Summary
The specificity of blood coagulation proteinases for substrate, inhibitor, and effector recognition is mediated by exosites on the surfaces of the catalytic domains, physically separated from the catalytic site. Some thrombin ligands bind specifically to either exosite I or II, while others engage both exosites. The involvement of different, overlapping constellations of exosite residues enables binding of structurally diverse ligands. The flexibility of the thrombin structure is central to the mechanism of complex formation and the specificity of exosite interactions. Encounter complex formation is driven by electrostatic ligand–exosite interactions, followed by conformational rearrangement to a stable complex. Exosites on some zymogens are in low affnity proexosite states and are expressed concomitant with catalytic site activation. The requirement for exosite expression controls the specificity of assembly of catalytic complexes on the coagulation pathway, such as the membrane-bound factor Xa•factor Va (prothrombinase) complex, and prevents premature assembly. Substrate recognition by prothrombinase involves a two-step mechanism with initial docking of prothrombin to exosites, followed by a conformational change to engage the FXa catalytic site. Prothrombin and its activation intermediates bind prothrombinase in two alternative conformations determined by the zymogen to proteinase transition that are hypothesized to involve prothrombin (pro)exosite I interactions with FVa, which underpin the sequential activation pathway. The role of exosites as the major source of substrate specificity has stimulated development of exosite-targeted anticoagulants for treatment of thrombosis.
Keywords: blood coagulation, exosites, factor Va, factor Xa, prothrombin, serine proteinases, zymogens
Introduction
Blood coagulation proteinases have evolved versatile and sensitive regulatory mechanisms for controlling the specificity of protein substrate recognition mediated by exosites. Exosites are surface sites on proteinases physically separated from the active site residues that determine the primary S1–S41 peptide substrate and inhibitor specificity. The concept of exosites in coagulation proteinases was introduced by John Fenton et al. around 1977 [2], and in molecular modeling of coagulation proteinase domains by Furie, Bing et al. in 1982 [3]. An extended substrate recognition site on thrombin, proposed to be intimately involved in its specificity for cleaving fibrinogen [4], was subsequently called the fibrinogen recognition exosite, or anion-binding exosite I due to its apparent affinity for negatively charged ligands [4]. Molecular modeling of the catalytic domains of thrombin, factor (F) Xa, and FIXa with chymotrypsin as a template, led to the prescient conclusion that substrate specificity is not determined primarily by the active site, but by surface insertion loops absent in chymotrypsin and unique to each coagulation proteinase, surrounding their catalytic sites [3]. After about 30 years of intensive research and numerous crystal structures of thrombin and its exosite ligand complexes, thrombin has emerged as the prototypical example of an exosite-regulated allosteric proteinase.
Many excellent reviews of the roles of exosite interactions in coagulation proteinase substrate specificity have been published previously, including many focused on the best studied thrombin exosites [5–11]. This review will focus on less covered aspects, such as: (i) structural and functional properties of thrombin exosites with a consideration of the dynamics of exosite-mediated protein–protein interactions; (ii) the role of low affinity exosite precursor forms (proexosites) and their expression via the zymogen-proteinase transition in controlling blood coagulation substrate specificity, in the context of prothrombin (ProT) activation; (iii) regulation of the substrate specificity of membrane-assembled proteinase–protein cofactor complexes, for the mechanism of substrate recognition by the prothrombinase complex; (iv) the role of FXa and (pro)thrombin exosites in kinetic control of the ProT activation pathway; and (v) the potential of developing anticoagulant therapy by targeting exosites.
Thrombin exosites
Two electropositive exosites, in near opposition on the thrombin surface, play crucial roles in the recognition of specific macromolecular substrates, effectors, and inhibitors, and their properties have been characterized by extensive crystallographic, mutagenesis, biophysics, and enzymology studies. Exosite I, composed of insertion loops 30–40 and 70–80 [12,13], mediates binding of fibrinogen, fibrin, PAR-1 and -4 substrate recognition, staphylocoagulase binding, and binding of the C-terminal 55–65 residue sequence of the thrombin-specific inhibitor, hirudin [5–8,11,14,15]. Exosite II, a positively charged cluster of residues, mediates different interactions specifically, including heparin [16], a specific monoclonal antibody [17], and the fragment 2 domain of ProT (F2) [18]. A variety of inhibitors from snake venoms and the saliva of hematophagous organisms also bind to thrombin exosites [11,19].
The thrombin catalytic site and the affinity of exosite I ligands are allosterically regulated by binding of Na+ [9,20,21]. Na+ binding switches thrombin from a ‘slow’ to a ‘fast form’ with higher specificity (kcat/Km) for the procoagulant substrates, fibrinogen, and PAR-1, greater reactivity toward antithrombin, and increased peptide substrate activity [8,20,22]. The Na+-free, slow form has higher relative specificity in protein C activation [22]. Thus, Na+ regulates the procoagulant and anticoagulant activities of thrombin. The structure of the slow form has been controversial, in that inactive thrombin conformations have also been concluded to represent the slow form [23–25]. Crystal packing interactions that influenced interpretation of previous structures of the fast, slow, and inactive forms are absent from the structure of thrombin D102N, which is thought to represent more closely the inactive slow form [26]. The thrombin D102N structure, and kinetic studies of Na+ binding [26–28], indicate that the catalytically active slow form is in equilibrium with a low level of inactive thrombin in which the catalytic and Na+ sites are blocked. Rapid-reaction kinetic studies of Na+ binding indicate that at 37 °C the inactive slow form is insignificantly populated [28]. Structures of the fast, slow, and inactive forms demonstrate dramatically the flexibility of thrombin.
Alanine scanning studies of thrombin exosite complexes provide a basis for interpreting the mechanisms involved in specific binding of structurally diverse protein substrates with strict specificities for exosite I or II. The structures illustrate a mechanism of specific ligand recognition in which the conformational flexibility of thrombin is necessary to accommodate diverse ligands with high specificity for each exosite. Figure 1 illustrates the structure of thrombin with exosite I residues that are most important for binding of particular ligands. It is apparent that exosite I-specific ligands bind competitively through overlapping constellations of distinctly different residues critical to each interaction. The epitope for Y63-sulfated hirudin-(55–65) involves exosite I extensively [15,29]. Fibrinogen [30,31] and PAR-1 [32] binding to exosite I overlap with the hirudin peptide and include many of the same residues. The extensive overlap in exosite I epitopes accounts for competitive binding of the hirudin peptide, fibrinogen, and PAR-1.
Fig. 1.
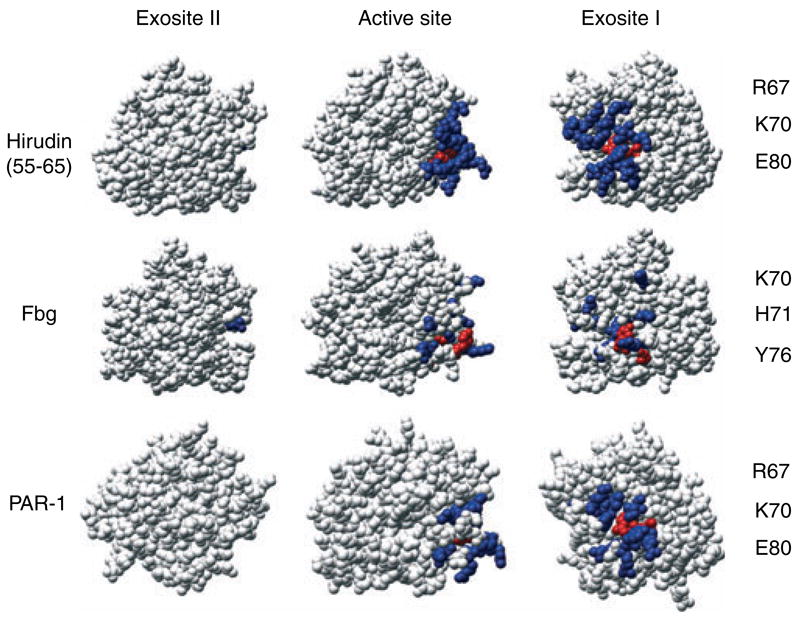
Functional mapping of thrombin exosite I residues involved in recognition of protein substrates. Thrombin structures are shown in standard Bode orientation [13] (middle) with the active site in the center, facing the reader. Two additional views are shown: c. 90° counter-clockwise rotation for exosite I (right), and c. 90° clockwise rotation for exosite II (left). Residues that cause at least a 2.3-fold effect by alanine-scanning mutagenesis are shown for exosite I (blue). For Y63-sulfated hirudin-(55–65): F34, K36, S36a, P37, Q38, L65, R67, K70, H71, R73, T74, R75, R77a, E80, E81, I82, M84, Y76, and E77a. For fibrinogen fragment E (Fbg): K36, R67, R73, K70, H71, Y76, R73, R77a, K81, and K110. For PAR-1: F34, R67, K70, R73, R75, Y76, R77a, E80, and I82. Residues that displayed the largest fold effect of alanine substitution are shown in red and listed on the right. The structures shown are those determined from thrombin complexes with Y63-sulfated hirudin peptide-(55–65) (1AFE), fibrinogen fragment E (1QVH), and PAR-1 (1NRN).
Rapid-reaction kinetic studies of hirudin and hirudin-(54–65) binding to thrombin support electrostatic steering of anionic exosite I ligands, such as the peptide, which mediates initial weak encounter complex formation, followed by establishing direct ionic interactions, and final conformational rearrangement of the exosite and ligand by engagement of hydrophobic interactions [33–35]. Thermodynamic studies of thrombin–hirudin interactions also support significant changes in the 70–80 loop of exosite I and structural ordering of the peptide on complex formation [36,37]. It has been suggested that all exosite I ligands follow a similar model, in which complementary asymmetric electrostatic fields enhance the rate of complex formation [35]. In agreement with this idea, analysis of the ionic strength dependence of the kinetics of fibrinogen cleavage by thrombin similarly supports diffusion-controlled electrostatic steering that facilitates initial formation of the transition state [38].
Exosite II interactions are stabilized mainly by electrostatic interactions, whereas exosite I interactions involve a greater contribution from hydrophobic interactions [6]. Exosite II-specific interactions with heparin [16] and ProT F2 [39] overlap to some extent (Fig. 2), but with only one common residue. As part of the GpIb-IX-V platelet receptor complex, GpIbα acts as a thrombin cofactor, producing a 5- to 7-fold enhancement in PAR-1 activation and 6- to tenfold acceleration of GpV cleavage, resulting in platelet activation, and a five thousandfold enhancement of FXI activation [40]. Two divergent crystal structures of GpIbα–thrombin complexes showed two thrombin molecules bound, one through exosite I and the other exosite II [41,42], which continued the controversy concerning the mode of thrombin binding. Mutagenesis studies with eight alanine substitutions of exosite II residues and two charge reversal mutants demonstrated decreased thrombin affinity [43,44], indicating exosite II binding (Fig. 2), whereas more limited exosite I mutagenesis showed no effect on GpIba binding [43–45].
Fig. 2.
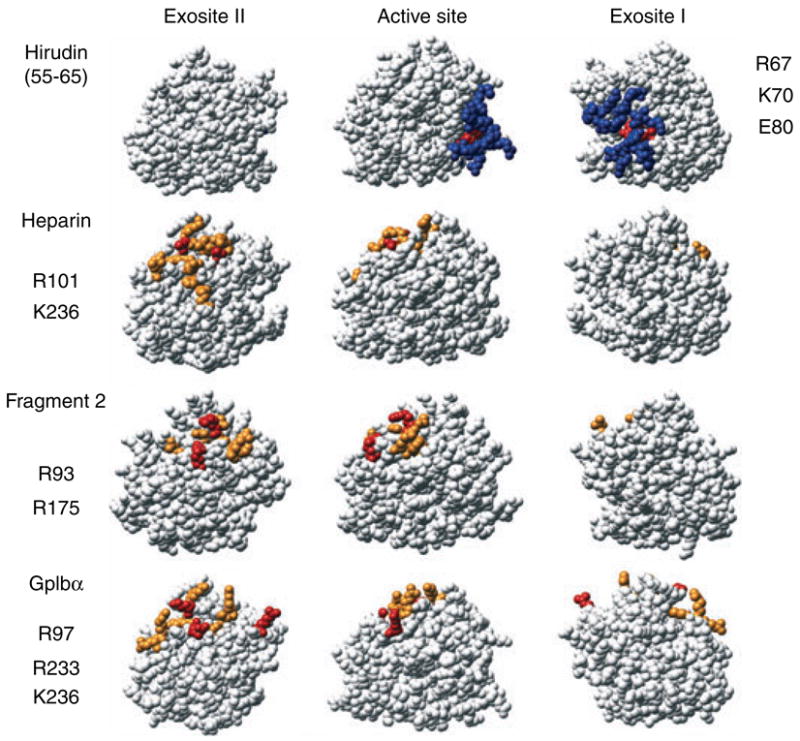
Functional mapping of thrombin exosite II residues involved in cofactor binding. Thrombin structures as described in Fig. 1, with the residues for interaction of Y63-sulfated hirudin-(55–65) shown as a reference. Residues identified by alanine-scanning mutagenesis as described in Fig. 1 for exosite I (blue) and exosite II (orange), with the exception of F2, where residues shown are those with the largest number of contacts in the crystal structure. For heparin: H91, R93, R101, R126, R165, H230, R233, K236, W237, and K240. For F2: P92, R93, Y94, W96, R97, R101, R175, and D178. For GpIbα, mutagenesis data indicated: R93, R97, R 101, R233, K236, and K240 [43], while the Dumas structure [40] also indicated direct contacts with R126, E127, N179, and K235. Red residues indicate the largest effect of alanine substitution or the largest number of contacts for F2, and are listed for exosite I (right) and exosite II (left). The structures of thrombin shown were those determined for the complexes with Y63-sulfated hirudin-(55–65) (1AFE), heparin (1XMN), F2 (2HPQ), and GpIbα (1P8V).
Some interactions make use of both exosites in determining binding specificity, presumably enhancing affinity and providing more strict control over the orientation of the bound thrombin for specific cleavages (Fig. 3). Binding of individual ligands to each exosite is typically accompanied by changes in tripeptide substrate specificity, and distinct fluorescence spectral changes reporting each binary interaction with thrombin active site-specifically labeled with tripeptide chloromethyl ketone-tethered fluorescence probes [46–50]. It had been suggested that exosite I and II binding was subject to extremely negative thermodynamic linkage, such that simultaneous interactions with both exosites was prevented allosterically [51]. An evaluation of linkage between binding of the exosite I ligands, fibrinogen, and hirudin peptide, and exosite II binding of a monoclonal antibody and F2 demonstrated no detectable inter-exosite linkage [17,50]. These studies concluded instead that binding of ligands to each exosite is linked to changes in the environment of the catalytic site. Ternary complexes formed with non-interacting ligands for both exosites produce additive or non-additive fluorescence changes, reporting changes in the active site environment [48,50]. These studies illustrate the sensitive linkage between the thrombin active site and exosite binding.
Fig. 3.
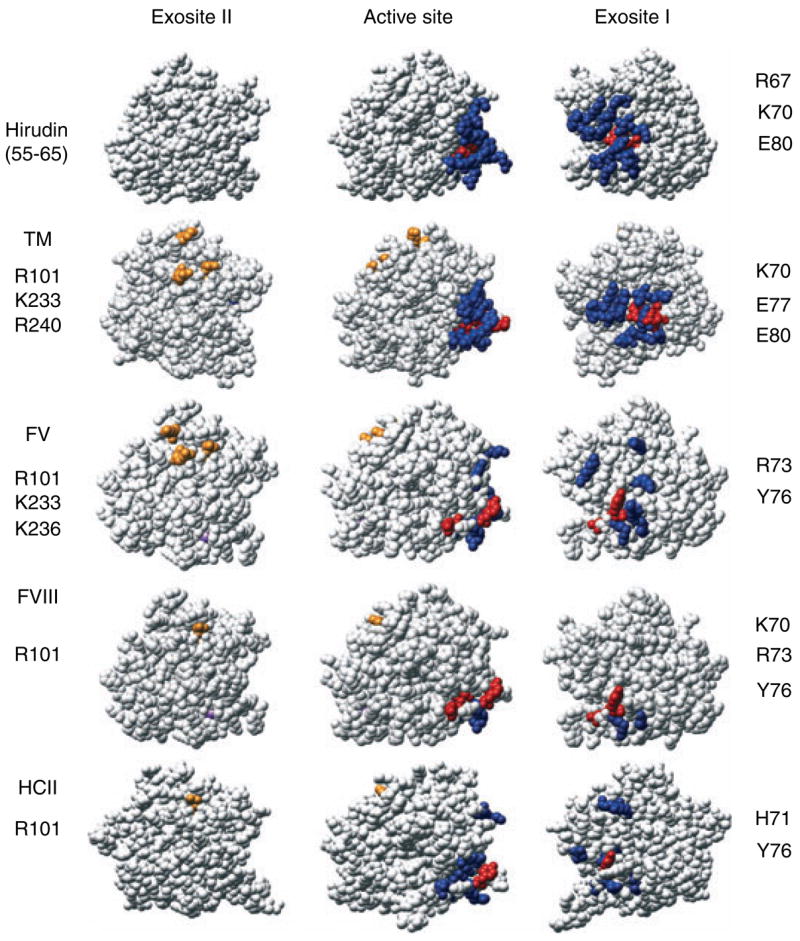
Functional mapping of thrombin ligands that use both exosites. Thrombin structures are shown in the same orientations as in Fig. 1, with residues involved in the complex with Y63-sulfated hirudin-(55–65) shown as a reference. Residues identified by alanine substitution for exosite I (blue) and exosite II (orange). For TM exosite I: F34, K36, P37, Q38a, L65, R67, K70, H71, R73, T74, R75, Y76, E77, R77a, E80, K81, I82, and K109, and in exosite II residues that interact with the chondroitin sulfate moiety: R101, R233, and K240. For factor V (FV) exosite I: K36, H71, R73, R75, Y76, R77a, K81, and exosite II: R101, R233, and K236. For factor VIII (FVIII) exosite I: K36, K70, H71, R73, R75, Y76 and exosite II: R101. For heparin cofactor II (HCII) exosite I: Q38, R67, K70, H71, R73, R75, Y76, K109, and K110, and in exosite II R101 is involved in heparin acceleration. Red residues are those displaying the largest effects of alanine substitution, which are listed for exosite I (right) and exosite II (left). The structures of thrombin shown were those determined for Y63-sulfated hirudin-(55–65) (1AFE), TM epidermal growth factor domain fragment-3,4,5 (1DX5), and heparin cofactor II (HCII) (1JMO). For FV and FVIII, where structures are unavailable, the mutagenesis data are shown on the fast form of thrombin with Na+ in purple (1SFQ).
Thrombomodulin interacts with exosite I residues [30,52] that also overlap those involving the hirudin peptide [29], fibrinogen [30], and PAR-1 binding [32], and with exosite II via its chondroitin sulfate moiety (Fig. 3). Factor V [53] and FVIII [54] substrate recognition are mediated primarily by a distinct group of exosite I residues, which are subsets of those involving the hirudin peptide. The epitopes overlap, but a smaller cluster of residues contributes to FVIII activation. Three important exosite II residues are involved in FV activation, while mutation of only one residue in exosite II has significant effects on FVIII activation. The differences between these interactions explain the primary involvement of exosite I and the secondary role of exosite II in FV and VIII activation [47,55–57]. Alanine scanning of heparin cofactor II (HCII) inactivation of thrombin showed interactions with exosite I to be important in the reaction, while only one residue in exosite II could be implicated in heparin acceleration [58].
The observations concerning the dynamics and electrostatic steering of thrombin protein–protein interactions parallel recent studies of the roles of flexibility and overlapping sites in the general mechanisms of protein–protein interactions. A good example is the binding of Enzyme I to the phosphocarrier protein of a bacterial signaling system [59,60]. Intermolecular paramagnetic relaxation enhancement studies using paramagnetic probe-labeled phosphocarrier protein were performed to detect sparsely populated complexes. Molecular modeling was used to generate a probability distribution structure of the non-specific encounter complexes. Many structurally distinct alternative encounter complexes are formed at low concentrations initially through electrostatic interactions. This is coupled to intramolecular rearrangement and water desolvation to the most stable complex observed crystallographically. This electrostatic searching mechanism of protein–protein interactions undoubtedly plays a major role in regulating the sensitivity and specificity of coagulation proteinase exosite binding. Molecular dynamics studies of protein–protein interactions also support a similar general model based on the critical role of flexibility [61].
Other experimental approaches have extended the understanding of the extent of thrombin conformational plasticity. Mapping mobility of thrombin by amide hydrogen/deuterium exchange demonstrated decreases in exchange of exosite I peptides accompanying active site inactivation with D-FPR-CH2Cl [62]. Comparison of ProT and thrombin showed that exosite I was more flexible in thrombin, whereas the autolysis loop and Na+ binding site exchanged more slowly [63]. Binding of a peptide from the γ′-fibrinogen variant to exosite II demonstrated that the 70–80 loop of exosite I and the autolysis loop were slower exchanging in the complex, suggesting long-range exosite I and II communication [64]. The source of the changes in intrinsic protein fluorescence accompanying binding of Na+ has been mapped by Phe substitution of each of the nine Trp residues [28]. Surprisingly, perturbation of all Trp residues occurred with formation of the thrombin•Na+ complex, with the largest contributions from W215 and W141, but significant, non-additive increases or decreases in fluorescence for all other Trp residues. The fluorescence and hydrogen/deuterium exchange results may reflect low energy structural rearrangements that may not be revealed by loss of function mutagenesis. W215 and W141 made equivalent, dominant contributions to the fluorescence change, but substitution of these or any of the other Trp residues was not associated with a significant change in Na+ binding [28]. Because Trp residues are dispersed over the whole molecule, the results favor the idea that binding of Na+, and probably other ligands to thrombin is accompanied by global effects on thrombin structure [28], which may not be detected from the crystal structure or mutagenesis. Although the changes in flexibility have been demonstrated clearly, which structural differences are involved in thrombin function and the mechanisms are at present unclear.
Exosites in the specificity of other blood coagulation proteinases
Factors Xa, IXa, VIIa, and activated protein C (APC) have topologically similar exosites I or II in their catalytic domains, which differ significantly in sequence [8]. Exosite I in these proteinases is the site of Ca2+ binding, which is absent from thrombin due to insertion of the K70 side chain into this site. Consequently, these proteinases are regulated by Ca2+, which is linked to Na+ binding sites in similar locations as in thrombin [65–68]. Exosite II of FXa is important for ProT substrate recognition and for the critical interaction with FVa [69,70] (Fig. 4). Experimental results from a number of studies of FXa–FVa interactions are nicely summarized in the context of a molecular model of the complex [71]. In FIXa and Xa, exosite II also overlaps the heparin binding site, which mediates enhancement of the rate of their inactivation by the antithrombin•heparin complex [72,73]. Exosite II of FIXa mediates its interaction with FVIIIa, which is linked to Ca2+ binding [65,74] (Fig. 4). APC, on the other hand, binds heparin through exosite I. The cationic exosite I of APC directs the interaction of APC with FVa, promoting initial cleavage at R506, and accommodating heparin binding that facilitates its inactivation by protein C inhibitor [75–79] (Fig. 4). Unlike thrombin, FXa, IXa, VIIa, and APC contain γ-carboxyglutamic acid domains required for calcium-dependent phospholipid membrane binding, which serves to localize their reactions to physiologic membranes. They also contain two epidermal growth factor-homologous domains, which mediate protein–protein interactions and participate in substrate recognition. Molecular modeling studies of the FXa•Va, FIXa•VIIIa, and APC•FVa complexes summarize the experimental evidence for these interactions [71,75,80,81]. The auxiliary domains contribute greatly to substrate specificity by controlling the assembly of membrane-bound productive enzyme–cofactor–substrate complexes, which enhance the rates of product formation 1000- to 300, 000-fold to the rates required for normal hemostasis [82–84]. Membrane assembly of coagulation complexes involves specific protein–protein interactions that provide the optimum orientation of the complex on the membrane surface and conformational changes due to exosite-mediated interactions critical to regulation of substrate specificity.
Fig. 4.
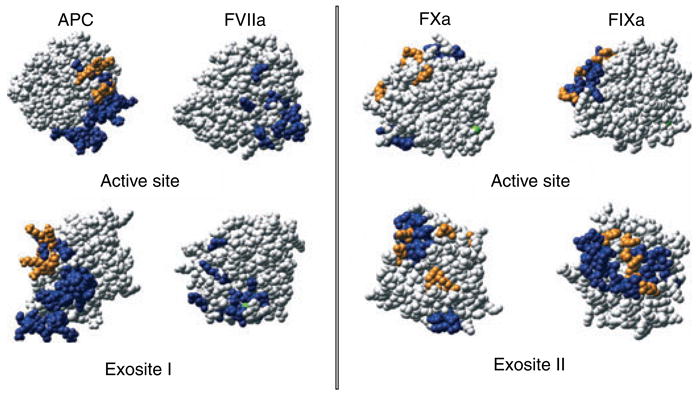
Comparison of exosite I and II locations on homologous proteinases, thrombin, factor Xa, APC, factor IXa, and factor VIIa. Structures are shown in standard Bode orientation (above) or turned to the relevant exosite, as indicated (below). Residues are colored blue for cofactor or substrate interactions and orange for the heparin binding site. For factor Xa (FXa): orange residues are R93, K96, R125, R165, K169, K236, and R240 [72]. Factor Xa residues involved in factor Va binding include all orange residues and blue residues D185-D189 [152] and V231-R245 [153]. For APC: orange heparin binding residues are K37, K38, K39, E60a, S61, K62, and K63 [77]. Blue residues involved in factor Va substrate recognition [154,155] are, S36-K39 (37 loop), D60-R67 (60 loop), E70-E80 (70–80 loop), and R147-R153 (autolysis loop) [156]. For factor IXa (FIXa): orange residues are R126, R165, R170, K230, and R233 and blue residues involved in factor VIIIa binding are K126-G134 (126-helix), L162-R170 (162-helix), F174-N178 [79,80]. For factor VIIa (FVIIa): blue residues involved in factor X substrate recognition are V21, K24, Q40, K60a, V67, E70, D72, L73, S74, E75, D79, E80, R148, T151, L153, E154, M156, T165, and K192 [105] determined by alanine scanning, excluding the residues that may directly impair substrate cleavage and Na+ binding [157]. The structures are the catalytic domains of FXa (1LQG), APC (1AUT), FIXa (1RFN), and FVIIa (1DVA).
Proexosite activation
Exosite I and the Na+ binding site on ProT are in low affinity, proexosite states that are expressed with the conformational changes that accompany catalytic site activation induced by insertion of the I16 N-terminus into the N-terminal binding pocket and formation of the critical salt bridge with D194 [46,85–89]. There is evidence that proexosite expression also plays a role in FXa, IXa and VIIa interactions. Activation of FX is apparently required for interaction of its exosite II with FVa and for expression of the Na+ site. These conclusions derive partly from Y225P FXa, which does not bind Na+, and exhibits weakened N-terminal insertion that allows it to assume a zymogen-like conformation [90]. The mutant has lower affinity for FVa, suggesting that FX binds weakly to FVa, and cofactor affinity is positively linked to Na+ binding [90]. Molecular dynamics studies of a FX model and FXa structure found major differences between the zymogen and proteinase catalytic domains in the Na+ and Ca2+ binding sites and a segment that participates in FVa binding, indicating that these sites are expressed by FX activation [91]. Direct studies of FX and FXa binding to FVa in solution and on platelets support proexosite expression in the observation that FX activation results in a >60-fold increase in affinity [92,93]. Based on structural studies of prethrombin 2 (Pre 2) [94] and FVII and VIIa [68,95] indicating the lack of formation of the Na+ binding site on both zymogens, and the Ca2+ binding site in the FVII proteinase domain [96], it was suggested that activation of other coagulation factors may be accompanied by formation of functional Na+ binding sites [90]. This suggests further that exosites on these enzymes linked to Na+ binding may also be present in proexosite states on the zymogens, with expression accompanying formation of the active proteinase.
The case of FIX appears similar to that of FX in that the heparin binding site (exosite II) overlaps segments mediating FVIIIa binding [73,97,98] (Fig. 4). Molecular modeling studies of the FIXa•VIIIa complex summarize the experimental evidence for these interactions [80]. Factor IXa binds tightly to platelets in the presence of FX and FVIIIa, but this is not diminished by a large excess of FIX, suggesting that the FVIIIa binding site is expressed on activation of FIX [99]. This is supported further by the demonstration that FIX activation is associated with a c. 28-fold increase in affinity for the FVIIIa light chain [100]. Similar to FX, disabling of the Na+ site by Y225P substitution weakens N-terminal insertion, affinity for FVIIIa, and catalytic efficiency, indicating that these functions are linked to the zymogen-proteinase transition [65]. There is scant evidence concerning whether exosite I of APC is in a proexosite state in the zymogen. Na+ binding to protein C could not be detected in intrinsic fluorescence studies [101], suggesting that this site is not formed. Using an inserted Trp residue as a reporter, however, Rezaie and co-workers [102] found that Ca2+ and Na+ binding were linked in the zymogen, with affinities for Na+ reduced 5- to 8-fold and Ca2+ increased 4-fold compared with APC [74,102]. Factor VIIa is an incompletely activated proteinase due to weak N-terminal insertion, which is induced by the cofactor, tissue factor (TF), through extended exosite interactions, including the FVIIa proteinase domain [96,103]. Mutations of certain residues in the D194 pocket of FVIIa that stabilize insertion increase TF affinity for FVIIa but not the zymogen, suggesting that the activating conformational change is required for allosteric regulation of TF affinity [104]. Factor VIIa has an electronegative cluster of residues in the catalytic domain in a position similar to the electropositive exosite I of thrombin, which play a key role in recognition of FX as a substrate [103,105,106] (Fig. 4). It has been proposed that TF stabilization of the inserted N-terminus is linked to restructuring of surface exosite residues for optimum substrate interactions [103]. A monoclonal antibody raised against the FVII zymogen and directed at the substrate-binding exosite binds with lower affinity to FVIIa compared with the zymogen, possibly reflecting expression of the exosite [103,106]. The kinetic pathway of blood coagulation zymogen activation is governed by exosite-driven assembly of coagulation complexes. Assembly of the FXa•Va and FIXa•VIIIa catalytic complexes are doubly regulated by the requirement for the activated protein cofactor as well as the active proteinase. The requirement for exosite expression coupled to catalytic activity ensures that the proteinase, activated cofactor and substrate are in the optimum conformations to catalyze the activation reaction specifically, and restricts premature activation of blood coagulation.
Thrombin exosite expression during ProT activation
ProT activation is accompanied by expression of exosites I and II on thrombin, as well as linkage between Na+ binding and exosite I, and the catalytic site. ProT is activated by sequential cleavage of two sites by the FXa•Va•membrane (prothrombinase) complex (described in an excellent review by Krishnaswamy [107]) (Fig. 5). Initial cleavage occurs at R320 in the catalytic domain, activating the catalytic site in the formation of meizothrombin (MzT). This is followed by cleavage at R271 between the catalytic and F2 domains, generating the free thrombin and fragment 1.2 (F1.2) activation products. In the absence of FVa, cleavage occurs in the alternative order, with slow R271 cleavage resulting in the inactive Pre 2•F1.2 non-covalent complex, followed by cleavage of R320 to form active thrombin. When thrombin is generated in vitro in the absence of thrombin inhibitors it cleaves ProT at R155, releasing fragment 1 (F1) and generating the zymogen form, prethrombin 1 (Pre 1). Pre 1 does not bind to membranes but is cleaved by prothrombinase to the corresponding alternative activation intermediates, meizothrombin-des-fragment 1 (MzT(-F1)) and the Pre 2•F2 complex, and subsequently to thrombin and F2.
Fig. 5.
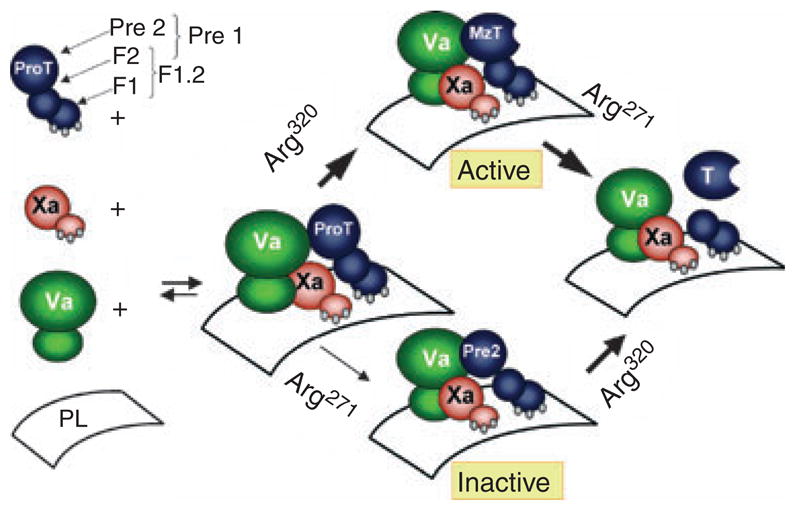
Mechanism of prothrombin activation. ProT activation is initiated by assembly of ProT, factor Xa (Xa), and factor Va (Va) on a phospholipid membrane surface. In the presence of saturating factor Va, cleavage at R320 in the ProT catalytic domain occurs first, yielding the active MzT reaction intermediate. Subsequent cleavage of MzT at R271 between the catalytic and F2 domains yields the products, thrombin (T) and F1.2. In the absence of factor Va, the alternative cleavage pathway predominates, with slow cleavage of R271 to form the inactive Pre 2•F1.2 non-covalent complex, followed by cleavage at R320 to form the products.
Evidence for the presence of proexosite I on ProT was obtained initially from studies of a fluorescein-labeled exosite I-specific hirudin-(53–64) peptide, demonstrating low affinity for ProT and expression of higher affinity on the activation intermediates and thrombin [87]. NMR studies indicated a very low, but detectable affinity (KD c. 500 μM) of ProT for N-acetyl-hirudin-(55–65) [108]. An antiexosite I monoclonal antibody and TM also demonstrated no detectable binding to ProT compared with thrombin, which was not changed on the reaction intermediates [88]. The pathway of exosite I expression was defined in equilibrium binding studies employing a fluorescein-labeled derivative of Y63-sulfated hirudin-(54–65) to quantitate the status of (pro)exosite I on ProT, Pre 1, and their activation intermediates [46,85,86,109]. The results of these studies are summarized in Fig. 6. The affinity of ProT for the hirudin peptide (KD c. 3 μM) is c. hundredfold lower than that of free thrombin, demonstrating overall expression of the exosite. The exosite activation pathway occurs with an increase in affinity accompanied by cleavage at R320 and activation of the catalytic site in MzT, whereas the zymogen intermediate, Pre 2•F1.2 binds with low affinity (KD c.1.3 μM). On this basis, the conformational changes accompanying the zymogen to proteinase transition result in expression of exosite I. Parallel studies comparing ProT with Pre 1 and its activation intermediates demonstrated an unexpected effect of the F1 domain in modulating proexosite I affinity (Fig. 6) [85]. Pre 1 binds the hirudin peptide with c. 7-fold higher affinity than does ProT, suggesting an interaction between F1 and the catalytic domain that attenuates affinity for the peptide [85,86]. Pre 1 and Pre 2•F2 have the same affinity for the peptide, which is enhanced c. twentyfold on formation of MzT(-F1) and thrombin [86]. These two effects account for the overall c. hundredfold increase in exosite I affinity accompanying ProT activation.
Fig. 6.
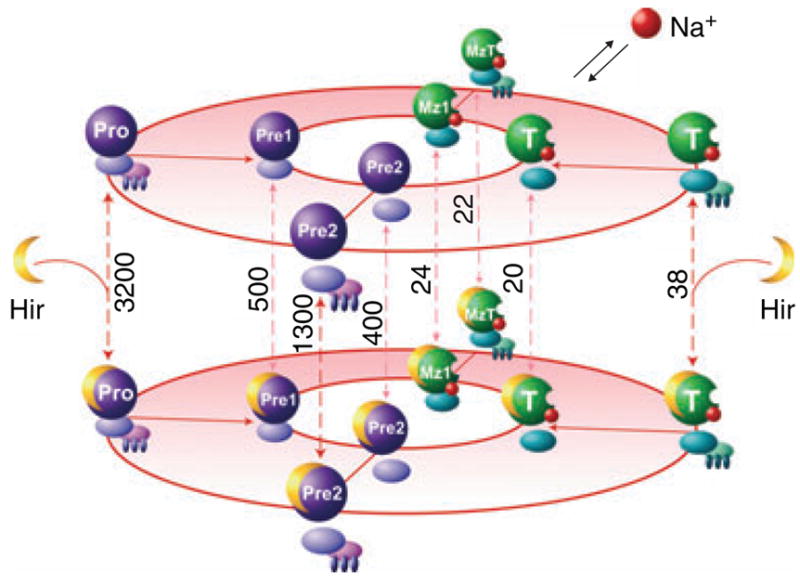
Pathways of exosite I and II expression and expression of the Na+ binding site during ProT and Pre 1 activation. Pathways of ProT (Pro) activation through the Pre 2•F1.2 or MzT alternative intermediates to the products, T + F1.2 (outer oval) and Pre 1 activation through Pre 2•F2 or MzT(-F1), to T + F2 (inner oval) are shown. Expression of the Na+ binding site is illustrated by association of Na+ (red spheres) with the intermediates and products. Purple proteins indicate inactive species, while green proteins indicate active forms. Fluorescein-labeled Y63-sulfated hirudin-(54-65) (Hir) binding equilibria are represented vertically (red dashed lines), with the dissociation constants for each species in nM. Modified with permission from Anderson and Bock [85].
Expression of exosite II was characterized from the binding of F2 and F1.2 to Pre 2 and thrombin [84,85]. Exosite II is covered by the F2 domain and is inaccessible on ProT, Pre 1, MzT, and MzT(-F1). Unlike exosite I, the relatively low affinity of exosite II on Pre 2 for F2 (KD c. 5 μM) is not affected significantly by the zymogen to proteinase transition, indicating its exposure accompanying dissociation of F2 (Fig. 6). Additional evidence for a modulatory role of the F1 domain was the c. sixteenfold higher affinity of F1.2 for Pre 2 compared with F2, and an associated c. 3-fold lower affinity of exosite I for the hirudin peptide [85]. The effect of F1 is specific for the zymogen as shown by the normalization of the affinities of F1.2 and F2 for thrombin. This is in contrast to exosite I expression on catalytic domain activation, in that the modulating effect of F1 on exosite II-mediated F1.2 binding is lost on activation of the catalytic site.
To determine whether (pro)exosite I on ProT zymogen species is allosterically regulated by Na+, as it is for thrombin, the effect of Na+ on (pro)exosite I affinity for the fluorescein-labeled hirudin peptide was quantitated for ProT and Pre 1 and their activation products (J Biol Chem, in press). Thermodynamic linkage between Na+ and (pro)exosite I affinity is undetectable for the zymogen forms, ProT, Pre 1, and Pre 2, and is expressed in concert with formation of the catalytic site on MzT, MzT(-F1), and thrombin (Fig. 6). The disrupted structure of the Na+ binding site in the crystal structure of the Pre 2–hirudin peptide complex demonstrates that the lack of linkage in the zymogens is due to the Na+ site not being formed, while it is activated as part of the conformational change in the zymogen to proteinase transition [94,110]. The implications of the finding of Na+-exosite I hirudin peptide linkage and enhancements of catalytic activity for all of the active species is that the substrate specificity of MzT for membrane-dependent FV and FVIII activation [57,111] and accelerated protein C activation when bound to TM [112,113] are also likely to be Na+ regulated, perhaps physiologically. The studies of exosite and regulatory site expression for ProT species demonstrate that all such sites are expressed as part of the zymogen to proteinase transition, tightly coupled to the generation of catalytic activity. An exception is that the intramolecular interaction of the F1 domain that attenuates proexosite I affinity is lost on formation of the catalytic site.
Exosites in recognition of substrates by coagulation proteinase–cofactor complexes
The formation of membrane-bound catalytic complexes among coagulation proteinases, their protein cofactors, and substrates is a central mechanism controlling blood coagulation substrate specificity [82–84]. Membrane complex assembly enhances specific substrate recognition by promoting the protein–protein interactions. Rapid-reaction kinetic studies of prothrombinase assembly on phospholipid vesicles showed that FXa and FVa bind rapidly and independently to the membrane, followed by formation of the complex by diffusion on the surface [114]. In the context of the electrostatic searching mechanism of protein–protein interactions, binding of the proteinase, cofactor and substrate orthogonally to the membrane surface reduces greatly the number of non-specific interactions required to form the FXa•Va•ProT productive complex.
The most intensively studied exosite-driven mechanism of substrate recognition by coagulation complexes is that of prothrombinase-catalyzed thrombin formation. Krishnaswamy et al. have amassed compelling thermodynamic, kinetic and structural evidence for a two-step substrate recognition mechanism [107,115–122]. ProT initially docks to exosites on the FXa–Va-membrane complex, forming a complex in which the active site of FXa remains accessible to small molecule substrates and inhibitors. This is followed by a conformational change in the ternary complex that engages the catalytic site of FXa for cleavage. The significance of this mechanism is that the apparent kcat and Km for the individual cleavages of ProT activation can depend on both the exosite binding and conformational change steps [107]. This mimics the well-established fact that FVa affects primarily kcat and not Km for thrombin formation. Analysis of the kinetics of the four individual ProT cleavages reveals that all substrates, ProT, Pre 2•F1.2, and MzT, have similar Km values and compete for the same exosite on prothrombinase [107,117,120,122] (see Fig. 5). Three of the four reactions also have indistinguishable kcat values, whereas cleavage of ProT at R271 proceeds with a c. thirtyfold lower kcat. This is a consequence of the locking conformational change being highly unfavorable for cleavage at R320 in Pre 2•F1.2, whereas the conformational equilibrium for MzT cleavage at R271 is favorable [107].
While there is ample evidence that exosites expressed on FXa contribute substantially to substrate recognition, the role of FVa in the mechanism is to induce their expression, without direct participation of the well-established ProT interactions with FVa in substrate recognition. ProT and its activation products bind specifically to the heavy chain subunit of FVa [47,56,123,124], but the role of this interaction in substrate recognition is not yet clear. Thrombin and MzT(-F1) bind to FVa in interactions that are exosite I-dependent based on their competitive displacement with the hirudin peptide [47,56] (and unpublished results). This originated the proposed role for ProT–FVa interactions in substrate specificity through (pro)exosite I interactions with FVa in the assembled complex. In the absence of membranes, binding of the hirudin peptide specifically and completely inhibits FVa-acceleration of ProT and Pre 1 activation through a mechanism in which the peptide binds to the free substrates in competition with their productive interactions with the FXa•FVa complex [109]. The simplest explanation in the context of the two-step model is that proexosite I on ProT either mediates directly initial binding to sites on FVa or modulates the conformational equilibrium engaging the FXa catalytic site. In agreement with this hypothesis, mutations in proexosite I residues of Pre 1 impair prothrombinase-catalyzed Pre 1 activation in parallel with loss of inhibition by hirudin peptide [125]. Additional residues on ProT contributing to FVa interactions were identified adjacent to proexosite I [126]. The two sulfated hirudin peptide-homologous sequences between residues 680 and 709 at the C-terminus of the FVa heavy chain were postulated to be the site of proexosite I interaction based on its specificity [46,47,109,127,128]. This is supported by studies of recombinant FVa with substitutions in the heavy chain and inhibition of FVa-accelerated ProT activation by other exosite I-specific ligands [127,129]. This interpretation is somewhat controversial, however, because an independent investigation of FVa mutants showed no effect of deletion of the hirudin-like sequences on the kinetics of prothrombinase-catalyzed ProT activation [130]. The initial studies of inhibition of FVa activity by the hirudin peptide were done in the absence of phospholipid membranes, which counteracted inhibition by the peptide [109]. Our subsequent unpublished fluorescence studies of FVa binding to active site-fluorescently labeled FXa on lipid vesicles characterized the binding of native ProT to the labeled prothrombinase complex to form the inactive enzyme–cofactor–substrate complex. ProT binds to the assembled FXa•Va complex and its binding is competitively inhibited by the hirudin peptide. These unpublished results provide evidence that (pro)exosite I plays an important role in substrate recognition by the membrane-bound prothrombinase complex. This mechanism is compatible with the substrate recognition mechanism of Krishnaswamy involving exosites on FXa, with the addition of ProT species interacting with FVa in proexosite I-dependent events that also contribute to substrate recognition and catalysis.
As pointed out by Krishnaswamy [107,118], the role of proexosite I in binding of all substrates and products to a common exosite on prothrombinase is apparently incompatible with one previous observation. Thrombin acts as an exosite-directed competitive inhibitor of FVa-accelerated Pre 2 activation. The hirudin peptide inhibits greatly the activation of Pre 2 and blocks competitive inhibition by thrombin, supporting the proexosite mechanism [118]. However, studies with chymotryptic fragments, ζ-1 and ζ-2, containing exosite I and most of exosite II [131], respectively, demonstrated that ζ-2, lacking exosite I, inhibited Pre 2 activation with the same affinity as thrombin, and the ζ-1 fragment did not inhibit Pre 2 activation [118]. These results argue that proexosite I is not required for Pre 2 activation or product inhibition of FVa-accelerated Pre 2 activation. This discrepancy between thrombin product inhibition and hirudin peptide inhibition was suggested to reflect indirect conformational changes in Pre 2 or thrombin by peptide binding that participate in substrate–prothrombinase interactions, rather than competition for exosites that directly mediate binding [118]. Although inhibition by exosite I ligands is clearly established in a number of studies, the role of FVa–ProT interactions and (pro)exosite I in mediating exosite docking and/or affecting the conformational locking step of the substrate recognition mechanism remains to be clarified.
The two-step docking and locking mechanism has been shown to be fairly general, accounting for the kinetics of FX activation by the FVIIa•TF complex [132] and activation of FIX by FXIa [133]. An exception is protein C activation by the thrombin•TM complex, which exhibits a major role for active site interactions in substrate recognition [134].
Exosite switching in regulation of the ProT activation pathway
Recent studies of the source of the ordered cleavages of ProT by prothrombinase have revealed the predominant role of substrate interactions in directing the pathway [107,115]. The pathway through cleavage at R320 to MzT is favored at the expense of Pre 2•F1.2 formation by a thirtyfold lower kcat for the latter reaction [122]. Cleavage of the identical site in MzT to form thrombin proceeds with a thirtyfold higher kcat. Studies of the activation pathway support a mechanism in which substrates in the zymogen and proteinase states bind in alternate conformations that direct sequential cleavage at the two activation sites by FXa [107,115]. The ProT zymogen is positioned for optimal cleavage at R320, whereas the product, MzT in the proteinase conformation ratchets to a different bound conformation for cleavage at R271. Transition of MzT into the active proteinase state is required for optimal cleavage at R271, as shown in studies of a ProT mutant in which the N-terminal sequence generated by cleavage at R320 was replaced by TAT, which prevents the N-terminal insertion required for activity [115]. The mutant ProT was cleaved normally at R320, whereas its subsequent conversion to thrombin was decreased c. twentyfold, and the rate of cleavage at R271 was rescued by stabilization of the active form with a reversible active site inhibitor. Covalent labeling of the catalytic site of a ProT mutant that could only be cleaved at R271 increased the rate of cleavage c. twelvefold [115]. This is thought to be due to occupation of the catalytic site by the covalent transition state analog changing the conformation of the zymogen catalytic domain toward a more active-like conformation. In agreement with this mechanism, active site labeling of ProT alters the ProT activation pathway, inhibiting accumulation of MzT and enhancing formation of the inactive Pre 2•F1.2 intermediate (unpublished results).
The activating conformational change to form MzT is accompanied by expression of increased exosite I affinity and Na+-exosite I linkage. The evidence supporting a role for (pro)exosite I in FVa interactions as part of substrate recognition suggests that the proexosite I to exosite I transition may control the bound substrate conformation, ratcheting it from initial cleavage at R320 in the zymogen to R271 cleavage in the active intermediate. This mechanism is compatible with the current model of substrate recognition and ratcheting, with the additional feature that (pro)exosite I-dependent FVa interactions are postulated to determine the alternate bound substrate conformations.
Exosite-targeted anticoagulants
The emergent predominance of exosite-mediated interactions in driving coagulation proteinase substrate specificity has led to several strategies for exosite-targeted anticoagulants. Inhibitors of ProT or thrombin (pro)exosite I, such as the hirudin peptide, an exosite I-specific DNA aptamer, and DYDYQ have been speculated to be potentially useful [57,127,129,135]. Based on the properties of exosites, and their multifunctionality for binding of various structurally different ligands via spatially overlapping residues, it has not yet been possible to identify a ligand that inhibits one but not all interactions of each exosite. Toward this goal, new computational approaches are being developed to identify hot spots and potential binding clefts, and docking approaches that address explicitly the flexibility and water-mediated interactions inherent in protein–protein interactions [136].
Exosite inhibitors derived from snake venoms and hematophagous organisms have been identified as possible anticoagulants [7,19,137]. Bothrojaracin does not block the catalytic site but interacts with both thrombin exosites and with high affinity to proexosite I on ProT, resulting in inhibition of thrombin generation [138,139]. NAPc2 is a flexible protein from the nematode Ancylostoma caninum that binds very specifically to FXa as an exosite-directed inhibitor of ProT activation, which is unaffected by incorporation of FXa into the prothrombinase complex [140,141]. The NAPc2•FXa complex is a potent inhibitor of the FVIIa•TF complex [142], which is the basis for its assessment in clinical trials as a promising new exosite-directed anticoagulant.
As a consequence of the different sets of residues involved in overlapping exosite interactions, site-directed mutation of one or two exosite residues can knock out particular interactions while preserving others. Substitution K60fE reduces fibrinogen clotting activity c. 6-fold while accelerating protein C activation in the presence of TM c. 2.5-fold [31]. R75E substitution of thrombin on the other hand has the converse effects, preserving activity with fibrinogen and not protein C. W215A substitution selectively inhibits Na+ binding and fibrinogen clotting activity, while having smaller effects on protein C activation and PAR-1 cleavage [143]. Similarly, thrombin E217A and E217K have no fibrinogen clotting activity but retain 50% activity in protein C activation [144,145], and E217A thrombin has anticoagulant activity in vivo [144]. The structure of thrombin E217K shows that the source of its low activity is disruption of the Na+ site and resultant collapse into an inhibited conformation, which is apparently restored by TM binding [23]. As a result of these findings, a two-residue substitution mutant (W215A/E217A) of thrombin was developed that has thirty-five thousandfold reduced catalytic activity toward certain chromogenic substrates, twenty thousandfold reduced activity with fibrinogen, and thousandfold reduced activity toward PAR-1, but develops normal activity in activation of protein C induced by binding to TM [146,147]. The structure of this mutant also demonstrated a disordered Na+ site and occluded S1 site in addition to an inactive arrangement of catalytic site residues [147,148]. This mutant has substantial anticoagulant and antithrombotic activity in vivo [149]. In a related approach, the murine ProT mutant R157A/R268A, which can only be cleaved by prothrombinase to a stable form of MzT, was developed based on the intrinsically low activity of MzT toward fibrinogen and platelet activation in contrast to its potency as a protein C activator when bound to TM [150]. This mutant has significant anticoagulant activity in a mouse model of thrombosis. Such ProT and thrombin mutants with selective functional inactivation show considerable promise as sources of novel exosite-based anticoagulants.
Active site-blocked coagulation proteinases have been proposed as exosite-directed anticoagulants. Active site-blocked FXa and FIXa, and an inactive TF mutant, are antithrombotic in vivo, with minimal effects on normal hemostasis compared with heparin [151]. In this vein, ProT labeled at the catalytic site with FPR-CH2Cl has anticoagulant and antithrombotic activity (unpublished results). In a mouse model of thrombosis, FPR-ProT decreases the rate of thrombus growth and size. FPR-ProT and S195A ProT have similar properties as competitive inhibitors of thrombin generation in a system of purified proteins. In platelet-rich and platelet-poor plasma, however, FPR-ProT increases the lag time in FVIIa-TF-initiated thrombin generation and reduces the maximum level of thrombin formed, whereas S195A ProT has a much smaller effect. As a result of active site labeling, the pathway of FPR-ProT cleavage by prothrombinase is shifted toward generation of the inactive Pre 2•F1.2 intermediate and away from the MzT pathway, whereas S195A ProT is cleaved normally. The mechanism of inhibition is thought to reflect FPR-ProT acting as a competing alternate substrate of prothrombinase. This illustrates an anticoagulant mechanism based on perturbation of the exosite-directed pathway of ProT activation.
Acknowledgments
Studies in the author’s laboratories were supported by NIH Heart, Lung and Blood Institute grant HL038779 to P. E. B. and HL080018 to I. M. A. V. A grant to P. E. B. from the Edmond Hustinx Foundation supported studies during sabbatical at the Cardiovascular Research Institute Maastricht, Department of Biochemistry, University Maastricht.
Footnotes
Schechter-Berger [1] notation referring to the residues of a substrate (from the N-terminal end) as …P4-P3-P2-P1-P1′-P2′… with the scissile bond at P1-P1′. The corresponding specificity subsites on the proteinase are designated…S4-S3-S2-S1-S1′-S2′…
Disclosure of conflict of interest
The authors state that they have no conflict of interests.
References
- 1.Schechter I, Berger A. On the size of the active site in proteases: I. Papain Biochem Biophys Res Commun. 1967;27:157–62. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 2.Bing DH, Cory M, Fenton JW., II Exosite affinity labeling of human thrombins. Similar labeling on the A chain and B chain/fragments of clotting alpha- and nonclotting gamma/beta-thrombins. J Biol Chem. 1977;252:8027–34. [PubMed] [Google Scholar]
- 3.Furie B, Bing DH, Feldmann RJ, Robison DJ, Burnier JP, Furie BC. Computer-generated models of blood coagulation factor Xa, factor IXa, and thrombin based upon structural homology with other serine proteases. J Biol Chem. 1982;257:3875–82. [PubMed] [Google Scholar]
- 4.Fenton JW, II, Olson TA, Zabinski MP, Wilner GD. Anion-binding exosite of human alpha-thrombin and fibrin(ogen) recognition. Biochemistry. 1988;27:7106–12. doi: 10.1021/bi00418a066. [DOI] [PubMed] [Google Scholar]
- 5.Lane DA, Philippou H, Huntington JA. Directing thrombin. Blood. 2005;106:2605–12. doi: 10.1182/blood-2005-04-1710. [DOI] [PubMed] [Google Scholar]
- 6.Huntington JA. Molecular recognition mechanisms of thrombin. J Thromb Haemost. 2005;3:1861–72. doi: 10.1111/j.1538-7836.2005.01363.x. [DOI] [PubMed] [Google Scholar]
- 7.Bode W. Structure and interaction modes of thrombin. Blood Cells Mol Dis. 2006;36:122–30. doi: 10.1016/j.bcmd.2005.12.027. [DOI] [PubMed] [Google Scholar]
- 8.Page MJ, Macgillivray RT, Di Cera E. Determinants of specificity in coagulation proteases. J Thromb Haemost. 2005;3:2401–8. doi: 10.1111/j.1538-7836.2005.01456.x. [DOI] [PubMed] [Google Scholar]
- 9.Di Cera E, Dang QD, Ayala YM. Molecular mechanisms of thrombin function. Cell Mol Life Sci. 1997;53:701–30. doi: 10.1007/s000180050091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Cera E, Cantwell AM. Determinants of thrombin specificity. Ann N Y Acad Sci. 2001;936:133–46. doi: 10.1111/j.1749-6632.2001.tb03502.x. [DOI] [PubMed] [Google Scholar]
- 11.Bode W. The structure of thrombin, a chameleon-like proteinase. J Thromb Haemost. 2005;3:2379–88. doi: 10.1111/j.1538-7836.2005.01356.x. [DOI] [PubMed] [Google Scholar]
- 12.Bode W, Turk D, Karshikov A. The refined 1. 9-A X-ray crystal structure of D-Phe-Pro-Arg chloromethylketone-inhibited human alpha-thrombin: structure analysis, overall structure, electrostatic properties, detailed active-site geometry, and structure-function relationships. Protein Sci. 1992;1:426–71. doi: 10.1002/pro.5560010402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bode W, Mayr I, Baumann U, Huber R, Stone SR, Hofsteenge J. The refined 1.9 A crystal structure of human alpha-thrombin: interaction with D-Phe-Pro-Arg chloromethylketone and significance of the Tyr-Pro-Pro-Trp insertion segment. EMBO J. 1989;8:3467–75. doi: 10.1002/j.1460-2075.1989.tb08511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedrich R, Panizzi P, Fuentes-Prior P, Richter K, Verhamme I, Anderson PJ, Kawabata S, Huber R, Bode W, Bock PE. Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation. Nature. 2003;425:535–9. doi: 10.1038/nature01962. [DOI] [PubMed] [Google Scholar]
- 15.Skrzypczak-Jankun E, Carperos VE, Ravichandran KG, Tulinsky A, Westbrook M, Maraganore JM. Structure of the hirugen and hirulog 1 complexes of alpha-thrombin. J Mol Biol. 1991;221:1379–93. [PubMed] [Google Scholar]
- 16.Carter WJ, Cama E, Huntington JA. Crystal structure of thrombin bound to heparin. J Biol Chem. 2005;280:2745–9. doi: 10.1074/jbc.M411606200. [DOI] [PubMed] [Google Scholar]
- 17.Colwell NS, Blinder MA, Tsiang M, Gibbs CS, Bock PE, Tollefsen DM. Allosteric effects of a monoclonal antibody against thrombin exosite II. Biochemistry. 1999;38:2610. doi: 10.1021/bi995066p. [DOI] [PubMed] [Google Scholar]
- 18.Arni RK, Padmanabhan K, Padmanabhan KP, Wu TP, Tulinsky A. Structure of the non-covalent complex of prothrombin kringle 2 with PPACK-thrombin. Chem Phys Lipids. 1994;68:59–66. doi: 10.1016/0009-3084(94)90124-4. [DOI] [PubMed] [Google Scholar]
- 19.Zingali RB. Interaction of snake-venom proteins with blood coagulation factors: mechanisms of anticoagulant activity. Toxin Rev. 2006;25:413–34. [Google Scholar]
- 20.Di Cera E. Thrombin: a paradigm for enzymes allosterically activated by monovalent cations. C R Biol. 2004;327:1065–76. doi: 10.1016/j.crvi.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 21.Wells CM, Di Cera E. Thrombin is a Na(+)-activated enzyme. Biochemistry. 1992;31:11721–30. doi: 10.1021/bi00162a008. [DOI] [PubMed] [Google Scholar]
- 22.Dang OD, Vindigni A, Di Cera E. An allosteric switch controls the procoagulant and anticoagulant activities of thrombin. Proc Natl Acad Sci U S A. 1995;92:5977–81. doi: 10.1073/pnas.92.13.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carter WJ, Myles T, Gibbs CS, Leung LL, Huntington JA. Crystal structure of anticoagulant thrombin variant E217 K provides insights into thrombin allostery. J Biol Chem. 2004;279:26387–94. doi: 10.1074/jbc.M402364200. [DOI] [PubMed] [Google Scholar]
- 24.Huntington JA, Esmon CT. The molecular basis of thrombin allostery revealed by a 1.8 A structure of the ‘slow’. Structure. 2003;11:469–79. doi: 10.1016/s0969-2126(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 25.Johnson DJ, Adams TE, Li W, Huntington JA. Crystal structure of wild-type human thrombin in the Na +-free state. Biochem J. 2005;392:21–8. doi: 10.1042/BJ20051217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pineda AO, Chen ZW, Bah A, Garvey LC, Mathews FS, Di Cera E. Crystal structure of thrombin in a self-inhibited conformation. J Biol Chem. 2006;281:32922–8. doi: 10.1074/jbc.M605530200. [DOI] [PubMed] [Google Scholar]
- 27.Lai MT, Di Cera E, Shafer JA. Kinetic pathway for the slow to fast transition of thrombin. Evidence of linked ligand binding at structurally distinct domains. J Biol Chem. 1997;272:30275–82. doi: 10.1074/jbc.272.48.30275. [DOI] [PubMed] [Google Scholar]
- 28.Bah A, Garvey LC, Ge J, Di Cera E. Rapid kinetics of Na(+) binding to thrombin. J Biol Chem. 2006;281:40049–56. doi: 10.1074/jbc.M608600200. [DOI] [PubMed] [Google Scholar]
- 29.Priestle JP, Rahuel J, Rink H, Tones M, Grutter MG. Changes in interactions in complexes of hirudin derivatives and human alpha-thrombin due to different crystal forms. Protein Sci. 1993;2:1630–42. doi: 10.1002/pro.5560021009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsiang M, Jain AK, Dunn KE, Rojas ME, Leung LL, Gibbs CS. Functional mapping of the surface residues of human thrombin. J Biol Chem. 1995;270:16854–63. doi: 10.1074/jbc.270.28.16854. [DOI] [PubMed] [Google Scholar]
- 31.Wu QY, Sheehan JP, Tsiang M, Lentz SR, Birktoft JJ, Sadler JE. Single amino acid substitutions dissociate fibrinogen-clotting and thrombomodulin-binding activities of human thrombin. Proc Natl Acad Sci U S A. 1991;88:6775–9. doi: 10.1073/pnas.88.15.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ayala YM, Cantwell AM, Rose T, Bush LA, Arosio D, Di Cera E. Molecular mapping of thrombin-receptor interactions. Proteins. 2001;45:107–16. doi: 10.1002/prot.1130. [DOI] [PubMed] [Google Scholar]
- 33.Jackman MP, Parry MA, Hofsteenge J, Stone SR. Intrinsic fluorescence changes and rapid kinetics of the reaction of thrombin with hirudin. J Biol Chem. 1992;267:15375–83. [PubMed] [Google Scholar]
- 34.Karshikov A, Bode W, Tulinsky A, Stone SR. Electrostatic interactions in the association of proteins: an analysis of the thrombin-hirudin complex. Protein Sci. 1992;1:727–35. doi: 10.1002/pro.5560010605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myles T, Le Bonniec BF, Betz A, Stone SR. Electrostatic steering and ionic tethering in the formation of thrombin-hirudin complexes: the role of the thrombin anion-binding exosite-I. Biochemistry. 2001;40:4972–9. doi: 10.1021/bi0023549. [DOI] [PubMed] [Google Scholar]
- 36.Ayala YM, Vindigni A, Nayal M, Spolar RS, Record MT, Jr, Di Cera E. Thermodynamic investigation of hirudin binding to the slow and fast forms of thrombin: evidence for folding transitions in the inhibitor and protease coupled to binding. J Mol Biol. 1995;253:787–98. doi: 10.1006/jmbi.1995.0591. [DOI] [PubMed] [Google Scholar]
- 37.Malkowski MG, Martin PD, Guzik JC, Edwards BF. The co-crystal structure of unliganded bovine alpha-thrombin and prethrombin-2: movement of the Tyr-Pro-Pro-Trp segment and active site residues upon ligand binding. Protein Sci. 1997;6:1438–48. doi: 10.1002/pro.5560060708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vindigni A, Di Cera E. Release of fibrinopeptides by the slow and fast forms of thrombin. Biochemistry. 1996;35:4417–26. doi: 10.1021/bi952834d. [DOI] [PubMed] [Google Scholar]
- 39.Arni RK, Padmanabhan K, Padmanabhan KP, Wu TP, Tulinsky A. Structures of the noncovalent complexes of human and bovine prothrombin fragment 2 with human PPACK-thrombin. Biochemistry. 1993;32:4727–37. doi: 10.1021/bi00069a006. [DOI] [PubMed] [Google Scholar]
- 40.Adams TE, Huntington JA. Thrombin-cofactor interactions: structural insights into regulatory mechanisms. Arterioscler Thromb Vasc Biol. 2006;26:1738–45. doi: 10.1161/01.ATV.0000228844.65168.d1. [DOI] [PubMed] [Google Scholar]
- 41.Dumas JJ, Kumar R, Seehra J, Somers WS, Mosyak L. Crystal structure of the GpIbalpha-thrombin complex essential for platelet aggregation. Science. 2003;301:222–6. doi: 10.1126/science.1083917. [DOI] [PubMed] [Google Scholar]
- 42.Celikel R, McClintock RA, Roberts JR, Mendolicchio GL, Ware J, Varughese KI, Ruggeri ZM. Modulation of alpha-thrombin function by distinct interactions with platelet glycoprotein Ibalpha. Science. 2003;301:218–21. doi: 10.1126/science.1084183. [DOI] [PubMed] [Google Scholar]
- 43.De Cristofaro R, De Candia E, Landolfi R, Rutella S, Hall SW. Structural and functional mapping of the thrombin domain involved in the binding to the platelet glycoprotein Ib. Biochemistry. 2001;40:13268–73. doi: 10.1021/bi010491f. [DOI] [PubMed] [Google Scholar]
- 44.Li CQ, Vindigni A, Sadler JE, Wardell MR. Platelet glycoprotein Ib alpha binds to thrombin anion-binding exosite II inducing allosteric changes in the activity of thrombin. J Biol Chem. 2001;276:6161–8. doi: 10.1074/jbc.M004164200. [DOI] [PubMed] [Google Scholar]
- 45.Yun TH, Baglia FA, Myles T, Navaneetham D, Lopez JA, Walsh PN, et al. Thrombin activation of factor XI on activated platelets requires the interaction of factor XI and platelet glycoprotein Ib alpha with thrombin anion-binding exosites I and II, respectively. J Biol Chem. 2003;278:48112–9. doi: 10.1074/jbc.M306925200. [DOI] [PubMed] [Google Scholar]
- 46.Anderson PJ, Nesset A, Dharmawardana KR, Bock PE. Characterization of proexosite I on prothrombin. J Biol Chem. 2000;275:16428–34. doi: 10.1074/jbc.M001254200. [DOI] [PubMed] [Google Scholar]
- 47.Dharmawardana KR, Olson ST, Bock PE. Role of regulatory exosite I in binding of thrombin to human factor V, factor Va, factor Va subunits, and activation fragments. J Biol Chem. 1999;274:18635–43. doi: 10.1074/jbc.274.26.18635. [DOI] [PubMed] [Google Scholar]
- 48.Hogg PJ, Jackson CM, Labanowski JK, Bock PE. Binding of fibrin monomer and heparin to thrombin in a ternary complex alters the environment of the thrombin catalytic site, reduces affinity for hirudin, and inhibits cleavage of fibrinogen. J Biol Chem. 1996;271:26088–95. doi: 10.1074/jbc.271.42.26088. [DOI] [PubMed] [Google Scholar]
- 49.Panizzi P, Friedrich R, Fuentes-Prior P, Kroh HK, Briggs J, Tans G, Bode W, Bock PE. Novel fluorescent prothrombin analogs as probes of staphylocoagulase-prothrombin interactions. J Biol Chem. 2006;281:1169–78. doi: 10.1074/jbc.M507955200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verhamme IM, Olson ST, Tollefsen DM, Bock PE. Binding of exosite ligands to human thrombin. Re-evaluation of allosteric linkage between thrombin exosites I and II. J Biol Chem. 2002;277:6788–98. doi: 10.1074/jbc.M110257200. [DOI] [PubMed] [Google Scholar]
- 51.Fredenburgh JC, Stafford AR, Weitz JI. Evidence for allosteric linkage between exosites 1 and 2 of thrombin. J Biol Chem. 1997;272:25493–9. doi: 10.1074/jbc.272.41.25493. [DOI] [PubMed] [Google Scholar]
- 52.Pineda AO, Cantwell AM, Bush LA, Rose T, Di Cera E. The thrombin epitope recognizing thrombomodulin is a highly cooperative hot spot in exosite I. J Biol Chem. 2002;277:32015–9. doi: 10.1074/jbc.M205009200. [DOI] [PubMed] [Google Scholar]
- 53.Myles T, Yun TH, Hall SW, Leung LL. An extensive interaction interface between thrombin and factor V is required for factor V activation. J Biol Chem. 2001;276:25143–9. doi: 10.1074/jbc.M011324200. [DOI] [PubMed] [Google Scholar]
- 54.Myles T, Yun TH, Leung LL. Structural requirements for the activation of human factor VIII by thrombin. Blood. 2002;100:2820–6. doi: 10.1182/blood-2002-03-0843. [DOI] [PubMed] [Google Scholar]
- 55.Esmon CT, Lollar P. Involvement of thrombin anion-binding exosites 1 and 2 in the activation of factor V and factor VIII. J Biol Chem. 1996;271:13882–7. doi: 10.1074/jbc.271.23.13882. [DOI] [PubMed] [Google Scholar]
- 56.Dharmawardana KR, Bock PE. Demonstration of exosite I-dependent interactions of thrombin with human factor V and factor Va involving the factor Va heavy chain: analysis by affinity chromatography employing a novel method for active-site-selective immobilization of serine proteinases. Biochemistry. 1998;37:13143–52. doi: 10.1021/bi9812165. [DOI] [PubMed] [Google Scholar]
- 57.Bukys MA, Orban T, Kim PY, Beck DO, Nesheim ME, Kalafatis M. The structural integrity of anion binding exosite I of thrombin is required and sufficient for timely cleavage and activation of factor V and factor VIII. J Biol Chem. 2006;281:18569–80. doi: 10.1074/jbc.M600752200. [DOI] [PubMed] [Google Scholar]
- 58.Fortenberry YM, Whinna HC, Gentry HR, Myles T, Leung LL, Church FC. Molecular mapping of the thrombin-heparin cofactor II complex. J Biol Chem. 2004;279:43237–44. doi: 10.1074/jbc.M406716200. [DOI] [PubMed] [Google Scholar]
- 59.Tang C, Iwahara J, Clore GM. Visualization of transient encounter complexes in protein-protein association. Nature. 2006;444:383–6. doi: 10.1038/nature05201. [DOI] [PubMed] [Google Scholar]
- 60.Blundell TL, Fernandez-Recio J. Cell biology: brief encounters bolster contacts. Nature. 2006;444:279–80. doi: 10.1038/nature05306. [DOI] [PubMed] [Google Scholar]
- 61.Grunberg R, Leckner J, Nilges M. Complementarity of structure ensembles in protein-protein binding. Structure. 2004;12:2125–36. doi: 10.1016/j.str.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 62.Croy CH, Koeppe JR, Bergqvist S, Komives EA. Allosteric changes in solvent accessibility observed in thrombin upon active site occupation. Biochemistry. 2004;43:5246–55. doi: 10.1021/bi0499718. [DOI] [PubMed] [Google Scholar]
- 63.Koeppe JR, Komives EA. Amide H/2H exchange reveals a mechanism of thrombin activation. Biochemistry. 2006;45:7724–32. doi: 10.1021/bi060405h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sabo TM, Farrell DH, Maurer MC. Conformational analysis of gamma’ peptide (410–427) interactions with thrombin anion binding exosite II. Biochemistry. 2006;45:7434–45. doi: 10.1021/bi060360k. [DOI] [PubMed] [Google Scholar]
- 65.Schmidt AE, Stewart JE, Mathur A, Krishnaswamy S, Bajaj SP. Na + site in blood coagulation factor IXa: effect on catalysis and factor VIIIa binding. J Mol Biol. 2005;350:78–91. doi: 10.1016/j.jmb.2005.04.052. [DOI] [PubMed] [Google Scholar]
- 66.Schmidt AE, Padmanabhan K, Underwood MC, Bode W, Mather T, Bajaj SP. Thermodynamic linkage between the S1 site, the Na + site, and the Ca2 + site in the protease domain of human activated protein C (APC). Sodium ion in the APC crystal structure is coordinated to four carbonyl groups from two separate loops. J Biol Chem. 2002;277:28987–95. doi: 10.1074/jbc.M201892200. [DOI] [PubMed] [Google Scholar]
- 67.Rezaie AR, He X. Sodium binding site of factor Xa: role of sodium in the prothrombinase complex. Biochemistry. 2000;39:1817–25. doi: 10.1021/bi992006a. [DOI] [PubMed] [Google Scholar]
- 68.Bajaj SP, Schmidt AE, Agah S, Bajaj MS, Padmanabhan K. High resolution structures of p-aminobenzamidine- and benzamidine-VIIa/soluble tissue factor: unpredicted conformation of the 192–193 peptide bond and mapping of Ca2 +, Mg2 +, Na +, and Zn2 + sites in factor VIIa. J Biol Chem. 2006;281:24873–88. doi: 10.1074/jbc.M509971200. [DOI] [PubMed] [Google Scholar]
- 69.Rezaie AR. Identification of basic residues in the heparin-binding exosite of factor Xa critical for heparin and factor Va binding. J Biol Chem. 2000;275:3320–7. doi: 10.1074/jbc.275.5.3320. [DOI] [PubMed] [Google Scholar]
- 70.Rudolph AE, Porche-Sorbet R, Miletich JP. Definition of a factor Va binding site in factor Xa. J Biol Chem. 2001;276:5123–8. doi: 10.1074/jbc.M006961200. [DOI] [PubMed] [Google Scholar]
- 71.Autin L, Steen M, Dahlback B, Villoutreix BO. Proposed structural models of the prothrombinase (FXa-FVa) complex. Proteins. 2006;63:440–50. doi: 10.1002/prot.20848. [DOI] [PubMed] [Google Scholar]
- 72.Rezaie AR. Heparin-binding exosite of factor Xa. Trends Cardiovasc Med. 2000;10:333–8. doi: 10.1016/s1050-1738(01)00070-6. [DOI] [PubMed] [Google Scholar]
- 73.Yang L, Manithody C, Rezaie AR. Localization of the heparin binding exosite of factor IXa. J Biol Chem. 2002;277:50756–60. doi: 10.1074/jbc.M208485200. [DOI] [PubMed] [Google Scholar]
- 74.Mathur A, Zhong D, Sabharwal AK, Smith KJ, Bajaj SP. Interaction of factor IXa with factor VIIIa. Effects of protease domain Ca2 + binding site, proteolysis in the autolysis loop, phospholipid, and factor X. J Biol Chem. 1997;272:23418–26. doi: 10.1074/jbc.272.37.23418. [DOI] [PubMed] [Google Scholar]
- 75.Pellequer JL, Gale AJ, Getzo3 ED, Griffin JH. Three-dimensional model of coagulation factor Va bound to activated protein C. Thromb Haemost. 2000;84:849–57. [PubMed] [Google Scholar]
- 76.Mather T, Oganessyan V, Hof P, Huber R, Foundling S, Esmon C, Bode W. The 2.8 A crystal structure of Gla-domainless activated protein C. EMBO J. 1996;15:6822–31. [PMC free article] [PubMed] [Google Scholar]
- 77.Friedrich U, Blom AM, Dahlback B, Villoutreix BO. Structural and energetic characteristics of the heparin-binding site in antithrombotic protein C. J Biol Chem. 2001;276:24122–8. doi: 10.1074/jbc.M011567200. [DOI] [PubMed] [Google Scholar]
- 78.Friedrich U, Nicolaes GA, Villoutreix BO, Dahlback B. Secondary substrate-binding exosite in the serine protease domain of activated protein C important for cleavage at Arg-506 but not at Arg-306 in factor Va. J Biol Chem. 2001;276:23105–8. doi: 10.1074/jbc.M103138200. [DOI] [PubMed] [Google Scholar]
- 79.Gale AJ, Tsavaler A, Grifin JH. Molecular characterization of an extended binding site for coagulation factor Va in the positive exosite of activated protein C. J Biol Chem. 2002;277:28836–40. doi: 10.1074/jbc.M204363200. [DOI] [PubMed] [Google Scholar]
- 80.Autin L, Miteva MA, Lee WH, Mertens K, Radtke KP, Villoutreix BO. Molecular models of the procoagulant factor VIIIa-factor IXa complex. J Thromb Haemost. 2005;3:2044–56. doi: 10.1111/j.1538-7836.2005.01527.x. [DOI] [PubMed] [Google Scholar]
- 81.Schmidt AE, Bajaj SP. Structure-function relationships in factor IX and factor IXa. Trends Cardiovasc Med. 2003;13:39–45. doi: 10.1016/s1050-1738(02)00210-4. [DOI] [PubMed] [Google Scholar]
- 82.Mann KG, Jenny RJ, Krishnaswamy S. Cofactor proteins in the assembly and expression of blood clotting enzyme complexes. Annu Rev Biochem. 1988;57:915–56. doi: 10.1146/annurev.bi.57.070188.004411. [DOI] [PubMed] [Google Scholar]
- 83.Mann KG, Krishnaswamy S, Lawson JH. Surface-dependent hemostasis. Semin Hematol. 1992;29:213–26. [PubMed] [Google Scholar]
- 84.Mann KG, Nesheim ME, Church WR, Haley P, Krishnaswamy S. Surface-dependent reactions of the vitamin K-dependent enzyme complexes. Blood. 1990;76:1–16. [PubMed] [Google Scholar]
- 85.Anderson PJ, Bock PE. Role of prothrombin fragment 1 in the pathway of regulatory exosite I formation during conversion of human prothrombin to thrombin. J Biol Chem. 2003;278:44489–95. doi: 10.1074/jbc.M306916200. [DOI] [PubMed] [Google Scholar]
- 86.Anderson PJ, Nesset A, Bock PE. Effects of activation peptide bond cleavage and fragment 2 interactions on the pathway of exosite I expression during activation of human prethrombin 1 to thrombin. J Biol Chem. 2003;278:44482–8. doi: 10.1074/jbc.M306917200. [DOI] [PubMed] [Google Scholar]
- 87.Liu LW, Ye J, Johnson AE, Esmon CT. Proteolytic formation of either of the two prothrombin activation intermediates results in formation of a hirugen-binding site. J Biol Chem. 1991;266:23633–6. [PubMed] [Google Scholar]
- 88.Wu Q, Picard V, Aiach M, Sadler JE. Activation-induced exposure of the thrombin anion-binding exosite. Interactions of recombinant mutant prothrombins with thrombomodulin and a thrombin exosite-specific antibody. J Biol Chem. 1994;269:3725–30. [PubMed] [Google Scholar]
- 89.Huber R, Bode W. Structural basis of the activation and action of trypsin. Acc Chem Res. 1978;11:114–22. [Google Scholar]
- 90.Camire RM. Prothrombinase assembly and S1 site occupation restore the catalytic activity of FXa impaired by mutation at the sodium-binding site. J Biol Chem. 2002;277:37863–70. doi: 10.1074/jbc.M203692200. [DOI] [PubMed] [Google Scholar]
- 91.Venkateswarlu D, Perera L, Darden T, Pedersen LG. Structure and dynamics of zymogen human blood coagulation factor X. Biophys J. 2002;82:1190–206. doi: 10.1016/S0006-3495(02)75476-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Miletich JP, Jackson CM, Majerus PW. Properties of the factor Xa binding site on human platelets. J Biol Chem. 1978;253:6908–16. [PubMed] [Google Scholar]
- 93.Persson E, Hogg PJ, Stenflo J. Effects of Ca2 + binding on the protease module of factor Xa and its interaction with factor Va. Evidence for two Gla-independent Ca(2 + )-binding sites in factor Xa. J Biol Chem. 1993;268:22531–9. [PubMed] [Google Scholar]
- 94.Zhang E, Tulinsky A. The molecular environment of the Na + binding site of thrombin. Biophys Chem. 1997;63:185–200. doi: 10.1016/s0301-4622(96)02227-2. [DOI] [PubMed] [Google Scholar]
- 95.Eigenbrot C, Kirchhofer D, Dennis MS, Santell L, Lazarus RA, Stamos J, Ultsch MH. The factor VII zymogen structure reveals reregistration of beta strands during activation. Structure. 2001;9:627–36. doi: 10.1016/s0969-2126(01)00624-4. [DOI] [PubMed] [Google Scholar]
- 96.Eigenbrot C, Kirchhofer D. New insight into how tissue factor allosterically regulates factor VIIa. Trends Cardiovasc Med. 2002;12:19–26. doi: 10.1016/s1050-1738(01)00139-6. [DOI] [PubMed] [Google Scholar]
- 97.Mertens K, Celie PH, Kolkman JA, Lenting PJ. Factor VIII-factor IX interactions: molecular sites involved in enzyme-cofactor complex assembly. Thromb Haemost. 1999;82:209–17. [PubMed] [Google Scholar]
- 98.Mathur A, Bajaj SP. Protease and EGF1 domains of factor IXa play distinct roles in binding to factor VIIIa. Importance of helix 330 (helix 162 in chymotrypsin) of protease domain of factor IXa in its interaction with factor VIIIa. J Biol Chem. 1999;274:18477–86. doi: 10.1074/jbc.274.26.18477. [DOI] [PubMed] [Google Scholar]
- 99.Ahmad SS, Rawala-Sheikh R, Walsh PN. Comparative interactions of factor IX and factor IXa with human platelets. J Biol Chem. 1989;264:3244–51. [PubMed] [Google Scholar]
- 100.Lenting PJ, ter Maat H, Clijsters PP, Donath MJ, van Mourik JA, Mertens K. Cleavage at arginine 145 in human blood coagulation factor IX converts the zymogen into a factor VIII binding enzyme. J Biol Chem. 1995;270:14884–90. doi: 10.1074/jbc.270.25.14884. [DOI] [PubMed] [Google Scholar]
- 101.De Cristofaro R, Picozzi M, Morosetti R, Landolfi R. Effect of sodium on the energetics of thrombin-thrombomodulin interaction and its relevance for protein C hydrolysis. J Mol Biol. 1996;258:190–200. doi: 10.1006/jmbi.1996.0242. [DOI] [PubMed] [Google Scholar]
- 102.Yang L, Prasad S, Di Cera E, Rezaie AR. The conformation of the activation peptide of protein C is influenced by Ca2 + and Na + binding. J Biol Chem. 2004;279:38519–24. doi: 10.1074/jbc.M407304200. [DOI] [PubMed] [Google Scholar]
- 103.Ruf W, Dickinson CD. Allosteric regulation of the cofactor-dependent serine protease coagulation factor VIIa. Trends Cardiovasc Med. 1998;8:350–6. doi: 10.1016/s1050-1738(98)00031-0. [DOI] [PubMed] [Google Scholar]
- 104.Petrovan RJ, Ruf W. Role of zymogenicity-determining residues of coagulation factor VII/VIIa in cofactor interaction and macromolecular substrate recognition. Biochemistry. 2002;41:9302–9. doi: 10.1021/bi0202169. [DOI] [PubMed] [Google Scholar]
- 105.Dickinson CD, Kelly CR, Ruf W. Identification of surface residues mediating tissue factor binding and catalytic function of the serine protease factor VIIa. Proc Natl Acad Sci U S A. 1996;93:14379–84. doi: 10.1073/pnas.93.25.14379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dickinson CD, Shobe J, Ruf W. Influence of cofactor binding and active site occupancy on the conformation of the macromolecular substrate exosite of factor VIIa. J Mol Biol. 1998;277:959–71. doi: 10.1006/jmbi.1998.1639. [DOI] [PubMed] [Google Scholar]
- 107.Krishnaswamy S. Exosite-driven substrate specificity and function in coagulation. J Thromb Haemost. 2005;3:54–67. doi: 10.1111/j.1538-7836.2004.01021.x. [DOI] [PubMed] [Google Scholar]
- 108.Ni F, Ning Q, Jackson CM, Fenton JW., II Thrombin exosite for fibrinogen recognition is partially accessible in prothrombin. J Biol Chem. 1993;268:16899–902. [PubMed] [Google Scholar]
- 109.Anderson PJ, Nesset A, Dharmawardana KR, Bock PE. Role of proexosite I in factor Va-dependent substrate interactions of prothrombin activation. J Biol Chem. 2000;275:16435–42. doi: 10.1074/jbc.M001255200. [DOI] [PubMed] [Google Scholar]
- 110.Vijayalakshmi J, Padmanabhan KP, Mann KG, Tulinsky A. The isomorphous structures of prethrombin2, hirugen-, and PPACK-thrombin: changes accompanying activation and exosite binding to thrombin. Protein Sci. 1994;3:2254–71. doi: 10.1002/pro.5560031211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tans G, Nicolaes GA, Thomassen MC, Hemker HC, van Zonneveld AJ, Pannekoek H, Rosing J. Activation of human factor V by meizothrombin. J Biol Chem. 1994;269:15969–72. [PubMed] [Google Scholar]
- 112.Doyle MF, Mann KG. Multiple active forms of thrombin: IV Relative activities of meizothrombins. J Biol Chem. 1990;265:10693–701. [PubMed] [Google Scholar]
- 113.Cote HC, Stevens WK, Bajzar L, Banfield DK, Nesheim ME, MacGillivray RT. Characterization of a stable form of human meizothrombin derived from recombinant prothrombin (R155A, R271A, and R284A) J Biol Chem. 1994;269:11374–80. [PubMed] [Google Scholar]
- 114.Krishnaswamy S, Jones KC, Mann KG. Prothrombinase complex assembly. Kinetic mechanism of enzyme assembly on phospholipid vesicles. J Biol Chem. 1988;263:3823–34. [PubMed] [Google Scholar]
- 115.Bianchini EP, Orcutt SJ, Panizzi P, Bock PE, Krishnaswamy S. Ratcheting of the substrate from the zymogen to proteinase conformations directs the sequential cleavage of prothrombin by prothrombinase. Proc Natl Acad Sci U S A. 2005;102:10099–104. doi: 10.1073/pnas.0504704102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Krishnaswamy S, Betz A. Exosites determine macromolecular substrate recognition by prothrombinase. Biochemistry. 1997;36:12080–6. doi: 10.1021/bi970979+. [DOI] [PubMed] [Google Scholar]
- 117.Boskovic DS, Krishnaswamy S. Exosite binding tethers the macromolecular substrate to the prothrombinase complex and directs cleavage at two spatially distinct sites. J Biol Chem. 2000;275:38561–70. doi: 10.1074/jbc.M006637200. [DOI] [PubMed] [Google Scholar]
- 118.Betz A, Krishnaswamy S. Regions remote from the site of cleavage determine macromolecular substrate recognition by the prothrombinase complex. J Biol Chem. 1998;273:10709–18. doi: 10.1074/jbc.273.17.10709. [DOI] [PubMed] [Google Scholar]
- 119.Wilkens M, Krishnaswamy S. The contribution of factor Xa to exosite-dependent substrate recognition by prothrombinase. J Biol Chem. 2002;277:9366–74. doi: 10.1074/jbc.M110848200. [DOI] [PubMed] [Google Scholar]
- 120.Boskovic DS, Troxler T, Krishnaswamy S. Active site-independent recognition of substrates and product by bovine prothrombinase: a fluorescence resonance energy transfer study. J Biol Chem. 2004;279:20786–93. doi: 10.1074/jbc.M400469200. [DOI] [PubMed] [Google Scholar]
- 121.Orcutt SJ, Pietropaolo C, Krishnaswamy S. Extended interactions with prothrombinase enforce a3nity and specificity for its macromolecular substrate. J Biol Chem. 2002;277:46191–6. doi: 10.1074/jbc.M208677200. [DOI] [PubMed] [Google Scholar]
- 122.Orcutt SJ, Krishnaswamy S. Binding of substrate in two conformations to human prothrombinase drives consecutive cleavage at two sites in prothrombin. J Biol Chem. 2004;279:54927–36. doi: 10.1074/jbc.M410866200. [DOI] [PubMed] [Google Scholar]
- 123.Guinto ER, Esmon CT. Loss of prothrombin and of factor Xa-factor Va interactions upon inactivation of factor Va by activated protein C. J Biol Chem. 1984;259:13986–92. [PubMed] [Google Scholar]
- 124.Luckow EA, Lyons DA, Ridgeway TM, Esmon CT, Laue TM. Interaction of clotting factor V heavy chain with prothrombin and prethrombin 1 and role of activated protein C in regulating this interaction: analysis by analytical ultracentrifugation. Biochemistry. 1989;28:2348–54. doi: 10.1021/bi00431a055. [DOI] [PubMed] [Google Scholar]
- 125.Chen L, Yang L, Rezaie AR. Proexosite-1 on prothrombin is a factor Va-dependent recognition site for the prothrombinase complex. J Biol Chem. 2003;278:27564–9. doi: 10.1074/jbc.M302707200. [DOI] [PubMed] [Google Scholar]
- 126.Yegneswaran S, Mesters RM, Fernandez JA, Griffin JH. Prothrombin residues 473–487 contribute to factor Va binding in the prothrombinase complex. J Biol Chem. 2004;279:49019–25. doi: 10.1074/jbc.M406645200. [DOI] [PubMed] [Google Scholar]
- 127.Beck DO, Bukys MA, Singh LS, Szabo KA, Kalafatis M. The contribution of amino Acid region asp695-tyr698 of factor v to procofactor activation and factor va function. J Biol Chem. 2004;279:3084–95. doi: 10.1074/jbc.M306850200. [DOI] [PubMed] [Google Scholar]
- 128.Kalafatis M, Beck DO, Mann KG. Structural requirements for expression of factor Va activity. J Biol Chem. 2003;278:33550–61. doi: 10.1074/jbc.M303153200. [DOI] [PubMed] [Google Scholar]
- 129.Kretz CA, Stafford AR, Fredenburgh JC, Weitz JI. HD1, a thrombin-directed aptamer, binds exosite 1 on prothrombin with high affinity and inhibits its activation by prothrombinase. J Biol Chem. 2006;281:37477–85. doi: 10.1074/jbc.M607359200. [DOI] [PubMed] [Google Scholar]
- 130.Toso R, Camire RM. Role of hirudin-like factor Va heavy chain sequences in prothrombinase function. J Biol Chem. 2006;281:8773–9. doi: 10.1074/jbc.M511419200. [DOI] [PubMed] [Google Scholar]
- 131.Lewis SD, Brezniak DV, Fenton JW, II, Shafer JA. Catalytically competent human and bovine zeta-thrombin and chimeras generated from unfolded polypeptide chains. Protein Sci. 1992;1:998–1006. doi: 10.1002/pro.5560010805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Baugh RJ, Dickinson CD, Ruf W, Krishnaswamy S. Exosite interactions determine the affinity of factor X for the extrinsic Xase complex. J Biol Chem. 2000;275:28826–33. doi: 10.1074/jbc.M005266200. [DOI] [PubMed] [Google Scholar]
- 133.Ogawa T, Verhamme IM, Sun MF, Bock PE, Gailani D. Exosite-mediated substrate recognition of factor IX by factor XIa. The factor XIa heavy chain is required for initial recognition of factor IX. J Biol Chem. 2005;280:23523–30. doi: 10.1074/jbc.M500894200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lu G, Chhum S, Krishnaswamy S. The affinity of protein C for the thrombin·thrombomodulin complex is determined in a primary way by active site-dependent interactions. J Biol Chem. 2005;280:15471–8. doi: 10.1074/jbc.M500881200. [DOI] [PubMed] [Google Scholar]
- 135.Bukys MA, Kim PY, Nesheim ME, Kalafatis M. A control switch for prothrombinase: characterization of a hirudin-like pentapeptide from the COOH terminus of factor Va heavy chain that regulates the rate and pathway for prothrombin activation. J Biol Chem. 2006;281:39194–204. doi: 10.1074/jbc.M604482200. [DOI] [PubMed] [Google Scholar]
- 136.Gonzalez-Ruiz D, Gohlke H. Targeting protein-protein interactions with small molecules: challenges and perspectives for computational binding epitope detection and ligand finding. Curr Med Chem. 2006;13:2607–25. doi: 10.2174/092986706778201530. [DOI] [PubMed] [Google Scholar]
- 137.Richardson JL, Kroger B, Hoeffken W, Sadler JE, Pereira P, Huber R, Bode W, Fuenter-Prior P. Crystal structure of the human alpha-thrombin-haemadin complex: an exosite II-binding inhibitor. EMBO J. 2000;19:5650–60. doi: 10.1093/emboj/19.21.5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Monteiro RQ, Zingali RB. Bothrojaracin, a proexosite I ligand, inhibits factor Va-accelerated prothrombin activation. Thromb Haemost. 2002;87:288–93. [PubMed] [Google Scholar]
- 139.Monteiro RQ, Bock PE, Bianconi ML, Zingali RB. Characterization of bothrojaracin interaction with human prothrombin. Protein Sci. 2001;10:1897–904. doi: 10.1110/ps.09001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Buddai SK, Toulokhonova L, Bergum PW, Vlasuk GP, Krishnaswamy S. Nematode anticoagulant protein c2 reveals a site on factor Xa that is important for macromolecular substrate binding to human prothrombinase. J Biol Chem. 2002;277:26689–98. doi: 10.1074/jbc.M202507200. [DOI] [PubMed] [Google Scholar]
- 141.Murakami MT, Rios-Steiner J, Weaver SE, Tulinsky A, Geiger JH, Arni RK. Intermolecular interactions and characterization of the novel factor Xa exosite involved in macromolecular recognition and inhibition: crystal structure of human Gladomainless factor Xa complexed with the anticoagulant protein NAPc2 from the hematophagous nematode Ancylostoma caninum. J Mol Biol. 2007;366:602–10. doi: 10.1016/j.jmb.2006.11.040. [DOI] [PubMed] [Google Scholar]
- 142.Stassens P, Bergum PW, Gansemans Y, Jespers L, Laroche Y, Huang S, Maki S, Messens J, Lauwereys M, Cappello M, Hotez PJ, Lasters I, Vlasuk GP. Anticoagulant repertoire of the hookworm Ancylostoma caninum. Proc Natl Acad Sci U S A. 1996;93:2149–54. doi: 10.1073/pnas.93.5.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Arosio D, Ayala YM, Di Cera E. Mutation of W215 compromises thrombin cleavage of fibrinogen, but not of PAR-1 or protein C. Biochemistry. 2000;39:8095–101. doi: 10.1021/bi0006215. [DOI] [PubMed] [Google Scholar]
- 144.Gibbs CS, Coutre SE, Tsiang M, Li WX, Jain AK, Dunn KE, Law VS, Mao CT, Matsumura SY, Mejza SY, Paborsky LR, Leung LLK. Conversion of thrombin into an anticoagulant by protein engineering. Nature. 1995;378:413–6. doi: 10.1038/378413a0. [DOI] [PubMed] [Google Scholar]
- 145.Leung LL, Hall SW. Dissociation of thrombin’s substrate interactions using site-directed mutagenesis. Trends Cardiovasc Med. 2000;10:89–92. doi: 10.1016/s1050-1738(00)00047-5. [DOI] [PubMed] [Google Scholar]
- 146.Cantwell AM, Di Cera E. Rational design of a potent anticoagulant thrombin. J Biol Chem. 2000;275:39827–30. doi: 10.1074/jbc.C000751200. [DOI] [PubMed] [Google Scholar]
- 147.Di Cera E. Anticoagulant thrombins. Trends Cardiovasc Med. 1998;8:340–50. doi: 10.1016/s1050-1738(98)00030-9. [DOI] [PubMed] [Google Scholar]
- 148.Pineda AO, Chen ZW, Caccia S, Cantwell AM, Savvides SN, Waksman G, Matthews FS, Di Cera E. The anticoagulant thrombin mutant W215A/E217A has a collapsed primary specificity pocket. J Biol Chem. 2004;279:39824–8. doi: 10.1074/jbc.M407272200. [DOI] [PubMed] [Google Scholar]
- 149.Gruber A, Fernandez JA, Bush L, Marzec U, Griffin JH, Hanson SR, Di Cera E. Limited generation of activated protein C during infusion of the protein C activator thrombin analog W215A/E217A in primates. J Thromb Haemost. 2006;4:392–7. doi: 10.1111/j.1538-7836.2006.01760.x. [DOI] [PubMed] [Google Scholar]
- 150.Shim K, Zhu H, Westfield LA, Sadler JE. A recombinant murine meizothrombin precursor, prothrombin R157A/R268A, inhibits thrombosis in a model of acute carotid artery injury. Blood. 2004;104:415–9. doi: 10.1182/blood-2004-02-0478. [DOI] [PubMed] [Google Scholar]
- 151.Himber J, Refino CJ, Burcklen L, Rouix S, Kirchhofer D. Inhibition of arterial thrombosis by a soluble tissue factor mutant and active site-blocked factors IXa and Xa in the guinea pig. Thromb Haemost. 2001;85:475–81. [PubMed] [Google Scholar]
- 152.Rezaie AR, Kittur FS. The critical role of the 185–189-loop in the factor Xa interaction with Na + and factor Va in the prothrombinase complex. J Biol Chem. 2004;279:48262–9. doi: 10.1074/jbc.M409964200. [DOI] [PubMed] [Google Scholar]
- 153.Yegneswaran S, Mesters RM, Griffin JH. Identification of distinct sequences in human blood coagulation factor Xa and prothrombin essential for substrate and cofactor recognition in the prothrombinase complex. J Biol Chem. 2003;278:33312–8. doi: 10.1074/jbc.M305906200. [DOI] [PubMed] [Google Scholar]
- 154.Gale AJ, Griffin JH. Characterization of a thrombomodulin binding site on protein C and its comparison to an activated protein C binding site for factor Va. Proteins. 2004;54:433–41. doi: 10.1002/prot.10627. [DOI] [PubMed] [Google Scholar]
- 155.Dahlback B, Villoutreix BO. Regulation of blood coagulation by the protein C anticoagulant pathway: novel insights into structure-function relationships and molecular recognition. Arterioscler Thromb Vasc Biol. 2005;25:1311–20. doi: 10.1161/01.ATV.0000168421.13467.82. [DOI] [PubMed] [Google Scholar]
- 156.Gale AJ, Heeb MJ, Griffin JH. The autolysis loop of activated protein C interacts with factor Va and differentiates between the Arg506 and Arg306 cleavage sites. Blood. 2000;96:585–93. [PubMed] [Google Scholar]
- 157.Silva FP, Jr, Antunes OA, de Alencastro RB, De Simone SG. The Na + binding channel of human coagulation proteases: novel insights on the structure and allosteric modulation revealed by molecular surface analysis. Biophys Chem. 2006;119:282–94. doi: 10.1016/j.bpc.2005.10.001. [DOI] [PubMed] [Google Scholar]


