Abstract
A quantitative algorithm was developed and applied to predict target genes of microRNAs encoded by herpesviruses. Although there is almost no conservation among microRNAs of different herpesvirus subfamilies, a common pattern of regulation emerged. The algorithm predicts that herpes simplex virus 1, human cytomegalovirus, Epstein–Barr virus, and Kaposi's sarcoma-associated herpesvirus all employ microRNAs to suppress expression of their own genes, including their immediate-early genes. In the case of human cytomegalovirus, a virus-coded microRNA, miR-112-1, was predicted to target the viral immediate-early protein 1 mRNA. To test this prediction, mutant viruses were generated that were unable to express the microRNA, or encoded an immediate-early 1 mRNA lacking its target site. Analysis of RNA and protein within infected cells demonstrated that miR-UL112-1 inhibits expression of the major immediate-early protein. We propose that herpesviruses use microRNA-mediated suppression of immediate-early genes as part of their strategy to enter and maintain latency.
Keywords: miRNAs, reactivation, immune evasion
MicroRNAs (miRNAs) are 20–23-nucleotide RNA molecules that bind to mRNA targets, generally within their 3′ untranslated region (3′ UTR), and interfere with their translation (1, 2). Viruses have co-evolved with cellular miRNAs and many encode their own miRNAs (3). Every herpesvirus genome that has been examined has been found to encode multiple miRNAs, including Epstein–Barr virus (EBV) (4–6), human cytomegalovirus (HCMV) (4, 7, 8), herpes simplex virus 1 (HSV-1) (9, 10), and Kaposi's sarcoma-associated herpesvirus (KSHV) (4, 6, 11, 12). These miRNAs can potentially function during lytic replication and latency. Lytic replication proceeds in a coordinated three-phase cascade: immediate-early (IE), early and late. IE products prepare the cell for infection and propagate the cascade. Early gene products support replication of viral DNA, and DNA replication is, in turn, a prerequisite for full activation of the late genes that encode the structural proteins of the virus. During latency, the virus is quiescent. A limited subset of the viral genome is expressed, but, importantly, the virus has the potential to reactivate and reenter the lytic cycle. Although the molecular mechanisms of reactivation are not understood, it is widely assumed that the lytic cascade is reinitiated with the expression of IE genes. Ectopic expression of a single IE protein has been shown to reactivate HSV-1 (13, 14), EBV (15), or KSHV (16, 17) in cell culture models of latency.
Whereas none or only a few protein-coding genes are expressed, multiple miRNAs are transcribed during latency. The HSV-1 miR-LAT lies within one of the latency associated transcripts (LATs), the only viral RNAs known to be expressed in latency. EBV and KSHV miRNAs also are expressed during latent infection. Because they are nonimmunogenic, miRNAs should be optimal agents for suppression of anti-viral responses and to modify behaviors of latently infected cells, and recent reports suggest this is the case. The HSV-1 miR-LAT has been reported to inhibit apoptosis in neurons by inhibiting translation of TGF-β and SMAD3 (10); the HCMV miR-UL112-1 represses expression of MICB, a ligand of the natural killer cell activating receptor NKG2D (18); and KSHV miRNAs down-regulate thrombospondin 1 expression, a regulator of cell adhesion, migration and angiogenesis (19).
Virus-coded miRNAs also could act to antagonize the expression of IE genes. This could contribute to down-regulation of IE expression during later phases of the lytic replication cycle when miRNAs have accumulated to relatively high levels, and it could help to establish latency and subsequently antagonize reactivation of latent virus, given the critical role of IE gene products in the initiation of the lytic cascade. Motivated by this reasoning, we developed an miRNA-target-predicting algorithm and applied it to all 3′ UTRs of four herpesviruses. The high probability targets predicted by this algorithm included IE transactivators for all four viruses: ICP0 of HSV-1; BZLF1 and BRLF1 of EBV; IE1 of HCMV; Rta and Zta of KSHV. We experimentally confirmed the ability of an HCMV-coded miRNA to reduce IE1 expression. Because these genes are necessary for efficient initiation of the lytic program, we propose that herpesviruses use a common miRNA-based strategy for maintaining latency.
Results
For further details, please see supporting information (SI) Materials and Methods and SI Tables 3–8.
Predicting Targets of HCMV-Coded miRNAs Within the HCMV Genome.
To test our hypothesis that herpesvirus miRNAs might inhibit expression of viral genes needed for efficient lytic replication and thereby favor latency, we asked whether viral miRNAs had potential to target viral 3′ UTRs. Instead of listing all conserved potential miRNA binding sites or computing scores based on various empirical rules, our algorithm uses a combination of analytical expressions and Monte Carlo simulations to determine exact probabilities that predicted miRNA targets would occur by chance. We use the standard assumption that the 3′ UTR sequence has coevolved with the sequence of the miRNA (20, 21) and the experimental observation that miRNA binding requires a perfect complementarity of a “seed” sequence near the 5′ end of the miRNA to a sequence in the 3′ UTR (2). This seed is usually a heptamer at positions 2–8 from the 5′ end of the miRNA. As a result of coevolution, the number of actual seed oligomers present in the 3′ UTR of a targeted gene will be higher than the number that would appear by chance in a random sequence with similar composition. The algorithm predicts functional miRNA targets in two steps.
First, for each miRNA-3′ UTR pair, we compute an approximate probability PVSH (P value for single hypothesis testing) that it would appear by chance in the random sequence; the smaller PVSH is, the more likely the given pair is to be biologically functional. (Probability PVSH is very nearly exact: The only approximation is that we assume independence between consecutive oligomers.) This procedure alone allows testing whether a given miRNA is likely to target a given 3′ UTR and is analogous to that used by Robins and Press (21).
Second, if we are interested in finding functional targets of multiple miRNAs among multiple 3′ UTRs, we need to take into account multiple hypothesis testing. We do this by performing a Monte Carlo simulation in which we compute the probability PVMH (P value for multiple hypothesis testing) that the top, say 10, miRNA-target pairs in a randomly generated genome with similar properties would have their PVSH lower than the corresponding top 10 miRNA-target pairs in the real genome. We used this approach instead of the now standard False Discovery Rate analysis (FDR) of Benjamini and Hochberg (22) because of the discrete nature of our data. In our case, most PVSH values are 1, and so FDR analysis is not applicable because it requires a fairly uniform distribution of PVSH except a small overrepresentation at values close to 0.
Details and various extensions of the algorithm, such as the generation of the random background sequence based on the Markov model (MM), the calculation of the single and multiple hypothesis P values, the use of conservation (21) among strains of the virus, and the choice of the seed sequence, are described in Materials and Methods.
We applied the algorithm to all 3′ UTRs of the HCMV genome, and Table 1 shows the 10 most probable miRNA-target pairs of the 4896 total possible miRNA-3′ UTR pairs. For each pair, the table shows the score PVSH and the statistical significance PVMH of all predictions up to this one. For instance, the 10th miRNA-target prediction, miR-UL112-1 targeting the IE transactivator protein 1 mRNA (IE1, encoded by the UL123 ORF, highlighted), has a score PVSH = 10−2.21 = 0.0062 and PVMH = 0.125, meaning only 12.5% of randomly generated genomes have top ten P values better or equal to PVSH = 10−2.21. For top 25 most probable miRNA-target pairs, see SI Table 5C. In fact, the data set in that table suggests that the most significant predictions are the top 10 listed in Table 1 because there is a sharp increase in PVMH from the 10th to 11th prediction: PVMH (10) = 0.125 and PVMH (11) = 0.309. Naturally, PVMH (k) increases toward 1 for larger k. In our analysis, we required that a target be conserved in six sequenced strains of HCMV. If conservation among strains is not taken into account, PVMH suggests that there are many more significant targets (35 with PVMH < 0.20, see SI Table 5C). Finally, the PVMH values listed in Table 1 are conservative upper bounds because we considered all published sequences of detected potential miRNAs although several are only slight variations of each other and some others are perhaps not real miRNAs.
Table 1.
Top 10 predicted miRNA-target pairs in HCMV when sorted by PVSH score
| 3′UTR |
hcmv-miR | Act.† | First-order local MM |
|||
|---|---|---|---|---|---|---|
| HCMV ORF | Length* | Exp.‡ | Log10 PVSH | PVMH | ||
| UL103 | 21 | US5-2 | 1 | 0.000 | −4.11 | 0.036 |
| UL112–113 | 67 | UL54-1 | 1 | 0.001 | −2.96 | 0.155 |
| RL10 | 57 | US5-1 | 1 | 0.003 | −2.52 | 0.273 |
| UL31 | 62 | UL112-1 | 1 | 0.003 | −2.46 | 0.229 |
| UL80 | 34 | UL70-5p | 1 | 0.004 | −2.42 | 0.187 |
| UL34 | 14 | UL112-1 | 1 | 0.004 | −2.41 | 0.155 |
| UL3 | 57 | UL70-5p | 1 | 0.005 | −2.33 | 0.155 |
| UL69 | 253 | US33-1 | 1 | 0.005 | −2.29 | 0.144 |
| UL57 | 426 | US25-2-5p | 2 | 0.108 | −2.27 | 0.127 |
| UL123(IE1) | 92 | UL112-1 | 1 | 0.006 | −2.21 | 0.125 |
The table shows the top 10 of 4,896 possible miRNA-3′ UTR pairs for the HCMV genome. The statistical significance of the top targets is measured by the multiple hypothesis P value PVMH. The random background used is the first-order local MM. IE1 (UL123) is shown in bold.
*Length denotes the total number of all conserved heptamers in the 3′UTR.
†Act. denotes the actual count (in the 3′ UTR) of conserved heptamers complementary to the miRNA seed.
‡Exp. denotes the count expected in the random sequence.
HCMV IE1 Protein Synthesis Is Suppressed by miR-UL-112-1.
Inhibition of any of the genes in Table 1 could potentially favor latency, but we considered IE1 to be a prime target, given its central role at the start of the HCMV transcriptional cascade. IE1 is one of two main products of the HCMV major IE locus, the other being IE2. IE1 and IE2 are required to execute the virus' transcriptional program (23), and they almost certainly influence the choice between latency and lytic replication. A mutant virus unable to produce a functional IE1 protein replicates efficiently only after infection at a high input multiplicity (24); at lower multiplicities, it fails to accumulate normal levels of early mRNAs (25). It activates transcription at least in part by controlling histone modifications (26).
The algorithm predicted a single binding site for miR-UL112-1 within the 99 nucleotide 3′ UTR of the IE1 mRNA. To test the prediction that miR-UL112-1 inhibits translation of IE1 protein, we prepared two reporter constructs. The first contained the wild-type IE1 3′ UTR downstream of the luciferase coding region and the second contained a derivative of the 3′ UTR lacking the 7-nucleotide seed sequence predicted to be the target of the miRNA (Fig. 1A, shaded sequence). HEK293T cells were cotransfected with set amounts of the reporter plasmids and increasing amounts of an effector plasmid expressing the miR-UL112-1 precursor hairpin sequence. The miRNA induced a statistically significant reduction in luciferase expression from the reporter with a wild-type IE1 3′ UTR (maximum repression = ≈60%) but not from the modified 3′ UTR lacking the seed sequence (Fig. 1B), arguing that miR-UL112-1 targets the seed sequence within the IE1 3′ UTR to reduce translation or degrade the RNA.
Fig. 1.
miR-UL112-1 inhibits expression from a reporter mRNA containing the IE1 3′ UTR. (A) The predicted miR-UL112-1-binding site within the HCMV major IE locus. At the top of the diagram, the spliced mRNAs that encode IE1 and IE2 are depicted with the noncoding exon 1 (Ex1) shown as an open box and the coding exons (Ex2–5) depicted as gray boxes. IE1 and IE2 share Ex2 and Ex3. The PolyA sites and the location of the miR-UL112-1 binding site in the 3′ UTR (gray pinhead) are shown. At the bottom of the diagram, the IE1 3′ UTR sequence is expanded and the putative miRNA/mRNA base pairing is depicted. The gray box denotes nucleotides within the miRNA seed sequence. (B) Reporter assay for miR-UL112-1 function. 293T cells were cotransfected with firefly luciferase expression plasmids containing either the wild-type (light gray) or mutant IE1 3′ UTR (dark gray) as well as a Renilla luciferase internal control. Cells were additionally cotransfected with the indicated amounts of a miR-UL112-1 expressing plasmid, and transfection mixtures were balanced with the expression plasmid lacking an insert. Firefly luciferase units were normalized to Renilla luciferase. The luciferase units are shown relative to the amount of luciferase from the reporter construct in the absence of miRNA expression plasmids. Asterisks denote P values <0.05 as determined by the Student's t test.
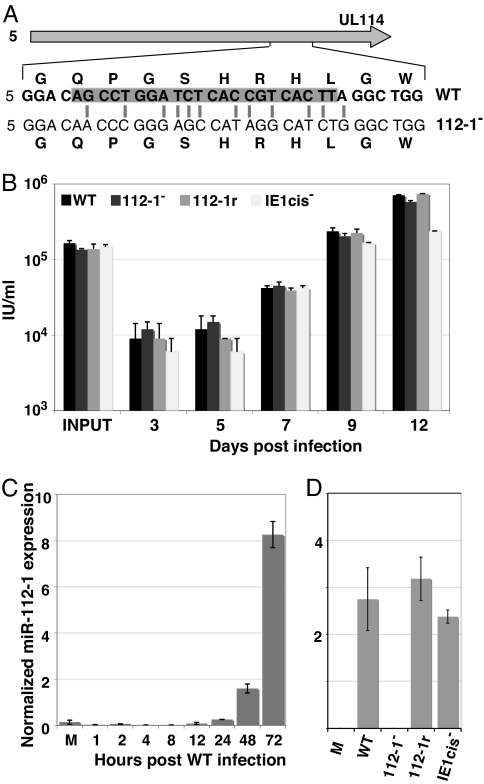
We next generated three viruses to test whether miR-UL112-1 targets IE1 expression within an HCMV-infected cell. The first, BFXdlIE1cis−, lacks the 7-nt seed sequence within the IE1 3′ UTR that is targeted by the miRNA. The second, BFXsub112-1−, is unable to express the miRNA. The miR-UL112-1 precursor is encoded on the DNA strand opposite UL114, and disruption of this ORF inhibits virus replication (27, 28). Consequently, we substituted 12 nucleotides within the miR-UL112-1 precursor sequence while maintaining the coding sequence of the UL114 ORF (Fig. 2A). The miR-UL112-1 mutation was repaired in the final virus, BFXsub112-1r, to control for potential off-target mutations. The viruses grew normally in fibroblasts (Fig. 2B). We also monitored accumulation of miR-UL112-1 by quantitative RT-PCR. The miRNA accumulated to a detectable level between 8–12 h after infection with wild-type virus and then increased as the infection progressed (Fig. 2C). No miR-UL112-1 was detected at 48 h after infection with BFXsub112-1−, a time at which the miRNA was readily detected in cells infected with the other viruses (Fig. 2D).
Fig. 2.
BFXsubUL112-1− and BFXdlIE1cis− grow with wild-type kinetics. (A) The nucleotides mutated in the BFXsubUL112-1− (112-1−) relative to BFXwt (WT) are shown with gray lines and the complement of the mature miRNA is shaded within the WT sequence. The substituted nucleotides in 112-1−conserves the amino acid sequence (shown above and below nucleotide sequences) of the UL114 gene. (B) MRC-5 fibroblasts were infected at a multiplicity of 0.1 pfu/cell with WT, 112-1−, BFXsub112-1r (112-1r, a revertant of 112-1−) or BFXdlIE1cis− (IE1cis−, lacks the 7-nucleotide seed sequence in IE1 mRNA). Virus (IU, infectious units) was assayed at the indicated times. (C) qRT-PCR with a miR-UL112-1 specific probe was used to detect the accumulation of the miRNA in WT-infected MRC5 fibroblasts over time. miR-UL112-1 levels were normalized to the expression of a cellular small nucleolar RNA, hsn6B. M, mock infected. (D) Accumulation of miR-112-1 was assayed by qRT-PCR at 48 hpi with the indicated viruses and normalized to hsn6B expression. The 112-1− virus had miRNA levels similar to mock infected cells.
To determine whether IE1 protein levels were affected by the expression of miR-UL112-1, we prepared extracts from infected cells after a 1-h 35S-labeling period at 6, 24 and 48 hpi with wild-type or mutant viruses. We did not monitor cells later than 48 hpi, even though the miRNA accumulated to higher levels at 72 hpi (Fig. 2A), because infected cells show severe cytopathic effect at the later time. We first examined the steady state levels of several proteins by Western blot assay (Fig. 3A Top). Tubulin levels, which are not altered by infection, provided a precise measure of the amount of cellular protein analyzed in each sample; and the accumulation of the late HCMV protein, pp28, confirmed that all infections progressed normally. We monitored IE1 steady state levels, but little difference was evident after infection with wild-type and mutant viruses. This was not surprising, because IE1 protein has a >20-h half life, and it accumulates to a high level before the miRNA is available.
Fig. 3.
Viruses that lack miR-UL112-1 or its binding site synthesize more IE1 protein. (A) MRC5 fibroblasts were mock-infected (M) or infected with BFXwt (WT), BFXsub112-1− (112-1−), BFXsub112-1r (112-1r), or BFXdlIE1cis− (IE1cis−). Cells were 35S-labeled for 1 h before harvesting at the indicated times after infection. Lysates were prepared and analyzed by Western blot for IE1, the late virus-coded pp28 or tubulin (Upper) or immunoprecipitation followed by electrophoresis for 35S-labeled IE1 (Lower). The experiment shown is a representative of 6 independent immunoprecipitations. (B Top) Quantification of 35S-labeled IE1 relative to tubulin. IE1 protein levels were quantified by phosphorimager analysis of immunoprecipated complexes from two independent experiments, each of which was analyzed by three independent immunoprecipitations, such as that displayed in A Lower. The levels of IE1 protein were normalized to tubulin levels from the Western blot in A. The mutant and revertant viruses are normalized to WT levels for each time point. P values were determined by the Student's t test. (B Middle) Quantification of IE1 RNA relative to UL37 RNA by qRT-PCR. Mutant and repaired viruses are normalized to WT levels for each time point. (Lower) ratio IE1 protein (from Top) to IE1 RNA (from Middle).
Next, IE1 was immunoprecipitated from extracts and subjected to electrophoresis to identify protein synthesized during each 1-h labeling period (Fig. 3A Bottom). The rate of IE1 synthesis was substantially greater at 6 hpi than at later times for all viruses, and this is expected because the promoter responsible for the production of IE1 mRNA is repressed late after infection (29, 30). Radioactivity in the IE1-specific band was quantified relative to the level of tubulin, and Fig. 3B Top presents the results of two independent experiments, each analyzed by performing three independent immunoprecipitations. At 6 and 24 hpi, we did not observe an effect attributable to miR-UL112-1 activity, consistent with the observation that the miRNA is not detected at 6 hpi and relatively little is present at 24 hpi. In contrast, at 48 hpi, when the miRNA has accumulated to higher levels, the miR-UL112-1-deficient and the IE1 target site-deficient mutants exhibited statistically significant increases (≈2-fold) in IE1 protein synthesis relative to the wild-type and revertant viruses.
At each time protein extracts were prepared, total RNA was isolated from a duplicate sample, and the amount of IE1 RNA was determined relative to the level of an independent IE RNA (UL37) by quantitative RT-PCR. IE1 RNA levels varied little among the viruses (Fig. 3B Middle), arguing that the miRNA does not significantly alter the stability of IE1 mRNA and supporting the conclusion that the changes in IE1 protein levels result from the inhibition of translation. The ratio of IE1 protein to RNA was calculated (Fig. 3B Lower), confirming a significant increase in protein synthesis when either the miRNA or its target site is disrupted.
Predictions of Targets for miRNAs Coded by Other Herpesviruses.
Encouraged by the experimental verification of our prediction in HCMV, we applied the algorithm to an analysis of three additional human herpesviruses. HSV-1, EBV, and KSHV each proved to encode miRNAs predicted to inhibit the expression of viral proteins, including IE proteins. Table 2 displays the rank of the IE-targeting miRNAs among all possible miRNA–3′ UTR pairs (the total number is equal to the number of 3′ UTRs times the number of miRNAs). The rank is again based on the P value PVSH computed according to the local first order MM or the global third order MM. ICP0 in HSV-1, BZLF1 and BRLF1 in EBV, and Zta and Rta in KSHV are among the virus-specific targets most likely to be targeted by virus-coded miRNAs (top 0.5–2% of virus-specific targets). The BZLF1/BRLF1 3′ UTR of EBV is predicted to be targeted by two miRNAs.
Table 2.
Whole genome ranks for predicted miRNA-IE target pairs in four herpesviruses
| Virus | 3′ UTR* | Length | miRNA | Seed | Count | Rank A† | Percentile | Rank B |
|---|---|---|---|---|---|---|---|---|
| HSV-1 | ICP0 | 186 | hsv1-miR-LAT | 3–8 | 1 | 4 of 154 | 97.40 | 12 of 154 |
| EBV | BZLF1, BRLF1 | 53 | ebv-miR-BART15 | 2–8 | 1 | 3 of 2,720 | 99.89 | 4 of 2,720 |
| EBV | BZLF1, BRLF1 | 53 | ebv-miR-BHRF1-3 | 2–8 | 1 | 4 of 2,720 | 99.85 | 3 of 2,720 |
| HCMV | IE1 | 92 | hcmv-miR-UL112-1 | 2–8 | 1 | 10 of 4,896 | 99.80 | 9 of 4,896 |
| KSHV | Zta, Rta | 1,144 | kshv-miR-K12-6-3p | 3–8 | 4 | 1 of 1,394 | 99.93 | 1 of 1,394 |
The table reports the top miRNA-IE target pairs for HSV-1, EBV, KSHV, and HCMV after sorting by PVSH score.
*BZLF1 and BRLF1 as well as Zta and Rta give rise to 3′ coterminal transcripts and therefore genes in each pair have the same 3′UTRs.
†Rank A (resp. rank B) denotes the rank among all possible miRNA—3′UTR pairs sorted by P values computed for the random sequence based on the first-order local (resp. the third-order global) MM. Percentile corresponds to Rank A.
SI Tables 3–6 show three additional pieces of information for each virus: First, there is a list (SI Tables 3A–6A) for each miRNA of the total actual and predicted number of binding sites across all 3′ UTRs with associated P value. miRNAs with smaller P value are more likely to regulate some (unspecified) viral genes. The total number of functional binding sites for miRNAs can be estimated from the difference of the total numbers of actual and predicted seed binding sites (21). It is reassuring that this number is always positive, regardless of the virus or the MM used. Second, there is a list (SI Tables 3B–6B) of the top 25 3′ UTR targets, sorted according to the P value based on the total actual and predicted binding-site counts across all miRNAs. 3′ UTRs with small P value are likely to be regulated by some combination of viral miRNAs. Third, there is a list (SI Tables 3C–6C) of the top 25 miRNA-3′ UTR pairs. Pairs with small P value are most likely to be functional pairs. The ranks of the IE genes in Table 2 are derived from this list.
Besides the IE genes, the top predicted miRNA targets include many genes involved in viral DNA replication as well as several inhibitors of apoptosis and other genes involved in immune evasion. Brief descriptions of the predicted targets in these functional groups are summarized in SI Table 7.
Discussion
Our computational analysis predicts that miRNAs encoded by members of three herpesvirus subfamilies target viral IE genes. HSV-1 (α-herpesvirus) miR-LAT likely targets ICP0. HCMV (β-herpesvirus) miR-UL112-1 targets IE1. EBV (γ-herpesvirus) miR-BHRF1–3 and miR-BART15 likely target BZLF1 and BRLF1, which share their 3′ UTR (Table 1 and SI Table 4C). If the effect of all EBV miRNAs is combined, this 3′ UTR is in fact the top target in the whole genome (SI Table 4B). In addition, the seed binding is actually longer than the heptamer 2–8 used in the analysis; it is an octamer 1–8 for miR-BHRF1-3, and if noncanonical binding U-G is allowed, a 9-mer 210 for miR-BART15. KSHV (γ-herpesvirus) Rta (ORF 50) and Zta (ORF K8) are homologues of EBV BRLF1 and BZLF1, respectively, although functionally BZLF1 seems to correspond more to the Rta. The 3′ UTR shared by Zta and Rta is likely targeted by the KSHV miR-K10-6-3p, which is numerically the strongest prediction of this algorithm (99.93 percentile). Finally, besides the IE genes, the analysis (SI Tables 3–6) suggests numerous other likely miRNA targets, e.g., the HSV-1 neurovirulence factor γ34.5, whose RNA contains an octamer seed binding site for miR-LAT.
We confirmed the predicted inhibition of HCMV IE1 translation by miR-UL112-1 within transfected cells by using reporter constructs (Fig. 1) and within virus-infected fibroblasts by analyzing mutant viruses (Figs. 2 and 3). As we were writing this manuscript, Gray et al. (31) reported the same conclusion. They demonstrated that addition of miR-UL112-1 RNA to cells before infection inhibited IE1 expression and viral DNA replication.
Given the broad range of predicted targets (SI Tables 3–6), we anticipate that herpesvirus-coded miRNAs exert regulatory effects directly on viral gene expression during replication and spread within infected hosts. This regulation could have many consequences, e.g., down-regulating viral genes as the infectious cycle progresses to avoid toxicity and helping to modulate viral gene expression to optimize replication in a variety of different cell types. Most importantly, however, our results also suggest that virus-coded miRNAs could play a central role in the establishment and maintenance of latency. Because they target IE products that act at the top of the lytic cascade, miRNAs expressed in cells destined for a latent infection can potentially antagonize the cascade and thereby favor entry into latency. Furthermore, miRNAs expressed during latency could help to prevent reactivation by inhibiting translation of IE transactivators.
There are several indirect justifications of this model. First, although expression of HCMV-coded miRNAs during latency has not yet been evaluated, HSV-1, EBV, and KSHV miRNAs are transcribed and available to function during latency. Second, if miRNAs are indeed central agents responsible for the maintenance of latency, this would explain the mysterious failure to detect proteins in the latent stage of HSV-1. Finally, our algorithm predicts that numerous early genes involved in DNA replication are targeted by viral miRNAs. In fact, the EBV miR-BART2 cleaves the mRNA of the DNA polymerase BALF5 (32). Early genes are normally activated by IE products, and their activity in turn would be expected to importantly influence the choice between lytic replication and latency.
In sum, antagonism of IE genes, which form a control node at the top of the herpesvirus lytic cascade, could contribute importantly to the establishment of latency and seems an optimal strategy for maintenance of latency.
Materials and Methods
Computation.
To determine the likelihood that a particular miRNA-3′ UTR pair is functional, we computed the corresponding probability PVSH. Let c denote the actual count of seed n-mers in the 3′ UTR of length l and p the probability (based on the MM described in SI Materials and Methods) that any given n-mer in the random background sequence is the seed n-mer. Then our P value PVSH gives the probability of finding at least c seed n-mers in a background sequence of length l which is equal to the P value of the binomial distribution,
 |
In practice, l is of the order of 100 or 1,000. For a hexamer seed sequence (n = 6), a typical p is 1/46 = 1/4,096 (exactly if all hexamers were equally likely) and therefore a typical c is zero, making the equation above impractical. An alternative exact expression for PVSH, which is numerically efficient is
where B(x,a,b) is the incomplete beta function and B(a,b) is the usual beta function,
 |
The statistical significance of the top miRNA-target pairs was evaluated by calculating probability PVMH. Because the majority of P value PVSH is equal to 1, we cannot use the standard method of estimating the false discovery rate (22). Instead, we use the following Monte Carlo procedure: First generate n = 1,000 random genomes analogous to the actual genome of interest. This means that each genome will have exactly the same number of 3′ UTRs as the genome of interest and each generated 3′ UTR will be of the same length as the corresponding real 3′ UTR. Each random 3′ UTR is generated by using the kth order MM based on the composition of the corresponding 3′ UTR in the real genome.
For each of the N randomly generated genomes, we repeat the same analysis of computing PVSH as we did for the real genome: i.e., we compute the score PVSH for each miRNA-3′ UTR and sort them. Now, evaluate the statistical significance of the top t miRNA-target pairs for the actual genome by counting the number Nt of the randomly generated genomes in which the tth top microRNA-3′ UTR pair has PVSH smaller than the tth pair in the actual genome. For each t, compute the P value PVMH(t) corrected for multiple hypothesis testing by
PVMH(t) is the probability of finding better scores for the top t potential microRNA–3′ UTR pairs in a random genome with similar properties as the actual genome. The smaller PVMH(t), the higher the chance that the predicted targets are real targets.
Additional details relevant to computation are described in SI Materials and Methods, including sequences used in the analysis, choice of the seed sequence, construction of random background sequence, use of conservation between strains or species.
Cells, Viruses, and Plasmids.
MRC5 and HEK293T cells were propagated in medium with 10% FBS or 10% newborn calf serum, respectively.
The wild-type virus used in these studies is BFXwt-GFP. It is a derivative of a bacterial artificial chromosome (BAC) clone of the HCMV VR1814 clinical isolate (33) in which a green fluorescent protein (GFP) expression cassette has been inserted upstream of the US7 ORF. Three derivatives of BFXwt-GFP were produced by using galK selection and counter selection to modify BAC DNAs (34) as described in SI Materials and Methods. BFXdlIE1cis− lacks the 7-nt seed sequence for miR-112-1 within the IE1 3′ UTR, BFXsub112-1− contains 12 single base pair substitutions that block expression of miR-112-1, BFXsub112-1r is a repaired derivative of BFXsub112-1−. Virus was generated by electroporation of MRC5 cells with BAC DNA (20 μg) plus an HCMV pp71-expressing plasmid (pCGNpp71). Virions were purified by centrifugation through a 20% sorbitol cushion. Virus titers were calculated by infecting fibroblasts and counting IE2-positive foci at 24 hpi.
The construction of plasmids is described in SI Materials and Methods.
mRNA and miRNA Quantification.
Real-time RT-PCR was performed on total RNA isolated from the cells by using the mirVana miRNA isolation kit (Ambion), which isolates total RNA while preserving the miRNA population. DNA was removed by using the DNA-free reagent kit (Ambion). Equal aliquots of total RNA were reverse transcribed by using the Taqman Reverse Transcription kit with random hexamers according to the manufacture's protocol (Applied Biosystems). To measure mRNA levels, real-time PCR was performed with SYBR green PCR master mix (Applied Biosystems) and primers specific to exon 4 of IE1 (5′-GCCTTCCCTAAGACCACCAAT-3′ and 5′-ATTTTCTGGGCATAAGCCATAATC-3′) or exon 1 of UL37 (5′-GACGAAGTCCGATGAGGAGGATG-3′ and 5′-TGGGACACTGGGCGTTGTTG-3′).
To measure levels of miR-UL112-1, a modified TaqMan-based stem loop RT-PCR was performed (35). TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems) was used according to the manufacturer's protocol with the following stem-loop oligonucleotide: 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGCCTG-3′. A 1:15 dilution of the product from the reverse transcriptase reaction was used in a TaqMan quantitative PCR along with 1.5 mM of forward primer (5′-GCCGCAAGTFACGGTGAGAT-3′), 0.7 mM of reverse primer (5′-GTGCAGGGTCCGAGGT-3′), 0.2 mM of TaqMan probe (5′-6FAM-TACGACAGCCTGGAT-mgbNFQ) and 1× Universal TaqMan PCR Master mix (Applied Biosystems). The results were normalized by quantifying the levels of human U6B small nuclear RNA using the RNU6B Taqman control assay (Applied Biosystems).
Protein Quantification.
MRC5 cells were infected at a multiplicity of 3 pfu per cell. Cells were starved for methionine and cystine before labeling by incubating for 1 h in medium with 10% dialyzed FBS. EasyTag Express Protein Labeling mix (100 μCi; PerkinElmer) was added to the cells for 1 h, after which the labeling medium was replaced with medium containing 10% FCS for 10 min to allow stalled translation to complete. Cells were washed in PBS and then lysed in buffer containing 20 mM Tris acetate pH 7.5, 0.27 M sucrose, 1 mM EDTA, 1 mM EGTA, 1 mM sodium orthovanadate, 10 mM sodium β-glycerophosphate, 50 mM sodium floride, 5 mM sodium pyrophosphate, and 1% Triton X-100. One tablet of Complete Mini Protease inhibitor (Roche Applied Science) was added per 10 ml of lysis buffer. Protein concentration was calculated by Bradford assay.
Aliquots (10 μg) were subjected to Western blot assay using monoclonal antibodies specific for HCMV IE1 (1B12), HCMV UL99 (10B4) and monoclonal anti-tubulin antibody (Sigma–Aldrich). An anti-mouse HRP conjugated antibody was used along with the ECL plus detection kit (Amersham) to detect specific bands. Chemiluminescence was analyzed by using a PhosphorImager and ImageQuant TL software (GE Healthcare Life Sciences, Piscataway, NJ).
For immunoprecipitation assays, aliquots of lysate (5 or 10 μg protein) were precleared with Protein A/G Plus Agarose beads (Santa Cruz Biotechnology) for 4 h at 4°C. Anti-IE1 monoclonal antibody (1B12) and Protein A/G Plus Agarose were added to the supernatant, which was incubated overnight at 4°C with shaking. Immunoprecipitated complexes were washed three times with RIPA buffer (50 mM Tris·HCl pH 7.4, 1% Nonidet P-40, 0.25% Na-deoxycholate, 150 mM NaCl, 1 mM EDTA) supplemented with Complete Mini Protease inhibitor. Beads were boiled in 2× SDS loading buffer and run on an 8% SDS/PAGE gel to separate the immunoprecipitated complexes. Gels were dried and exposed to a phosphor screen, which was analyzed by using a PhosphorImager and ImageQuant TL software.
Supplementary Material
Acknowledgments.
We thank M. Krasnitz and G. Atwal for useful discussions. This work was supported by National Cancer Institute Grant CA085786. E.M. was supported by a postdoctoral fellowship from the American Cancer Society (PF-0427101-MBC).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711910105/DC1.
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Cullen BR. Viruses and microRNAs. Nat Genet. 2006;38(Suppl):S25–S30. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- 4.Pfeffer S, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 5.Cai X, et al. Epstein–Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA. 2006;12:733–750. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grey F, Antoniewicz A, Allen E, Saugstad J, McShea A, Carrington JC, Nelson J. Identification and characterization of human cytomegalovirus-encoded microRNAs. J Virol. 2005;79:12095–12099. doi: 10.1128/JVI.79.18.12095-12099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunn W, et al. Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cell Microbiol. 2005;7:1684–1695. doi: 10.1111/j.1462-5822.2005.00598.x. [DOI] [PubMed] [Google Scholar]
- 9.Cui C, et al. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J Virol. 2006;80:5499–5508. doi: 10.1128/JVI.00200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta A, Gartner JJ, Sethupathy P, Hatzigeorgiou AG, Fraser NW. Anti-apoptotic function of a microRNA encoded by the HSV-1 latency-associated transcript. Nature. 2006;442:82–85. doi: 10.1038/nature04836. [DOI] [PubMed] [Google Scholar]
- 11.Cai X, et al. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci USA. 2005;102:5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samols MA, Hu J, Skalsky RL, Renne R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J Virol. 2005;79:9301–9305. doi: 10.1128/JVI.79.14.9301-9305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris RA, Everett RD, Zhu XX, Silverstein S, Preston CM. Herpes simplex virus type 1 immediate-early protein Vmw110 reactivates latent herpes simplex virus type 2 in an in vitro latency system. J Virol. 1989;63:3513–3515. doi: 10.1128/jvi.63.8.3513-3515.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu XX, Chen JX, Young CS, Silverstein S. Reactivation of latent herpes simplex virus by adenovirus recombinants encoding mutant IE-0 gene products. J Virol. 1990;64:4489–4498. doi: 10.1128/jvi.64.9.4489-4498.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Countryman J, Miller G. Activation of expression of latent Epstein–Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc Natl Acad Sci USA. 1985;82:4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun R, et al. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1998;95:10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukac DM, Renne R, Kirshner JR, Ganem D. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology. 1998;252:304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- 18.Stern-Ginossar N, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samols MA, et al. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 2007;3:e65. doi: 10.1371/journal.ppat.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 21.Robins H, Press WH. Human microRNAs target a functionally distinct population of genes with AT-rich 3′ UTRs. Proc Natl Acad Sci USA. 2005;102:15557–15562. doi: 10.1073/pnas.0507443102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 23.Mocarski ES, Shenk T, Pass RF. In: Fields Virology. Knipe DM, Howley PM, editors. Philadelphia, PA: Lippincott, Williams and Wilkins; 2007. pp. 2702–2772. [Google Scholar]
- 24.Mocarski ES, Kemble GW, Lyle JM, Greaves RF. A deletion mutant in the human cytomegalovirus gene encoding IE1(491aa) is replication defective due to a failure in autoregulation. Proc Natl Acad Sci USA. 1996;93:11321–11326. doi: 10.1073/pnas.93.21.11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gawn JM, Greaves RF. Absence of IE1 p72 protein function during low-multiplicity infection by human cytomegalovirus results in a broad block to viral delayed-early gene expression. J Virol. 2002;76:4441–4455. doi: 10.1128/JVI.76.9.4441-4455.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nevels M, Paulus C, Shenk T. Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc Natl Acad Sci USA. 2004;101:17234–17239. doi: 10.1073/pnas.0407933101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu D, Silva MC, Shenk T. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc Natl Acad Sci USA. 2003;100:12396–12401. doi: 10.1073/pnas.1635160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prichard MN, Duke GM, Mocarski ES. Human cytomegalovirus uracil DNA glycosylase is required for the normal temporal regulation of both DNA synthesis and viral replication. J Virol. 1996;70:3018–3025. doi: 10.1128/jvi.70.5.3018-3025.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macias MP, Huang L, Lashmit PE, Stinski MF. Cellular or viral protein binding to a cytomegalovirus promoter transcription initiation site: effects on transcription. J Virol. 1996;70:3628–3635. doi: 10.1128/jvi.70.6.3628-3635.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reeves M, et al. Autorepression of the human cytomegalovirus major immediate-early promoter/enhancer at late times of infection is mediated by the recruitment of chromatin remodeling enzymes by IE86. J Virol. 2006;80:9998–10009. doi: 10.1128/JVI.01297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grey F, Meyers H, White EA, Spector DH, Nelson J. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS pathogens. 2007;3:e163. doi: 10.1371/journal.ppat.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfeffer S, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 33.Hahn G, et al. The human cytomegalovirus ribonucleotide reductase homolog UL45 is dispensable for growth in endothelial cells, as determined by a BAC-cloned clinical isolate of human cytomegalovirus with preserved wild-type characteristics. J Virol. 2002;76:9551–9555. doi: 10.1128/JVI.76.18.9551-9555.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen C, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.