Abstract
The outer membrane (OM) of most Gram-negative bacteria contains lipopolysaccharide (LPS) in the outer leaflet. LPS, or endotoxin, is a molecule of important biological activities. In the host, LPS elicits a potent immune response, while in the bacterium, it plays a crucial role by establishing a barrier to limit entry of hydrophobic molecules. Before LPS is assembled at the OM, it must be synthesized at the inner membrane (IM) and transported across the aqueous periplasmic compartment. Much is known about the biosynthesis of LPS but, until recently, little was known about its transport and assembly. We applied a reductionist bioinformatic approach that takes advantage of the small size of the proteome of the Gram-negative endosymbiont Blochmannia floridanus to search for novel factors involved in OM biogenesis. This led to the discovery of two essential Escherichia coli IM proteins of unknown function, YjgP and YjgQ, which are required for the transport of LPS to the cell surface. We propose that these two proteins, which we have renamed LptF and LptG, respectively, are the missing transmembrane components of the ABC transporter that, together with LptB, functions to extract LPS from the IM en route to the OM.
Keywords: ABC transporter, bioinformatics, endosymbiont, membrane biogenesis
The hallmark of Gram-negative bacteria is the presence of two extracytoplasmic membranes: the inner and outer membranes. The inner membrane (IM), which surrounds the cytoplasm, is separated from the outer membrane (OM) by an aqueous compartment known as the periplasm (1, 2). The IM is composed of phospholipids, integral transmembrane (TM) proteins that span the IM with α-helical TM domains, and lipoproteins (3, 4). In contrast, the OM is typically an asymmetric lipid bilayer where the inner leaflet is composed of phospholipids and the outer leaflet is composed mainly of lipopolysaccharide (LPS) (5). In addition, the OM contains lipoproteins and integral outer membrane proteins (OMPs), most of which span the OM via antiparallel β-sheets that fold into β-barrels (4, 6). Although some Gram-negative bacteria lack LPS and in others LPS is not essential (7), LPS is essential in Escherichia coli, Salmonella, and probably many other bacteria.
In E. coli, the main function of the OM is to serve as a selective permeability barrier against many toxic chemicals, such as detergents and antibiotics (1). Porin proteins in the OM control permeability to hydrophilic molecules, but unlike typical phospholipid bilayers, the OM is quite impermeable to hydrophobic molecules, mainly because of LPS (1). For the OM to serve as a barrier, all OM components must be assembled properly. None of the components of the OM are synthesized in situ, so they must be transported to the OM from their site of synthesis. All OMPs and lipoproteins are made in the cytoplasm and cross the IM through the Sec translocon (3). After their signal sequence is removed, OMPs are thought to travel across the periplasm aided by chaperones that deliver them to the OM Bam complex (β-barrel assembly machine, formerly known as the YaeT complex) for assembly (8–12). Lipoproteins are modified at the periplasmic face of the IM by signal sequence cleavage and the addition of lipid moieties at the N terminus. OM lipoproteins interact with the ABC transporter LolCDE, which extracts them from the IM and passes these proteins to the periplasmic chaperone LolA for delivery to LolB at the OM (4).
Both phospholipids and LPS are synthesized at the cytoplasmic face of the IM and are flipped across the bilayer by the ABC transporter MsbA (13, 14). In addition, phospholipids can be flipped by an ATP-independent mechanism that involves α-helical TM peptides (15). How phospholipids are transported to the OM is unknown. In contrast, several factors required for LPS transport to the cell surface have recently been identified and a transport model analogous to the aforementioned Lol system has emerged (16–19). The IM-associated cytoplasmic ATPase LptB (for LPS transport) is thought to be the nucleotide-binding domain (NBD) component of an LPS ABC transporter (18), but its TM components are unknown. The bitopic IM protein LptC (formerly YrbK) is also required for LPS extraction from the IM (A. Polissi, personal communication); however, its simple topology suggests that it cannot serve as the integral TM component of an ABC transporter (20). At the OM, the LptDE (formerly Imp–RlpB) complex is required for the assembly of LPS at the cell surface (16, 17, 19). Periplasmic LptA could be the chaperone that transports LPS across the periplasm (18); however, in vivo experiments question the existence of a soluble periplasmic intermediate (21).
There are still OM biogenesis factors yet to be identified, such as those involved in phospholipid traffic to the OM. More than 40% of the proteins in E. coli K-12 are of unknown function (22), so to limit the number of potential candidates for OM biogenesis factors, we applied a reductionist bioinformatic approach. This led to the discovery that the essential IM proteins YjgP and YjgQ are required for LPS transport to the cell surface in E. coli. We propose that YjgP and YjgQ are the TM component of the ABC transporter that, together with the NBD LptB (18), extracts LPS from the IM. Therefore, YjgP and YjgQ are renamed LptF and LptG, respectively.
Results
Rationale.
Our goal was to identify novel OM assembly factors in E. coli. To do so, we filtered the large body of published bioinformatic data through a series of criteria until we obtained a small number of candidates suitable for experimental testing. This goal was attained after applying the following consecutive filters, which are based on properties we predict unknown OM biogenesis factors to have. First, an OM assembly factor should be an envelope protein. Second, an OM assembly factor should be present in Gram-negative endosymbiotic bacteria possessing an OM similar in composition to that of E. coli's. We applied this filter because evolution of endosymbionts is marked by extensive gene loss (23). Thus, their genomes are among the smallest ones known; still, they encode all of the machinery required for building a bacterial cell. Third, we focused on those remaining proteins that are predicted to be essential in E. coli because the OM is an essential organelle, and of unknown function—that is, they are encoded by “y” genes.
Identification of LptFG as Potential OM Biogenesis Factors.
To employ the strategy described above, we first obtained a list of the 1,479 envelope proteins in E. coli from EchoLOCATION (24). We next searched for published genomes of endosymbiotic bacteria that encode enzymes for the biosynthesis of LPS because such bacteria would likely have an OM resembling that of E. coli. Particularly, we were interested in endosymbionts phylogenetically related to E. coli.
We chose Blochmannia floridanus because its genome, which encodes 583 proteins (25), is the smallest one among endosymbionts that, like E. coli, belong to the Enterobacteriaceae family. Despite the fact that the Bl. floridanus proteome is only 14% the size of E. coli's, BLAST searches revealed that it contains most of the major OM biogenesis factors known in E. coli; only SecG, DegP, BamB (formerly YfgL), and LolB were absent in Bl. floridanus [supporting information (SI) Table S1]. In E. coli, LolB is essential (26) but SecG, DegP, and BamB are not under normal growth conditions (27–29). In contrast, Bl. floridanus encodes all of the known proteins required for LPS assembly in E. coli. Overall, this comparative analysis reveals the Bl. floridanus proteome as a great candidate for our studies.
We then used GeneVenn (30) to find that 109 of the 1,479 E. coli envelope proteins are present in the Bl. floridanus. We filtered this list through that of essential proteins provided by Baba et al. (22) using GeneVenn (30). This yielded 40 proteins, 8 of which are encoded by y genes. Of these eight Y proteins, three lack published demonstration of their function: YrbK, YjgP, and YjgQ. However, as stated above, Polissi and collaborators (A. Polissi, personal communication) have shown that YrbK (now LptC) is required for LPS transport across the cell envelope, validating our approach. Therefore, we sought to determine whether YjgP and YjgQ function in OM biogenesis in E. coli. These two proteins are the focus of our work, and we have renamed them LptF and LptG, respectively, for reasons stated below.
In both E. coli and Bl. floridanus, lptF and lptG are located in a two-gene operon and their expression must be cotranslationally coupled because the stop codon of lptF overlaps with the start codon of lptG. Both LptF and LptG are homologous to each other (23.457% identity overall; FASTA analysis) and they belong to the Pfam family PF03739, which has been annotated as the “predicted permease YjgP/YjgQ family” (31). This motif encompasses their entire ORF, but there is no experimental evidence to support permease function. However, prediction and experimental studies show that both LptF and LptG have a topology typical of the TM domain (TMD) component of ABC transporters (20) because they are IM proteins containing six TM segments with cytoplasmic N and C termini (32).
Links Between LptF/LptG and LPS Biogenesis.
Searches to investigate the conservation of the lptFG operon among Gram-negative bacteria revealed linkage with lpt genes, which are required for LPS transport. In Caulobacter crescentus CB15, the 3′ end of lptG has a 7-bp overlap with the gene encoding the ortholog of the OMP LptD. In Synechococcus sp. CC9311, the start codon of lptG overlaps with the last stop codon in the lptAB operon. We also found in Thermatoga maritima MSB8 a 1,074-aa hypothetical protein (TM1735) with an N-terminal domain belonging to the aforementioned Pfam YjgP/YjgQ family that is followed by an OstA domain (Pfam: PF03968), which, in E. coli, constitutes most of LptA and the N terminus of LptD. Together, these findings suggest that LptFG could participate in LPS biogenesis. Furthermore, because of their abovementioned topology, we hypothesized that LptF and LptG could be the missing TMD of the ABC transporter that, with the NBD LptB, releases LPS from the IM.
Both lptF and lptG Are Essential.
lptFG are predicted to be essential in E. coli (22, 33). In agreement with these predictions, we could not delete either one or both genes unless we provided in trans a copy of the lptFG operon under the control of the PBAD promoter and grew strains in the presence of its inducer arabinose (34). We therefore constructed three E. coli strains in which we could deplete LptF, LptG, or both by inserting an arabinose-inducible copy of the lptFG operon at the λatt site and deleting one or both native chromosomal genes (35). In the resulting strains, NR1139, NR1141, and NR1113, we can stop expression of lptF, lptG, or both, respectively, by omitting the inducer arabinose from the growth medium.
The growth of all three depletion strains is arabinose-dependent (Fig. S1), demonstrating that both lptF and lptG are essential in E. coli. In the presence of arabinose, all three strains, NR1139 (LptF-depletion), NR1141 (LptG-depletion), and NR1113 (LptFG-depletion), grow similarly to the parent lptFG diploid strain. However, when all three depletion strains are diluted in media without arabinose, growth ceases after approximately four generations, and a low level of lysis is evidenced by a slight decrease in cell density and an increased accumulation of cell debris. Light microscopy also showed that all three depletion strains form filaments composed of 10–20 cells with constricted septa when grown without arabinose (data not shown). In agreement with our hypothesis that LptF and LptG are involved in LPS transport, similar phenotypes have been reported for cells where each of the lpt gene products LptABDE have been depleted (17–19).
Low Levels of LptF and/or LptG Cause Increased OM Permeability and Abnormal Membrane Structures.
Strains with defects in LPS biogenesis exhibit increased sensitivity to many hydrophobic antibiotics and detergents (36–38). We found that in media containing low levels of arabinose (0.0067%), all three depletion strains, NR1139, NR1141, and NR1113, exhibit increased sensitivity to hydrophobic antibiotics (Table S2), demonstrating that low levels of these IM proteins compromise the barrier function of the OM.
Depletion of each of the lpt gene products LptABDE causes striking alterations in envelope structure (18, 19). TEM (Fig. S2) revealed that in the presence of inducer, the cell envelope of the LptFG double-depletion strain appears normal, but depletion of LptFG results in defective envelopes that include extra membranous material in the periplasm. This phenotype is characteristic of cells where LPS transport has been compromised (18, 19).
Depletion of LptF and/or LptG Leads to Altered LPS but Does Not Affect OMP Assembly.
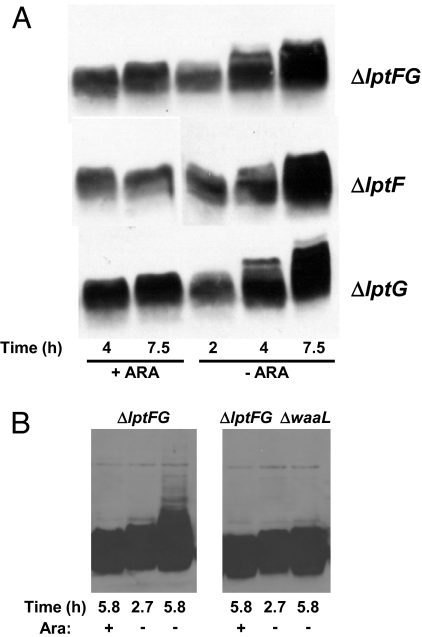
Depletion of LptAB results in an anomalous form of LPS that migrates as a ladder in gel electrophoresis (18). We observe a similar LPS ladder that increases in size with depletion time in each of our three depletion strains, NR1139 (LptF-depletion), NR1141 (LptG-depletion), and NR1113 (LptFG-depletion). The appearance of this ladder is a consequence of LptF and/or LptG depletion because it is absent when these strains are grown with arabinose (Fig. 1A).
Fig. 1.
Depletion of LptF and/or LptG leads to altered LPS. (A) Samples from depletion strains NR1113, NR1139, and NR1141 grown for the time indicated below panels in the presence (+ARA) or absence (−ARA) of arabinose were subjected to LPS immunoblotting. Samples correspond to the growth curve presented in Fig. S1B. (B) The ladder-like mobility of LPS in LptFG-depletion strain NR1113 grown in the absence of arabinose is WaaL-dependent.
The LPS molecule can be divided into three structural portions: lipid A, core, and O-antigen (39). Ligation of O-antigen to core by WaaL (RfaL) occurs at the periplasmic face of the IM (14). Despite the fact that E. coli K-12 lacks O-antigen (40), WaaL is present and can ligate colanic acid to the LPS core, and the resulting LPS migrates in gel electrophoresis as a ladder resembling those reported here (41). Moreover, colanic acid ligation to LPS occurs upon depletion of LptABDE (A. Polissi, personal communication). Therefore, we tested whether WaaL was required for the appearance of the LPS ladder upon depletion of LptFG in NR1113. As Fig. 1B shows, deletion of waaL blocks the production of the LPS ladder that appears in the absence of arabinose. Because WaaL-mediated ligation occurs at the periplasmic face of the IM, these results demonstrate that LptFG function after MsbA translocates LPS across the IM.
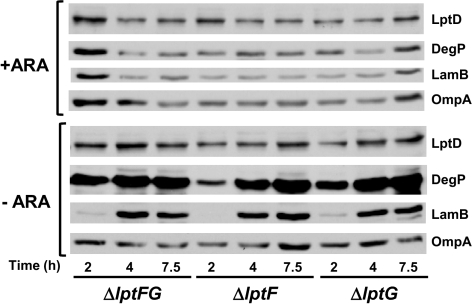
LptDE are required for the transport of LPS to the cell surface, but their depletion does not affect OMP biogenesis (19). If LptFG function in LPS transport, we would expect the same phenotype. Defects in OMP assembly lead to decreased levels of OMPs primarily because the mistargeted molecules are degraded by the periplasmic protease DegP (10). Thus, we compared the levels of three OMPs (LamB, OmpA, and LptD) in each of our three depletion strains grown with or without arabinose. As shown in Fig. 2, in the presence of arabinose, all depletion strains contain normal levels of OMPs. Upon depletion of LptF and/or LptG, levels of LptD and OmpA remain constant in all strains, whereas LamB levels actually increase (Fig. 2). Such increase is probably caused by the loss of catabolite repression of lamB synthesis upon removal of arabinose from the medium.
Fig. 2.
Depletion of LptF and/or LptG does not affect OMP biogenesis but increases DegP levels. Samples from depletion strains NR1113, NR1139, and NR1141 grown for the time indicated below panels in the presence (+ARA) or absence (−ARA) of arabinose were subjected to immunoblotting for OMPs (LptD, LamB, and OmpA) and the periplasmic protease DegP. Samples correspond to the growth curve presented in Fig. S1B.
Although LptFG are clearly not involved in OMP biogenesis, their depletion causes a significant increase in DegP levels (Fig. 2). Likely, DegP levels increase during depletion of LptF and/or LptG because the σE response is induced by defects in LPS transport to the OM, as LPS defects are known to induce this stress response (42).
Depletion of LptFG Leads to Modification of LPS by PagP.
LPS defects caused either by defective LPS biogenesis or by the release of LPS molecules from the OM during EDTA treatment result in the translocation of phospholipids to the outer leaflet of the OM (16, 19, 43). This translocation activates the OM enzyme PagP, which transfers a palmitate group from a phospholipid to lipid A (44), converting wild-type, hexa-acyl lipid A to hepta-acyl lipid A (45). Therefore, we can use the appearance of hepta-acyl lipid A as a reporter of defects in LPS transport (16, 19).
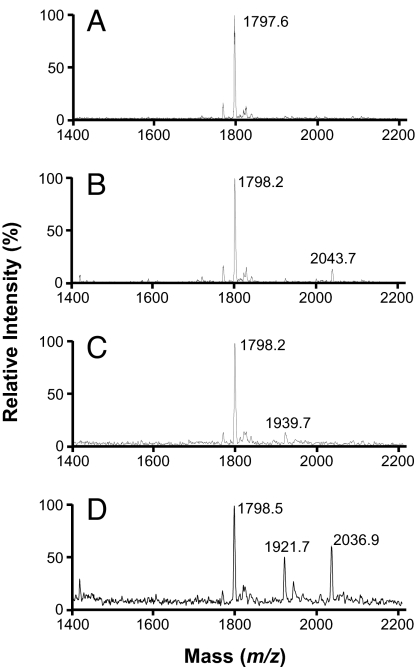
We analyzed the different species of lipid A by MALDI-TOF MS (19, 46) and saw that, as expected, the spectrum from the lptFG diploid strain NR1111 (Fig. 3A) contains a peak that corresponds to hexa-acyl lipid A (m/z 1,797.6) but not one corresponding to hepta-acyl lipid A (m/z ≈2,040). However, treatment of this diploid strain with EDTA causes the appearance of hepta-acyl lipid A (m/z 2,043.7; Fig. 3B) because it causes translocation of phospholipids to the cell surface and activation of PagP (43, 44).
Fig. 3.
Depletion of LptFG leads to PagP-modified LPS. Shown are MALDI-TOF profiles of LPS isolated from lptFG diploid strain NR1111 grown with arabinose (A) and EDTA (B), and from LptFG-depletion strain NR1113 grown with (C) or without arabinose (D).
Samples from all three depletion strains NR1113, NR1139, and NR1141 grown with arabinose also each showed a peak corresponding to hexa-acyl lipid A but not one corresponding to hepta-acyl lipid A (Fig. 3C and data not shown). We also found minor peaks (m/z 1,921–1,940; Fig. 3C and data not shown) indicative of additional modifications that could correspond to phosphoethanolamine- or aminodeoxypentose-modified lipid A, molecules that appear when the OM is damaged (39, 47). Therefore, even under inducing conditions, our depletion strains have slight defects in LPS biogenesis.
Depletion of LptF and/or LptG significantly alters the MALDI-TOF spectra. In all three depletion strains, growth without arabinose leads to the appearance of hepta-acyl lipid A (Fig. 3D and data not shown), similarly to what is observed upon depletion of either LptD or LptE (19). In addition, depletion of LptF and/or LptG causes an increase in the intensity of peaks with an m/z 1,921–1,940 value (Fig. 3D and data not shown). Thus, depletion of LptF and/or LptG allows translocation of phospholipids to the outer leaflet of the OM. In addition, in every test conducted, depletion of LptF, LptG, or both confers the same phenotypes, suggesting that they both function in the same pathway.
LptFG Are Required for the Transport of LPS to the Cell Surface.
Based on their topology (32), we hypothesized that LptFG constitute the missing TMD of the ABC transporter that releases LPS from the IM. If they are indeed required for LPS transport to the OM, their depletion should stop this essential step and LPS synthesized after depletion should not reach the cell surface.
We can exploit the fact that only LPS located at the cell surface is modified by PagP (44) to determine whether de novo-synthesized LPS reaches the cell surface, using a pulse–chase assay (19). As stated above, depletion of LptF and/or LptG causes the appearance of PagP-modified LPS. LPS that had been transported to the cell surface before depletion can be easily modified by PagP, but we wanted to know whether LPS that was synthesized after depletion could also be modified by PagP. Absence of modification of the LPS made after depletion would indicate that LPS cannot reach the outer leaflet of the OM without LptFG.
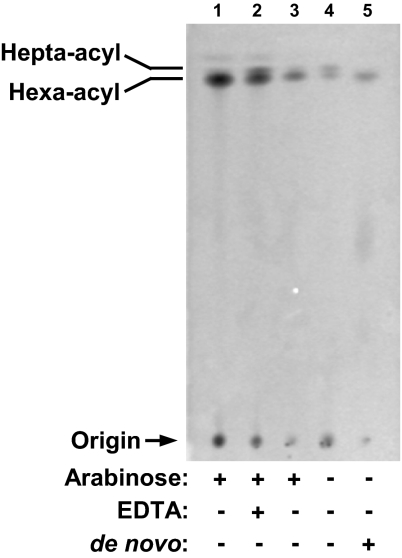
We monitored the modification status of newly synthesized LPS by labeling with [1-14C]acetate and separating LPS species, using TLC (19). As controls, lptFG diploid strain NR1111 grown with arabinose was either treated with EDTA to induce modification of LPS by PagP or not treated (44). In agreement with Fig. 3 A and B, most of the LPS in NR1111 contains hexa-acyl lipid A, but EDTA treatment greatly increases the levels of hepta-acyl LPS (Fig. 4, lanes 1 and 2). Also as expected, the main LPS species in LptFG double-depletion strain NR1113 grown with arabinose contains hexa-acyl lipid A (Fig. 4, lane 3). If we monitor steady-state LPS species by labeling at the time of subculturing in the absence of arabinose, we see that depletion of LptFG significantly increases the amounts of hepta-acyl LPS (Fig. 4, lane 4). In contrast, if we label LPS after depletion, we can only detect hexa-acyl radiolabeled LPS (Fig. 4, lane 5), demonstrating that without LptFG, de novo LPS cannot be modified by PagP. We conclude that, under these conditions, LPS does not reach the cell surface.
Fig. 4.
De novo-synthesized LPS requires LptFG to reach the cell surface. LptFG-diploid strain NR1111 grown in the presence of arabinose (lane 1) was treated with EDTA (lane 2) to serve as negative and positive controls, respectively, for the position of hepta-acyl lipid A. LptFG-depletion strain NR1113 was grown with (lane 3) or without (lanes 4 and 5) arabinose. [1-14C]Acetate was added either at the beginning of subculturing to label steady-state LPS (lanes 1–4) or after LptFG depletion to label de novo-synthesized LPS (lane 5). Equal amounts of radioactive material were spotted for each sample.
Discussion
Here, we have applied a bioinformatic approach that takes advantage of the small genome of bacterial endosymbionts to identify two essential IM proteins, LptF and LptG, required for the transport of LPS from the periplasmic face of the IM to the OM in E. coli. LptFG are encoded by a two-gene operon, and they function in the same pathway, because depletion of one or both proteins results in indistinguishable phenotypes. Given that both are IM proteins with the typical topology of the IM components of ABC transporters (i.e., 6-TM segments with cytoplasmic N and C termini) (20, 32), we propose that LptF and LptG are the missing TM components of the ABC transporter whose function is to extract LPS from the outer leaflet of the IM en route to the OM. Likely, the NBD component of this ABC transporter is LptB (18).
The recently discovered envelope factors required for LPS transport (16–19) support a model that is analogous to the Lol system, which shuttles OM lipoproteins from the IM to the OM (4, 18). According to this model, LPS transport requires an ABC transporter at the IM (LptBFG), a periplasmic protein (LptA), and an OM assembly site (LptDE). It is logical to envision that once MsbA translocates LPS to the periplasmic face of the IM, LPS interacts with LptFG, the TMD of the LptBFG ABC transporter. Then, LptB provides, through ATP hydrolysis, the energy required for extraction of LPS from the IM. LptA could serve as a periplasmic chaperone that delivers LPS to the OM LptDE complex for assembly at the cell surface.
The Lpt and Lol systems are strikingly similar in composition, but we do not know the extent of their similarity at the mechanistic level. For example, the Lol system lacks LptC and LptD equivalents. In addition, although the Lol pathway includes a soluble periplasmic intermediate composed of LolA and its cargo (4), it is not clear whether the Lpt system does. LptA, when overexpressed, can be detected in the periplasmic fraction (18). Yet, in vivo data show that LPS transport can occur at IM-OM contact sites without the release of soluble intermediates (21). Although these data do not distinguish whether such sites are membrane or protein bridges, they are consistent with the idea that a single functional machine that spans the periplasm transports LPS across the envelope without a soluble intermediate. Notably, LptFG and LptC possess large periplasmic domains that could mediate physical interactions even with components at the OM.
Having identified most, if not all, of the essential components required for LPS assembly, the focus of future research must be to clarify the mechanistic role played by each of these envelope proteins. While it seems likely that LptBFG are involved in providing the energy to extract LPS from the IM, it is less clear how the hydrophobic LPS molecule transits the periplasm to reach the OM machinery (LptDE) required for its insertion in the outer leaflet of the OM. Is there a soluble intermediate, or is there a physical structure that effectively connects the two membranes? Available evidence does not clearly distinguish these two models. What is clear is that if a physical bridge is required for transport, new biochemical methods will need to be developed to study this process. However, developing these tools will be well worth the effort given the pressing need for new antibiotics that can target Gram-negative bacteria and the great potential of proteins in the LPS pathway as antibiotic targets.
Materials and Methods
Bacterial Strains.
Except DY378 (48), all strains are derivatives of NR754, an araD+ revertant of MC4100 (49). Alleles were transferred using P1 transduction (50). The ΔrfaL::kan allele from the Keio collection (22) was introduced into NR1113 to yield NR1118.
Growth Conditions.
Luria–Bertani (LB) broth and agar were prepared as described in ref. 50. Except for recombineering (48), all liquid cultures were grown under aeration at 37°C, and their growth was monitored by optical density at 600 nm (OD600). When appropriate, kanamycin (25 μg/ml), ampicillin (125 μg/ml for plasmid maintenance and 25 μg/ml for strains with InCh alleles), and arabinose (0.2% wt/vol) were added. Unless indicated otherwise, for depletions, 1 ml of overnight cultures grown with arabinose was pelleted, washed once with LB broth, and resuspended in 1 ml of LB broth. Fresh LB broth with or without arabinose was inoculated with a 1:500 dilution of washed cultures, except with NR1139, for which a 1:1,000 dilution was used.
Contruction of lptFG Diploid Strain.
We amplified lptFG (from 26 bp upstream of lptF to 71 bp downstream of lptG) from MC4100 using primers 5YjgP4484215EcoRI (5′-CGGAATTCGACGAGTTTTTACGGGCG) and 3yjgQ4486494XbaI (5′-GCTCTAGACAGGAATGAACGAAGCACAC). The PCR product and plasmid pBAD18 (34) were digested with EcoRI and XbaI and ligated to produce pBADyjgPQ1. pBADyjgPQ1 was integrated into the λ-att site of NR754 using the λInCh procedure (35) to yield NR1111.
Construction of LptF and LptG Depletion Strains.
To construct a ΔlptFG::kan allele where the lptFG locus was deleted from the −53 bp position of lptF ORF to 70 bp downstream of the lptG ORF, we used primers yjgP4484119P1 (5′-GTGAATCCGTTGAGTATAATTATCTTAGCGACGATTTCGACGACTCAAGAGAATAAATGACGTTTAAGCCGTGTAGGCTGGAGCTGCTTC) and yjgQ4486494P2 (5′-GCCTGATGCGACGCTGGCGCGTCTTATCATGCCCACCCCACTGCAATATATTGAATTTTAATTATTTTTCCATATGAATATCCTCCTTA) to amplify from pKD4 (51) a PCR product that contains a kanamycin-resistance cassette flanked by 70 bp of homology to the target sequences in the lptG locus. This PCR product was electroporated into DY378 (pBADyjgPQ1) for recombineering (48). Transformants were selected in media with kanamycin and arabinose. The resulting ΔlptFG::kan allele was introduced into NR1111 using P1 transduction by selecting on media containing kanamycin and arabinose to yield NR1112. The kan cassette was excised (51) to generate the LptFG-depletion strain NR1113. The same procedures were used to construct LptF- and LptG-depletion strains NR1139 and NR1141, respectively, except with the following modifications. Primer pairs yjgPDEL1P1 (5′-ATAAATGACGTTTAAGCCATGAAACAAGCTAAAATCCTGCAAAAGACGAGTTTTTACGGGCGTATTTAAAGTGTAGGCTGGAGCTGCTTC) and yjgQDEL1P2 (5′-ACAGTGTCATCATGATGGTGGTGAAAATAGTTTTACCGATATAGCGGTCAAGTACGCCAAAAGGTTGCATCATATGAATATCCTCCTTa) and yjgQDEL1P1 (5′- CCTTTGGGACACCGTGCCGGTCCGCCGCCTGCGCGCCAGTTTTTCGCGTAAAGGAGCGGTGTGATGCAACGTGTAGGCTGGAGCTGCTTC) and yjgQ4486494P2 were used to construct the ΔlptF::kan and ΔlptG::kan alleles, respectively, in DY378 (pBADyjgPQ1). These single-depletion alleles were moved into NR1111 to yield NR1137 and NR1140, respectively, and strains NR1139 and NR1141 were obtained after their kan cassette was excised (51).
Immunoblotting.
One-milliliter samples from cultures were pelleted (16,000 × g, 2 min). To standardize samples, pellets were resuspended in a volume (ml) of SDS sample buffer equal to OD600/10. Samples were boiled for 10 min, and equal volumes were subjected to electrophoresis. For OMP and DegP detection, we used SDS/10% PAGE and immunoblotting was performed using LptD (1:7,000 dilution), DegP, LamB, and OmpA (1:30,000 dilution) rabbit polyclonal sera as described in ref. 38. For LPS immunoblots, we used SDS/15% PAGE with a Tris-Tricine SDS buffer system (National Diagnostics), mouse monoclonal antiserum against LPS core (1:5,000 dilution; HyCult Biotechnology), and goat anti-mouse horseradish peroxidase conjugate (Bio-Rad) (1:10,000 dilution). Bands were visualized using the ECL antibody detection kit (Amersham Pharmacia Biotech) and Hyblot CL film (Denville Scientific).
Isolation of Lipid A and MS Characterization.
Overnight cultures of NR1111, NR1113, NR1139, and NR1141 were diluted 1:1,000 into 25 ml of LB broth with or without arabinose and grown until growth of the depleted and nondepleted cultures diverged (≈3.5 h for NR111, NR1113, and NR1141, and ≈4 h for NR1139). Cells were treated and lipid A was extracted as described in ref. 19. Spectra were acquired in the negative reflector mode using a time-of-flight MALDI Voyager DE Pro mass spectrometer (Applied Biosystems) equipped with a 337-nm nitrogen laser and set at a 20-kV extraction voltage. Each spectrum was the average of 100 shots.
Labeling LPS and TLC Analysis.
Two 25-ml cultures of NR1111 and NR1113 and a 50-ml culture of NR1113 were inoculated from overnight cultures by 1:1,000 dilution into LB broth. Twenty-five microliters of 1 mCi/ml sodium [1-14C]acetate was added to all but the 50-ml culture. One 25-ml NR1113 culture was grown with arabinose; the other two NR1113 cultures lacked arabinose. Cells were grown until growth of the depleted cultures started to diverge. At this point, the 50-ml NR1113 culture was pulse-labeled with 50 μl of 1 mCi/ml sodium [1-14C]acetate, and all cultures were grown for an additional 30 min. Ten minutes before harvesting, 25 mM EDTA was added to one of the NR1111 cultures. Samples were processed and subjected TLC as described in ref. 19.
For details on compound sensitivity and electron microscopy, see SI Text.
Acknowledgments.
We thank Peggy Bisher for processing TEM samples. This work was supported by National Institute of General Medical Sciences Grants GM34821 (to T.J.S.) and GM66174 (to D.K.) and a National Science Foundation Graduate Research Fellowship (to L.S.G.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801196105/DCSupplemental.
References
- 1.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruiz N, Kahne D, Silhavy TJ. Advances in understanding bacterial outer-membrane biogenesis. Nat Rev Microbiol. 2006;4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- 3.Dalbey RE, Chen M. Sec-translocase mediated membrane protein biogenesis. Biochim Biophys Acta. 2004;1694:37–53. doi: 10.1016/j.bbamcr.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 4.Tokuda H, Matsuyama S. Sorting of lipoproteins to the outer membrane in E. coli. Biochim Biophys Acta. 2004;1694:IN1–IN9. [PubMed] [Google Scholar]
- 5.Kamio Y, Nikaido H. Outer membrane of Salmonella typhimurium: Accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry. 1976;15:2561–2570. doi: 10.1021/bi00657a012. [DOI] [PubMed] [Google Scholar]
- 6.Koebnik R, Locher KP, Van Gelder P. Structure and function of bacterial outer membrane proteins: Barrels in a nutshell. Mol Microbiol. 2000;37:239–253. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- 7.Steeghs L, et al. Meningitis bacterium is viable without endotoxin. Nature. 1998;392:449–450. doi: 10.1038/33046. [DOI] [PubMed] [Google Scholar]
- 8.Malinverni J, et al. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol. 2006;61:151–164. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- 9.Sklar JG, et al. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:6400–6405. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sklar JG, Wu T, Kahne D, Silhavy TJ. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007;21:2473–2484. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- 12.Wu T, et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 13.Doerrler WT. Lipid trafficking to the outer membrane of Gram-negative bacteria. Mol Microbiol. 2006;60:542–552. doi: 10.1111/j.1365-2958.2006.05130.x. [DOI] [PubMed] [Google Scholar]
- 14.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kol MA, et al. Phospholipid flop induced by transmembrane peptides in model membranes is modulated by lipid composition. Biochemistry. 2003;42:231–237. doi: 10.1021/bi0268403. [DOI] [PubMed] [Google Scholar]
- 16.Bos MP, Tefsen B, Geurtsen J, Tommassen J. Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc Natl Acad Sci USA. 2004;101:9417–9422. doi: 10.1073/pnas.0402340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braun M, Silhavy TJ. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol Microbiol. 2002;45:1289–1302. doi: 10.1046/j.1365-2958.2002.03091.x. [DOI] [PubMed] [Google Scholar]
- 18.Sperandeo P, et al. Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J Bacteriol. 2007;189:244–253. doi: 10.1128/JB.01126-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu T, et al. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2006;103:11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linton KJ, Higgins CF. Structure and function of ABC transporters: The ATP switch provides flexible control. Pflügers Arch. 2007;453:555–567. doi: 10.1007/s00424-006-0126-x. [DOI] [PubMed] [Google Scholar]
- 21.Tefsen B, Geurtsen J, Beckers F, Tommassen J, de Cock H. Lipopolysaccharide transport to the bacterial outer membrane in spheroplasts. J Biol Chem. 2005;280:4504–4509. doi: 10.1074/jbc.M409259200. [DOI] [PubMed] [Google Scholar]
- 22.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silva FJ, Latorre A, Moya A. Genome size reduction through multiple events of gene disintegration in Buchnera APS. Trends Genet. 2001;17:615–618. doi: 10.1016/s0168-9525(01)02483-0. [DOI] [PubMed] [Google Scholar]
- 24.Misra RV, Horler RS, Reindl W, Goryanin II, Thomas GH. EchoBASE: An integrated post-genomic database for Escherichia coli. Nucleic Acids Res. 2005;33:D329–D333. doi: 10.1093/nar/gki028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gil R, et al. The genome sequence of Blochmannia floridanus: Comparative analysis of reduced genomes. Proc Natl Acad Sci USA. 2003;100:9388–9393. doi: 10.1073/pnas.1533499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka K, Matsuyama SI, Tokuda H. Deletion of lolB, encoding an outer membrane lipoprotein, is lethal for Escherichia coli and causes accumulation of lipoprotein localization intermediates in the periplasm. J Bacteriol. 2001;183:6538–6542. doi: 10.1128/JB.183.22.6538-6542.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eggert US, et al. Genetic basis for activity differences between vancomycin and glycolipid derivatives of vancomycin. Science. 2001;294:361–364. doi: 10.1126/science.1063611. [DOI] [PubMed] [Google Scholar]
- 28.Flower AM, Hines LL, Pfennig PL. SecG is an auxiliary component of the protein export apparatus of Escherichia coli. Mol Gen Genet. 2000;263:131–136. doi: 10.1007/s004380050039. [DOI] [PubMed] [Google Scholar]
- 29.Lipinska B, Fayet O, Baird L, Georgopoulos C. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J Bacteriol. 1989;171:1574–1584. doi: 10.1128/jb.171.3.1574-1584.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pirooznia M, Nagarajan V, Deng Y. GeneVenn—A web application for comparing gene lists using Venn diagrams. Bioinformation. 2007;1:420–422. doi: 10.6026/97320630001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finn RD, et al. Pfam: Clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daley DO, et al. Global topology analysis of the Escherichia coli inner membrane proteome. Science. 2005;308:1321–1323. doi: 10.1126/science.1109730. [DOI] [PubMed] [Google Scholar]
- 33.Gerdes SY, et al. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol. 2003;185:5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyd D, Weiss DS, Chen JC, Beckwith J. Towards single-copy gene expression systems making gene cloning physiologically relevant: Lambda InCh, a simple Escherichia coli plasmid-chromosome shuttle system. J Bacteriol. 2000;182:842–847. doi: 10.1128/jb.182.3.842-847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sampson BA, Misra R, Benson SA. Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics. 1989;122:491–501. doi: 10.1093/genetics/122.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sperandeo P, Pozzi C, Deho G, Polissi A. Non-essential KDO biosynthesis and new essential cell envelope biogenesis genes in the Escherichia coli yrbG-yhbG locus. Res Microbiol. 2006;157:547–558. doi: 10.1016/j.resmic.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 38.Ruiz N, Falcone B, Kahne D, Silhavy TJ. Chemical conditionality: A genetic strategy to probe organelle assembly. Cell. 2005;121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 39.Raetz CR, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu D, Reeves PR. Escherichia coli K12 regains its O antigen. Microbiology. 1994;140:49–57. doi: 10.1099/13500872-140-1-49. [DOI] [PubMed] [Google Scholar]
- 41.Meredith TC, et al. Modification of lipopolysaccharide with colanic acid (M-antigen) repeats in Escherichia coli. J Biol Chem. 2007;282:7790–7798. doi: 10.1074/jbc.M611034200. [DOI] [PubMed] [Google Scholar]
- 42.Ruiz N, Silhavy TJ. Sensing external stress: Watchdogs of the Escherichia coli cell envelope. Curr Opin Microbiol. 2005;8:122–126. doi: 10.1016/j.mib.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 43.Leive L. Release of lipopolysaccharide by EDTA treatment of E. coli. Biochem Biophys Res Commun. 1965;21:290–296. doi: 10.1016/0006-291x(65)90191-9. [DOI] [PubMed] [Google Scholar]
- 44.Jia W, et al. Lipid trafficking controls endotoxin acylation in outer membranes of Escherichia coli. J Biol Chem. 2004;279:44966–44975. doi: 10.1074/jbc.M404963200. [DOI] [PubMed] [Google Scholar]
- 45.Bishop RE, et al. Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. EMBO J. 2000;19:5071–5080. doi: 10.1093/emboj/cdd507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Z, Lin S, Cotter RJ, Raetz CR. Lipid A modifications characteristic of Salmonella typhimurium are induced by NH4VO3 in Escherichia coli K12. Detection of 4-amino-4-deoxy-L-arabinose, phosphoethanolamine and palmitate. J Biol Chem. 1999;274:18503–18514. doi: 10.1074/jbc.274.26.18503. [DOI] [PubMed] [Google Scholar]
- 47.Brabetz W, Muller-Loennies S, Holst O, Brade H. Deletion of the heptosyltransferase genes rfaC and rfaF in Escherichia coli K-12 results in an Re-type lipopolysaccharide with a high degree of 2-aminoethanol phosphate substitution. Eur J Biochem. 1997;247:716–724. doi: 10.1111/j.1432-1033.1997.00716.x. [DOI] [PubMed] [Google Scholar]
- 48.Yu D, et al. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Casadaban MJ. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 50.Silhavy TJ, Berman ML, Enquist LW. Experiments with Gene Fusions. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1984. [Google Scholar]
- 51.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]