Abstract
The regulation of gene expression by the hormone auxin is a crucial mechanism in plant development. We have shown that the Arabidopsis F-box protein TIR1 is a receptor for auxin, and our recent structural work has revealed the molecular mechanism of auxin perception. TIR1 is the substrate receptor of the ubiquitin–ligase complex SCFTIR1. Auxin binding enhances the interaction between TIR1 and its substrates, the Aux/IAA repressors, thereby promoting the ubiquitination and degradation of Aux/IAAs, altering the expression of hundreds of genes. TIR1 is the prototype of a new class of hormone receptor and the first example of an SCF ubiquitin-ligase modulated by a small molecule. Here, we describe the design, synthesis, and characterization of a series of auxin agonists and antagonists. We show these molecules are specific to TIR1-mediated events in Arabidopsis, and their mode of action in binding to TIR1 is confirmed by x-ray crystallographic analysis. Further, we demonstrate the utility of these probes for the analysis of TIR1-mediated auxin signaling in the moss Physcomitrella patens. Our work not only provides a useful tool for plant chemical biology but also demonstrates an example of a specific small-molecule inhibitor of F-box protein–substrate recruitment. Substrate recognition and subsequent ubiquitination by SCF-type ubiquitin ligases are central to many cellular processes in eukaryotes, and ubiquitin-ligase function is affected in several human diseases. Our work supports the idea that it may be possible to design small-molecule agents to modulate ubiquitin-ligase function therapeutically.
Keywords: chemical biology, TIR1, ubiquitin ligase, SCF, plant hormones
The plant hormone auxin is a core regulator of plant growth and development. By controlling cell division and elongation and triggering specific differentiation events, indole-3-acetic acid (IAA), the predominant naturally occurring auxin, regulates developmental phenomena as diverse as embryo polarization, vascular differentiation, tropic response to light and gravity, and apical dominance, among many others (1). Auxin's influence over these manifold events stems from its ability to regulate differentially the expression of hundreds of genes. Central to this transcriptional control is the auxin-enhanced and ubiquitin-mediated degradation of a family of transcriptional repressor proteins called Aux/IAAs (1). Recent work has identified TIR1 and related AFB proteins as auxin receptors for this response (2–4). TIR1 is an F-box protein and a component of the E3 ubiquitin ligase complex SCFTIR1. E3 ubiquitin ligases are responsible for catalyzing the ubiquitination of target proteins, which in most cases results in the rapid degradation of the target proteins in the 26S proteasome. SCF-type E3s consists of three comportments, a SKP subunit (termed ASK proteins in Arabidopsis), and CULLIN subunit and the F-box protein (5). It is the F-box protein component, of which there are >700 in Arabidopsis, that is responsible for the recruitment of specific protein substrates (6). In the case of TIR1, the Aux/IAA targets are recruited via an interaction between the leucine-rich repeat (LRR) domain of TIR1 and a short so-called “degron” motif within domain II of the Aux/IAA (7). This interaction is enhanced by the direct binding of auxin to TIR1 make the SCFTIR1–auxin–Aux/IAA complex the hub of the fundamental derepression model for auxin-regulated control of gene expression: When auxin levels are low, Aux/IAAs are relatively stable and able to exert repression on target genes. As auxin levels rise, TIR1-mediated proteolysis of Aux/IAAs relieves this repression, and genes are expressed (5).
Recent structural analysis of the receptor protein TIR1 in complex with auxin and the Aux/IAA domain II peptide has revealed the molecular mechanism of auxin perception. Both auxin and Aux/IAA bind to the same pocket formed by the LRRs on the surface of TIR1. Auxin nestles on the floor of this common pocket, whereas the Aux/IAA binds on top of auxin, trapping it underneath. In this way, auxin is thought to act as a “molecular glue” by increasing the extent of hydrophobic interaction among the three components (8).
The vast majority of the molecular genetic research on which our understanding of auxin signaling is based has come from work on the model plant Arabidopsis. Beyond this dicot model, the role of auxin in the growth and development of diverse plant species is unclear. Assessing the physiological role of auxin in these species is often hampered by lack of genetic tools to dissect auxin response. To address this problem and to complement molecular genetic approaches to auxin study, we have taken a chemical biology approach, by generating specific auxin signaling probes that can be used to dissect auxin signal transduction (9, 10).
Many structural and biological investigations of auxin compounds have led to the development of several synthetic auxins, including 2,4-dichlorophenoxy acetic acid (2,4-D) and 1-napthalene acetic acid (NAA). In contrast, there are few reports on antiauxinic compounds. p-Chlorophenoxy isobutylic acid (PCIB) is used as an antiauxin although it does not antagonize all auxin responses, and its mode of action is not clear (11, 12).
Here, we report a series of systematically designed small-molecule agonists and antagonists of TIR1 receptor function. By introducing different alkyl chains to the α-position of IAA, we have generated specific TIR1 angonists and antagonists that modulate all tested TIR1-mediated auxin responses in Arabidopsis, from the molecular to the whole-plant level. The mode of action of these small-molecule probes is demonstrated by x-ray crystallography of the TIR1–probe complexes. We then use these probes to provide preliminary evidence of an ancient functional TIR1-mediated auxin response mechanism in the lower plant P. patens (moss). Our studies demonstrate not only the power of chemical biology in dissecting the auxin response beyond model plant species but also provide an example of a specific small-molecule inhibitor of substrate recognition by an F-box protein containing ubiquitin ligase.
Results
α-Alkyl IAA Probes Are Specific to TIR1.
Although extensive structure-activity analysis of modifications to the indole ring of IAA has failed to identify experimentally useful antiauxin and auxin derivatives, the effect of substitutions at the α-position of IAA on auxin activity has not been investigated systematically. To examine the effect of such modifications, a series of α-alkyl chains were introduced to the α-position of IAA (Fig. 1A). Synthesis procedures are described in supporting information (SI) Appendix. The α-alkyl IAAs were prepared as racemic mixtures at α-position, and the racemate was used for biological evaluation because of the difficulty of separating each enantiomer.
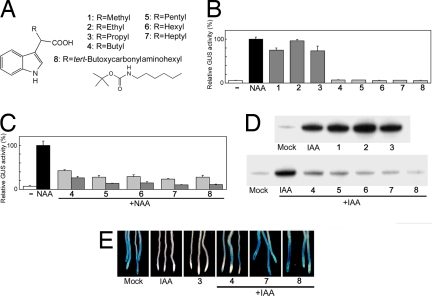
Fig. 1.
α-Alkyl IAAs act on TIR1-mediated auxin signaling. (A) The chemical structures of probes 1–8. (B and C) Effects of the probes on auxin-responsive gene expression. Arabidopsis DR5::GUS reporter line was treated with/without chemicals for 5 h. The induced GUS activity is expressed relative to 1 μM NAA treatment (100%). Error bars, mean +/− SD of three independent experiments. (B) Auxin activity of 1–3 (10 μM) and 4–8 (50 μM). (C) Antiauxin activity of 4–8 (light gray, 20 μM; dark gray, 50 μM). (D) The effect of probes on the Aux/IAA–TIR1 interaction. c-myc-tagged TIR1 was pulled-down using biotinylated IAA7 domain II peptide in the presence of chemicals (100 μM probes and/or 0.5 μM IAA), and recovery of TIR1-myc was monitored by Western blot analysis with anti-c-Myc antibody. (E) The effect of probes on Aux/IAA stability. The HS::AXR3NT-GUS line was incubated with chemicals (2 μM 3, 1 μM IAA, and/or 50 μM 4–8) after heat-shock induction. GUS expression was visualized by histochemical GUS staining.
To examine the auxin and antiauxin activity of the probes, we used the Arabidopsis auxin response reporter line DR5::GUS, which has a synthetic auxin responsive promoter driving GUS expression (13). The IAAs with methyl to propyl chain substitutions (probes 1–3) at the α-position induced DR5::GUS expression demonstrating that these molecules retain auxin activity in this assay (Fig. 1B). In contrast, the introduction of butyl or longer chains (probes 4–8) to IAA abolished auxin activity. Further, these probes inhibited NAA-induced DR5::GUS expression (Fig. 1C), indicating that the introduction of butyl or longer chain (4–8) confers an antiauxin activity to these new molecules. The potency of the antiauxin activity of 4–8 was approximately proportional to chain length.
To test the idea that these probes act by affecting the binding of Aux/IAAs to the receptor TIR1, we performed in vitro pulldown assays in the presence and absence of both IAA and each of the probes using a biotinylated Aux/IAA domain II degron peptide and c-myc-tagged TIR1 (14). Fig. 1D shows that probes 1–3 have auxinic activity in this assay, enhancing the interaction between TIR1 and Aux/IAA domain II peptides. In contrast, probes 4–8 (butyl or longer chains) blocked the IAA-enhanced interaction, indicating that these molecules are antiauxins blocking the TIR1-Aux/IAA interaction (Fig. 1D). Consistent with DR5::GUS reporter analysis, this inhibitory activity was also proportional to alkyl chain length, with probe 8 exhibiting the most potent antiauxin activity. To confirm the effects of the probes on TIR1–Aux/IAA interaction in vivo, Aux/IAA stability was monitored by using the Arabidopsis HS::AXR3NT-GUS line, in which a translational fusion between domains I and II of the Aux/IAA AXR3 and the GUS reporter protein is expressed under the control of a heat-shock promoter (15). HS::AXR3NT-GUS seedlings were treated with the probes in the presence or absence of IAA after heat induction. After a 20-min incubation, probe 3 enhanced the degradation of the fusion, whereas probes 4, 7, and 8 blocked IAA-enhanced degradation (Fig. 1E). Together, these data indicate that these auxin and antiauxin molecules act at the level of TIR1–Aux/IAA interaction by mimicking or interfering with auxin activity.
Antiauxin Probes Block Auxin Response via the SCFTIR1–Aux/IAA Pathway.
To examine further the biological activity of the antiauxin probes, we evaluated their effects on typical auxin responses in the Arabidopsis root. Auxin inhibits primary root growth while promoting root hair and lateral root formation (1). Consistent with its activity at the molecular level, probe 8 antagonized these root responses to auxin, suppressing the inhibition of primary root (Fig. 3 A e and f and B) and the promotion of root hair and lateral root formation (Fig. 2 A and B and Fig. S2). Probes 4 and 7 similarly antagonized these three root auxin responses (Figs. S1 and S2 in SI Appendix). For further detailed analysis of antiauxin action, probe 8 was used because of its higher potency. In the same bioassays, probe 3 continued to exhibit auxin activity (Figs. S1 and S2). Auxin is an essential regulator of the root gravitropic response and is also required for the proper development of columella cells in which the gravity is perceived (1). To test the effects of 8 on gravitropism, 5-day-old Arabidopsis seedlings were transferred onto agar plates with or without both IAA and 8. The plates were then rotated 135° and cultured for another 2 days in the dark. As shown in Fig. 2C, probe 8 perturbed gravitropic response of Arabidopsis roots although this effect could be rescued by exogenous IAA.
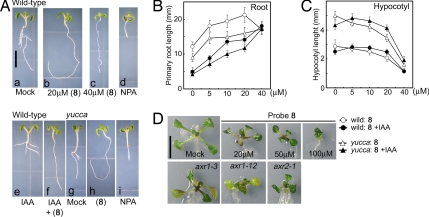
Fig. 3.
The effects of probe 8 on the growth and development of Arabidopsis. (A–C) The effects of 8 on the elongation of hypocotyl and root in wild-type and auxin overproducing yucca mutants. The seedlings were grown for 6 days in the presence of chemicals. Five μM NPA and 50 nM IAA were used for assays unless otherwise stated. (A) Photos of Arabidopsis seedlings grown in the presence or absence of chemicals (a–f, wild-type; g–i, yucca mutant). Plants grown with 8 (b, c, and h), with NPA (d and i), or with IAA and/or 20 μM 8 (e and f). (B and C) Root and hypocotyl length was measured after cultivating with 8 and with and without 50 nM IAA. Values are the mean +/− SD of two independent experiments. (D) Wild-type plants treated with 8 phenocopy auxin-insensitve mutants, axr1–3, axr1–12, and axr2–1. Wild type were grown for 14 days with or without 8. Mutants were grown for 14 days without 8. (Scale bars, 5 mm in A and 10 mm in D.)
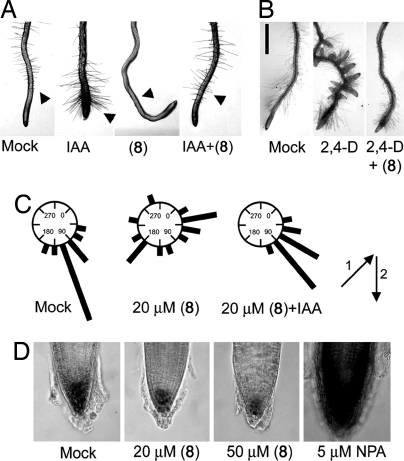
Fig. 2.
The antiauxin probe 8 blocks typical auxin responses in the Arabidopsis root. (A) Probe 8 blocks auxin-induced root hair formation. Five-day-old seedlings were cultured with chemicals (0.1 μM IAA and/or 20 μM 8) for 36 h. Arrows indicate root hair. (B) Effect of 8 on auxin-induced lateral root formation. Five-day-old seedlings were cultured with chemicals for another 4 days. Left, untreated root; Center, treated with 0.2 μM 2,4-D; Right, treated with 0.2 μM 2,4-D and 20 μM 8. Scale bar, 10 mm. (C) Effects of 8 on root gravitropic response. Five-day-old seedlings (n = 15) were grown in the dark for 2 days after rotating seedling 135° angle along their growth axis. The arrows indicate the vector of gravity before (i) and after (ii) the start of gravistimulation. The root angles were plotted on circular histograms at 20° intervals. Assays were performed in duplicate. (D) Root tip grown for 5 days in the presence of 8 and NPA at the concentrations indicated. Lugol staining was used to show starch granules in columella cells.
Although auxin transport inhibitors such as 1-N-naphthylphthalamic acid (NPA) can elicit similar gravitropic defects (16), other developmental abnormalities arising from inhibition of auxin transport are very different (Fig. 3 A a and d). For example, NPA induces abnormal columella cell positioning by perturbing auxin transport, whereas probe 8 affects columella cells by reducing their number but not their positioning (Fig. 2D), suggesting probe 8 action is distinct from that typical of auxin transport inhibitors such as NPA. The antiauxin activity of probe 8 is not confined to root responses to auxin. The etiolation response of Arabidopsis hypocotyls is strongly inhibited by auxin but could be fully recovered by cotreatment with 8 (Fig. S3).
To assess the effects of probe 8 on responses to endogenous auxin in Arabidopsis, wild-type and yucca mutant plants (which have an auxin overproduction phenotype) (17) were grown in the presence of 8, with or without exogenous IAA. yucca mutants have shorter root and longer hypocotyls than wild-type because of elevated endogenous auxin levels (Fig. 3 Ag, B, and C). Probe 8 promoted root elongation in wild-type and yucca in a dose-dependent manner, indicating that endogenous auxin action is being antagonized (Fig. 3 A b, c, and h, and B). Exogenous IAA (50 nM) counteracted this effect of probe 8 on root elongation in both WT and yucca (Fig. 3B). In contrast to root response, probe 8 blocked endogenous auxin action to inhibit WT and yucca hypocotyl elongation in a dose-dependent manner (Fig. 3 A b, c, and h, and C). The control of hypocotyl growth with respect to auxin is complex (18). Although yucca mutants have longer hypocotyls than WT, both genotypes exhibit reduced hypocotyl elongation in response to exogenous auxin (Fig. 3C), suggesting that in each background, there is an optimal level of auxin with compensatory changes in response in the yucca mutant. This is supported by the observation that the dose–response curve of hypocotyls treated with 8 and exogenous IAA displayed a bell-shaped response in both WT and yucca mutant, but higher concentrations of 8 (40 μM) inhibited hypocotyl growth to the same extent in all conditions (Fig. 3C). Probe 8 (20 μM) was sufficient to almost fully rescue the yucca phenotypes under our conditions (Fig. 3Ah). Together, these experiments support the idea that the antiauxin 8 antagonizes responses to endogenous auxin.
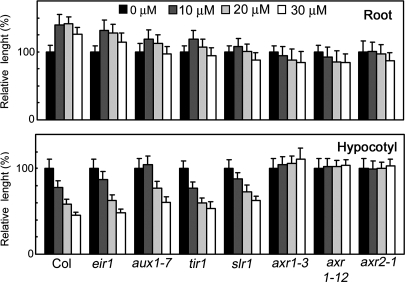
Several Arabidopsis mutants with specific defects in the TIR1-Aux/IAA pathway have been identified (1, 19). These range from the strongly auxin-resistant mutants axr1–3, axr1–12, and axr2–1 to the more moderately resistant mutants tir1–1 and slr-1. The strong auxin resistance phenotypes of axr1–3, axr1–12, and axr2–1 mutants were phenocopied by the treatment of WT plants with 20–50 μM probe 8 (Fig. 3D) consistent with the idea that probe 8 blocks TIR1 function in vivo. This is further supported by the finding that strongly auxin resistant mutants such as axr1–12 and axr2–1 were also highly insensitive to 8, whereas tir1–1 and slr-1 were more moderately insensitive (Fig. 4) presumably because the effect of 8 on the growth of wild-type hypocotyls and roots depends on the response to endogenous IAA, and this response is attenuated in each mutant. In contrast, auxin transport mutations affecting either auxin influx (aux1–7) or auxin efflux (eir1) displayed only weak or no resistance to 8, suggesting that probe 8 acts to disrupt auxin signaling principally by affecting the initial auxin perception event.
Fig. 4.
Effects of 8 on the growth of Arabidopsis auxin mutants. Arabidopsis wild type and mutants were grown for 6 days. Upper shows the effect of increasing concentrations of 8 on root length relative to untreated control (100%). (Lower) Hypocotyl length relative to untreated control (100%). Values are the mean +/− SD of two independent experiments.
Molecular Mechanism of Auxin Probe Action in Auxin Perception.
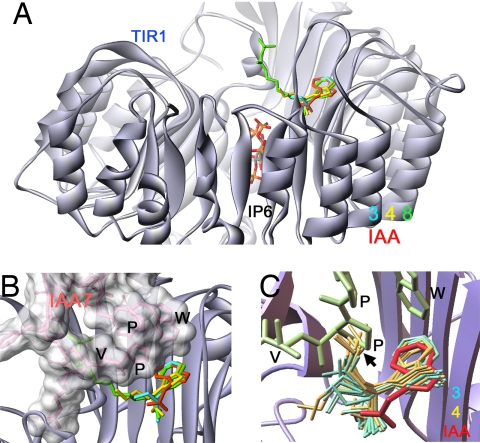
To understand the molecular mechanism of the α-alkyl probes, we solved the crystal structures of TIR1 (and ASK1) in complex with probes 3, 4, and 8 (Table S1 in SI Appendix) and also carried out molecular docking calculations using the program AutoDock (20). In the crystal structures of the TIR1 complexed with 3 (auxin), 4, or 8 (antiauxin), the indole 3-acetic acid moiety of three probes sits in the auxin-binding pocket of TIR1 in the same way as IAA (8), whereas the alkyl chain of the probes is oriented to the Aux/IAA-binding cavity (Fig. 5 A and B and Fig. S4). We could not determine the exact position of alkyl chains beyond β-carbon in alkyl chain (the one next to α-position) because of weak electron density of alkyl carbons (Fig. S4). The coordinates of alkyl chain are represented as one of the most likely chain conformations (Fig. 5 A and B). IAA and IAA7 molecules were superimposed into experimentally determined coordinates (8). Interestingly, all bound probes had S-form stereochemistry even though racemic probes were used for crystallization, indicating that it is the S-form, which binds, and is thus more potent than the R-form. These structures indicate that the alkyl chains is significantly disordered and oriented into Aux/IAA binding cavity. Consistent with crystal structure of auxin-bound TIR1, no conformational change in TIR1 was observed among three complexes (with auxin 3 or antiauxin 4 and 8) supporting the molecular glue hypothesis of Tan et al. (8).
Fig. 5.
Crystal structure and molecular docking analysis of TIR1–probe complexes. (A and B) Crystal structure of TIR1–probe complexes. TIR1 is shown as silver ribbon. Probes 3, 4, and 8 are shown as blue, yellow, and green, respectively. IAA7 degron peptide (pink, surface-filled model) and IAA (red) were superimposed on the coordinates in the crystal structure of the TIR1-IAA-IAA7 complex. (C) Molecular docking of TIR1 probe. Predicted binding conformers of 3 (blue) and 4 (yellow) to TIR1 auxin-binding site. Fifty possible binding conformers were predicted by the program AutoDock. Ten representative conformers were shown based on rmsd values to the coordinates of IAA moiety in 3 and 4 in crystal structure.
Molecular docking calculation analysis complemented the crystallographic analysis of the TIR1–probe complexes. Docking analysis predicted the possible conformers of the three probes in the TIR1 auxin/Aux/IAA-binding site. The predicted binding conformers of 3 and 4 are indicated as blue and yellow colored molecules, and red- and green-colored molecules represent IAA and IAA7 superimposed into the coordinates in crystal (Fig. 5C). In the predicted binding position of 3 and 4, the IAA moiety of both probes sits in the TIR1 groove in a manner similar to IAA. Consistent with crystal structures, alkyl chains are disordered (Fig. 5C). The butyl chains of 4 (antiauxin) were directed to the Aux/IAA-binding cavity, where they would prevent access of the domain II Aux/IAA degron in the binding cavity. The butyl chain conflicts with the second proline in the IAA7 core degron sequence GWPPV, which is critical for Aux/IAA recognition by TIR1. In contrast, the propyl chain of 3 (auxin) fits into a small vacant cavity without hindering subsequent binding of the Aux/IAA (Fig. 5C). Thus, the addition of a methyl group in the butyl chain of 4 is all that is required to convert an auxin to an antiauxin. This was further confirmed by auxinic and antiauxinic activity of another two types of α-alkyl auxins, α-alkyl NAA, and 2,4-D. Previous structural work has demonstrated that NAA and 2,4-D bind to TIR1 with a binding conformation similar to IAA (8). Therefore, it is predicted that the introduction of α-alkyl chain to two different types of auxins would confer similar auxin/antiauxin properties to the α-alkyl IAA derivatives. Consistent with our structural analysis, α-methyl to ethyl NAA and α-methyl to propyl 2,4-D retained auxin activity in DR5::GUS expression assays, whereas the introduction of over butyl chain showed antiauxin activity as observed with probes 4–8 (Figs. S5 and S6). The activity of propyl NAA was more complex showing both weak auxin activity in the absence of NAA and weak antiauxin activity in the presence of NAA. This reflects the fact that some conformers of propyl NAA with the disordered chain are predicted to allow the molecule to occupy the auxin-binding site without blocking the Aux/IAA pocket, whereas other conformers do not.
The crystal structure of TIR1 complexed with 8 shows clearly that the probe would block Aux/IAA access (Fig. 5B). The predicted binding conformers of 8 are shown in Fig. S7. The IAA moiety of the conformers is consistent with experimental coordinates of 8. The alkyl chain is more disordered than probe 4 because of a longer chain length and would hinder the access of Aux/IAA more effectively (Fig. S7). These data explain, at molecular level, both why 3 and 4 have opposite activities (agonist/antagonist), and why probe 8 is more potent than 4.
Auxin-binding protein 1 (ABP1) is a candidate auxin receptor of unknown function, distinct from the TIR1/AFB family. The crystal structure of ABP1–auxin complex (21) has demonstrated that auxin binds to small pocket in ABP1 (Fig. S8A). Molecular modeling analysis revealed that probe 8 could not bind to ABP1 because of the bulky alkyl chain of the probe (Fig. S8B), suggesting that probe 8 could not block ABP1 function and that probe 8 is specific to TIR1 and TIR1-related AFB receptors.
SCFTIR1 Pathway Is Conserved in Lower Land Plants.
To confirm the utility of the probes for studying auxin response in other plant species, we examined the effect of 8 on auxin responses in the monocot grass Oryza sativa (rice) and the moss P. patens. As expected from the high homology of the rice TIR1 orthologue to Arabidopsis TIR1, probe 8 blocked typical auxin responses in rice, such as lateral root and root hair formation (Fig. S9). The moss P. patens is a lower-land plant, and available sequence data suggests that a functional TIR1-Aux/IAA pathway could be conserved in moss. Moss is therefore an attractive model in which to assess the early evolution of auxin signaling mechanisms. P. patens shows two distinct developmental stages, the protonema, a filamentous network of the chloronemata and caulonemata; and the gametophore, a leafy shoot. The gametophore is developed from a subapical cell of the caulonema (22). In P. patens, exogenous auxin is known to elongate the gametophore and promote the formation of rhizoids and caulonemata (23). The P. patens genome contains a TIR1 orthologue (PpTIR1) with high homology to TIR1. Homology modeling indicates auxin-binding site of TIR1 is conserved in PpTIR1 (Fig. 10A). Molecular docking calculation between PpTIR1 and 8 indicated that 8 could bind to PpTIR1 in similar way to TIR1 (Fig. S10B).
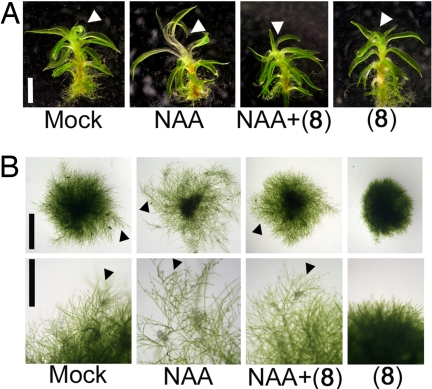
We investigated the effects of 8 on P. patens auxin responses as a potential means of probing the existence of functional PpTIR1-mediated auxin signaling pathway in moss. Probe 8 antagonized NAA-induced gametophore elongation (Fig. 6A). Additionally, probe 8 inhibited the elongation of chloronema cell and the formation of gametophore by blocking the transition from chloronema to caulonema (Fig. 6B and Fig. S11). Probe 8 also antagonized the enhancement by NAA of this developmental transition from chloronema to caulonema cells (Fig. 6B and Fig. S11). Auxin application promotes the transition gametophore bud cells to rhizoid-like bud structure and probe 8 also blocked this process (Fig. S11). Taken together with homology modeling and molecular docking calculation, these findings support the idea thus that auxin response mediated by TIR1 and Aux/IAA orthologues is an ancient mechanism. Interestingly, this contrasts with the lack of a functional gibberellin receptor-DELLA GA response pathway in moss, despite the existence of apparent homologs for both components (24). The developmental role of auxin and its means of perception and signal transduction in other lower land plants such as liverworts and ferns is unknown, and our probes offer useful tools for the study of auxin-regulated development across diverse plant species.
Fig. 6.
The TIR1/AFB specific probe 8 blocks auxin responses of moss P. patens. (A) Effects of 8 on NAA-induced elongation of P. patens gametophores. The juvenile gametophore was incubated for 60 h with chemicals (2 μM NAA and/or 20 μM 8). Arrows indicate the elongation zone in response to NAA. (Scale bar, 10 mm.) (B) Effects of 8 and NAA on the development of chloronemata. Chloronema cells were cultured on a BCDATG medium for 10 days in the presence of 0.5 μM NAA and/or 10 μM 8. Arrows indicate caulonemata. [Scale bars, 2.5 mm (Upper) and 1 mm (Lower).]
Discussion
We have designed a set of molecules that modulate, either positively or negatively, the interaction between TIR1 (and AFBs) and Aux/IAAs and hence transcriptional responses to auxin. These auxin agonists and antagonists affect all assayed TIR1-mediated responses to auxin. By x-ray crystallography of TIR1 in complex with the probes, we show how binding events at the receptor relate to these in vitro and in vivo responses. Further, we demonstrate the utility of these molecules for probing auxin response in species in which the role of auxin and the mechanism of its perception are unclear.
The probe concentrations required to block IAA activity are relatively high (10–50 μM probe 8 against 0.1–2 μM IAA). There are three possible reasons for this. First, the long and disordered alkyl-chain of probes such as 8 would probably decrease the affinity of the probe to TIR1 compared with IAA. Second, the structural analysis of TIR1–probe complex revealed that the S-form of the probes are the predominant binding enantiomer, suggesting that pure S-form probes would show higher activity than the racemates assayed here. Finally, there are no affinity data available for the binding of IAA to TIR1 in the absence of Aux/IAA protein. A prediction of the molecular glue hypothesis would be that without Aux/IAA binding on top of the TIR1-bound IAA, the TIR1-IAA affinity would be relatively low (8). Because the alkyl chains of the antiauxins such as 8 preclude Aux/IAA binding, much higher concentrations of antiauxin would be required to compete effectively with IAA, which is retained more readily in the tight complex of Aux/IAA-auxin-TIR1.
Given that auxin-induced interactions between the TIR1-related AFB proteins and Aux/IAAs have also been demonstrated, we predict that our probes will also act on these related receptor proteins (2, 5). In addition to binding to the TIR1/AFB receptors, there is a possibility that our probes could act in other ways, for example, by interacting with other receptors or affecting auxin transport. The evidence presented here suggests that this is not the case: structural modeling of the only other candidate auxin receptor, ABP1, clearly shows that our antiauxin probes cannot bind to its auxin-binding site. Also, although some of the effects of probe 8 are similar to those of auxin transport inhibitors such as NPA, most effects are different. For example, NPA enhances DR5 response in the presence of auxin, whereas probe 8 has the opposite effect. Together, the molecular and structural data, these observations suggest that these α-alkyl IAA molecules are effective TIR1/AFB-specific probes.
Our work also has implications beyond plant biology. The F-box protein TIR1 is the first example of an entirely new class of receptor in which the activity of an otherwise generic mechanism for protein ubiquitination, common throughout eukaryotes, is regulated by the binding of a small molecule (in this case auxin). The vast majority of the hundreds of F-box proteins identified remain uncharacterized and their target proteins unknown (6). A second example of an F-box protein acting as a small-molecule receptor has already been identified (25), suggesting it is likely that this mechanism will be more widespread. Our results thus provide not only a useful tool for plant chemical biology but also demonstrate an example of a specific small-molecule inhibitor of F-box protein-substrate recruitment. Substrate recognition and subsequent ubiquitination by SCF-type ubiquitin ligases is central to many cellular processes in eukaryotes and ubiquitin-ligase function is affected in several human diseases including Parkinson's disease and certain cancers (26, 27). Our work substantiates the idea that it may be possible to design small-molecule agents to ameliorate ubiquitin ligase function in human diseases. Such drugs could be formulated for maximal specificity and offer an exciting opportunity for pharmaceutical research.
Materials and Methods
Materials and Plant Growth Conditions.
Full details of synthetic procedures and the spectroscopic data of probes are described in SI Appendix. Arabidopsis thaliana Columbia and Physcomitrella patens were cultured at 24°C under continuous light in this study. Arabidopsis plants were grown on a dish containing germination (GM) medium (9) with 0.1% gellan gum (Sigma) containing the indicated hormone and/or chemicals for the indicated time. P. patens was maintained as protonemata on a plate containing BCDATG medium (28). To observe the auxin response of gametophores, the gametophores were cultured on a plate with BCD medium (28) with 0.1 mM CaCl2 and 0.1% gellan gum for 60 h.
Assays.
For the β-glucuronidase (GUS) reporter assay, 5-day-old DR5::GUS seedlings (n = 10−12) were incubated in liquid GM medium for 5 h with or without chemicals. Induced GUS activity was determined fluorometrically. For pulldown assay with Aux/IAA and c-myc tagged TIR1, pulldown assays with the biotinylated Aux/IAA degron peptide were performed as described (14). The Aux/IAA protein degradation assay was assayed as described in ref. 15. Briefly, 8-day-old HS::AXR3NT-GUS transgenic seedlings after heat shock (2 h, 37°C and then 10 min, 23°C) were treated with chemicals for another 20 min. GUS activity was histochemically stained. Root gravitropic response assay were performed as described (10). For hypocotyl and root assays (n = 25–40), the length of hypocotyl and root was recorded by digital camera and analyzed by the program NIH Image (National Institutes of Health) after 6-day cultivation.
Crystallographic Study and Molecular Docking Analysis.
TIR1–ASK1 complex purification and crystallization were performed as described (8). Different probes were soaked into the crystals by placing the crystals in the crystallization well buffer supplemented with 0.5 mM probes for 3 days. Diffraction datasets were collected at the BL5.0.2 beamlines at the Advanced Light Source in Berkeley, CA, using crystals flash-frozen in crystallization buffers supplemented with 15–20% glycerol at −170°C. The structures were determined as described (8). The program AutoDock 3.0 (20) was used for the molecular docking calculation. The grid box was set to cover the whole active site of TIR1 (56′56′56′ grid with a grid spacing of 0.375 Å). In each case, 50 docking runs were performed by using the Lamarckian genetic algorithm. A composite file of all possible conformers was analyzed by AutoDock Tools (29).
Acknowledgments.
We thank Dr. Yunde Zhao (University of California, San Diego) for the yucca mutant, Dr. Mitsuyasu Hasebe and Yusuke Hiwatashi (National Institute of Basic Biology, Okazaki, Japan) for helpful suggestions for the P. patens culture, and Dr. Mark Estelle (Indiana University, Bloomington) for helpful suggestions. This work was supported by Japan Society for the Promotion of Science Grant-in-Aid for scientific research (Grant 18510197, to K.-i.H.), a Royal Society Research Grant (S.K.), and the Pew Scholarship (N.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The structure coordinates and structural factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3C6N, 3C60, and 3C6P).
This article contains supporting information online at www.pnas.org/cgi/content/full/0711146105/DCSupplemental.
References
- 1.Woodward AW, Bartel B. Auxin: regulation, action, and interaction. Ann Bot (London) 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 3.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 4.Dharmasiri N, et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 5.Kepinski S. The anatomy of auxin perception. BioEssays. 2007;29:953–956. doi: 10.1002/bies.20657. [DOI] [PubMed] [Google Scholar]
- 6.Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA. 2002;99:11519–11524. doi: 10.1073/pnas.162339999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dharmasiri N, Dharmasiri S, Jones AM, Estelle M. Auxin action in a cell-free system. Curr Biol. 2003;13:1418–1422. doi: 10.1016/s0960-9822(03)00536-0. [DOI] [PubMed] [Google Scholar]
- 8.Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi K, et al. Yokonolide B, a novel inhibitor of auxin action, blocks degradation of AUX/IAA factors. J Biol Chem. 2003;278:23797–23806. doi: 10.1074/jbc.M300299200. [DOI] [PubMed] [Google Scholar]
- 10.Yamazoe A, Hayashi K, Kepinski S, Leyser O, Nozaki H. Characterization of terfestatin A, a new specific inhibitor for auxin signaling. Plant Physiol. 2005;139:779–789. doi: 10.1104/pp.105.068924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oono Y, et al. p-Chlorophenoxyisobutyric acid impairs auxin response in Arabidopsis root. Plant Physiol. 2003;133:1135–1147. doi: 10.1104/pp.103.027847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biswas KK, et al. Genetic Characterization of Mutants Resistant to the Antiauxin p-chlorophenoxyisobutyric Acid (PCIB) Reveals that AAR3, a Gene Encoding DCN1-Like Protein, regulates responses to the synthetic auxin 2,4-dichlorophenoxyacetic acid in Arabidopsis roots. Plant Physiol. 2007;145:773–785. doi: 10.1104/pp.107.104844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kepinski S, Leyser O. Auxin-induced SCFTIR1-Aux/IAA interaction involves stable modification of the SCFTIR1 complex. Proc Natl Acad Sci USA. 2004;101:12381–12386. doi: 10.1073/pnas.0402868101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature. 2001;414:271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- 16.Rashotte AM, Brady SR, Reed RC, Ante SJ, Muday GK. Basipetal auxin transport is required for gravitropism in roots of Arabidopsis. Plant Physiol. 2000;122:481–490. doi: 10.1104/pp.122.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Y, et al. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- 18.Collett CE, Harberd NP, Leyser O. Hormonal interactions in the control of Arabidopsis hypocotyl elongation. Plant Physiol. 2000;124:553–562. doi: 10.1104/pp.124.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liscum E, Reed JW. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol. 2002;49:387–400. [PubMed] [Google Scholar]
- 20.Morris GM, et al. Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. J Comput Chem. 1988;19:1639–1662. [Google Scholar]
- 21.Woo EJ, et al. Crystal structure of auxin-binding protein 1 in complex with auxin. EMBO J. 2002;21:2877–2885. doi: 10.1093/emboj/cdf291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cove D. The moss Physcomitrella patens. Annu Rev Genet. 2005;39:339–358. doi: 10.1146/annurev.genet.39.073003.110214. [DOI] [PubMed] [Google Scholar]
- 23.Decker EL, Frank W, Sarnighausen E, Reski R. Moss systems biology en route: phytohormones in Physcomitrella development. Plant Biol. 2006;8:397–405. doi: 10.1055/s-2006-923952. [DOI] [PubMed] [Google Scholar]
- 24.Yasumura Y, Crumpton-Taylor M, Fuentes S, Harberd NP. Step-by-step acquisition of the gibberellin-DELLA growth-regulatory mechanism during land-plant evolution. Curr Biol. 2007;17:1225–1230. doi: 10.1016/j.cub.2007.06.037. [DOI] [PubMed] [Google Scholar]
- 25.Thines B, et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka K, Suzuki T, Hattori N, Mizuno Y. Ubiquitin, proteasome and parkin. Biochim Biophys Acta. 2004;1695:235–247. doi: 10.1016/j.bbamcr.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 27.Strohmaier H, et al. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- 28.Nishiyama T, Hiwatashi Y, Sakakibara I, Kato M, Hasebe M. Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res. 2000;7:9–17. doi: 10.1093/dnares/7.1.9. [DOI] [PubMed] [Google Scholar]
- 29.Sanner MF. Python: A programming language for software integration and development. J Mol Graphics Mod. 1999;17:57–61. [PubMed] [Google Scholar]