4C). Hence our data suggest that the PCNA–Ub fusion does not recapitulate the effects seen for K164–Ub–PCNA.
Monoubiquitination of PCNA Prevents Polδ from Replacing Polη.
The above experiments mimic the exchange between the replicative and the lesion bypass Pols when Polδ holoenzyme is stalled by a DNA lesion. The exchange process recruits Polη to the DNA damage site for lesion bypass DNA synthesis. After the synthesis a second Pol exchange presumably happens to replace Polη with the normal replicative Polδ. To address this process we designed an experiment to mimic the reverse Pol exchange step (see Fig. 5A). After the loading of either unmodified PCNA or K164–Ub–PCNA by RFC onto the singly primed M13 DNA substrate, the full-length Polη was introduced to form a complex with the processivity factor on DNA. Then Polδ and dNTPs were added to initiate the processive DNA synthesis. If Polδ can effectively replace Polη and regain control of PCNA, a processive DNA synthesis should be observed. Our results indicate that when unmodified PCNA was used, the presence of Polη had little effect on DNA synthesis by Polδ–PCNA holoenzyme (Fig. 5 B and C). However, when K164–Ub–PCNA was used, the presence of Polη dramatically reduced the DNA synthesis by Polδ with the largest decrease of 60% at the highest Polη concentration tested. Our results suggest that monoubiquitination of PCNA prevents the back exchange of Polδ.
Fig. 5.
The reverse exchange between Polδ and Polη in the presence of either PCNA or K164–Ub–PCNA. (A) Reaction sequence of reverse exchange between Polη and Polδ is shown. (B) Alkaline agarose gel electrophoresis of the DNA synthesis product by Polδ in the presence of increasing concentration of Polη is shown. (C) The quantification of DNA product as shown in B is depicted.
Discussion
Monoubiquitination of PCNA Does Not Appreciably Reduce the Stability of Polδ Holoenzyme.
In addressing the possible role of PCNA monoubiquitination in TLS we first considered whether the covalent modification of PCNA destabilizes the Polδ–PCNA holoenzyme, and thus facilitates the dissociation of Polδ from DNA. We first compared the processivity of Polδ holoenzyme assembled with unmodified PCNA versus monoubiquitinated PCNA. Under our assay condition monoubiquitination of PCNA does not reduce the processivity of the Polδ holoenzyme, which is in accord with a previous study (21). The similar replicative properties displayed by holoenzyme assembled with either PCNA or monoubiquitin-modified PCNA indicate that the covalent modification of PCNA does not adversely affect the stability of Polδ holoenzyme.
To quantify this notion we used a catalytically impaired Polδ mutant (PolδAA) as a dominant negative trap to probe the stability of Polδ holoenzyme. PolδAA has a double mutation (D762A, D764A) in the active site of the catalytic subunit Pol3. This mutant was purified following the same protocol used for the WT Polδ as a ternary complex of Pol3, Pol31, and Pol32. Because the mutation is localized in the enzyme active site it is unlikely that it alters the enzyme's affinity for PCNA. This notion was confirmed by the experiment showing that PolδAA–PCNA holoenzyme assembled at the DNA primer end prohibited the DNA synthesis by the WT Polδ.
We first demonstrated that the PolδAA trap does not adversely affect the processive DNA synthesis by the Polδ holoenzyme, which is in marked difference from the T4 DNA Pol holoenzyme. This distinction may be understood in view of the structural difference between yeast Polδ and the T4 gp43. Although the processivity factors PCNA and gp45 are highly similar in their 3D structures (both are toroids with a diameter of ≈60 Å), the yeast Polδ (220 kDa) is twice as large as gp43 (100 kDa) and probably has more sites of interactions with its cognate clamp protein than the T4 Pol (24). The bulkiness of Polδ may exclude the simultaneous binding of two copies of Polδ to PCNA, which could be an essential intermediate for the active Pol exchange process as suggested by a previous study (9).
Therefore we were able to use PolδAA as a passive trap protein to probe the stability of the Polδ–PCNA holoenzyme in the presence or absence of PCNA monoubiquitination. If monoubiquitination of PCNA reduces the stability of the Polδ holoenzyme, the presence of PolδAA would disrupt the DNA synthesis by competing with WT Polδ for rebinding to the processivity factor PCNA. However, prolonged incubation of Polδ holoenzyme formed with either PCNA or monoubiquitinated PCNA with the PolδAA trap showed no difference in their time-dependent activity profiles. Thus we conclude that monoubiquitination of PCNA does not appreciably reduce the stability of the Polδ–PCNA holoenzyme.
The Exchange Process Is Regulated by the Movement of Holoenzyme and the Monoubiquitination of PCNA.
Most known TLS Pols, including yeast Polη, are low-fidelity copiers. Therefore it is imperative to restrict the access of TLS Pols to the vicinity of the DNA lesion. The current study identified two molecular events, namely the stalling of holoenzyme and the monoubiquitination of PCNA, as major regulating factors for the Pol exchange step in TLS. Our conclusion partially mirrors the findings from E. coli replication system, showing that low-fidelity Pol IV only gains access to the primer/template DNA when Pol III is stalled (12). Intriguingly, the eukaryotic DNA replication seems to possess another level of complexity in regulating TLS, namely the posttranslational modification of PCNA by Ub. Although previous studies have pointed to monoubiquitination of PCNA as an important step in eukaryotic TLS (14, 15, 29), the current study provides direct biochemical support of this notion in a reconstituted system.
The Role of Polη PIP Motif in Promoting Pol Exchange.
In the current study we used Polη mutants that harbor defects in the PIP motif or carry a C-terminal deletion in which both the PIP and the UBZ domain have been removed. We found that when the PCNA–Ub fusion was used to assemble holoenzyme, alteration in the PIP motif by either mutating the essential hydrophobic residues (Phe-627–Ala, Phe-628–Ala) or truncation of PIP only attenuates Polη's ability to promote Pol exchange, whereas the loss of both the PIP motif and the UBZ domain in Polη completely abolished Pol exchange. However, when the native K164 monoubiquitinated PCNA was used in combination with the Polη PIP mutants, no significant Pol exchange was observed. This observation agrees with a previous genetic study that examined the ability of the same rad30 (residues 1–624) and rad30 FF627,628AA mutant genes to complement the UV sensitivity of the rad5Δ rad30Δ double mutant (25). It was found that both mutant genes are highly defective in complementing the UV sensitivity of rad30Δ mutation. Hence, the binding of PCNA by Polη through its PIP motif is essential for Pol exchange to occur. Thus, although both the PCNA–Ub and K164–Ub–PCNA fusions can promote Pol exchange, our results suggest that they differ in the underlying mechanisms; as, for example, with the PCNA–Ub fusion, the binding of Polη to the Ub moiety apparently overrides the absolute requirement for the PIP motif as is seen for the K164–Ub–PCNA.
At present the interaction between full-length Polη and monoubiquitinated PCNA has not been quantified. A recent NMR study measured a modest binding affinity (≈73–81 μM) between the Polη UBZ domain and Ub moiety (30). However, despite the poor affinity of UBZ for Ub, because the covalently attached Ub moiety is located at the outer rim of PCNA toroid the interaction of Polη with the Ub moiety on PCNA could serve as an initial step in recruiting Polη to the site of DNA damage, followed by the PIP–PCNA interaction.
The Reverse Pol Exchange Step in TLS Requires Deubiquitination of PCNA.
The TLS across a damaged DNA site requires more than one Pol exchange step. After the synthesis past a lesion, a second Pol exchange is needed to restore the replicative DNA Pol. Our results suggest that a complex formed between monoubiquitinated PCNA and Polη masks the DNA primer-template end and blocks Polδ binding to DNA. In contrast, Polδ can readily reform holoenzyme with unmodified PCNA in the presence of Polη. A binary complex between S. cerevisiae Polη and PCNA was detected by size exclusion chromatography (25). Using a FRET approach we measured a dissociation constant of 100 ± 20 nM between Polη and PCNA (Z.Z., unpublished results). Although both experiments were done in the absence of DNA, the physical interaction between Polη and PCNA should be retained on DNA. The stimulation of Polη synthesis activity by PCNA observed on primed M13 DNA is in accord with this notion (25). This stimulation is specific because the alteration of the Polη PIP motif abolished the stimulation. Thus we conclude that even in the absence of monoubiquitin a ternary complex of Polη–PCNA–DNA is readily formed. But in the absence of the Ub modification of PCNA, the Polη–PCNA complex does not exclude rebinding by Polδ.
Although a cocrystal structure of Polη and PCNA is not available, we can gain useful insight from the E. coli Pol IV little finger domain-β clamp costructure. Pol IV is a low-fidelity Pol that shares significant sequence similarity with Polη. The little finger domains of Pol IV and the PAD domain of Polη are highly conserved in their structures. The Pol IV little finger-β clamp structure reveals a nonproductive conformation of Pol IV binding to the β clamp with Pol IV active site angled away from the primer-template DNA (12, 31). Thus it is possible that Polη binds to PCNA in two different conformations, i.e., unproductive versus productive conformations. Monoubiquitination of PCNA may modulate the binding of Polη to PCNA by favoring a productive conformation for Polη. As a result Polη is able to access the primer-template DNA and may effectively compete with the replicative DNA Polδ for the DNA 3′ end.
Monoubiquitination of PCNA Plays Multiple Roles in the Regulation of TLS.
PCNA monoubiquitination could contribute to TLS in multiple ways. First, it might attract a TLS Pol to the site of DNA lesion. Second, monoubiquitination of PCNA may favor a productive replication conformation for Polη. Third, after lesion bypass synthesis the removal of Ub moiety will reduce the affinity of Polη to the DNA end, thus facilitating the return of Polδ. At present we do not know whether Polδ and Polη bind to PCNA simultaneously during the TLS process, similar to what has been demonstrated for E. coli Pol IV and Pol III. However, given the trimeric structure of PCNA and the finding that the exchange depends on the identity of the trapping Pol a tool-belt mechanism may be applicable. If this is indeed the case the exchange between various Pols is determined by which Pol has access to the DNA end. Therefore a conformational change will play an essential role in switching between participating Pols.
Our results also suggest a deubiquitination step is necessary after the successful lesion-bypass DNA synthesis. Although in S. cerevisiae the enzyme that catalyzes such deubiquitination reaction remains elusive, in human USP1 is able to deubiquitinate PCNA (32). The regulatory role of USP1 in damage tolerance has been demonstrated by showing that UV irradiation of cells results in the autocleavage of USP1, which favors the monoubiquitinated form of PCNA (32).
To date, Pol exchange has been observed in T4 bacteriophage, E. coli, and S. cerevisiae. Although a common paradigm regarding Pol exchange can be found in all three systems, the hierarchic nature of its regulation is evident. In T4 phage the Pol exchange between Pol gp43 occurs readily during the whole genomic DNA replication and a regulatory mechanism seems to be lacking. In E. coli the switch between Pol IV and Pol III is regulated by the movement of the holoenzyme that prevents the low-fidelity Pol from introducing unwanted mutation. The eukaryotic replication machinery has adapted a more complex “double-safety” mechanism that involves both the movement of the holoenzyme and the monoubiquitination state of PCNA. The tighter regulation of TLS is essential to maintain the fitness of eukaryotic organism by regulating the extent of error-prone DNA repair.
Materials and Methods
[α-32P]dGTP and [α-32P]dCTP were purchased from PerkinElmer. Unlabeled deoxynucleotides were purchased from Roche Biochemicals. The S. cerevisiae proteins were purified as described (25, 33). To overexpress the WT and catalytically inactive forms of the yeast Polδ holoenzyme, the genes encoding the Pol3 catalytic subunit and the accessory subunits Pol31 and Pol32 were cloned downstream from a GAL–PGK promoter as described (34). GST-tagged WT or the catalytically inactive Polδ complex was expressed and purified from yeast strain YRP654 harboring the plasmids pBJ1244 (GST-Pol31), pBJ1180 (Pol32), and either pBJ1231 (Pol3) or pBJ1259 (Pol3 D762D764 AA) as described (34). The K164–Ub–PCNA and PCNA–Ub fusion proteins were prepared following published procedures (16, 20).
Pol Exchange Assayed Under Stalled Condition.
A 46-mer DNA primer (5′-TCT GAC CTG AAA GCG TAA GAA TAC GTG GCA CAG ACA ATA TTT TTG A-3′) was annealed to M13mp18 ssDNA (positions 5017 to 5062). A typical reaction was carried out in a solution containing 2.3 nM singly primed M13 ssDNA, 6 nM Polδ, 60 nM RFC, 70 nM PCNA or K164–Ub–PCNA, 1.4 μM E. coli SSB, 1 mM ATP, and 100 μM dNTPs. E. coli SSB can be interchanged for yeast RPA in processive DNA synthesis by Polδ–PCNA holoenzyme (35). The reaction solution also contained 25 mM Tris (pH 7.5), 5 mM MgCl2, 10% glycerol, 25 mM NaCl, 1 mM DTT, and 0.1 mg/ml BSA. The Pol exchange was found to be insensitive to different salt concentration (25, 50, 75, and 100 mM NaCl). The reactions were carried out at 37°C. The reaction solution containing primed M13 DNA, Polδ, RFC, PCNA, or K164–Ub–PCNA was incubated with ATP, dATP, and dTTP for 2 min to assemble the Polδ holoenzyme. Either Polη or PolδAA was then added and the reaction solution was incubated for varied times. Lastly, dNTPs with radioactive nucleotide were introduced to initiate the DNA synthesis. The DNA synthesis was allowed to proceed for 20 s before it was stopped by rapid addition of quench solution (500 mM EDTA, pH 8.0). DNA synthesis products were separated by 1.2% alkaline agarose gel electrophoresis and quantified by PhosphorImager (Storm; GE Healthcare Bioscience). The amount of DNA product was reported as cpm.
In another set of experiments, Polη mutants, including Polη (1–624), Polη FF627,628AA, and Polη (1–513), at varied concentrations were added into the reaction solution containing the assembled Polδ holoenzyme. The reaction solution was incubated for 1 min. Then dNTPs with radioactive nucleotide were introduced to initiate the DNA synthesis for 20 s before being stopped by rapid addition of quench solution (500 mM EDTA).
Pol Exchange Assayed Under Moving Condition.
The Polδ holoenzyme was assembled on the singly primed M13 DNA substrate as described above. A mixture of dNTPs with radioactive nucleotide was added to initiate the processive DNA synthesis by Polδ holoenzyme. Ten seconds after the addition of dNTPs Polη with varied concentration was added into reaction solution. The reaction was then allowed to proceed for another 30 s before being quenched by the addition of 0.5 M EDTA solution.
The Reverse Pol Exchange.
A solution containing 25 nM PCNA or K164–Ub–PCNA, 40 nM RFC, 2.3 nM DNA, 450 nM SSB, 1 mM ATP, 25 μM dATP, and 25 μM dTTP were incubated for 1 min to load the PCNA or K164–Ub–PCNA onto DNA. Polη at increasing concentrations (0–80 nM) was then added, and the reaction solution was incubated for 1 min. Lastly, 8 nM Polδ was added with the full set of dNTPs (50 μM) with radioactive nucleotide to initiate the DNA synthesis (2 min).
Acknowledgments.
We thank Dr. Peter M. Burgers (Washington University, St. Louis) for furnishing the RFC sample and critically reading the manuscript. This work was supported by National Institutes of Health Grants GM13306 and CA107650.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801310105/DCSupplemental.
References
- 1.Garg P, Burgers PM. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit Rev Biochem Mol Biol. 2005;40:115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 2.Kraus E, Leung WY, Haber JE. Break-induced replication: A review and an example in budding yeast. Proc Natl Acad Sci USA. 2001;98:8255–8262. doi: 10.1073/pnas.151008198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox MM. The nonmutagenic repair of broken replication forks via recombination. Mutat Res. 2002;510:107–120. doi: 10.1016/s0027-5107(02)00256-7. [DOI] [PubMed] [Google Scholar]
- 4.Broomfield S, Hryciw T, Xiao W. DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat Res. 2001;486:167–184. doi: 10.1016/s0921-8777(01)00091-x. [DOI] [PubMed] [Google Scholar]
- 5.Torres-Ramos CA, Prakash S, Prakash L. Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:2419–2426. doi: 10.1128/MCB.22.7.2419-2426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blastyak A, et al. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 8.Friedberg EC, Lehmann AR, Fuchs RP. Trading places: How do DNA polymerases switch during translesion DNA synthesis? Mol Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 9.Yang J, Zhuang Z, Roccasecca RM, Trakselis MA, Benkovic SJ. The dynamic processivity of the T4 DNA polymerase during replication. Proc Natl Acad Sci USA. 2004;101:8289–8294. doi: 10.1073/pnas.0402625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson DE, Takahashi M, Hamdan SM, Lee SJ, Richardson CC. Exchange of DNA polymerases at the replication fork of bacteriophage T7. Proc Natl Acad Sci USA. 2007;104:5312–5317. doi: 10.1073/pnas.0701062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamdan SM, et al. Dynamic DNA helicase–DNA polymerase interactions assure processive replication fork movement. Mol Cell. 2007;27:539–549. doi: 10.1016/j.molcel.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 12.Indiani C, McInerney P, Georgescu R, Goodman MF, O'Donnell M. A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol Cell. 2005;19:805–815. doi: 10.1016/j.molcel.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 13.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 14.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 15.Haracska L, Torres-Ramos CA, Johnson RE, Prakash S, Prakash L. Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:4267–4274. doi: 10.1128/MCB.24.10.4267-4274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bienko M, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 17.Wood A, Garg P, Burgers PM. A ubiquitin-binding motif in the translesion DNA polymerase Rev1 mediates its essential functional interaction with ubiquitinated proliferating cell nuclear antigen in response to DNA damage. J Biol Chem. 2007;282:20256–20263. doi: 10.1074/jbc.M702366200. [DOI] [PubMed] [Google Scholar]
- 18.Plosky BS, et al. Controlling the subcellular localization of DNA polymerases ι and η via interactions with ubiquitin. EMBO J. 2006;25:2847–2855. doi: 10.1038/sj.emboj.7601178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe K, et al. Rad18 guides pol η to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haracska L, Unk I, Prakash L, Prakash S. Ubiquitylation of yeast proliferating cell nuclear antigen and its implications for translesion DNA synthesis. Proc Natl Acad Sci USA. 2006;103:6477–6482. doi: 10.1073/pnas.0510924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garg P, Burgers PM. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases η and REV1. Proc Natl Acad Sci USA. 2005;102:18361–18366. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker JL, Bielen AB, Dikic I, Ulrich HD. Contributions of ubiquitin- and PCNA-binding domains to the activity of polymerase η in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:881–889. doi: 10.1093/nar/gkl1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhuang Z, Yoder BL, Burgers PM, Benkovic SJ. The structure of a ring-opened proliferating cell nuclear antigen-replication factor C complex revealed by fluorescence energy transfer. Proc Natl Acad Sci USA. 2006;103:2546–2551. doi: 10.1073/pnas.0511263103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansson E, Garg P, Burgers PM. The Pol32 subunit of DNA polymerase δ contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J Biol Chem. 2004;279:1907–1915. doi: 10.1074/jbc.M310362200. [DOI] [PubMed] [Google Scholar]
- 25.Haracska L, Kondratick CM, Unk I, Prakash S, Prakash L. Interaction with PCNA is essential for yeast DNA polymerase η function. Mol Cell. 2001;8:407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- 26.Acharya N, Brahma A, Haracska L, Prakash L, Prakash S. Mutations in the ubiquitin binding UBZ motif of DNA polymerase η do not impair its function in translesion synthesis during replication. Mol Cell Biol. 2007;27:7266–7272. doi: 10.1128/MCB.01196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kondratick CM, Washington MT, Prakash S, Prakash L. Acidic residues critical for the activity and biological function of yeast DNA polymerase η. Mol Cell Biol. 2001;21:2018–2025. doi: 10.1128/MCB.21.6.2018-2025.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trincao J, et al. Structure of the catalytic core of S. cerevisiae DNA polymerase η: Implications for translesion DNA synthesis. Mol Cell. 2001;8:417–426. doi: 10.1016/s1097-2765(01)00306-9. [DOI] [PubMed] [Google Scholar]
- 29.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase η with monoubiquitinated PCNA: A possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 30.Bomar MG, Pai MT, Tzeng SR, Li SS, Zhou P. Structure of the ubiquitin-binding zinc finger domain of human DNA Y-polymerase η. EMBO Rep. 2007;8:247–251. doi: 10.1038/sj.embor.7400901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bunting KA, Roe SM, Pearl LH. Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the β-clamp. EMBO J. 2003;22:5883–5892. doi: 10.1093/emboj/cdg568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang TT, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 33.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Polη. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 34.Johnson RE, Prakash L, Prakash S. Yeast and human translesion DNA synthesis polymerases: Expression, purification, and biochemical characterization. Methods Enzymol. 2006;408:390–407. doi: 10.1016/S0076-6879(06)08024-4. [DOI] [PubMed] [Google Scholar]
- 35.Burgers PM. Saccharomyces cerevisiae replication factor C. II. Formation and activity of complexes with the proliferating cell nuclear antigen and with DNA polymerases δ and ϵ. J Biol Chem. 1991;266:22698–22706. [PubMed] [Google Scholar]