Abstract
The specific ability of the major human fungal pathogen Candida albicans, as well as many other pathogenic fungi, to extend initial short filaments (germ tubes) into elongated hyphal filaments is important for a variety of virulence-related processes. However, the molecular mechanisms that control hyphal extension have remained poorly understood for many years. We report the identification of a novel C. albicans transcriptional regulator, UME6, which is induced in response to multiple host environmental cues and is specifically important for hyphal extension. Although capable of forming germ tubes, the ume6Δ/ume6Δ mutant exhibits a clear defect in hyphal extension both in vitro and during infection in vivo and is attenuated for virulence in a mouse model of systemic candidiasis. We also show that UME6 is an important downstream component of both the RFG1-TUP1 and NRG1-TUP1 filamentous growth regulatory pathways, and we provide evidence to suggest that Nrg1 and Ume6 function together by a negative feedback loop to control the level and duration of filament-specific gene expression in response to inducing conditions. Our results suggest that hyphal extension is controlled by a specific transcriptional regulatory mechanism and is correlated with the maintenance of high-level expression of genes in the C. albicans filamentous growth program.
INTRODUCTION
Candida albicans, the most common human fungal pathogen, is responsible for a wide variety of mucosal and systemic infections. Mucosal infections include oral and vaginal thrush, whereas systemic infections can occur in nearly every organ and tissue of the human body (Odds, 1988; Dupont, 1995; Weig et al., 1998). Candida spp. are the fourth-leading cause of hospital-acquired bloodstream infections in the United States, with an attributable mortality rate approaching 35% (Edmond et al., 1999). In this country approximately $1 billion per year is spent on antifungal treatments for patients with hospital-acquired Candida infections (Miller et al., 2001). Immunocompromised individuals, including AIDS patients, organ transplant recipients, cancer patients on chemotherapy, and recipients of artificial joints and prosthetic devices are especially vulnerable to infection (for reviews see Shepherd et al., 1985; Dupont, 1995; Weig et al., 1998).
C. albicans possesses several virulence properties including the ability to undergo a reversible morphological conversion from single round budding yeast cells (blastospores) to elongated cells attached end-to-end (filaments) (for reviews see Mitchell, 1998; Brown, 2002; Calderone and Gow, 2002). C. albicans filaments are known to occur in two distinct forms: pseudohyphae and hyphae. Pseudohyphal cells are elliptical in shape and have constrictions at cell junctions, whereas hyphal cells have parallel sides and true septa (lacking constrictions); for a more complete description of the differences between these two forms see Sudbery et al. (2004). Tissues infected with C. albicans typically contain a mixture of blastospores, pseudohyphae, and hyphae (Odds, 1988).
The C. albicans blastospore-to-filament transition is required for virulence and is known to occur in response to a wide variety of inducing conditions present in host tissues including serum, body temperature, N-actetylglucosamine, neutral pH, amino acids, and certain human hormones (Odds, 1988; Lo et al., 1997; Brown, 2002; Saville et al., 2003). When C. albicans cells encounter these conditions, they initially form small projections, termed “germ tubes.” Subsequent cell division at the apical tip of the germ tube allows C. albicans to form extended filaments (Odds, 1988; Sudbery et al., 2004).
The ability to form and extend hyphal filaments has been associated with a number of virulence-related processes in C. albicans as well as several other pathogenic fungi. Studies on clinical isolates have shown a clear correlation between extensive invasion of oral epithelial mucosal surfaces and increased number of hyphal filaments (Bartie et al., 2004). Other studies have reported that although C. albicans hyphae can be found within epithelial cells, blastospores are generally found either between these cells or on the epithelial cell surface, strongly suggesting that extended hyphae are the invasive form (Scherwitz, 1982; Ray and Payne, 1988; Filler and Sheppard, 2006). Indeed, epithelial cell invasion is believed to be caused by the mechanical force exerted by hyphal extension (Kumamoto and Vinces, 2005b). C. albicans hyphal filament extension is critical for the process of thigmotropism (guidance of hyphal growth along ridges of the substratum, which is believed to be an adaptation for tissue invasion) and also plays an important role in the ability of C. albicans to breach endothelial cells and lyse macrophages and neutrophils when endocytosed (Zink et al., 1996; Lo et al., 1997; Jong et al., 2001; Korting et al., 2003; Kumamoto and Vinces, 2005a,b). In the widespread fungal pathogen Aspergillus fumigatus, as well as Zygomycete fungi such as Rhizopus oryzae, hyphal filament extension is specifically important for the process of angioinvasion (Filler and Sheppard, 2006). Although the importance of hyphal extension to virulence in C. albicans and other pathogenic fungi has been well-established, little is known about the molecular mechanisms that control this process or the specific molecular properties of C. albicans hyphal filaments that are associated with enhanced virulence.
To address these questions, we previously identified, by DNA microarray analysis, a transcriptional program of 61 genes that is induced in the presence of one of the strongest filament-inducing conditions, serum at 37°C (Kadosh and Johnson, 2005). Although certain genes in this program do not appear to be involved in filamentation per se, they do carry out several known virulence functions (e.g., adherence to host cells and degradation of host cell membranes), suggesting that filament formation and hyphal extension are coregulated with other virulence properties. In this study we examine the basis for hyphal filament extension by focusing on the mechanisms that are important for controlling induction and maintaining expression of the C. albicans filamentous growth program. We have previously demonstrated that approximately half of all genes in this program are under negative control in the blastospore form by three key transcriptional repressors: Rfg1, Nrg1, and Tup1 (Kadosh and Johnson, 2005). Strains deleted for any of these regulators are filamentous in the absence of filament-inducing conditions and are highly attenuated for virulence in a mouse model of systemic candidiasis (Braun and Johnson, 1997; Braun et al., 2000, 2001; Kadosh and Johnson, 2001; Khalaf and Zitomer, 2001; Murad et al., 2001). In Saccharomyces cerevisiae, the RFG1 and NRG1 homologues encode DNA-binding proteins, which are known to direct transcriptional repression of target genes by recruitment of the Tup1 corepressor (Keleher et al., 1992; Deckert et al., 1995; Tzamarias and Struhl, 1995; Park et al., 1999; Smith and Johnson, 2000). Numerous lines of evidence have suggested that in C. albicans these regulators function in a similar manner to direct repression of filament- and virulence-specific target genes (Braun et al., 1997, 2000, 2001; Kadosh and Johnson, 2001; Khalaf and Zitomer, 2001; Murad et al., 2001). Previous studies have demonstrated that the C. albicans NRG1 transcript is down-regulated in response to serum at 37°C (Braun et al., 2001; Murad et al., 2001). Down-regulation of NRG1 appears to be an important step in the blastospore to filament transition because strains expressing NRG1 at high constitutive levels are locked in the blastospore form and are highly attenuated for virulence in a mouse model of systemic candidiasis (Braun et al., 2001; Saville et al., 2003).
Here, we report the identification of a novel filament-specific transcriptional regulator of C. albicans hyphal extension and virulence, UME6, which is important for controlling the level and duration of NRG1 down-regulation as well as maintaining expression of filament-specific genes in response to inducing conditions. We examine the functional relationships between UME6 and both the RFG1-TUP1 and NRG1-TUP1 filamentous growth regulatory pathways. The identification of UME6 also provides us with a unique opportunity to examine the specific transcriptional regulatory mechanisms that control the important, but poorly understood, C. albicans virulence trait of hyphal filament extension.
MATERIALS AND METHODS
Strains and DNA Constructions
Wild-type (CAF2-1), rfg1Δ/rfg1Δ, nrg1Δ/nrg1Δ, and tup1Δ/tup1Δ strains have been described previously (Fonzi and Irwin, 1993; Braun and Johnson, 1997; Braun et al., 2001; Kadosh and Johnson, 2001), and the genotypes of all strains used in this study are listed in Table 1. The ume6Δ/ume6Δ strain was generated using a fusion PCR strategy previously described by Noble and Johnson (Noble and Johnson, 2005). Briefly, 5′ and 3′ flanking sequences immediately outside of the UME6 coding region were generated using primers 1 and 2 for the upstream flank and primers 3 and 4 for the downstream flank (see Table S1 for a listing of primers used in this study). These flanks were then used in a second round of fusion PCR with LEU2 and HIS1 markers (obtained using primers 5/6 and plasmids pSN40 and pSN52) to generate ume6Δ::LEU2 and ume6Δ::HIS1 knockout PCR products. To construct the strain MBY1 (ume6Δ/+), SN152 (leu2−, his1−, arg4−; Noble and Johnson, 2005) was first transformed with ume6Δ::LEU2, and wild-type alleles of C. albicans ARG4 (generated using primers 33/34) and HIS1 (generated using the primers 35/36) were also subsequently added back to this strain. Our wild-type control strain, DK318, was constructed by adding back wild-type alleles of C. albicans ARG4 and HIS1 to strain SN95 (his1−, arg4−; Noble and Johnson, 2005) using the PCR products described above. The ume6Δ/ume6Δ strain (DK312) was generated by transforming SN152 with ume6Δ::LEU2, followed by ume6Δ::HIS1 and finally by a wild-type allele of C. albicans ARG4. To construct the ume6Δ/ume6Δ::UME6 add-back strain, a 4.3-kb BamHI-XhoI fragment containing Candida dubliniensis ARG4 was cloned from pSN69 to pSN75. Roche Expand High Fidelity Plus polymerase (Roche Diagnostics, Basel, Switzerland) was next used to generate a 4.3-kb BamHI-BamHI PCR fragment containing UME6 (using primers 7/8), which was subsequently cloned into pSN75-ARG4 cut with BamHI. The resulting construct was linearized by digestion with SmaI and integrated at the UME6 promoter of a ume6Δ/ume6Δ (arg4−) strain (DK244). The nrg1Δ/nrg1Δ ume6Δ/ume6Δ (MBY61) and rfg1Δ/rfg1Δ ume6Δ/ume6Δ (MBY79) strains were constructed as follows: first, rfgΔ::ARG4 and nrg1Δ::ARG4 PCR knockout products were generated by fusion PCR (using primers 9/10 and 11/12 for NRG1 flanks, primers 17/18 and 19/20 for RFG1 flanks and primers 5/6 for plasmid pSN69 containing the ARG4 marker) and were used to transform a ume6Δ/ume6Δ (arg4−) strain (DK244). To delete the second copy of each gene, PCR was used to generate 5′ and 3′ flanks for RFG1 (with primers 21/22 and 23/24) and NRG1 (with primers 13/14 and 15/16). The 5′ RFG1 and NRG1 flanks were digested with KpnI and XhoI, and the 3′ flanks were digested with NotI and SacII; the resulting fragments were cloned into vector pSFS2 (Reuss et al., 2004) digested with the appropriate restriction enzymes. Finally, the resulting constructs were digested with SacII and KpnI to release rfg1Δ::SAT1 and nrg1Δ::SAT1 fragments (5.2 kb each), which were used to transform rfg1Δ/+ ume6Δ/ume6Δ and nrg1Δ/+ ume6Δ/ume6Δ strains, respectively; the SAT marker was subsequently looped out of the final double mutant strains as described previously (Reuss et al., 2004). The tup1Δ/tup1Δ ume6Δ/ume6Δ strain (MBY93) was generated as follows: PCR was used to generate UME6 5′ and 3′ flanks (with primers 25/26 and 27/28). The 5′ UME6 flank was digested with BamHI and PstI, and the 3′ UME6 flank was digested with BglII and KpnI; the resulting fragments were cloned into pBB510 (Braun and Johnson, 2000) digested with the appropriate restriction enzymes. A 4.9-kb PstI-KpnI fragment containing the 5′ and 3′ UME6 flanking regions and the URA3 marker was then released from this construct and used to disrupt the first copy of UME6 in strain BCa2-5 (tup1Δ/tup1Δ, ura3-; Braun and Johnson, 1997). To delete the second copy of UME6 in this strain, 5′ and 3′ UME6 flanks (obtained by PCR with primers 29/30 and 31/32) were digested with KpnI-XhoI and NotI-SacII, respectively, and cloned into pSFS2 cut with the appropriate restriction enzymes. The resulting construct was digested with KpnI and SacII to release a 5.2-kb ume6Δ::SAT1 fragment, which was used to generate the final tup1Δ/tup1Δ ume6Δ/ume6Δ strain (MBY93). Correct integration of all disruption constructs was verified by whole cell PCR across the 5′ and 3′ disruption junctions and absence of the open reading frame (ORF) in homozygous deletion strains was confirmed by PCR using internal ORF primers.
Table 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| CAF2-1 | ura3Δ::imm434/URA3 iro1Δ::imm434/IRO1 | Fonzi and Irwin (1993) |
| SN95 | arg4Δ/arg4Δ his1Δ/his1Δ ura3Δ::imm434/URA3 iro1Δ::imm434/IRO1 | Noble and Johnson (2005) |
| SN152 | arg4Δ/arg4Δ leu2Δ/leu2 Δ his1Δ/his1Δ ura3Δ::imm434/URA3 iro1Δ::imm434/IRO1 | Noble and Johnson (2005) |
| DK318 (WT) | arg4Δ::ARG4/arg4Δ his1Δ::HIS1/his1Δ ura3Δ::imm434/URA3 iro1Δ::imm434/IRO1 | This study |
| MBY1 (ume6Δ/+) | ume6Δ::C.m.LEU2/UME6 arg4Δ::ARG4/arg4Δ leu2Δ/leu2Δ his1Δ::HIS1/his1Δ ura3Δ ::imm434/URA3 iro1Δ ::imm434/IRO1 | This study |
| DK244 | ume6Δ::C.m.LEU2/ume6Δ::C.d.HIS1 arg4Δ/arg4Δ leu2Δ/leu2Δ his1Δ/his1Δ ura3Δ::imm434/URA3 iro1Δ::imm434/IRO1 | This study |
| DK312 (ume6Δ/Δ) | ume6Δ::C.m.LEU2/ume6Δ::C.d.HIS1 arg4Δ::ARG4/arg4Δ leu2Δ/leu2Δ his1Δ/his1Δ ura3Δ::imm434/URA3 iro1Δ::imm434/IRO1 | This study |
| MBY16 (ume6Δ/Δ::UME6) | ume6Δ::UME6::C.d.ARG4::C.m.LEU2/ume6Δ::C.d.HIS1 arg4Δ/arg4Δ leu2Δ/leu2Δ his1Δ/his1Δ ura3Δ::imm434/URA3 iro1Δ::imm434/IRO1 | This study |
| MBY79 (ume6Δ/Δ rfg1Δ/Δ) | ume6Δ::C.m.LEU2/ume6Δ::C.d.HIS1 rfg1Δ::ARG4/rfg1Δ::frt arg4Δ/arg4Δ leu2Δ/leu2Δ his1Δ/his1Δ ura3Δ::imm434/URA3 iro1Δ::imm434/IRO1 | This study |
| MBY61 (ume6Δ/Δ nrg1Δ/Δ) | ume6Δ::C.m.LEU2/ume6Δ::C.d.HIS1 nrg1Δ::ARG4/nrg1Δ::frt arg4Δ/arg4Δ leu2Δ/leu2Δ his1Δ/his1Δ ura3Δ::imm434/URA3 iro1Δ::imm434/IRO1 | This study |
| MBY93 (ume6Δ/Δ tup1Δ/Δ) | ume6Δ::URA3/ume6Δ::SAT1 tup1Δ/tup1Δ ura3Δ::imm434/ura3Δ::imm434 iro1Δ::imm434/iro1Δ::imm434 | This study |
| BCa2-10 (tup1Δ/Δ) | tup1Δ/tup1Δ::URA3 ura3Δ::imm434/ura3Δ::imm434 iro1Δ::imm434/iro1Δ::imm434 | Braun and Johnson, (1997) |
| BCa23-3 (nrg1Δ/Δ) | nrg1Δ/nrg1Δ::URA3 ura3Δ::imm434/ura3Δ::imm434 iro1Δ::imm434/iro1Δ::imm434 | Braun et al. (2001) |
| DK129 (rfg1Δ/Δ) | rfg1Δ/rfg1Δ::URA3 ura3Δ::imm434/ura3Δ::imm434 iro1Δ::imm434/iro1Δ::imm434 | Kadosh and Johnson (2001) |
C.d., C. dubliniensis; C.m., C. maltosa.
Media and Growth Conditions
Standard non–filament-inducing growth conditions for C. albicans were yeast extract-peptone-dextrose (YEPD) medium at 30°C (Guthrie and Fink, 1991). Serum medium consisted of YEPD plus 10% fetal calf serum (FCS). Spider and Lee's media were prepared as described previously (Lee et al., 1975; Liu et al., 1994). Liquid Spider-, Lee's pH 6.8-, serum- and temperature-induction experiments were carried out, with certain modifications, using previously described protocols (Kadosh and Johnson, 2005). For DNA microarray experiments, saturated overnight cultures of both wild-type (DK318) and ume6Δ/ume6Δ (DK312) strains were diluted into 2 L of YEPD medium and grown at 30°C overnight until cells reached an OD600 of ∼8.5 (first experiment) or ∼10.5 (second experiment). At this point (the zero time point) for each strain an aliquot of cells was harvested for RNA preparation, and additional 200-ml aliquots were diluted into 2 L of fresh pre-warmed YEPD medium at 30°C or YEPD medium plus 10% serum at 37°C. Cells were harvested at 1-, 2-, 3- and 5-h time points for RNA preparation (please note that this protocol is similar to our previously described optimized protocol for serum and temperature induction (Kadosh and Johnson, 2005)). A similar procedure was used to prepare cells for the Nrg1-Ume6 kinetics experiment (Figure 6) except that cells were grown overnight in YEPD medium to an OD600 of ∼ 4.0 and diluted 1:10 into 150 ml of pre-warmed YEPD at 30°C or YEPD plus 10% serum at 37°C, and 5-ml aliquots were harvested for RNA preparation (we have noticed that the optimal OD600 for serum and temperature induction can vary depending on culture volume). For the experiment testing the effect of various filament-inducing conditions on UME6 transcript levels (Figure 5), cells were grown overnight in YEPD to an OD600 of ∼4.0, and diluted 1:25 into 150 ml of each inducing medium (pre-warmed), and 40-ml aliquots were harvested for RNA preparation at 30 min after induction. In the Nrg1-Tup1-Ume6 epistasis experiment (Figure 8B), strains were grown overnight under non–filament-inducing conditions (YEPD medium at 30°C) to an OD600 of ∼1.0, at which point 5-ml aliquots of cells were collected to prepare RNA. For the Rfg1-Ume6 epistasis experiment (see Figure 8C), a saturated overnight culture of each strain was diluted in YEPD medium to an OD600 of 0.2 and grown at 30°C to an OD600 of ∼1.0. For both epistasis experiments 5-ml aliquots of cells were collected for RNA preparation (although data for one wild-type strain (CAF2-1) is shown for the epistasis experiments, we also obtained very similar results using the DK318 wild-type strain as well).
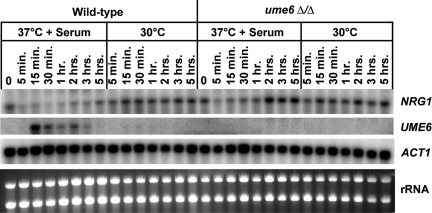
Figure 6.
Kinetics of NRG1 down-regulation and UME6 induction in response to serum at 37°C. Wild-type and ume6Δ/Δ strains were grown under non–filament-inducing conditions (YEPD medium at 30°C) and diluted into pre-warmed YEPD medium at 30°C or YEPD medium + 10% FCS at 37°C. Cells were harvested at the zero time point (immediately before induction) and at the indicated time points after induction for total RNA preparation. Northern analysis was carried out using 3 μg of RNA from each sample to assess transcript levels of the indicated genes. The ACT1 transcript, as well as ribosomal RNA (rRNA) are shown as loading controls.
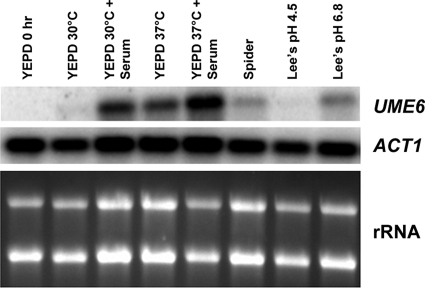
Figure 5.
Effect of a variety of filament-inducing conditions on induction of UME6. A wild-type strain was grown under non–filament-inducing conditions (YEPD medium at 30°C) and diluted into pre-warmed flasks containing either YEPD or the indicated filament-inducing media at 30°C (or 37°C where shown). Cells were harvested at the zero time point (immediately before induction) and at 30 min after induction for total RNA preparation. Northern analysis was carried out using 3 μg of RNA from each sample to assess transcript levels of the indicated genes. The ACT1 transcript, as well as ribosomal RNA (rRNA) are shown as loading controls.
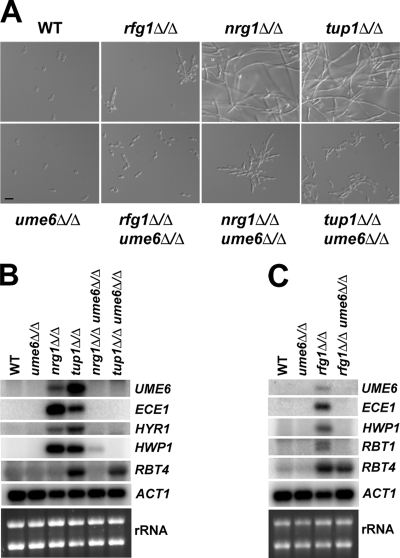
Figure 8.
Cell morphologies and filament-specific gene expression in rfg1Δ/Δ ume6Δ/Δ, nrg1Δ/Δ ume6Δ/Δ, and tup1Δ/Δ ume6Δ/Δ double mutants and respective single mutants. (A) Cell morphologies of the indicated strains are shown after growth in liquid YEPD medium at 30°C to an OD600 ∼ 1.0. Aliquots of cells were fixed in 4.5% formaldehyde, washed twice in 1× PBS, and then visualized by Nomarski/DIC optics (Bar, 10 μm). (B and C) Total RNA was prepared from the strains shown in A, and Northern analysis was carried using 3 μg RNA and probes to the indicated filament-specific transcripts. The ACT1 transcript and ribosomal RNA (rRNA) are included as loading controls.
RNA Preparation and Analysis
For both DNA microarray and Northern analysis, total RNA was prepared using hot acid phenol (as described previously by Ausubel et al., 1992) and quantified by spectrophotometer. For Northern analysis 3 μg of RNA from each sample was loaded onto a formaldehyde denaturing gel and transferred by capillary action to a GeneScreen Plus nylon membrane (Perkin Elmer, Waltham, MA). Probes for Northern analysis were generated by PCR, purified using a Qiaquick column and labeled with α-32P-dATP using a random priming kit (GE Healthcare Biosciences, Piscataway, NJ) (see Table S1 for a complete listing of primers used to generate the probes). Blots were hybridized overnight at 42°C, washed twice for 10 min at 59°C using the Church protocol (Church and Gilbert, 1984), and scanned using a Molecular Dynamics Typhoon 9400 phosphorimager (Sunnyvale, CA). Autoradiographs were visualized and quantified using Image Quant 2.0 software (Molecular Dynamics).
DNA Microarray Analysis
Construction of whole-genome C. albicans DNA microarrays, poly(A) RNA selection, cDNA preparation, and hybridization of coupled cDNA to microarrays as well as DNA microarray analysis were carried out as previously described (Bennett et al., 2003; Kadosh and Johnson, 2005). All data were normalized by NOMAD (http://ucsf-nomad.sourceforge.net) and filtered so as to include only spots with a median signal intensity ≥500 in at least one channel. The serum- and temperature-induction experiment was carried out twice using both wild-type and ume6Δ/ume6Δ strains (note that microarray experiment 2 (Supplementary Data) was performed using oligonucleotide-based arrays). Each hybridization contained a Cy5-labeled experimental sample and a Cy3-labeled universal reference sample, which has been previously described (Kadosh and Johnson, 2005). For both wild-type and ume6Δ/ume6Δ samples, signal ratios (ratio of the median signal intensity of each spot) for each time point versus reference were divided by the signal ratio of the zero time point (for each strain, respectively) versus reference. Signal ratio values for the zero time point versus reference (for both strains) represented median values from three independent DNA microarray experiments (using cDNAs from the same total RNA preparation). Gene annotation was based on the C. albicans orf19 assembly (Braun et al., 2005) and on the Candida Genome Database (Arnaud et al., 2005).
Virulence Experiments and Histology
Overnight cultures (grown in YEPD medium at 25°C) of wild-type (DK318), ume6Δ/ume6Δ and ume6Δ/ume6Δ::UME6 strains were harvested by centrifugation and washed three times in sterile pyrogen-free saline. Based on cell counts made using a hemocytometer, appropriate dilutions were carried out, and 200 μl of cell suspension from each strain was injected by lateral tail vein into individual 6–8-wk-old BALB/c mice (eight mice per group). Mice were monitored for survival for 21 d after infection (moribund mice were killed and were recorded as dying on the next day). The Kaplan-Meier log rank test was used to determine statistical differences between groups and analyses were carried out using Prism and GraphPad Software (San Diego, CA).
For all animals, one kidney was removed at the time of death for histological analysis. Briefly, these kidneys were fixed in 10% buffered Formalin, embedded in paraffin, sectioned, and stained with Grocot-Gomori methenamine-silver (GMS).
RESULTS
UME6 Is Important for C. albicans Filamentous Growth in Response to a Variety of Filament-inducing Conditions
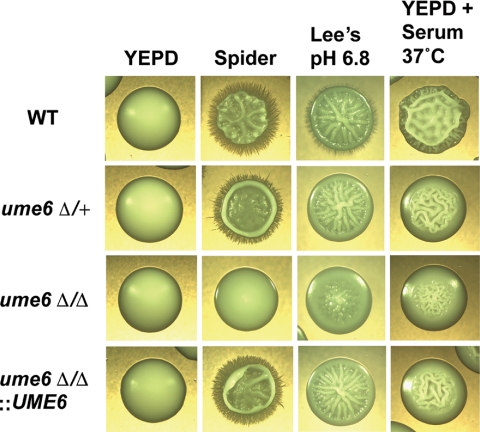
UME6 was first identified as a component of the C. albicans filamentous growth program that is induced in response to the host filament-inducing conditions of serum and body temperature, 37°C (Kadosh and Johnson, 2005). The S. cerevisiae UME6 homolog encodes a zinc-finger DNA-binding protein that is known to function as a key transcriptional regulator of early meiosis-specific genes and is also important for controlling arginine catabolism, peroxisomal function, and DNA repair (Strich et al., 1994; Bowdish et al., 1995; Einerhand et al., 1995; Steber and Esposito, 1995; Sweet et al., 1997). C. albicans UME6 (orf19.1822) encodes a 843-amino acid protein with a zinc-finger DNA-binding domain that is 41% identical to that of S. cerevisiae Ume6. Although C. albicans is not known to undergo meiosis, we hypothesized that CaUME6 may play a role in filamentous growth because this gene is induced by serum and 37°C and is repressed by the Nrg1-Tup1 pathway in blastospores under non–filament-inducing conditions (Kadosh and Johnson, 2005). To further explore this possibility, we generated both heterozygous and homozygous ume6 deletion mutants and tested for filamentation on a variety of solid filament-inducing media (wrinkled or rough colonies typically indicate filamentation). As shown in Figure 1 the ume6Δ/ume6Δ mutant is significantly defective for filamentation in response to Spider medium (nitrogen and carbon starvation), Lee's medium, pH 6.8 (neutral pH), and YEPD medium plus 10% serum at 37°C compared to a wild-type strain under these same growth conditions (microscopic examination revealed that under all filament-inducing conditions the ume6Δ/ume6Δ mutant formed filaments that were significantly shorter in length than those of the wild-type strain). In contrast, the ume6Δ/+ strain showed a milder filamentation defect, suggesting that the ume6 mutant phenotype is dosage-dependent. We also generated a strain in which a single wild-type copy of UME6 is added back to the ume6Δ/ume6Δ mutant. The ume6Δ/ume6Δ::UME6 “add-back” strain showed clear complementation of the ume6Δ/ume6Δ filamentation defect and appeared phenotypically similar to the ume6Δ/+ strain under all filament-inducing conditions tested (Figure 1), confirming that the original defect was due to a loss of UME6 function.
Figure 1.
ume6Δ/Δ mutants are defective for filamentous growth in response to a variety of filament-inducing conditions. Colony morphologies of wild-type (WT), ume6Δ/+, ume6Δ/Δ, and ume6Δ/Δ::UME6 (add-back) strains grown on solid non–filament-inducing medium (YEPD) and on various solid filament-inducing media. All colonies were grown at 30°C for 5 d, with the exception of cells grown on YEPD + 10% serum plates, which were grown at 37°C for 3 d, and photographed at ∼×20 magnification.
Because filamentation is known to be important for C. albicans biofilm formation (for reviews see Ramage et al., 2005; Blankenship and Mitchell, 2006), we also tested the strains described above for their ability to form biofilms on an inert substrate (polystyrene wells). The ume6Δ/ume6Δ mutant showed a partial defect in biofilm formation that was complemented in the ume6Δ/ume6Δ::UME6 “add-back” strain (Figure S1).
UME6 Is Specifically Important for Hyphal Filament Extension
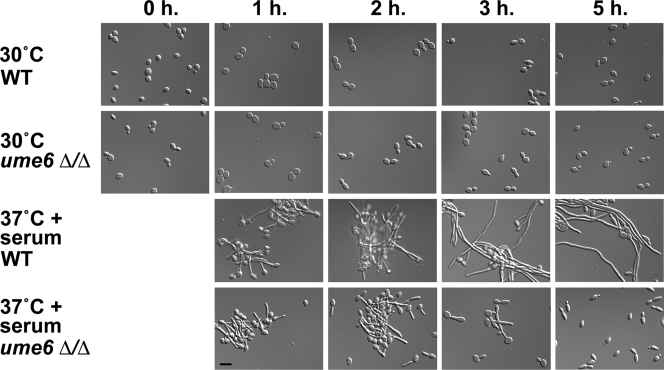
Because UME6 is induced by serum and temperature, we next sought to determine the extent to which this regulator is required for hyphal filament formation in liquid medium under these conditions. Using a previously optimized protocol in which nearly all cells are induced to form hyphal filaments (Kadosh and Johnson, 2005), we carried out a serum- and temperature-induction time course experiment to compare wild-type and ume6Δ/ume6Δ strains. Aliquots of cells from both strains were harvested at 0-, 1-, 2-, 3-, and 5-h time points, fixed, and examined by microscopy (Figure 2). In contrast to the wild-type strain, the ume6Δ/ume6Δ mutant exhibited a clear defect in the extension of hyphal filaments. Many cells formed germ tubes within the first 2 h of induction. At the 3-h time point many of these cells remained as germ tubes or very short filaments and by the 5-h time point, a large fraction of ume6Δ/ume6Δ cells appeared to take on an elongated blastospore-type morphology. The long extended hyphal filaments normally present in the wild-type strain at the 3- and 5-h time points were absent in the ume6Δ/ume6Δ mutant (Figure 2). Thus, although not required for initial germ tube formation, Ume6 appears to be specifically important for hyphal filament extension.
Figure 2.
Morphology of wild-type and ume6Δ/Δ cells undergoing the blastospore to filament transition. Wild-type (WT) and ume6Δ/Δ strains grown under non–filament-inducing conditions (YEPD medium at 30°C) were diluted into pre-warmed YEPD medium at 30°C or 37°C in the presence or absence of 10% fetal calf serum (FCS). Induction time (h) is shown on top. Aliquots of cells were fixed in 4.5% formaldehyde, washed twice in 1× PBS, and then visualized by Nomarski/DIC optics. Please note that the 0-h time point shows cells immediately before induction. Bar, 10 μm.
UME6 Controls the Level and Duration of Gene Expression in the C. albicans Filamentous Growth Program
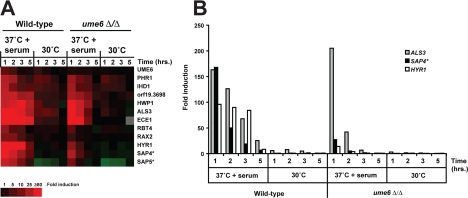
To examine the expression levels of filament-specific transcripts, both wild-type and ume6Δ/ume6Δ mutant cells were harvested at each time point during the serum- and temperature-induction experiment described above, and RNA was prepared for DNA microarray analysis. As indicated by the cluster diagram shown in Figure 3A, although nearly all of the top filament-specific target genes were induced in both wild-type and ume6Δ/ume6Δ strains, the level and duration of induction was significantly reduced in the ume6Δ/ume6Δ mutant. A transcriptional profile (Figure 3B) indicates that although some of the more highly induced genes in the wild-type strain, such as ALS3, are also strongly induced in the ume6Δ/ume6Δ mutant at the 1-h time point, others (such as SAP4 and HYR1) show reduced expression even at this early stage; most transcripts showed significantly reduced induction in the ume6Δ/ume6Δ strain at the 2-, 3- and 5-h time points. These results correlate well with the morphological differences observed between wild-type and ume6Δ/ume6Δ strains (Figures 1 and 2) and suggest that Ume6 is required to maintain full expression of the C. albicans filamentous growth program in response to serum at 37°C.
Figure 3.
Transcriptional profile of serum- and temperature-induced genes in wild-type and ume6Δ/Δ strains. (A) Cluster diagram indicating expression levels of several previously identified (Kadosh and Johnson, 2005) top serum- and temperature-induced transcripts (≥5-fold mean induction in the wild-type strain, n = 2, at the 37°C + 10% FCS 1-h time point). Only data from one serum- and temperature-induction experiment and only genes with greater than 93% of data present are shown. Red, increased expression; green, reduced expression; gray, no data available. Expression levels reflect fold induction relative to the zero time point for each strain. None of the genes showed a significant difference in absolute level of expression between wild-type and ume6Δ/Δ strains at the zero time point. (B) Histogram indicates fold induction, relative to the zero time point of wild-type or ume6Δ/Δ strains, for three serum- and temperature-induced genes: ALS3, an adhesin important for both epithelial and endothelial cell adhesion (Zhao et al., 2004), SAP4, a secreted aspartyl protease important for host tissue invasion and virulence (Sanglard et al., 1997; Schaller et al., 1999), and HYR1, a putative cell wall glycoprotein (Bailey et al., 1996) described in A. *Note that expression levels of SAP5 may, in part, reflect those of SAP4 or SAP6 (and SAP4 levels may reflect SAP5 and SAP6 levels) due to cross-hybridization on the microarray, since these genes have nearly identical DNA sequences.
The ume6 Δ/ume6Δ Mutant Is Attenuated for Virulence and Defective for Hyphal Extension in a Mouse Model of Systemic Candidiasis
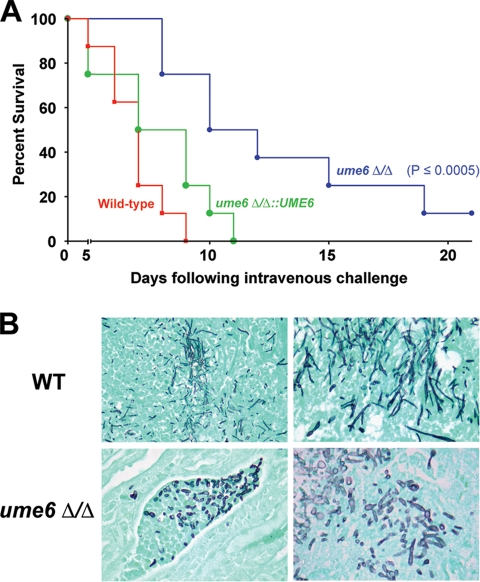
In an experiment to determine the effect of the ume6Δ/ume6Δ mutation on virulence, wild-type, ume6Δ/ume6Δ, and ume6Δ/ume6Δ::UME6 (add-back) strains were each injected by tail vein into eight 6–8-wk-old female BALB/c mice. Although mice injected with the wild-type strain all died by 5–9 d after infection, mice injected with the ume6Δ/ume6Δ strain generally died after a significantly longer time period (p ≤ 0.0005), indicating that the ume6Δ/ume6Δ mutant is attenuated for virulence. The ume6Δ/ume6Δ::UME6 strain did not show a statistically significant difference in virulence compared with the wild-type strain, indicating that the observed virulence defect of the ume6Δ/ume6Δ strain was specifically due to a loss of UME6 function (Figure 4A).
Figure 4.
The ume6Δ/Δ mutant is attenuated for virulence and defective for hyphal filament extension in a mouse model of systemic candidiasis. (A) Eight female BALB/c mice (6–8 wk old) were each injected with 2 × 105 CFUs of a wild-type, ume6Δ/Δ, or ume6Δ/Δ::UME6 (add-back) strain. Survival was monitored over the course of 21 d. A Kaplan-Meier test was performed to confirm that the difference in virulence between the wild-type and ume6Δ/Δ strains is statistically significant (p ≤ 0.0005) (the ume6Δ/Δ::UME6 strain did not show a statistically significant difference in virulence when compared with the wild-type strain). (B) Kidney tissues from infected mice were fixed, sectioned, and stained with Grocot- Gomori methenamine-silver to visualize fungal cells. Top panels, sections taken from mice infected with the wild-type (WT) strain; bottom panels, sections taken from mice infected with the ume6Δ/Δ strain.
We next carried out histopathology studies to determine whether the virulence defect of the ume6Δ/ume6Δ mutant was due to altered C. albicans morphology during infection. Mouse kidneys were fixed, sectioned, and stained to visualize fungal cells. In contrast to kidneys of mice infected with a wild-type strain, which showed a mixture of blastospores and extended hyphal and pseudohyphal filaments, kidneys of mice infected with the ume6Δ/ume6Δ mutant showed a mixture of blastospores and short, stubby filaments (Figure 4B). These findings are consistent with our previous results (Figure 2) and indicate that the ume6Δ/ume6Δ mutant is defective for hyphal filament extension both in vitro as well as during a systemic infection in vivo.
UME6 Is Induced in Response to a Variety of Filament-inducing Conditions
The results described above suggest that induction of UME6 appears to be critical for proper hyphal filament extension both during infection in vivo and in response to the combination of serum and 37°C in vitro. We next sought to determine the relative contribution of serum and temperature as well as a variety of other filament-inducing conditions present in host tissues to UME6 induction. We examined induction of UME6 by Northern analysis in response to YEPD plus 10% serum alone, YEPD at 37°C, YEPD plus 10% serum at 37°C, Spider medium (nitrogen and carbon starvation), and Lee's medium (at both pH 4.5 and 6.8) (Figure 5). We found that serum and 37°C both made separate contributions and showed an additive effect on UME6 induction. The UME6 transcript was also induced in the presence of both Spider and Lee's pH 6.8, but not Lee's pH 4.5 media. Interestingly, NRG1, which encodes a transcriptional regulator that has been shown to repress UME6 under non–filament-inducing conditions (Kadosh and Johnson, 2005), is down-regulated in response to serum and temperature as well as neutral pH (Braun et al., 2001; Murad et al., 2001; Lotz et al., 2004). Taken together, these results suggest that UME6 and NRG1 may function together in a coordinated manner to control induction of filament-specific target genes in response to a variety of specific inducing conditions and signaling pathways.
UME6 Controls the Level and Duration of NRG1 Down-Regulation in Response to Serum at 37°C
To further define the functional relationship between UME6 and NRG1, we carried out an experiment to determine the kinetics of UME6 induction and NRG1 down-regulation under inducing conditions. Both wild-type and ume6Δ/ume6Δ strains were induced to form filaments in response to serum at 37°C, and RNA was prepared from cells harvested at very early time points up to 5 h. We observed strong NRG1 down-regulation as early as our first time point, 5 min (Figure 6). NRG1 levels remained very low (about 5-fold repressed) through the 1-h time point and then began to rise from the 2- to 5-h time points. In contrast, in the ume6Δ/ume6Δ mutant, NRG1 was significantly down-regulated only at the 5-min time point and showed a clear reduction in down-regulation at the 15-min through 1-h time points; by 2 h after serum and temperature induction NRG1 levels in the ume6Δ/ume6Δ mutant were equivalent or even slightly greater than those under non-inducing conditions (Figure 6). These results suggest that Ume6 functions as negative regulator of NRG1 and plays an important role in controlling the level and duration of NRG1 down-regulation in response to serum at 37°C. We first observed induction of UME6 (∼40-fold) at the 15-min time point. In general, UME6 transcript levels gradually decline over the time course, although we did consistently observe a slight increase between the 1- and 2-h time points (Figure 6). Our results indicate that down-regulation of NRG1 precedes induction of UME6 in the series of regulatory events that mediate the filamentation response to serum at 37°C.
Epistatic Relationships between UME6 and RFG1, NRG1, and TUP1
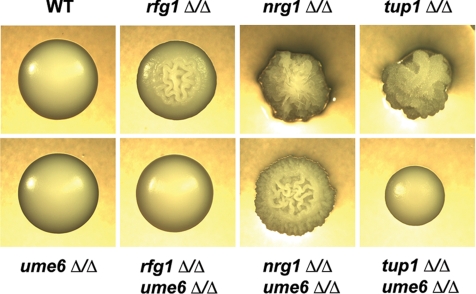
The results described above suggest a functional relationship between UME6 and the NRG1-TUP1 filamentous growth regulatory pathway. To define this relationship more precisely and to investigate whether UME6 also functions in the RFG1-TUP1 pathway, we generated rfg1Δ/rfg1Δ ume6Δ/ume6Δ, nrg1Δ/nrg1Δ ume6Δ/ume6Δ, and tup1Δ/tup1Δ ume6Δ/ume6Δ double mutant strains and examined both colony and cell morphologies (compared with the individual single mutants) when cells were grown in the absence of filament-inducing conditions. Under non–filament-inducing conditions the ume6Δ/ume6Δ mutant grows as blastospores, the rfg1Δ/rfg1Δ mutant is mildly filamentous and nrg1Δ/nrg1Δ and tup1Δ/tup1Δ mutants are highly filamentous. We observed that both rfg1Δ/rfg1Δ ume6Δ/ume6Δ and tup1Δ/tup1Δ ume6Δ/ume6Δ double mutants grew as smooth colonies on solid non–filament-inducing medium and that tup1Δ/tup1Δ ume6Δ/ume6Δ colonies were mildly defective for growth (Figure 7). The nrg1Δ/nrg1Δ ume6Δ/ume6Δ double mutant showed a wrinkled colony morphology, although this strain did not appear as wrinkled or as adherent to the solid agar as the nrg1Δ/nrg1Δ single mutant. In liquid non–filament-inducing medium (YEPD) the rfg1Δ/rfg1Δ ume6Δ/ume6Δ mutant grew as a combination of blastospores and short filaments (Figure 8A). Under these conditions both nrg1Δ/nrg1Δ ume6Δ/ume6Δ and tup1Δ/tup1Δ ume6Δ/ume6Δ double mutants were still mostly filamentous but lacked the extended filaments that are normally observed in the nrg1Δ/nrg1Δ and tup1Δ/tup1Δ single mutants. This defect was significantly more pronounced in the tup1Δ/tup1Δ ume6Δ/ume6Δ mutant and a significant number of individual cells of this strain also appeared to grow in an elongated oval-like morphology. These findings suggest that while the ume6Δ/ume6Δ mutation is not completely epistatic to the rfg1Δ/rfg1Δ, nrg1Δ/nrg1Δ, or tup1Δ/tup1Δ mutations with respect to filamentation, UME6 plays a specific important role in the ability of these mutants to generate extended filaments.
Figure 7.
Colony morphologies of rfg1Δ/Δ ume6Δ/Δ, nrg1Δ/Δ ume6Δ/Δ, and tup1Δ/Δ ume6 Δ/Δ double mutants and respective single mutants. Colony morphologies of the indicated strains are shown after growth on solid YEPD medium at 30°C (non–filament-inducing conditions) for 3 d. Colonies were visualized by light microscopy and photographed at ∼×20 magnification.
We also examined epistatic relationships between UME6 and RFG1, NRG1, TUP1 at the transcriptional level. Northern analysis was carried out using total RNA prepared from all the single and double mutant strains described above as well as a wild-type control strain grown under non–filament-inducing conditions. We examined several strongly induced filament-specific transcripts that have previously been shown to be derepressed in rfg1Δ/rfg1Δ, nrg1Δ/nrg1Δ, and/or tup1Δ/tup1Δ mutants (Braun and Johnson, 2000; Braun et al., 2001; Kadosh and Johnson, 2001, 2005; Murad et al., 2001). The majority of these transcripts failed to show derepression in the rfg1Δ/rfg1Δ ume6Δ/ume6Δ, nrg1Δ/nrg1Δ ume6Δ/ume6Δ, and tup1Δ/tup1Δ ume6Δ/ume6Δ double mutant strains under non–filament-inducing conditions (Figures 8, B and C). These results indicate that with respect to the transcriptional regulation of several filament-specific genes the ume6Δ/ume6Δ mutation is epistatic to rfg1Δ/rfg1Δ, nrg1Δ/nrg1Δ, and tup1Δ/tup1Δ mutations and suggest that Ume6 functions as a downstream component of the Rfg1, Nrg1, and Tup1 pathways. Interestingly, the HWP1 transcript was mildly derepressed in the nrg1Δ/nrg1Δ ume6Δ/ume6Δ double mutant and derepression of the RBT4 transcript did not appear to be affected to a significant degree in the rfg1Δ/rfg1Δ ume6Δ/ume6Δ and tup1Δ/tup1Δ ume6Δ/ume6Δ mutants. These results suggest that in the case of a few specific genes Rfg1, Nrg1, and Tup1 may direct repression to some extent by a Ume6-independent pathway.
In confirmation of our previous findings by DNA microarray analysis (Kadosh and Johnson, 2005), the UME6 transcript was derepressed in both the nrg1Δ/nrg1Δ and tup1Δ/tup1Δ mutant strains in the absence of filament-inducing conditions (Figure 8B). The level of UME6 derepression was greater in the tup1Δ/tup1Δ mutant compared with that in the nrg1Δ/nrg1Δ mutant, suggesting that UME6 is also regulated by a Nrg1-independent Tup1 repression pathway(s). Consistent with this finding, we observed that UME6 is mildly derepressed in the rfg1Δ/rfg1Δ mutant, indicating that UME6 is also under negative control by the Rfg1-Tup1 repression pathway (Figure 8C). We also note that under the growth conditions used in our experiment the ECE1 transcript appears to show greater derepression in the nrg1Δ/nrg1Δ versus tup1Δ/tup1Δ mutant (Figure 8B), suggesting that this gene may partially be under the control of a Tup1-independent Nrg1 repression pathway.
DISCUSSION
A Novel Transcriptional Regulator Specifically Controls Hyphal Filament Extension and Virulence
A wide variety of both pathogenic and nonpathogenic fungal species are capable of growing in a filamentous form. Extension of initial short filaments (germ tubes) to elongated hyphae is critical for a diverse array of processes including nutrient scavenging, mating, thigmotropism, and virulence (Odds, 1988; Collier et al., 1998; Madden and Snyder, 1998; Kumamoto and Vinces, 2005a). In pathogenic fungi hyphal extension has been shown to be specifically important for the virulence-related processes of tissue invasion (particularly invasion of mucosal epithelial cells), angioinvasion, breaching of endothelial cells and the lysis of macrophages and neutrophils (Lo et al., 1997; Korting et al., 2003; Kumamoto and Vinces, 2005b; Filler and Sheppard, 2006).
Despite the widespread importance of hyphal filament extension for a variety of fungal species, little is known about the regulatory mechanisms that control this process. In Neurospora crassa, maintenance of a Ca2+ gradient at the hyphal tip as well as interaction between two kinases (POD6 and COT1) is important for the extension of hyphal filaments (Silverman-Gavrila and Lew, 2003; Seiler et al., 2006). C. albicans also appears to utilize a Ca2+-dependent mechanism to orient hyphal growth (Brand et al., 2007), and calcineurin has been shown to be important for hyphal elongation in both A. fumigatus and Cryptococcus neoformans (Cruz et al., 2001; Steinbach et al., 2006). In the corn smut fungus Ustilago maydis, deletion of a kinesin affects both hyphal extension as well as pathogenesis and several kinesins are up-regulated in hyphae (Lehmler et al., 1997; Schuchardt et al., 2005).
The identification of UME6 represents a significant advance in our understanding of the molecular mechanisms that regulate hyphal filament extension. Because UME6 encodes a transcriptional regulator, it appears that expression of many of the components required for hyphal extension (such as those described above) is controlled in a coordinated manner by a central regulatory mechanism. There are several explanations for our finding that the ume6Δ/ume6Δ mutant is specifically defective for hyphal extension, but not germ tube formation. One possibility is that germ tube formation and hyphal extension represent distinct processes that are controlled by separate gene sets. A second more likely possibility, supported by our finding that filament-specific gene expression is still induced (albeit at lower levels) during germ tube formation in the ume6Δ/ume6Δ mutant, is that a common, or very similar, set of genes functions to direct both processes.
These studies also allow us to gain insight into the role of hyphal filament extension in virulence. Previous reports have shown that C. albicans strains locked in the blastospore form, such as the efg1Δ/efg1Δ mutant or a strain expressing high constitutive levels of NRG1, are completely attenuated for virulence in a mouse model of systemic candidiasis (Lo et al., 1997; Saville et al., 2003). Interestingly, although the ume6Δ/ume6Δ mutant, which can form filaments but is defective for hyphal extension both in vitro and during infection in vivo, is clearly attenuated for virulence, most of the mice eventually succumb to infection and die (albeit after a significantly longer time period than mice infected with a wild-type strain). Our results suggest that the attenuated virulence of this mutant can be at least partially ascribed to a defect in hyphal filament extension causing reduced and/or delayed tissue invasion and damage. Hyphal extension may therefore be important, but not absolutely required, for virulence in this infection model. Although our findings are consistent with previous studies implicating hyphal extension in a variety of virulence-related processes (Scherwitz, 1982; Ray and Payne, 1988; Zink et al., 1996; Lo et al., 1997; Jong et al., 2001; Kumamoto and Vinces, 2005b; Filler and Sheppard, 2006), we cannot exclude the possibility that the ume6Δ/ume6Δ mutant is attenuated for virulence as a result of the reduced expression of virulence factors that are not involved in filamentation per se.
Correlation of Hyphal Extension with the Level and Duration of Expression the C. albicans Filamentous Growth Program
An important observation is that UME6, itself a filament-induced transcript, plays a role in controlling the level and duration of expression of nearly all of the most highly induced genes in the C. albicans filamentous growth program. Consistent with this observation, the ume6Δ/ume6Δ defect in hyphal extension becomes most apparent toward the later time points (after 2 h) following serum and temperature induction. Interestingly, our results suggest that genes in the C. albicans filamentous growth program must be expressed at high levels for an extended period of time in order for cells to make the transition from germ tubes to extended hyphal filaments. It is important to note that our DNA microarray analysis did not detect the activation or repression of a novel set of genes that specifically correlates with the ume6Δ/ume6Δ hyphal extension defect. Thus, we hypothesize that hyphal filament extension is correlated with the level and duration of filament-specific gene expression rather than activation (or repression) of a separate gene set.
Do specific Ume6 target genes play key roles in carrying out the hyphal filament extension process? Although we do not yet have definitive evidence to answer this question, two hyphal-specific genes, encoding Eed1 and the Hgc1 cyclin-related protein, are transcriptionally regulated and have been shown to play important roles in hyphal extension (Zheng et al., 2004; Zakikhany et al., 2007). Sinha et al. (2007) have recently proposed a model for the initiation and maintenance of polarized hyphal growth that involves regulation of septin phosphorylation by Cdc28-cyclin complexes that can include Hgc1, and Zheng et al. (2007) have recently shown that phosphorylation of Rga2, a GTPase activating protein, by Cdc28/Hgc1 and localization of Rga2 away from the hyphal tip is important for extended hyphal cell development. These findings suggest that hyphal extension may be regulated not only at the transcriptional level but also at the level of posttranslational modification and intracellular protein localization.
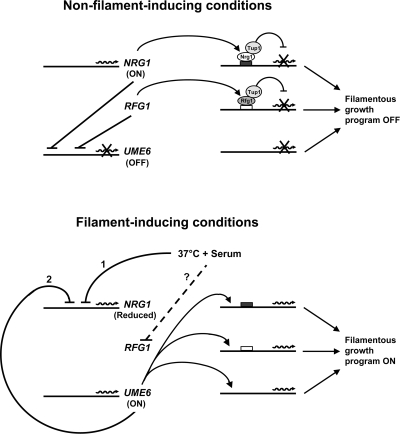
Model for Control of Induction and Maintenance of Expression of the C. albicans Filamentous Growth Program by Rfg1, Nrg1, Tup1, and Ume6
Our results suggest a model in which Nrg1 and Ume6 function together by a negative feedback loop to control the induction and maintained expression of the C. albicans filamentous growth program in response to filament-inducing conditions such as serum at 37°C (Figure 9). Under non–filament-inducing conditions the UME6 transcript would not be expressed (or expressed at very low levels) as a consequence of strong repression by the Nrg1-Tup1 pathway. NRG1 levels would be too high and UME6 levels would be too low to cause expression of filament-specific target genes. Initially, within 5 min after induction by serum at 37°C, the NRG1 transcript is strongly down-regulated; although the mechanism controlling NRG1 down-regulation is not entirely clear, another key transcriptional regulator of filamentous growth, Efg1 (the target of a cAMP-PKA signaling pathway; Ernst, 2000), has been shown to be important for this process (Braun et al., 2001). Rapid down-regulation of the NRG1 transcript would lead to a strong induction (or derepression) of UME6, which is subsequently observed at the 15-min time point. We believe that a certain threshold level of Ume6 expression is required to maintain repression of the NRG1 transcript. As the time course progresses Ume6 levels eventually fall below this threshold, causing NRG1 transcript levels to increase starting at ∼2 h after induction. The net result of this negative feedback loop would be a transient reduction of NRG1 levels and elevation in UME6 levels, which becomes most apparent at early time points. As a consequence of NRG1 down-regulation, Nrg1-Tup1–controlled target genes in the C. albicans filamentous growth program would be derepressed. In addition Ume6, when expressed at sufficiently high levels, would function independently as a strong activator of filament- and virulence-specific target genes. Rfg1 also appears to function as a repressor of UME6 under non–filament-inducing conditions, and we hypothesize that relief of Rfg1-Tup1–mediated repression, by an unknown mechanism, may also play a role in the induction of UME6. This model is supported by several lines of evidence: 1) UME6 is repressed by both the Rfg1-Tup1 and Nrg1-Tup1 pathways under non–filament-inducing conditions, and epistasis analysis indicates that Ume6 functions as a downstream component of these pathways to control expression of several filament-specific transcripts, 2) the NRG1 repressor is down-regulated by serum at 37°C starting at the 5-min time point, 3) transcription of UME6 is induced by serum and 37°C at the 15-min time point, after down-regulation of NRG1, 4) UME6 functions as a repressor of NRG1 only under inducing conditions and is important for controlling the level and duration of NRG1 down-regulation in response to serum and 37°C, 5) several genes in the filamentous growth program, which are not repressed by the Rfg1-Tup1 or Nrg1-Tup1 pathways (Kadosh and Johnson, 2005), show reduced serum and temperature induction in the ume6Δ/ume6Δ mutant (e.g., USO5, NBP35, ERV46), suggesting that Ume6 also functions independently as a strong activator of these transcripts, 6) both the level and duration of filament-specific gene induction are reduced in the ume6Δ/ume6Δ mutant, suggesting that a mechanism for amplifying the inducing signal (and consequently maintaining expression of filament-specific genes) is in place.
Figure 9.
Model for control of induction and maintenance of expression of the C. albicans filamentous growth program by Rfg1, Nrg1, Tup1, and Ume6. Nrg1 and Ume6 are believed to repress each other in a negative feedback loop. Under steady-state non–filament-inducing conditions (top) Nrg1 levels are high, and UME6 is not expressed (due to strong Nrg1-Tup1 repression). In turn, many filament-specific transcripts are not induced as a consequence of repression by the Nrg1-Tup1 pathway and/or lack of activation by Ume6. In addition, under non–filament-inducing conditions, UME6 as well as several filament-specific transcripts, are repressed by the Rfg1-Tup1 pathway. In the presence of filament-inducing conditions such as serum at 37°C (bottom) the NRG1 transcript undergoes a very rapid transient down-regulation (1) causing UME6 levels to rise. As a consequence of repression by Ume6 (2), the NRG1 transcript is maintained at a reduced level for a significantly longer time period. Filament-specific transcripts are subsequently induced as a result of relief of repression by the Nrg1-Tup1 and Rfg1-Tup1 pathways and/or activation by Ume6. Also, in the presence of serum at 37°C repression of UME6 by the Rfg1-Tup1 pathway is believed to be relieved by a mechanism not yet determined (dashed line). Please note that it is unclear at this point as to whether Ume6 and Nrg1 repress each other by direct or indirect effects.
It is important to bear in mind that this model may not apply to all genes in the C. albicans filamentous growth program. RBT4, for example, appears to be controlled by the Rfg1-Tup1 pathway but not by Ume6 (to a significant degree). We also cannot exclude the possibility of alternative models in which UME6 is induced by Rfg1-, Nrg1-, and Tup1-independent mechanisms (or by relief of repression by a Tup1 pathway that does not depend upon Rfg1 or Nrg1) or models in which down-regulation of NRG1 is maintained by additional Ume6-independent mechanisms.
How can UME6 simultaneously function as a repressor of NRG1 and activator of filament-specific transcripts? It is possible that at least one of these events occurs as the result of an indirect effect. For example, rather than directly repressing the NRG1 transcript, Ume6 may instead function to activate a strong repressor of NRG1. It is also unclear at this point whether Rfg1 represses UME6 under non–filament-inducing conditions by a direct or indirect mechanism.
Interestingly, although UME6 is most strongly induced at early time points after serum- and temperature-induction, this regulator plays an important role in promoting hyphal extension at later time points. We hypothesize that this “lag time” may be due to the continued presence of Ume6 protein levels after UME6 transcript levels have decreased. Consistent with this hypothesis, filament-specific transcript levels are reduced most significantly at the later time points following serum- and temperature-induction in the ume6Δ/ume6Δ mutant, suggesting that in a wild-type strain the Ume6 protein is normally present at sufficiently high levels to cause activation of these genes (we are currently in the process of generating Ume6 antibodies to address this hypothesis as well as carry out additional planned Ume6 protein studies). Alternatively, because many filament-specific transcripts are initially expressed at lower levels in the ume6Δ/ume6Δ mutant, these transcripts may sooner decline (due to degradation over the time course) below the threshold required to maintain extended hyphal growth.
The work presented in this article indicates that UME6 functions as an important regulator of C. albicans hyphal filament extension and virulence as well as a key downstream component of the Rfg1, Nrg1, and Tup1 filamentous growth regulatory pathways. Our observation that UME6 is induced by multiple filament-inducing conditions (in addition to serum and 37°C) suggests that a variety of filamentous growth signaling and regulatory pathways may utilize this mechanism to promote hyphal filament extension. Future studies on Ume6 should provide significantly greater insight into the molecular basis for this poorly understood virulence trait.
Supplementary Material
ACKNOWLEDGMENTS
We thank Brian Wickes for critical reading of this manuscript as well as members of the San Antonio Center for Medical Mycology for fruitful discussions and technical advice during the course of the experiments. We are grateful to Alexander Johnson (University of California, San Francisco) for providing whole-genome C. albicans DNA microarrays; Suzanne Noble (University of California, San Francisco), Joachim Morschhäuser (Universität Würzburg, Germany), and Stephen Saville (University of Texas at San Antonio) for providing plasmids and/or strains; and P. Dube for use of the differential interference contrast microscope in his laboratory. C. albicans sequence data were provided by the Stanford Genome Technology Center (http://www-sequence.stanford.edu/group/candida). We thank Alexander Johnson for supporting several of the initial experiments that are described in this article. This work was also supported by a University of Texas Health Science Center at San Antonio Executive Research Committee New Investigator Award to D.K. and by Grant RO1AI064562 from the National Institute of Allergy and Infectious Diseases (NIAID) to J.L.L.-R. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID or the National Institutes of Health.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-11-1110) on January 23, 2008.
REFERENCES
- Arnaud M. B., Costanzo M. C., Skrzypek M. S., Binkley G., Lane C., Miyasato S. R., Sherlock G. The Candida Genome Database (CGD), a community resource for Candida albicans gene and protein information. Nucleic Acids Res. 2005;33:D358–D363. doi: 10.1093/nar/gki003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K., editors. New York: Greene Publishing Associates and Wiley-Interscience; 1992. Current Protocols in Molecular Biology. [Google Scholar]
- Bailey D. A., Feldmann P. J., Bovey M., Gow N. A., Brown A. J. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J. Bacteriol. 1996;178:5353–5360. doi: 10.1128/jb.178.18.5353-5360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartie K. L., Williams D. W., Wilson M. J., Potts A. J., Lewis M. A. Differential invasion of Candida albicans isolates in an in vitro model of oral candidosis. Oral Microbiol. Immunol. 2004;19:293–296. doi: 10.1111/j.1399-302X.2004.00155.x. [DOI] [PubMed] [Google Scholar]
- Bennett R. J., Uhl M. A., Miller M. G., Johnson A. D. Identification and characterization of a Candida albicans mating pheromone. Mol. Cell. Biol. 2003;23:8189–8201. doi: 10.1128/MCB.23.22.8189-8201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship J. R., Mitchell A. P. How to build a biofilm: a fungal perspective. Curr. Opin. Microbiol. 2006;9:588–594. doi: 10.1016/j.mib.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Bowdish K. S., Yuan H. E., Mitchell A. P. Positive control of yeast meiotic genes by the negative regulator UME6. Mol. Cell. Biol. 1995;15:2955–2961. doi: 10.1128/mcb.15.6.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A., Shanks S., Duncan V. M., Yang M., Mackenzie K., Gow N. A. Hyphal orientation of Candida albicans is regulated by a calcium-dependent mechanism. Curr. Biol. 2007;17:347–352. doi: 10.1016/j.cub.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun B. R., Head W. S., Wang M. X., Johnson A. D. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics. 2000;156:31–44. doi: 10.1093/genetics/156.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun B. R., Johnson A. D. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science. 1997;277:105–109. doi: 10.1126/science.277.5322.105. [DOI] [PubMed] [Google Scholar]
- Braun B. R., Johnson A. D. TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics. 2000;155:57–67. doi: 10.1093/genetics/155.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun B. R., Kadosh D., Johnson A. D. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 2001;20:4753–4761. doi: 10.1093/emboj/20.17.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun B. R., et al. A human-curated annotation of the Candida albicans genome. PLoS Genet. 2005;1:36–57. doi: 10.1371/journal.pgen.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. J. Morphogenetic signaling pathways in Candida albicans. In: Calderone R. A., editor. Candida and Candidiasis. Washington, DC: ASM Press; 2002. pp. 95–106. [Google Scholar]
- Calderone R. A., Gow N. A. Host recognition by Candida species. In: Calderone R. A., editor. Candida and Candidiasis. Washington, DC: ASM Press; 2002. pp. 67–86. [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc. Natl. Acad. Sci. USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier L., Balons A., Sussman M., editors. New York: Oxford University Press; 1998. Microbiology and Microbial Infections. [Google Scholar]
- Cruz M. C., Fox D. S., Heitman J. Calcineurin is required for hyphal elongation during mating and haploid fruiting in Cryptococcus neoformans. EMBO J. 2001;20:1020–1032. doi: 10.1093/emboj/20.5.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckert J., Rodriguez Torres A. M., Simon J. T., Zitomer R. S. Mutational analysis of Rox1, a DNA-bending repressor of hypoxic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:6109–6117. doi: 10.1128/mcb.15.11.6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont P. F. Candida albicans, the opportunist. A cellular and molecular perspective. J. Am. Podiatr. Med. Assoc. 1995;85:104–115. doi: 10.7547/87507315-85-2-104. [DOI] [PubMed] [Google Scholar]
- Edmond M. B., Wallace S. E., McClish D. K., Pfaller M. A., Jones R. N., Wenzel R. P. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin. Infect. Dis. 1999;29:239–244. doi: 10.1086/520192. [DOI] [PubMed] [Google Scholar]
- Einerhand A. W., Kos W., Smart W. C., Kal A. J., Tabak H. F., Cooper T. G. The upstream region of the FOX3 gene encoding peroxisomal 3-oxoacyl-coenzyme A thiolase in Saccharomyces cerevisiae contains ABF1- and replication protein A-binding sites that participate in its regulation by glucose repression. Mol. Cell. Biol. 1995;15:3405–3414. doi: 10.1128/mcb.15.6.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J. F. Transcription factors in Candida albicans—environmental control of morphogenesis. Microbiology. 2000;146:1763–1774. doi: 10.1099/00221287-146-8-1763. [DOI] [PubMed] [Google Scholar]
- Filler S. G., Sheppard D. C. Fungal invasion of normally non-phagocytic host cells. PLoS Pathog. 2006;2:e129. doi: 10.1371/journal.ppat.0020129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzi W. A., Irwin M. Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Fink G. R. San Diego: Academic Press; 1991. Guide to Yeast Genetics and Molecular Biology. [Google Scholar]
- Jong A. Y., Stins M. F., Huang S. H., Chen S. H., Kim K. S. Traversal of Candida albicans across human blood-brain barrier in vitro. Infect. Immun. 2001;69:4536–4544. doi: 10.1128/IAI.69.7.4536-4544.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D., Johnson A. D. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol. Cell. Biol. 2001;21:2496–2505. doi: 10.1128/MCB.21.7.2496-2505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D., Johnson A. D. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell. 2005;16:2903–2912. doi: 10.1091/mbc.E05-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleher C. A., Redd M. J., Schultz J., Carlson M., Johnson A. D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- Khalaf R. A., Zitomer R. S. The DNA binding protein Rfg1 is a repressor of filamentation in Candida albicans. Genetics. 2001;157:1503–1512. doi: 10.1093/genetics/157.4.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korting H. C., Hube B., Oberbauer S., Januschke E., Hamm G., Albrecht A., Borelli C., Schaller M. Reduced expression of the hyphal-independent Candida albicans proteinase genes SAP1 and SAP3 in the efg1 mutant is associated with attenuated virulence during infection of oral epithelium. J. Med. Microbiol. 2003;52:623–632. doi: 10.1099/jmm.0.05125-0. [DOI] [PubMed] [Google Scholar]
- Kumamoto C. A., Vinces M. D. Alternative Candida albicans lifestyles: growth on surfaces. Annu. Rev. Microbiol. 2005a;59:113–133. doi: 10.1146/annurev.micro.59.030804.121034. [DOI] [PubMed] [Google Scholar]
- Kumamoto C. A., Vinces M. D. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell Microbiol. 2005b;7:1546–1554. doi: 10.1111/j.1462-5822.2005.00616.x. [DOI] [PubMed] [Google Scholar]
- Lee K. L., Buckley H. R., Campbell C. C. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia. 1975;13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- Lehmler C., Steinberg G., Snetselaar K. M., Schliwa M., Kahmann R., Bolker M. Identification of a motor protein required for filamentous growth in Ustilago maydis. EMBO J. 1997;16:3464–3473. doi: 10.1093/emboj/16.12.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Kohler J., Fink G. R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- Lo H. J., Kohler J. R., DiDomenico B., Loebenberg D., Cacciapuoti A., Fink G. R. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Lotz H., Sohn K., Brunner H., Muhlschlegel F. A., Rupp S. RBR1, a novel pH-regulated cell wall gene of Candida albicans, is repressed by RIM101 and activated by NRG1. Eukaryot. Cell. 2004;3:776–784. doi: 10.1128/EC.3.3.776-784.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden K., Snyder M. Cell polarity and morphogenesis in budding yeast. Annu. Rev. Microbiol. 1998;52:687–744. doi: 10.1146/annurev.micro.52.1.687. [DOI] [PubMed] [Google Scholar]
- Miller L. G., Hajjeh R. A., Edwards J. E., Jr. Estimating the cost of nosocomial candidemia in the United States. Clin. Infect. Dis. 2001;32:1110. doi: 10.1086/319613. [DOI] [PubMed] [Google Scholar]
- Mitchell A. P. Dimorphism and virulence in Candida albicans. Curr. Opin. Microbiol. 1998;1:687–692. doi: 10.1016/s1369-5274(98)80116-1. [DOI] [PubMed] [Google Scholar]
- Murad A.M.A., et al. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 2001:4742–4752. doi: 10.1093/emboj/20.17.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble S. M., Johnson A. D. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell. 2005;4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds F. C. Candida and Candidosis. London: Baillière Tindall; 1988. [Google Scholar]
- Park S. H., Koh S. S., Chun J. H., Hwang H. J., Kang H. S. Nrg1 is a transcriptional repressor for glucose repression in STA1 gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:2044–2050. doi: 10.1128/mcb.19.3.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage G., Saville S. P., Thomas D. P., Lopez-Ribot J. L. Candida biofilms: an update. Eukaryot. Cell. 2005;4:633–638. doi: 10.1128/EC.4.4.633-638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray T. L., Payne C. D. Scanning electron microscopy of epidermal adherence and cavitation in murine candidiasis: a role for Candida acid proteinase. Infect. Immun. 1988;56:1942–1949. doi: 10.1128/iai.56.8.1942-1949.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuss O., Vik A., Kolter R., Morschhauser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Sanglard D., Hube B., Monod M., Odds F. C., Gow N. A. A triple deletion of the secreted aspartyl proteinase genes SAP4, SAP5, and SAP6 of Candida albicans causes attenuated virulence. Infect. Immun. 1997;65:3539–3546. doi: 10.1128/iai.65.9.3539-3546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville S. P., Lazzell A. L., Monteagudo C., Lopez-Ribot J. L. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell. 2003;2:1053–1060. doi: 10.1128/EC.2.5.1053-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller M., Korting H. C., Schafer W., Bastert J., Chen W., Hube B. Secreted aspartic proteinase (Sap) activity contributes to tissue damage in a model of human oral candidosis. Mol. Microbiol. 1999;34:169–180. doi: 10.1046/j.1365-2958.1999.01590.x. [DOI] [PubMed] [Google Scholar]
- Scherwitz C. Ultrastructure of human cutaneous candidosis. J. Invest. Dermatol. 1982;78:200–205. doi: 10.1111/1523-1747.ep12506451. [DOI] [PubMed] [Google Scholar]
- Schuchardt I., Assmann D., Thines E., Schuberth C., Steinberg G. Myosin-V, Kinesin-1, and Kinesin-3 cooperate in hyphal growth of the fungus Ustilago maydis. Mol. Biol. Cell. 2005;16:5191–5201. doi: 10.1091/mbc.E05-04-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler S., Vogt N., Ziv C., Gorovits R., Yarden O. The STE20/germinal center kinase POD6 interacts with the NDR kinase COT1 and is involved in polar tip extension in Neurospora crassa. Mol. Biol. Cell. 2006;17:4080–4092. doi: 10.1091/mbc.E06-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd M. G., Poulter R. T., Sullivan P. A. Candida albicans: biology, genetics, and pathogenicity. Annu. Rev. Microbiol. 1985;39:579–614. doi: 10.1146/annurev.mi.39.100185.003051. [DOI] [PubMed] [Google Scholar]
- Silverman-Gavrila L. B., Lew R. R. Calcium gradient dependence of Neurospora crassa hyphal growth. Microbiology. 2003;149:2475–2485. doi: 10.1099/mic.0.26302-0. [DOI] [PubMed] [Google Scholar]
- Sinha I., Wang Y. M., Philp R., Li C. R., Yap W. H., Wang Y. Cyclin-dependent kinases control septin phosphorylation in Candida albicans hyphal development. Dev. Cell. 2007;13:421–432. doi: 10.1016/j.devcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Smith R. L., Johnson A. D. Turning genes off by Ssn6-Tup1, a conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci. 2000;25:325–330. doi: 10.1016/s0968-0004(00)01592-9. [DOI] [PubMed] [Google Scholar]
- Steber C. M., Esposito R. E. UME6 is a central component of a developmental regulatory switch controlling meiosis-specific gene expression. Proc. Natl. Acad. Sci. USA. 1995;92:12490–12494. doi: 10.1073/pnas.92.26.12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbach W. J., et al. Calcineurin controls growth, morphology, and pathogenicity in Aspergillus fumigatus. Eukaryot. Cell. 2006;5:1091–1103. doi: 10.1128/EC.00139-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strich R., Surosky R. T., Steber C., Dubois E., Messenguy F., Esposito R. E. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 1994;8:796–810. doi: 10.1101/gad.8.7.796. [DOI] [PubMed] [Google Scholar]
- Sudbery P., Gow N., Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Sweet D. H., Jang Y. K., Sancar G. B. Role of UME6 in transcriptional regulation of a DNA repair gene in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:6223–6235. doi: 10.1128/mcb.17.11.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzamarias D., Struhl K. Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8-Tup1 corepressor complex to differentially regulated promoters. Genes Dev. 1995;9:821–831. doi: 10.1101/gad.9.7.821. [DOI] [PubMed] [Google Scholar]
- Weig M., Gross U., Muhlschlegel F. Clinical aspects and pathogenesis of Candida infection. Trends Microbiol. 1998;6:468–470. doi: 10.1016/s0966-842x(98)01407-3. [DOI] [PubMed] [Google Scholar]
- Zakikhany K., Naglik J. R., Schmidt-Westhausen A., Holland G., Schaller M., Hube B. In vivo transcript profiling of Candida albicans identifies a gene essential for interepithelial dissemination. Cell Microbiol. 2007;9:2938–2954. doi: 10.1111/j.1462-5822.2007.01009.x. [DOI] [PubMed] [Google Scholar]
- Zhao X., Oh S. H., Cheng G., Green C. B., Nuessen J. A., Yeater K., Leng R. P., Brown A. J., Hoyer L. L. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology. 2004;150:2415–2428. doi: 10.1099/mic.0.26943-0. [DOI] [PubMed] [Google Scholar]
- Zheng X., Wang Y., Wang Y. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 2004;23:1845–1856. doi: 10.1038/sj.emboj.7600195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X. D., Lee R. T., Wang Y. M., Lin Q. S., Wang Y. Phosphorylation of Rga2, a Cdc42 GAP, by CDK/Hgc1 is crucial for Candida albicans hyphal growth. EMBO J. 2007;26:3760–3769. doi: 10.1038/sj.emboj.7601814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink S., Nass T., Rosen P., Ernst J. F. Migration of the fungal pathogen Candida albicans across endothelial monolayers. Infect. Immun. 1996;64:5085–5091. doi: 10.1128/iai.64.12.5085-5091.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.