Abstract
The disparity in species richness among groups of organisms is one of the most pervasive features of life on earth. A number of studies have addressed this pattern across higher taxa (e.g. ‘beetles’), but we know much less about the generality and causal basis of the variation in diversity within evolutionary radiations at lower taxonomic scales. Here, we address the causes of variation in species richness among major lineages of Australia's most diverse vertebrate radiation, a clade of at least 232 species of scincid lizards. We use new mitochondrial and nuclear intron DNA sequences to test the extent of diversification rate variation in this group. We present an improved likelihood-based method for estimating per-lineage diversification rates from combined phylogenetic and taxonomic (species richness) data, and use the method in a hypothesis-testing framework to localize diversification rate shifts on phylogenetic trees. We soundly reject homogeneity of diversification rates among members of this radiation, and find evidence for a dramatic rate increase in the common ancestor of the genera Ctenotus and Lerista. Our results suggest that the evolution of traits associated with climate tolerance may have had a role in shaping patterns of diversity in this group.
Keywords: diversification rates, phylogeny, key innovation, speciation, scincid lizards
1. Introduction
Why do some groups of organisms contain vastly greater numbers of species than other groups? A prominent explanation holds that variation in species richness among groups can be explained in part by differences in per-lineage rates of species origination and extinction (Slowinski & Guyer 1989; Mooers & Heard 1997). Many previous studies have tested for variation in diversification rates across large phylogenetic and taxonomic scales, such as angiosperms (Magallon & Sanderson 2001; Sims & McConway 2003; Davies et al. 2004) or passerine birds (Phillimore et al. 2006; Ricklefs 2006). By comparison, few studies have addressed the extent to which diversification rates vary within species-level radiations, and particularly within groups that do not represent classic adaptive radiations (Ruber & Zardoya 2005; Kozak et al. 2006; McKenna & Farrell 2006). Extending the analysis of diversification rates to species-level radiations of non-model groups is important, because recent work suggests that clade age and not diversification rate might be the dominant signal influencing the distribution of diversity among major animal clades (McPeek & Brown 2007). Understanding the contribution of diversification rate variation to differences in species richness requires more data from evolutionary radiations at lower taxonomic scales.
Australian lizards known as sphenomorphine skinks are a particularly appropriate system in which to test the extent to which diversification rates vary among lineages during continental evolutionary radiations. This group comprises at least 232 species that appear to have diverged in situ within mainland Australia (Cogger 2000; Reeder 2003; Skinner in press), making it by far the most diverse vertebrate radiation in Australia, and one of the most diverse continental radiations across all amniotes. Sphenomorphine skinks occur in virtually all terrestrial habitats in Australia, and body elongation and limb reduction have evolved repeatedly within the group (Greer 1989). Of the 15 Australian sphenomorphine genera, the genus Ctenotus alone includes nearly 100 described species (Cogger 2000). Although it has been suggested previously that Ctenotus may have experienced elevated diversification rates (Reeder 2003), there have been no formal tests for heterogeneity of diversification rates within the sphenomorphine skink radiation.
Here, we present the first molecular phylogenetic analysis of major lineages within Ctenotus and use these historical data to test whether it is necessary to invoke among-lineage variation in diversification rates to explain the high disparities in extant diversity among major groups of this radiation. We also test the monophyly of the genus Ctenotus to exclude the possibility that the high species diversity of this genus is simply a taxonomic artefact. Finally, we use a likelihood framework based on the birth–death process to test whether high species diversity within Ctenotus is the outcome of increased diversification rates within the genus, or whether it reflects a decline in diversification elsewhere in the radiation. Our results indicate a dramatic increase in diversification occurring in the lineage leading to Ctenotus and its sister taxon Lerista and suggest that arid-adapted lineages may have diversified explosively in response to the expansion of the Australian arid zone over the past 20 Myr.
2. Material and methods
(a) Taxon sampling
We obtained tissue samples for 34 species of Ctenotus, including multiple representatives of the 12 recognized ‘species groups’ (Storr et al. 1999) and most of the phenotypic diversity present in this high-diversity genus (Greer 1989; Cogger 2000). A summary of major morphotypes within Ctenotus is provided as electronic supplementary material (electronic supplementary material, table s1). Genomic DNA was extracted using DNeasy Tissue kits (Qiagen). We sequenced 718 bp of mitochondrial ND4 and 181 bp of three flanking tRNAs (tRNA-ser, tRNA-his, tRNA-leu) using primers and protocols described by Reeder (2003). In addition, we sequenced 671 bp of the nuclear ATP synthetase β-subunit intron, after Skinner (in press). We aligned new DNA sequences for Ctenotus to those used in a previous analysis of sphenomorphine skink relationships (Reeder 2003; Skinner in press). The full data matrix contained 81 species, including representatives of all Australian sphenomorphine genera, and 6 outgroup taxa. In addition to ND4, tRNA and ATP synthetase intron sequences, we included 1442 bp of mitochondrial 12S and 16S rRNA sequences for 48 of these species that were available from previous studies (Reeder 2003; Skinner in press). Alignment of ND4 did not require gap insertion and was straightforward. Ribosomal 12S and 16S alignments used secondary structure models from the European ribosomal database (Wuyts et al. 2001), tRNAs were aligned after Macey & Verma (1997) and ATP synthetase intron sequences were aligned using ClustalX (Thompson et al. 1997) with default parameters. GenBank accession numbers, museum voucher numbers and sampling localities for all sequences included in this study are available as electronic supplementary material (electronic supplementary material, table s2).
(b) Phylogenetic analysis
Bayesian phylogenetic analyses were performed on the combined dataset using separate partitions for intron (ATP synthetase), protein coding (ND4) and structural (tRNAs, rRNAs). Appropriate models of molecular evolution for the three partitions were determined using MrModeltest v. 2.2 (Nylander 2004). Recognition of additional data partitions (Brandley et al. 2005) did not change results, and we present analyses based on only these three partitions. We performed two simultaneous runs of Metropolis-coupled Markov chain Monte Carlo (MCMCMC) sampling using the parallel processor version of MrBayes v. 3.1.2 (Altekar et al. 2004), with four incrementally heated chains run simultaneously for 10 million generations and trees sampled every 1000th generation. We discarded the first 7.5 million generations as burn-in, and checked for convergence by (i) testing for stationarity of log L values, (ii) by plotting the posterior probabilities for nodes as a function of the number of generations, and (iii) examining standard deviations of split frequencies for independent runs (Huelsenbeck et al. 2001). We performed two additional runs using identical conditions to verify that results converged on the same posterior distribution of trees.
(c) Variation in diversification rates
We obtained an ultrametric tree for diversification analyses by applying penalized likelihood (PL; Sanderson 2002) to the Bayesian consensus phylogram using the software package r8s (Sanderson 2003). Prior to rate smoothing, we removed all non-Australian sphenomorphine taxa from the tree and applied cross-validation procedure implemented in r8s with an additive penalty to select an optimal smoothing parameter. The diversification rate analyses discussed below are robust to absolute age estimates and require only relative divergence times. However, to facilitate comparison with other Australian radiations for which approximate time scales are available (e.g. Jennings et al. 2003; Crisp et al. 2004), we inferred the age of the basal split among Australian sphenomorphine species. No fossil calibration points exist within either the sphenomorphine radiation or the closely related outgroup taxa (Martin et al. 2004), so we used published rates of squamate mtDNA sequence evolution (Zamudio & Greene 1997; Calsbeek et al. 2003; Richmond & Jokusch 2007). We determined mean pairwise maximum-likelihood (ML) distances between taxa in the two basal clades under the best-fit model and parameters inferred by MrModeltest for this data partition and estimated the timing of the split under an mtDNA divergence rate of 0.895% corrected sequence divergence per Myr (range: 0.47–1.32%; Zamudio & Greene 1997).
Diversification rate analyses were conducted on the genus-level tree created by pruning all but one lineage per genus from the PL tree; species totals were assigned to each terminal following Wilson & Swan (2003). We tested whether some groups of Australian skinks are characterized by exceptionally high or low diversification rates using the approach of Magallon & Sanderson (2001). We plotted the number of species within Australian sphenomorphine genera as a function of stem clade age, as inferred from the PL chronogram. The stem clade age is simply the age of the divergence between the crown group and its extant sister group, corresponding in this case to genus age; this assumes that genera are monophyletic. We do not have complete sampling of all sphenomorphine species, therefore we cannot distinguish between crown- and stem-group ages, and these divergence times thus represent the transition time from a single ancestral species to a clade with some level of standing diversity in the present.
We then considered the Australian sphenomorphine radiation as a whole and estimated the net diversification rate r or the difference between the speciation rate (λ) and the extinction rate (μ). We compared the extant diversity of sphenomorphine genera with the 95% CIs around expected species diversity for clades of similar age that have diversified with the net diversification rate observed for the overall radiation (Magallon & Sanderson 2001, eqn (10)).
Net diversification rates were estimated for the entire sphenomorphine radiation using two methods. First, we used the whole-clade estimator described by Magallon & Sanderson (2001, eqn (7)). This estimator is a function of three parameters: the number of species in the clade; the age of the clade; and the ratio of the extinction rate to the speciation rate μ/λ (denoted by a). Even with a constant net diversification rate, the expected outcome of a stochastic diversification process can vary with the extinction fraction a (e.g. Raup 1985). We therefore computed CIs around expected diversity under a=0 and 0.99; these values represent extremes on a continuum of relative extinction rates. We computed one-tailed probabilities of the observed diversity of each clade, given the estimated overall diversification rate and clade age, to test whether sphenomorphine skinks are characterized by an excess of species-rich or species-poor lineages; we combined individual p-values using the Z-transform method (Whitlock 2005; additional information in the electronic supplementary material, table s4).
We repeated these analyses using an estimator of r that uses both taxonomic and phylogenetic data, because our data consist of terminal taxa of known diversity (e.g. numbers of species within genera), as well as the phylogenetic relationships of these taxa. Our estimator generalizes the Yule process estimator of Sanderson & Wojciechowski (1996) to the birth–death process. The log likelihood of the observed diversity of T terminal taxa is given by
| (2.1) |
where
| (2.2) |
ni is the diversity of the ith terminal; ti is the stem age of the terminal; and a is the extinction fraction. The log likelihood that each of the N internal branches in the phylogeny will result in exactly two descendant species after some time t is given by
| (2.3) |
where xi is the birth time of the ith lineage in units of time before present. The log likelihood of the combined taxonomic and phylogenetic data is simply log L=log LP+log LT. ML estimates of r were found using the one-dimensional ‘optimize’ routine in R (http://cran.r-project.org/). A derivation of this estimator and its relationship to previous taxonomic/phylogenetic estimators is given in appendix A.
(d) Shifts in diversification rate
We tested for shifts in diversification rate within the Australian sphenomorphine radiation by contrasting the likelihood of the data under a model with equal diversification rates for all lineages to the likelihood under a model where an ancestral diversification rate r1 shifts to a new rate r2 along some branch in the tree (Sanderson 1994; Sanderson & Wojciechowski 1996). To compute likelihoods and estimate r, we used the birth–death estimator based on phylogenetic and taxonomic data described in this study. For this model, referred as the flexible-rate model, we sequentially split the tree at each branch and optimized r onto the resulting pair of subtrees. The node resulting in the maximum combined likelihood for the bipartite tree was the ML estimate of the shift point. Analyses were conducted under extinction fractions of a=0 and 0.99.
We predicted that the clade containing Ctenotus and Lerista would show an increase in diversification rates relative to other lineages simply owing to the greater diversity of these genera relative to other sphenomorphine genera. However, an alternative hypothesis is that this clade retained an ancestral but elevated diversification rate, while another clade or clades exhibited a decline in diversification. To distinguish between these possibilities, we repeated our flexible-rate analysis with the constraint that the subtree partition containing Ctenotus must include the root node. Thus, no rate shift was permitted along the path between the root node and Ctenotus. If the likelihood of the data under this constrained model is similar to that under the flexible-rate model, we cannot distinguish between an increase in diversification rates for Ctenotus and a decrease in diversification rates for some other clade. To avoid conditioning results on any particular tree topology and branch lengths, we sampled 1000 phylogenies from the set of post-burn-in topologies (1 tree every 5000 generations) and applied PL to each. We repeated our analyses on this set of trees to generate the posterior distribution of likelihood differences between these two variable-rate diversification models, with the expectation that the rate-decrease model would consistently provide a poorer fit to the data than the flexible-rate model. Functions for all diversification analyses described in this study have been included in the LASER library for the R programming language (Rabosky 2006a).
3. Results and discussion
(a) Phylogenetic relationships
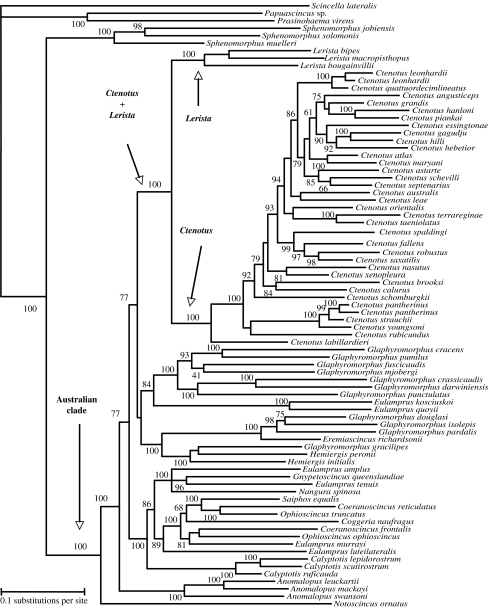
Bayesian phylogenetic analysis supports Ctenotus as monophyletic with a high level of confidence (figure 1), implying that the exceptional species diversity within Ctenotus cannot be explained as a taxonomic artefact. In addition, our results strongly support sister clade relationship between Ctenotus and Lerista, the other highly diverse genus of sphenomorphine skink (79 species). Although taxon sampling of Lerista is limited, morphological and molecular data (Greer 1989; A. Skinner 2007, personal communication) strongly suggest that the genus is monophyletic.
Figure 1.
Bayesian consensus phylogram for 75 Australian and 6 non-Australian sphenomorphine skink species based on combined analysis of mtDNA and nuclear intron sequences. Only posterior probabilities (×100) greater than or equal to 60 are shown. Branch lengths are means from the posterior distribution of sampled trees.
Our results corroborate those of several previous studies (Reeder 2003; Skinner in press) and provide further support for the monophyly of Australian sphenomorphine skinks, as well as a basal split between Notoscincus and all other Australian sphenomorphine lineages. These results were also supported by parsimony and likelihood bootstrap analyses (electronic supplementary material, table s3). As found previously (Reeder 2003), several sphenomorphine genera are not monophyletic (e.g. Eulamprus, Glaphyromorphus). Taken together, monophyly of Ctenotus and Ctenotus plus Lerista suggests higher diversification rates for these lineages relative to the remainder of the Australian sphenomorphine radiation, because 75% of the species in the radiation fall within these genera.
(b) Variation in diversification rates
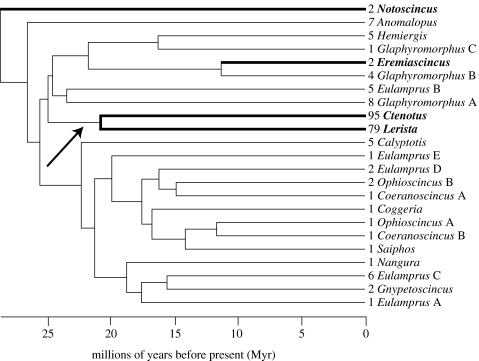
Cross-validation analysis of PL trees with different smoothing parameters and of a tree with branch lengths estimated under a molecular clock constraint indicated optimal performance of PL with a smoothing parameter of 100. We pruned the PL tree to include 23 lineages for which we could assign levels of taxonomic diversity (figure 2). We inferred a basal divergence time of 28.2 Myr BP (Myr ago; range: 19.1–53.6 Ma) using ML distances between Notoscincus and other Australian sphenomorphine skinks. In the absence of robust fossil calibration points, we prefer to treat this age estimate provisionally, but note that a recently published substitution rate for squamate reptile ND4 (1.6% Myr−1) would imply that the radiation has occurred even more recently (Feldman & Spicer 2006). With the exception of the absolute magnitude of inferred diversification rates, all results are independent of the proposed age of the sphenomorphine radiation.
Figure 2.
Penalized likelihood chronogram for Australian sphenomorphine diversification derived from Bayesian consensus phylogram, indicating taxonomic diversity of each terminal. Basal divergence of 28.2 Ma is based on estimated mtDNA rate of 0.895 substitutions Myr−1. The four paraphyletic genera (Coeranoscincus, Eulamprus, Glaphyromorphus and Ophioscincus) were assigned taxonomic diversity levels based on the full phylogeny (figure 1) or from prior information regarding morphological species groups. Results of diversification analyses are robust to species richness assigned to paraphyletic genera (electronic supplementary material, figure s1). ML estimate of rate-shift location inferred under two-rate diversification model is indicated by an arrow. Bold taxon names and branch lengths denote the four arid-adapted taxa.
We were able to assign taxonomic diversity levels to the four paraphyletic genera, because taxon sampling included most or all species within three of those genera (2/2 Coeranoscincus, 2/3 Ophioscincus, 11/13 Glaphyromorphus) and because major lineages within Eulamprus correspond to phenotypically and ecologically distinctive categories (O'Connor & Moritz 2003). To verify that results were robust to alternative species richness assignments, we generated 100 datasets by randomly partitioning generic diversity into paraphyletic lineages (e.g. the 15 recognized species of Eulamprus were divided at random among Eulamprus lineages A, B, C, D and E in each replicate dataset) and repeated the full complement of diversification analyses on each. Shuffling diversity levels within paraphyletic genera in this fashion did not change results described below (electronic supplementary material, figure s1).
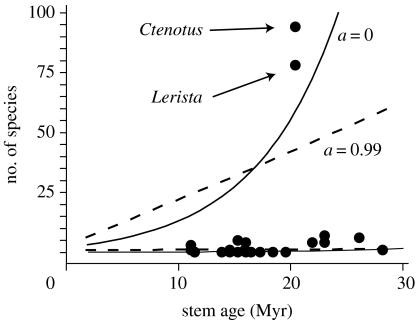
Comparisons of observed species diversity within terminal taxa relative to those expected under a homogeneous net diversification rate across all Australian sphenomorphine skinks reject the null hypothesis that net diversification rates have been constant among lineages (p<0.013, a=0; p<0.031, a=0.99; figure 3). Regardless of whether sphenomorphine diversification rates are estimated using the combined taxonomic/phylogenetic approach described in this study (figure 3) or the whole-clade estimator (p<0.001; electronic supplementary material, table s4), there is clearly an excess of species-poor or species-rich lineages. Although this test is conservative (see electronic supplementary material, table s4e), these results are robust to assumptions about the underlying model of extinction. Therefore, clade age alone does not explain the striking disparity in species richness among sphenomorphine skinks.
Figure 3.
Relationship between stem clade age and extant diversity for Australian sphenomorphine skinks. Lines indicate the 95% confidence envelope around the expected species diversity through time for a clade that diversifies with a rate equal to that observed across the Australian sphenomorphines considered as a whole under relative extinction rates of a=0 (solid line) and a=0.99 (dashed line). Ctenotus and Lerista have an excess of species and the remaining lineages have a deficit of species assuming a constant diversification rate for the entire sphenomorphine clade (a=0, p<0.013; a=0.99, p<0.032). Net diversification rate was estimated from combined taxonomic/phylogenetic data.
(c) Shifts in diversification rate
The data clearly reject the constant diversification model in favour of the flexible-rate model (p<0.001; table 1). The ML estimate of the shift point in the flexible-rate model is the node corresponding to the most recent common ancestor (MRCA) of Ctenotus and Lerista (figure 2) under extinction fractions of both a=0 and 0.99; ML estimates of r for Ctenotus/Lerista and all other Australian sphenomorphine lineages suggest that the net diversification rate within this clade has increased approximately 2.5-fold (a=0) to 15-fold (a=0.99) relative to that observed across the remainder of the tree. While palaeontological evidence supports the view that relative extinction rates have generally been high (e.g. Stanley 1979; Gilinsky 1994; Alroy 1996), we are aware of no evidence suggesting that extant clades have diversified in the absence of extinction. It is thus probable that the true magnitude of the rate increase for Ctenotus and Lerista exceeded that inferred under a=0. Rate shifts at other internal nodes are far less likely than those at the MRCA of Ctenotus/Lerista, as assessed by the difference in likelihood scores (ΔL) between the best-fit location of the rate shift and alternative nodes (figure 2): MRCACtenotus–Hemiergis, ΔL=4.06; MRCACtenotus–Anomalopus, ΔL=9.99; shift on Ctenotus branch only, ΔL=7.05; and shift on Lerista branch only, ΔL=8.36 (ΔL given for a=0, but results for a=0.99 are qualitatively similar).
Table 1.
Model-based analysis of diversification rates in Australian sphenomorphine skinks. (Flexible-rate model considers all possible bipartitions of tree and finds bipartition giving the highest likelihood when net diversification rates are optimized separately to each partition. Rate-decrease model assumes that Ctenotus and Lerista have retained the ancestral diversification rate present at the root node.)
| model | constant-rate | flexible-rate | rate-decrease |
|---|---|---|---|
| log L (ΔAICa) | log L (ΔAIC) | log L (ΔAIC) | |
| a=0 | −117.6 (19.0)b | −106.1 (0) | −111.6 (10.9) |
| parametersc | r=0.135 | r=0.086; rCL=0.218 | r=0.073; rCL=0.160 |
| a=0.99 | −136.3 (32.7)b | −117.9 (0) | − 124.7 (13.6) |
| parametersc | r=0.005 | r=0.002; rCL=0.030 | r=0.0003; rCL=0.005 |
ΔAIC is the difference in AIC scores between each model and the overall best-fit model.
The data reject the constant-rate model in favour of the flexible-rate model under a=0 and 0.99 (Χ2≥23; p<0.001). Rate-decrease model does not show a simple nested relationship with flexible-rate model, but AIC strongly favours the flexible-rate model.
Maximum-likelihood estimate of the net diversification rate r in lineages Myr−1. rCL is the net diversification rate of the subtree partition containing Ctenotus and Lerista.
The hypothesis that Lerista/Ctenotus have merely retained high diversification rates from the ancestral sphenomorphine lineage is not supported, as this scenario would require that diversification rates have decreased multiple times throughout the tree. The model specifying increased diversification at the Lerista/Ctenotus MRCA performs much better than a model where no rate increase is permitted between the root node and Lerista/Ctenotus (rate-decrease model; table 1), as assessed by the AIC. This result is not conditional on the topology and branch lengths shown in figure 2, because the posterior distribution of the difference in likelihood scores between the flexible-rate and rate-decrease models strongly favours the former model (electronic supplementary material, figure s2).
Because our analyses are partially dependent on taxonomy, our results could have been influenced by a failure to adequately account for true species diversity within genera. Very few intraspecific phylogeographic studies have been conducted on sphenomorphine skinks, and we predict that future investigations will reveal morphologically cryptic species within many taxa. However, this issue would only influence our results if Lerista and Ctenotus harbour proportionately fewer cryptic species than other taxa.
(d) Aridification of Australia and diversification rates
The progressive expansion of the Australian arid zone during the past 25 Myr appears to have catalysed increased diversification in a number of sclerophyllous plant clades, including eucalypts and casuarinas (Crisp et al. 2004). Likewise, some lineages of agamid lizards experienced rapid diversification during the aridification of Australia (Harmon et al. 2003; Hugall & Lee 2004; Rabosky 2006b). The majority of species within both Lerista and Ctenotus occur in the arid and semi-arid regions of Australia, suggesting a possible link between aridification and diversification rates in this group. Temporal calibration of the sphenomorphine tree (figure 2) yields dates for the radiations of Ctenotus and Lerista that accord well with the known chronology for the aridification of Australia and that show broad congruence with the timing of diversification in these other arid-adapted groups.
Given the diversity of these two genera in the Australian arid zone, it is striking that only a few other lineages of sphenomorphine skinks (figure 2) can be considered arid zone taxa—two species each of Notoscincus and Eremiascincus. Thus, while nearly half of all the species of sphenomorphine skinks occur in the arid and semi-arid regions of the continent, this diversity is concentrated within just 4 of 23 major lineages (figure 2). Yet the arid and semi-arid climatic regions of Australia are defined by a climatic zone that occupies more than three-fourths of the continent's surface area (James & Shine 2000). That so few major lineages have succeeded in entering a geographically vast region, which nonetheless comprises a small fraction of the total climatic diversity in Australia (James & Shine 2000), suggests the possibility that phylogenetic conservatism of climatic tolerances (Ricklefs & Latham 1992; Wiens & Graham 2005) may underlie the exceptional diversification of a few sphenomorphine lineages. Such phylogenetic conservatism of traits is hypothesized to limit the dispersal of lineages between tropical and temperate regions (Ricklefs & Latham 1992; Wiens et al. 2006) and among elevational zones in the montane tropics (Wiens et al. 2007); we similarly hypothesize that such constraints have influenced the ability of lineages to shift from mesic-to-arid environments.
Can these diversification patterns be explained by niche conservatism in conjunction with the larger area of the arid zone, relative to the more mesic regions of continental Australia? There is considerable evidence that the geographical area occupied by a clade exerts a strong effect on diversification rates (Losos & Schluter 2000; Ricklefs 2003; Davies et al. 2005). The mesic-to-arid gradient might act as an environmental filter, limiting the dispersal of lineages between those environments, and the larger area of the arid zone would thus provide an expanded theatre of diversification for clades that successfully made this transition. If arid Australia harbours a diversity of sphenomorphine species proportional to its geographical area, and if a restricted number of clades have been able to enter this climatic zone, we would expect to observe increased diversification rates in those clades as a consequence of the interaction between the geographical area and phylogenetic conservatism of climatic tolerance. Indeed, Ctenotus species richness within major climatic regions of Australia is proportional to the geographical area occupied by those zones (James & Shine 2000). This suggests that part of the answer to ‘why do so many Ctenotus species occur in the arid zone’ (Pianka 1972; Morton & James 1988; James & Shine 2000) may come from understanding why other sphenomorphine clades fail to show this pattern.
The phylogenetic conservatism/geographical area hypothesis might only be part of the explanation for the increased diversification of Ctenotus and Lerista relative to other sphenomorphine skinks. Assuming that the ancestral Australian sphenomorphine was not arid adapted, which is likely based on the general climatic tolerances of sphenomorphine outgroup taxa, only four major lineages have successfully made the transition to arid environments (if the putative shift in climatic tolerance occurred in the ancestor of Ctenotus and Lerista, then only three lineages have made this transition). Yet two of these lineages failed to radiate (Notoscincus and Eremiascincus). Previous studies have found that the Ctenotus lineage is characterized by a substantial increase in both critical thermal maximum temperatures and preferred active temperatures relative to other sphenomorphine species, including Eremiascincus (Benett & John-Alder 1986; Huey & Bennett 1987; Garland et al. 1991). Although data are needed for Lerista and Notoscincus, these results suggest the possibility that traits related to thermal physiology might underlie the dramatic radiation of these groups in the arid zone. However, despite a close phylogenetic relationship, Ctenotus and Lerista have followed radically different evolutionary paths: Ctenotus is diurnal, surface active and shows minimal interspecific variation in body shape, whereas Lerista is fossorial to cryptozoic, frequently nocturnal and shows a tremendous range of interspecific variation in limb reduction and body elongation (Greer 1989).
4. Conclusions
This study demonstrates exceptional heterogeneity in diversification rates within a major continental radiation. The high species richness of Ctenotus and Lerista relative to other genera of Australian skinks cannot be explained by clade age or as a taxonomic artefact.
Tremendous variation in diversification rates appears to have characterized several continental plant radiations (Hodges & Arnold 1995; Klak et al. 2004; Hughes & Eastwood 2006), but we are unaware of a comparable, sustained contrast in diversification rates within animal radiations restricted to a geographically contiguous region. The diversification of Australian sphenomorphine skinks has occurred against a background of apparently strong phylogenetic niche conservatism, and we speculate that a key physiological innovation may have catalysed diversification in some, but not all, arid-adapted lineages.
Acknowledgments
This research was supported by NSF-OSIE-0612855, the Mario Einaudi Center at Cornell University, and an EAPSI Fellowship to D. Rabosky administered jointly by NSF and the Australian Academy of Science. All research was conducted in accordance with the legal requirements of the USA and Australia. We thank A. Agrawal, M. Hutchinson, A. McCune and the CLO Evolutionary Biology Lab discussion group for comments on the manuscript and/or valuable discussion of these topics. We are especially grateful to A. Skinner for providing sequences and alignments from Skinner (in press).
Appendix A.
Here, we derive our estimator of the net diversification rate r and explain differences from previous estimators (Sanderson & Wojciechowski 1996; Paradis 2003). The taxonomic data consists of T terminals in a molecular phylogeny with branch length data. The length of each terminal branch represents the stem clade age of each taxon or the maximum elapsed time between a single ancestral species and n descendant species. Let λ and μ represent per-lineage probabilities of speciation and extinction, and define r=λ−μ and a=μ/λ. Under the birth–death process, the probability of observing n species in the present given a single ancestral species is given by (Kendall 1948)
| (A1) |
where
| (A2) |
| (A3) |
and the probability of observing zero descendant species is
| (A4) |
This is identical to Raup (1985, eqn (A 17)) for the case of a single ancestral species. A ML estimator of r based only on the taxonomic data can be obtained from the product of the probabilities of the taxonomic data
| (A5) |
This is the likelihood used by Paradis (2003). However, if a diversification process has left zero descendants in the present, it cannot be observed. Thus, we condition Pr(n|t) on the probability that the clade has survived to the present (Nee et al. 1994), which it does with a probability of 1−α. After conditioning,
| (A6) |
Multiplying the probabilities for each of the T terminals together and taking the log gives the log likelihood of the taxonomic data (equation (2.1)). The phylogenetic component consists of N internal branches, each beginning with a single lineage and ending with a speciation event. The probability density of each branching event, conditional upon each lineage surviving to the present, is given by Nee et al. (1994, eqn (17)). For the special case of a single ancestral lineage, we have
| (A7) |
where x is the birth time or branching time (Nee et al. 1994; Nee 2001) of the lineage in time units before present. Multiplying the probability densities for each of the N branching events and taking the log of both sides gives equation (2.3). This phylogenetic likelihood differs from Paradis (2003), in that Paradis (2003, eqn (2.5)) includes the likelihood that each of the T terminal lineages does not undergo a speciation event on the terminal branch. This is the case for a complete phylogeny, but we know that at least some of the T terminals have left more than one progeny in the present; furthermore, the likelihoods of the terminal branches have already been considered in the taxonomic portion of the expression (LT; equation (2.1)).
The log likelihood of phylogenetic data plus taxonomic/diversity data is given by the sum of equation (2.1) and equation (2.3). Under the special case of a=0, this expression is identical to the Yule process model used by Sanderson & Wojciechowski (1996). Numerical optimization techniques can be used to find the ML estimate of r under any assumed level of background extinction (μ<λ).
Supplementary Material
Detailed supplement to main text, including specimens used and sampling localities, summary of parsimony and likelihood phylogenetic analyses, and results from several sensitivity analyses
References
- Alroy J. Constant extinction, constrained diversification, and uncoordinated stasis in North American mammals. Palaeogeogr. Palaeoclim. Palaeoecol. 1996;127:285–311. doi:10.1016/S0031-0182(96)00100-9 [Google Scholar]
- Altekar G, Dwarkadas S, Huelsenbeck J.P, Ronquist F. Parallel metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics. 2004;20:407–415. doi: 10.1093/bioinformatics/btg427. doi:10.1093/bioinformatics/btg427 [DOI] [PubMed] [Google Scholar]
- Bennett A.F, John-Alder H. Thermal relations of some Australian skinks (Sauria, Scincidae) Copeia. 1986;198b:57–64. doi:10.2307/1444888 [Google Scholar]
- Brandley M.C, Schmitz A, Reeder T.W. Partitioned Bayesian analyses, partition choice, and the phylogenetic relationships of scincid lizards. Syst. Biol. 2005;54:373–390. doi: 10.1080/10635150590946808. doi:10.1080/10635150590946808 [DOI] [PubMed] [Google Scholar]
- Calsbeek R, Thompson J.N, Richardson J.E. Patterns of molecular evolution and diversification in a biodiversity hotspot: the California Floristic Province. Mol. Ecol. 2003;12:1021–1029. doi: 10.1046/j.1365-294x.2003.01794.x. doi:10.1046/j.1365-294X.2003.01794.x [DOI] [PubMed] [Google Scholar]
- Cogger H.G. Reed; Sydney, Australia: 2000. Reptiles and amphibians of Australia. [Google Scholar]
- Crisp M, Cook L, Steane D. Radiation of the Australian flora: what can comparisons of molecular phylogenies across multiple taxa tell us about the evolution of diversity in present-day communities? Phil. Trans. R. Soc. B. 2004;359:1551–1571. doi: 10.1098/rstb.2004.1528. doi:10.1098/rstb.2004.1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies T.J, Barraclough T.G, Chase M.W, Soltis P.S, Soltis D.E, Savolainen V. Darwin's abominable mystery: insights from a supertree of the angiosperms. Proc. Natl Acad. Sci. USA. 2004;101:1904–1909. doi: 10.1073/pnas.0308127100. doi:10.1073/pnas.0308127100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies T.J, Savolainen V, Chase M.W, Goldblatt P, Barraclough T.G. Environment, area, and diversification in the species-rich flowering plant family Iridaceae. Am. Nat. 2005;166:418–425. doi: 10.1086/432022. doi:10.1086/432022 [DOI] [PubMed] [Google Scholar]
- Feldman C.R, Spicer G.S. Comparative phylogeography of woodland reptiles in California: repeated patterns of cladogenesis and population expansion. Mol. Ecol. 2006;15:2201–2222. doi: 10.1111/j.1365-294X.2006.02930.x. doi:10.1111/j.1365-294X.2006.02930.x [DOI] [PubMed] [Google Scholar]
- Garland T, Jr, Huey R.B, Bennett A.F. Phylogeny and coadaptation of thermal physiology in lizards—a reanalysis. Evolution. 1991;45:1969–1975. doi: 10.1111/j.1558-5646.1991.tb02703.x. doi:10.2307/2409846 [DOI] [PubMed] [Google Scholar]
- Gilinsky N.L. Volatility and the Phanerozoic decline of background extinction intensity. Paleobiology. 1994;20:445–458. [Google Scholar]
- Greer A.E. Surrey Beatty & Sons; Chipping Norton, Australia: 1989. Biology and evolution of Australian lizards. [Google Scholar]
- Harmon L.J, Schulte J.A, Larson A, Losos J.B. Tempo and mode of evolutionary radiation in iguanian lizards. Science. 2003;301:961–964. doi: 10.1126/science.1084786. doi:10.1126/science.1084786 [DOI] [PubMed] [Google Scholar]
- Hodges S.A, Arnold M.L. Spurring plant diversification: are floral nectar spurs a key innovation? Proc. R. Soc. B. 1995;262:343–348. doi:10.1098/rspb.1995.0215 [Google Scholar]
- Huelsenbeck J.P, Ronquist F, Nielsen R, Bollback J.P. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–2314. doi: 10.1126/science.1065889. doi:10.1126/science.1065889 [DOI] [PubMed] [Google Scholar]
- Huey R.B, Bennett A.F. Phylogenetic studies of coadaptation—preferred temperatures versus optimal performance temperatures of lizards. Evolution. 1987;41:1098–1115. doi: 10.1111/j.1558-5646.1987.tb05879.x. doi:10.2307/2409194 [DOI] [PubMed] [Google Scholar]
- Hugall A.F, Lee M.S.Y. Molecular claims of Gondwanan age for Australian agamid lizards are untenable. Mol. Biol. Evol. 2004;21:2102–2110. doi: 10.1093/molbev/msh219. doi:10.1093/molbev/msh219 [DOI] [PubMed] [Google Scholar]
- Hughes C, Eastwood R. Island radiation on a continental scale: exceptional rates of plant diversification after uplift of the Andes. Proc. Natl Acad. Sci. USA. 2006;103:10 334–10 339. doi: 10.1073/pnas.0601928103. doi:10.1073/pnas.0601928103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James C.D, Shine R. Why are there so many coexisting species of lizards in Australian deserts? Oecologia. 2000;125:127–141. doi: 10.1007/PL00008884. doi:10.1007/PL00008884 [DOI] [PubMed] [Google Scholar]
- Jennings W.B, Pianka E.R, Donnellan S. Systematics of the lizard family pygopodidae with implications for the diversification of Australian temperate biotas. Syst. Biol. 2003;52:757–780. doi:10.1080/10635150390250974 [PubMed] [Google Scholar]
- Kendall D.G. On the generalized birth-and-death process. Ann. Math. Stat. 1948;19:1–15. [Google Scholar]
- Klak C, Reeves G, Hedderson T. Unmatched tempo of evolution in Southern African semi-desert ice plants. Nature. 2004;427:63–65. doi: 10.1038/nature02243. doi:10.1038/nature02243 [DOI] [PubMed] [Google Scholar]
- Kozak K.H, Weisrock D.W, Larson A. Rapid lineage accumulation in a non-adaptive radiation: phylogenetic analysis of diversification rates in eastern North American woodland salamanders (Plethodontidae: Plethodon) Proc. R. Soc. B. 2006;273:539–546. doi: 10.1098/rspb.2005.3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losos J.B, Schluter D. Analysis of an evolutionary species–area relationship. Nature. 2000;408:847–850. doi: 10.1038/35048558. doi:10.1038/35048558 [DOI] [PubMed] [Google Scholar]
- Macey J.R, Verma A. Homology in phylogenetic analysis: alignment of transfer RNA genes and the phylogenetic position of snakes. Mol. Phylogenet. Evol. 1997;7:272–279. doi: 10.1006/mpev.1997.0379. doi:10.1006/mpev.1997.0379 [DOI] [PubMed] [Google Scholar]
- Magallon S, Sanderson M.J. Absolute diversification rates in angiosperm clades. Evolution. 2001;55:1762–1780. doi: 10.1111/j.0014-3820.2001.tb00826.x. [DOI] [PubMed] [Google Scholar]
- Martin J.E, Hutchinson M.N, Meredith R, Case J.A, Pledge N.S. The oldest genus of scincid lizard (Squamata) from the tertiary etadunna formation of South Australia. J. Herpetol. 2004;38:180–187. doi:10.1670/25-03A [Google Scholar]
- McKenna D.D, Farrell B.D. Tropical forests are both evolutionary cradles and museums of leaf beetle diversity. Proc. Natl Acad. Sci. USA. 2006;103:10 947–10 951. doi: 10.1073/pnas.0602712103. doi:10.1073/pnas.0602712103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPeek M.A, Brown J.M. Clade age and not diversification rate explains species richness among animal taxa. Am. Nat. 2007;169:E97–E106. doi: 10.1086/512135. doi:10.1086/512135 [DOI] [PubMed] [Google Scholar]
- Mooers A.O, Heard S.B. Evolutionary process from phylogenetic tree shape. Q. Rev. Biol. 1997;72:31–54. doi:10.1086/419657 [Google Scholar]
- Morton S.R, James C.D. The diversity and abundance of lizards in arid Australia—a new hypothesis. Am. Nat. 1988;132:237–256. doi:10.1086/284847 [Google Scholar]
- Nee S. Inferring speciation rates from phylogenies. Evolution. 2001;55:661–668. doi: 10.1554/0014-3820(2001)055[0661:isrfp]2.0.co;2. doi:10.1554/0014-3820(2001)055[0661:ISRFP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Nee S, May R.M, Harvey P.H. The reconstructed evolutionary process. Phil. Trans. R. Soc. B. 1994;344:305–311. doi: 10.1098/rstb.1994.0068. doi:10.1098/rstb.1994.0068 [DOI] [PubMed] [Google Scholar]
- Nylander, J. A. A. 2004 MrModeltest See http://www.abc.se/∼nylander/
- O'Connor D, Moritz C. A molecular phylogeny of the Australian skink genera Eulamprus, Gnypetoscincus and Nangura. Aust. J. Zool. 2003;51:317–330. doi:10.1071/ZO02050 [Google Scholar]
- Paradis E. Analysis of diversification: combining phylogenetic and taxonomic data. Proc. R. Soc. B. 2003;270:2499–2505. doi: 10.1098/rspb.2003.2513. doi:10.1098/rspb.2003.2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillimore A.B, Freckleton R.P, Orme C.D.L, Owens I.P.F. Ecology predicts large-scale patterns of phylogenetic diversification in birds. Am. Nat. 2006;168:220–229. doi: 10.1086/505763. doi:10.1086/505763 [DOI] [PubMed] [Google Scholar]
- Pianka E.R. Zoogeography and speciation of Australian desert lizards—ecological perspective. Copeia. 1972;1972:127–145. doi:10.2307/1442789 [Google Scholar]
- Rabosky D.L. LASER: a maximum likelihood toolkit for inferring temporal shifts in diversification rates. Evol. Bioinform. Online. 2006a;2:257–260. [PMC free article] [PubMed] [Google Scholar]
- Rabosky D.L. Likelihood methods for detecting temporal shifts in diversification rates. Evolution. 2006b;60:1152–1164. [PubMed] [Google Scholar]
- Raup D.M. Mathematical models of cladogenesis. Paleobiology. 1985;11:42–52. [Google Scholar]
- Reeder T.W. A phylogeny of the Australian Sphenomorphus group (Scincidae: Squamata) and the phylogenetic placement of the crocodile skinks (Tribolonotus): Bayesian approaches to assessing congruence and obtaining confidence in maximum likelihood inferred relationships. Mol. Phylogenet. Evol. 2003;27:384–397. doi: 10.1016/s1055-7903(02)00448-7. doi:10.1016/S1055-7903(02)00448-7 [DOI] [PubMed] [Google Scholar]
- Richmond J.Q, Jokusch E.L. Body size evolution simultaneously creates and collapses species boundaries in a clade of scincid lizards. Proc. R. Soc. B. 2007;274:1701–1708. doi: 10.1098/rspb.2007.0364. doi:10.1098/rspb.2007.0364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklefs R.E. Global diversification rates of passerine birds. Proc. R. Soc. B. 2003;270:2285–2291. doi: 10.1098/rspb.2003.2489. doi:10.1098/rspb.2003.2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklefs R.E. Global variation in the diversification rate of passerine birds. Ecology. 2006;87:2468–2478. doi: 10.1890/0012-9658(2006)87[2468:gvitdr]2.0.co;2. doi:10.1890/0012-9658(2006)87[2468:GVITDR]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Ricklefs R.E, Latham R.E. Intercontinental correlation of geographical ranges suggests stasis in ecological traits of relict genera of temperate perennial herbs. Am. Nat. 1992;139:1305–1321. doi:10.1086/285388 [Google Scholar]
- Ruber L, Zardoya R. Rapid cladogenesis in marine fishes revisited. Evolution. 2005;59:1119–1127. [PubMed] [Google Scholar]
- Sanderson M.J. Reconstructing the evolution of genes and organisms using maximum likelihood. In: Fambrough D, editor. Molecular evolution of physiological processes. Rockefeller; New York, NY: 1994. pp. 13–26. [Google Scholar]
- Sanderson M.J. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol. Biol. Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- Sanderson M.J. r8s; inferring absolute rates of evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. doi:10.1093/bioinformatics/19.2.301 [DOI] [PubMed] [Google Scholar]
- Sanderson M.J, Wojciechowski M.F. Diversification rates in a temperate legume clade: are there “so many species”; of Astragalus (Fabaceae)? Am. J. Bot. 1996;83:1488–1502. doi:10.2307/2446103 [Google Scholar]
- Sims H.J, McConway K.J. Non-stochastic variation of species-level diversification rates within angiosperms. Evolution. 2003;57:460–479. doi: 10.1111/j.0014-3820.2003.tb01538.x. [DOI] [PubMed] [Google Scholar]
- Skinner, A. In press. Phylogenetic relationships and rate of early diversification of Australian Sphenomorphus group scincids (Scincoidea, Squamata). Biol. J. Linn. Soc
- Slowinski J.B, Guyer C. Testing the stochasticity of patterns of organismal diversity—an improved null model. Am. Nat. 1989;134:907–921. doi:10.1086/285021 [Google Scholar]
- Stanley S.M. Freeman; San Francisco, CA: 1979. Macroevolution: pattern and process. [Google Scholar]
- Storr G.M, Smith L.A, Johnstone R.E. Western Australian Museum; Perth, Australia: 1999. Lizards of Western Australia. I. Skinks. [Google Scholar]
- Thompson J.D, Gibson T.J, Plewniak F, Jeanmougin F, Higgins D.G. The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. doi:10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock M.C. Combining probability from independent tests: the weighted Z-method is superior to Fisher's approach. J. Evol. Biol. 2005;18:1368–1373. doi: 10.1111/j.1420-9101.2005.00917.x. doi:10.1111/j.1420-9101.2005.00917.x [DOI] [PubMed] [Google Scholar]
- Wiens J.J, Graham C.H. Niche conservatism: integrating evolution, ecology, and conservation biology. Annu. Rev. Ecol. Syst. 2005;36:519–539. doi:10.1146/annurev.ecolsys.36.102803.095431 [Google Scholar]
- Wiens J.J, Graham C.H, Moen D.S, Smith S.A, Reeder T.W. Evolutionary and ecological causes of the latitudinal diversity gradient in hylid frogs: treefrog trees unearth the roots of high tropical diversity. Am. Nat. 2006;168:579–596. doi: 10.1086/507882. doi:10.1086/507882 [DOI] [PubMed] [Google Scholar]
- Wiens J.J, Parra-Olea G, García-París M, Wake D.B. Phylogenetic history underlies elevational biodiversity patterns in tropical salamanders. Proc. R. Soc. B. 2007;274:919–928. doi: 10.1098/rspb.2006.0301. doi:10.1098/rspb.2006.0301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S, Swan G. Princeton University Press; Princeton, NJ: 2003. Reptiles of Australia. [Google Scholar]
- Wuyts J, De Rijk P, Van de Peer Y, Winkelmans T, De Wachter R. The European large subunit ribosomal RNA database. Nucleic Acids Res. 2001;29:175–177. doi: 10.1093/nar/29.1.175. doi:10.1093/nar/29.1.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamudio K.R, Greene H.W. Phylogeography of the bushmaster (Lachesis muta: Viperidae): implications for neotropical biogeography, systematics, and conservation. Biol. J. Linn. Soc. 1997;62:421–442. doi:10.1006/bijl.1997.0162 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed supplement to main text, including specimens used and sampling localities, summary of parsimony and likelihood phylogenetic analyses, and results from several sensitivity analyses