Abstract
An assembly of a catalytic unit for aminoacylation of an RNA microhelix is demonstrated here. This assembly may recapitulate a step in the historical development of tRNA synthetases. The class-defining domain of a tRNA synthetase is closely related to the primordial enzyme that catalyzed synthesis of aminoacyl adenylate. RNA binding elements are imagined to have been added so that early RNA substrates could be docked proximal to the activated amino acid. RNA microhelices that recapitulate the acceptor stem of modern tRNAs are potential examples of early substrates. In this work, we examined a fragment of Escherichia coli alanyl-tRNA synthetase, which catalyzes aminoacyl adenylate formation but is virtually inactive for catalysis of RNA microhelix aminoacylation. Fusion to the fragment of either of two unrelated nonspecific RNA binding domains activated microhelix aminoacylation. Although the fusion proteins lacked the RNA sequence specificity of the natural enzyme, their activity was within 1–2 kcal⋅mol−1 of a truncated alanyl-tRNA synthetase that has aminoacylation activity sufficient to sustain cell growth. These results show that, starting with an activity for adenylate synthesis, barriers are relatively low for building catalytic units for aminoacylation of RNA helices.
We report here an example of a pathway by which one component of the genetic code could be assembled in evolution. Aminoacyl-tRNA synthetases establish the genetic code by attaching amino acids to transfer RNAs that bear the corresponding anticodons defined by the code (1–3). Because they establish the code, these enzymes are believed to be amongst the earliest proteins (4). The enzymes activate amino acids by coupling them to ATP to form an aminoacyl adenylates and subsequently transfer the amino acid to the 3′ acceptor end of tRNA (5, 6). Extant aminoacyl-tRNA synthetases are divided into two classes based on the structures of their active sites. Class I enzymes are characterized by a Rossman nucleotide binding fold having an 11-aa signature sequence that ends in the tetrapeptide HIGH and a KMSKS pentapeptide (7–9). Class II enzymes have a distinct fold made up of an antiparallel β-sheet and three α-helices that encompass three highly degenerate sequence motifs known as motifs 1, 2, and 3 (5, 10). Members of the two classes of synthetases are thought to have evolved independently from two distinct primordial enzymes that catalyzed the formation of aminoacyl adenylate. Specificity for distinct transfer RNAs and amino acids evolved through changes within the ancestral core and through the incorporation of additional domains that are idiosyncratic to particular synthetases (11, 12).
Transfer RNAs also may have evolved in a piecemeal fashion. Modern tRNAs form an L-shaped structure made up of two domains. One domain is a 12-bp minihelix that is made up of the TΨC and acceptor stems and that terminates at the 3′-end in the single-stranded tetranucleotide N73CCA with the amino acid acceptance site. (N73 is any of the four nucleotides and is referred to as the “discriminator base.”) The second domain contains the anticodon loop at the end of a 10-bp helix comprised of the dihydrouridine and anticodon stems. The two domains segregate distinct functions of the tRNA. Thus, the anticodon-containing domain serves as a reading head that interacts with messenger RNAs whereas the acceptor-TΨC domain contains the site for amino acid attachment and interacts with the peptidyl transfer center in the ribosome.
Several lines of evidence suggested that the two domains of tRNAs arose independently. For example, each domain of tRNA interacts with a separate rRNA on the ribosome (13). In addition, the minihelix is an ancient tag for RNA genomes (14, 15) and is the part of contemporary tRNAs that is used as a primer for viral reverse transcription (16–18). Finally, at least 10 tRNA synthetases isolated across evolution from bacterial to human cells can aminoacylate helical RNA substrates corresponding to the acceptor-TΨC domain or to the acceptor stem alone (termed mini- and microhelices, respectively) (19–27). Aminoacylation of these truncated substrates generally depends on the identity of the N73 discriminator base and on one or two base pairs within the last four pairs of the acceptor stem. The relationship between acceptor stem nucleotides/structures and specific amino acids constitutes an “operational RNA code” for amino acids. This code may have predated the addition of the anticodon branch to the tRNA structure (28).
In one evolutionary scheme for tRNA synthetases, ancient “core enzymes” activated amino acids to form aminoacyl adenylates. The active sites are imagined to be the progenitors of either of the two structural formats that determine the two classes of modern tRNA synthetases. The reaction to form (from an adenylate) an aminoacyl ester linkage with the 3′-hydroxyl of an RNA is thermodynamically favorable. Thus, the challenge in evolution may have been to dock potential RNA substrates proximal to the bound aminoacyl adenylates. These early substrates could have been RNA minihelix-like because such structures are thought to be prevalent in the RNA world (14, 15, 29). When these early RNA substrates incorporated an anticodon-containing branch, the primitive synthetase then could incorporate additional RNA binding elements to make contacts with the second domain of the tRNA.
The dissection by deletions of a modern tRNA synthetase can be viewed as a form of retroevolution. For example, deletion of the C-terminal half of Escherichia coli alanyl-tRNA synthetase (AlaRS) yields a 461-aa N-terminal fragment (461N) that aminoacylates microhelix substrates with the same efficiency as the full length enzyme (30). Because the efficiency of aminoacylation of full-length tRNA substrates by fragment 461N is reduced ≈100-fold when compared with that of the full-length enzyme, this deletion is believed to remove parts of the enzyme that make contact with the second (anticodon-containing) domain of the L-shaped tRNA. (Contacts of AlaRS with the second domain of the tRNA do not include the anticodon triplet, which is unprotected from nuclease attack by the bound enzyme (31).) However, when expressed from a multicopy plasmid, the activity of fragment 461N is sufficient to maintain cell growth (32, 33). Thus, fragment 461N might be seen as an intermediate in the evolutionary development of AlaRS: that is, as a protein that efficiently aminoacylates the acceptor stem domain in isolation.
Further truncation of E. coli AlaRS to fragment 368N yields an enzyme that is fully competent for formation of alanyl adenylate but is inactive under standard conditions for aminoacylation of RNA substrates (32, 34). The fragment contains the three sequence motifs that are characteristic of the active sites of class II enzymes. Fragment 368N lacks RNA binding activity, with a Kd for tRNA at pH 5.2 that is reduced by more than three orders of magnitude compared with that of fragment 461N (35). Therefore, the deletion of the region from Thr369-Asp461 creates an enzyme that may resemble the functional progenitor in the evolutionary scheme described above: that is, one that catalyzes formation of aminoacyl adenylate but cannot effectively bind or aminoacylate RNA.
We imagined that a step in the evolution of aminoacylation activity might be the fusion of a nonspecific RNA binding domain to the catalytic unit for adenylate synthesis. The presence of nonspecific RNA binding domains in modern aminoacyl-tRNA synthetases such as E. coli isoleucyl-tRNA synthetase (36) and Saccharomyces cerevisiae glutaminyl-tRNA synthetase (37) is consistent with these domains playing a role in the development of tRNA synthetases. To test the feasibility of conferring aminoacylation activity on fragment 368N of AlaRS, we fused two different nonspecific RNA binding domains to the core fragment for adenylate synthesis. It was not obvious that a fusion of this sort would result in aminoacylation activity without further refinements by protein engineering.
Materials and Methods
Construction and Purification of Truncated and Fusion Proteins.
All plasmid constructs are based on pQE-AlaS (38), which encodes the E. coli alanyl-tRNA synthetase with a C-terminal His6 tag sequence under the control of an inducible T5 promoter. PCR mutagenesis (39) was used to produce silent mutations that introduce StuI and SacI restriction sites after the codons for Leu374 and Leu465, respectively, yielding the plasmid pAlaS-ESS. To construct a plasmid encoding fragment 465N with a C-terminal His6 tag, an oligonucleotide cassette encoding six His residues and a stop codon was inserted between the SacI and SpeI sites of pAlaS-ESS to give plasmid pESS-465. The N-terminal 227-residue appended domain of S. cerevisiae glutaminyl-tRNA synthetase was amplified by PCR from plasmid pEFW110, which encodes the complete sequence of yeast glutaminyl tRNA synthetase (40). For this purpose, the primers GCGCGCAGGCCTGATGTCTTCTGTAGAAGAATTGACTCAG and GCGCGCGAGCTCCAAATCACCTAGGAAACCTTCATTGAACATAGTCC (restriction sites in italics) introduced StuI and SacI sites, respectively, at the 5′- and 3′-ends of the gene for glutaminyl tRNA synthetase. Subcloning into pESS-465 gave plasmid pESS-AQ, which encodes the first 374 residues of E. coli AlaRS fused to the appended domain of S. cerevisiae glutaminyl tRNA synthetase, with the sequence ELHHHHHH at the C terminus of the fusion. The resulting protein was designated as fusion 374N-AdScQRS. Fusion of the C-terminal domain of Arc1p was achieved analogously by PCR from pCW119 [which encodes the complete Arc1p protein (37)] and ligation of the coding sequence of the C-terminal domain into pESS-465 to give plasmid pESS-AA. The protein expressed from this plasmid was designated as fusion 374N-CArc1p.
Fusion proteins and His6-tagged fragment 465N were purified by nickel affinity chromatography as described (38). Cells were grown in LB medium (supplemented with 50 mg/ml ampicillin) to OD600 of 0.8 and then were inoculated with isopropyl β-d-thiogalactoside to a final concentration of 1.5 mM. The cells were grown an additional 4 hours and then were harvested and lysed in a French press. The His6-tagged enzyme was purified on a Ni–nitrilotriacetic acid agarose (Qiagen, Santa Clarita, CA) column in 50 mM Na2HPO4 (pH 8.0), 300 mM NaCl, and 10% glycerol. Elution was with a gradient of 20–250 mM imidazole. Fragment 368N was prepared as described (41) by trypsin digestion and was purified by gel filtration chromatography on Superose-12.
RNA Substrates and Aminoacylation Assays.
Microhelix RNA substrates were chemically synthesized on a Pharmacia Gene Assembler synthesizer by using published procedures (42). Products of the syntheses were purified on denaturing 20% polyacrylamide gels.
Aminoacylation assays were carried out at ambient temperature (25°C) in 50 mM Hepes (pH 7.5), 20 mM alanine, 4 mM ATP, 20 mM KCl, 10 mM MgCl2, 20 mM β-mercaptoethanol, and 0.1 mg/ml BSA, as described (43). To insure proper base pairing, microhelices were heated to 80°C and were cooled in the absence of magnesium before addition to the aminoacylation reaction mixture. In assays in which different constructions were being directly compared, RNA was at a concentration of 2.5 or 10 μM whereas enzyme concentrations were 0.5 or 1 μM. [Enzyme concentrations were determined by the alanine-dependent ATP-pyrophosphate exchange activity (44).] Kinetic parameters were determined by direct fitting of data points to the Michaelis-Menten equation. Initial rates were determined at ambient temperature (25°C), with RNA concentrations from 10 to 400 μM (465N), 5 to 400 μM (374N-CArc1p), or 2 to 20 μM (374N-AdScQRS). Enzyme concentrations of 0.5 or 1 μM were used.
Results
Nonspecific RNA Binding Domains.
Two well-studied nonspecific RNA binding domains were chosen as fusion partners for this work. The 228 amino acid N-terminal appended domain of S. cerevisiae glutaminyl-tRNA synthetase binds RNA nonspecifically, with a KD for tRNA, pseudoknots, and poly-A RNA of 0.6–1.8 μM (37). Arc1p specifically binds methionyl-tRNA synthetase and glutamyl-tRNA synthetase in the yeast S. cerevisiae. It is thought to deliver tRNA substrates to these enzymes by enhancing the local effective concentration of the substrates around the proteins (45, 46). On the basis of secondary structure predictions, the protein has been divided into three domains. The N-terminal 132 residues are thought to interact with the aminoacyl-tRNA synthetases whereas the middle (133–201) and C-terminal domains (202–376) are responsible for RNA binding. A truncation composed of the only the middle and C-terminal domains nonspecifically binds tRNA with the same affinity as does the full-length protein, with a KD of 5–10 nM. The C-terminal domain alone also binds tRNA, but with a >50-fold reduction in affinity (46).
C-terminal His6-tagged fusions were constructed by attaching the appended domain of yeast GlnRS (AdScQRS) or the C-terminal domain of Arc1p (CArc1p) to the C terminus of fragment 374N of E. coli AlaRS (Fig. 1). (We chose fragment 374N instead of 368N for reasons of technical convenience in constructing the plasmids that encode the fusion proteins. Previous work showed that fragments 368N and 385N have the full adenylate synthesis activity of the native enzyme(35). For that reason we assumed that a fusion protein with the first 374 amino acids of AlaRS would have full activity for adenylate synthesis (see below).) Attempts to express fusions to full length Arc1p and the middle and C-terminal domains of Arc1p yielded no soluble protein and were not pursued further. The 374N-AdScQRS and 374N-CArc1p fusion proteins were isolated by nickel affinity chromatography to a purity of >90%.
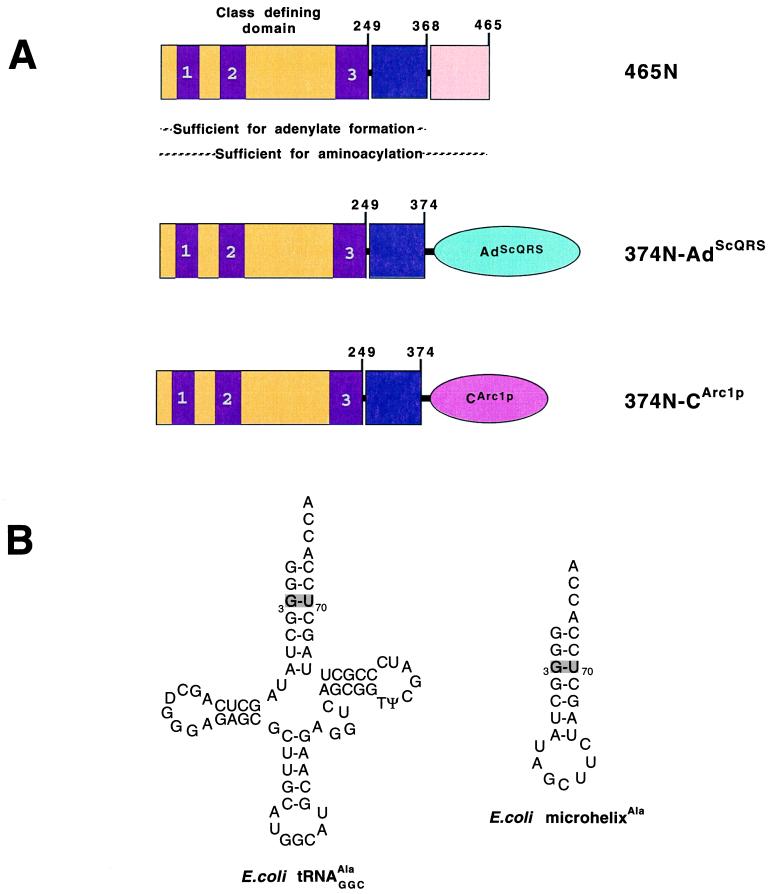
Figure 1.
(A) Schematic diagram of fragment 465N of E. coli alanyl-tRNA synthetase and the fusions to nonspecific RNA binding domains CArc1p and AdScQRS. The class-defining active site region extends to His249 whereas a fragment produced by extensive proteolysis terminates at Arg368. The nonspecific RNA binding domains are schematically shown as ellipsoids. Conserved class II motifs in the class defining domain are labeled as 1, 2, and 3. (B) Sequence and cloverleaf structure of E. coli tRNAAla and sequence and hairpin structure of microhelixAla. The G3:U70 base pair is indicated by a shaded box.
Concentrations of fusion proteins 374N-AdScQRS and 374N-CArc1p were determined by adenylate synthesis activity and by standard Bradford assays. Based on the agreement between these assays, the fusions had full adenylate synthesis activity, indicating that alanine activation is not affected by the addition of the heterologous RNA binding domains or of the His6-tag.
Aminoacylation of RNA Microhelices with Fusion Proteins.
For the aminoacylation assays, a hairpin microhelix comprised of the seven base pairs of the acceptor stem of E. coli tRNAAla was used as a substrate (Fig. 2). This substrate is efficiently aminoacylated by fragment 461N. Aminoacylation is highly specific with respect to the sequence of the microhelix. In particular, it depends on a G3:U70 base pair that marks the molecule for aminoacylation with alanine (47, 48) Our experiments were directed at determining whether the fusion constructions would charge the microhelix, and, if so, then we wanted to compare the specificity of charging with that of fragment 465N of AlaRS. The sensitivity of charging to changes in the essential G3:U70 base pair provided a simple way to test for specificity.
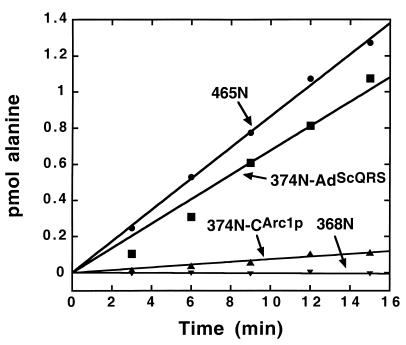
Figure 2.
Aminoacylation of microhelixAla by fusion constructs. Aminoacylation assays were at pH 7.5, 25°C, with 2.5 μM RNA and 500 nM enzyme.
In these experiments, fragment 465N of AlaRS with a 6-His tag appended to the C terminus was used as a standard for sequence-specific aminoacylation of the microhelixAla. Fragment 368N was used as a negative control. [Although fragment 374N could have been used in principle, the purification of this protein could have easily resulted in fragment 368N as a product of proteolysis by cellular proteases (34, 49).] Fragment 465N charged microhelixAla (Fig. 2). As expected, no charging under these conditions was seen with Fragment 368N.
In contrast, both the 374N-AdScQRS and 374N-CArc1p fusion proteins charged microhelixAla at rates clearly above the background of Fragment 368N (Fig. 2). Remarkably, the activity seen with fusion 374N-AdScQRS was comparable to that of fragment 465N. As for the 374N-CArc1p fusion construction, the efficiency of aminoacylation was ≈15-fold lower than that with fragment 465N.
Kinetic parameters were measured for each of the proteins over a range of at least 10-fold in the concentration of microhelixAla. This analysis showed that that both fusion proteins have kcat values ≈20-fold lower than that for fragment 465N. [The value of kcat for the His-tagged fragment 465N is ≈9-fold lower than that previously measured for aminoacylation of microhelixAla by full length AlaRS (50) at 37°C. The value of Km (100 μM) for fragment 465N is similar to that previously reported for full length AlaRS. Because fragment 461N aminoacylates truncated RNA substrates with the same efficiency as the full length enzyme (51), this lowering of kcat could be attributable to the difference in temperature (25°C versus 37°C) and, additionally or alternatively, to the presence of the His-tag appended to fragment 465N. It is possible that the tag also attenuates kcat for the fusion proteins, but this possibility has not been proven.] However, fusion 374N-AdScQRS has a Km of 7 μM for microhelixAla. This Km is ≈15-fold lower than that of fragment 465N. Consequently, the combined effect of the reduced kcat and Km for fusion 374N-AdScQRS results in kcat/Km being comparable to that of fragment 465N.
In the case of fusion 374N-CArc1p, the entire difference between it and fusion 374N-AdScQRS is explained by the difference in Km for microhelixAla. This difference is consistent with the previously measured apparent 10-fold higher tRNA binding affinity of AdScQRS when compared with a large fragment of Arc1p that includes the C-terminal domain (37).
Specificity of MicrohelixAla Aminoacylation by Fusion Proteins.
As stated above, alanyl-tRNA synthetases require a G3:U70 base pair for aminoacylation of tRNAAla and of RNA substrates based on the acceptor stem of tRNAAla. The unpaired exocyclic 2-amino group of G3 in the minor groove is a critical atomic determinant, as demonstrated by replacement of G3 by inosine (I) in duplex substrates that recapitulate the acceptor stem of tRNAAla. The only difference between the G:U and I:U pair is the presence or absence of the 2-amino group. E. coli AlaRS fails to charge substrates with an I3:U70 pair (52). Moreover, inserting a base pair comprised of 2-amino adenosine (2-AA) and isocytidine (isoC) at the 3:70 position can restore aminoacylation (53, 54). With the 2-AA3:isoC70 pair, major and minor groove atomic determinants are different from that of the G:U pair, but the presence of the unpaired exocyclic amino group is maintained. Microhelices incorporating the 2-AA3:U70 base pair are efficiently charged with alanine, further demonstrating the importance of the unpaired 2-amino group at the 3-position of the helix.
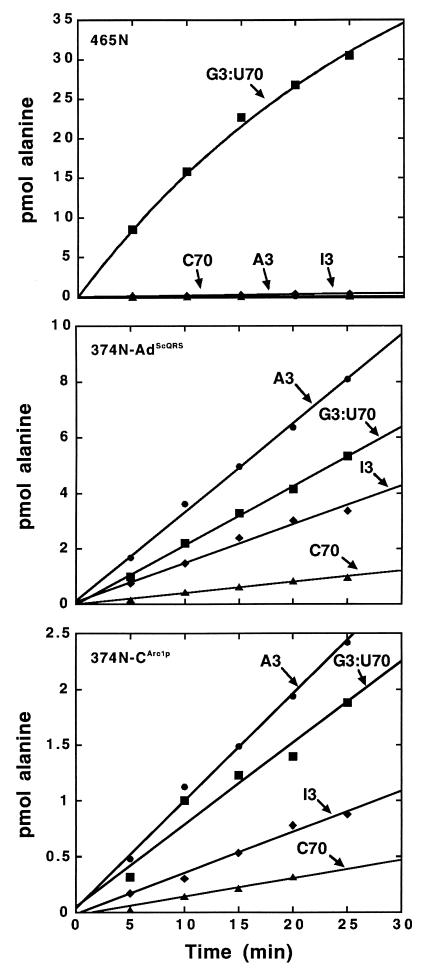
Thus, to test the RNA sequence specificity of the fusion proteins, the comparison of a microhelix containing the G3:U70 pair with one containing I3:U70 was considered essential. We confirmed that fragment 465N charged microhelixAla containing the G3:U70 base pair and that substitution with I3:U70 eliminated aminoacylation (Fig. 3 Top). In contrast, both fusion proteins charged microhelices containing the I3:U70 base pair, albeit with a relatively small decrease in efficiency (Fig. 3 Middle and Bottom). Thus, the fusion proteins do not recapitulate the critical specificity for the unpaired 2-amino group at the 3-position of the RNA helix. This result is consistent with previous work that demonstrated the importance of the region from Thr369 to Asp461 for recognition of G3:U70 (41).
Figure 3.
Specificity of aminoacylation for a G3:U70 base pair. Aminoacylation of microhelixAla containing the indicated nucleotides at the 3:70 position with fragment 465N (Top), fusion 374N-AdScQRS (Middle), and fusion 374N-CArc1p (Bottom). Aminoacylation assays were at pH 7.5, 25°C, with 10 μM RNA and 1 μM enzyme.
We also checked substrates containing an A3:U70 and a G3:C70 base pair. As expected, neither of these substrates was charged by fragment 465N. However, both variants were aminoacylated by the fusion constructions. Of the four substrates (G3:U70, I3:U70, A3:U70, and G3:C70 microhelixAla), the A3:U70 microhelixAla was the most efficiently and the G3:C70 microhelixAla the least efficiently charged by both fusion proteins (Fig. 3). These results demonstrate that, although the fusion constructions are active for aminoacylation and show some sensitivity to the base pair at the 3:70 position, they lack the high selectivity of AlaRS for G3:U70.
Discussion
The portion of AlaRS that extends to His249 contains the structural unit that defines the class II tRNA synthetases. This unit is thought to be the original, historic tRNA synthetase that contained the structural components needed to catalyze synthesis of the aminoacyl adenylate. RNA binding elements are imagined to have been added to this structural unit to give rise to the 10 distinct class II enzymes. These RNA binding elements are more or less specific to the synthetase; that is, enzymes in the same class often do not share common RNA binding elements. Thus, these elements may have been added in an idiosyncratic way. Any peptide or protein that docked the end of an RNA close to the bound aminoacyl adenylate should facilitate synthesis of the aminoacyl RNA. From this perspective, the key to success lies in the orientation in which the RNA substrate is docked onto the moiety that holds the aminoacyl adenylate.
In the present study, the success of the experiment was undoubtedly influenced by using a fragment of AlaRS that extended beyond His249 (to Leu374). This extension was necessary because C-terminal truncations of AlaRS significantly beyond Arg368 result in a loss of the capacity to synthesize alanyl adenylate. For example, fragment 257N is inactive for adenylate synthesis (32). By extending the fragment of AlaRS to Leu374, we incorporated elements that are important to recognize the discriminator base A73 and the first two base pairs of microhelixAla (41). In particular, Asp285 and Arg314 are especially important for binding microhelixAla to AlaRS (55). These highly conserved residues are not involved in adenylate synthesis but are part of a predicted two-helix pair that establishes a platform for binding RNA helices. However, as demonstrated by the lack of aminoacylation activity for fragment 368N (Fig. 2), this two-helix pair does not provide sufficient binding interactions to achieve productive docking of microhelixAla. Thus, the nonspecific RNA binding domains added here provided reinforcement for weak interactions—including those involving the platform around Asp285 and Arg314—that place the microhelix substrate in the proper orientation for aminoacylation.
The lack of specificity of aminoacylation with the fusion constructs provides further support for the earlier conclusion that determinants for recognition of G3:U70 lie between Arg368 and Asp461 (41). These fusion constructions have kcat− values that are ≈20-fold below that of fragment 465N. This difference corresponds to <2 kcal⋅mol−1 in transition state stabilization free energy. Possibly the bulk of this difference is an entropic component that arises from unproductive orientations of the 3′-end of the substrate when it is docked with the assistance of the nonspecific RNA binding domains from S. cerevisiae glutaminyl-tRNA synthetase and Arc1p. With specificity for G3:U70 (i.e., specific recognition of the 2-amino group of G3), the orientation of the 3′-end of the bound substrate undoubtedly is made precise. Thus, the energetic benefit of specific contacts with G3:U70 may be used to offset the entropic cost of fixing precisely the orientation of the 3′-end of the microhelix substrate.
In this connection, it is of interest that the Km parameters for the fusion proteins are equal to or less than that for fragment 465N (Table 1). This circumstance suggests that evolving (within the context of either nonspecific RNA binding domain) an energetically favorable specific contact with G3:U70 could offset the entropic cost of raising kcat without having to make a sacrifice in Km.
Table 1.
Kinetic parameters for aminoacylation of microhelixAla at pH 7.5, 25°C
| Enzyme | kcat, min−1 | KM, μM | kcat/KM (relative) |
|---|---|---|---|
| 465N | 1.9 | 100 | (1.0) |
| 374N-AdScQRS | 0.09 | 7 | 0.68 |
| 374N-CArc1p | 0.11 | 110 | 0.05 |
Kinetic parameters were determined over a microhelixAla range of 10−400 μM (465N), 5−400 μM (374N-CArc1p), or 2−20 μM (374N-AdScQRS).
Aminoacyl-tRNA synthetases must perform several distinct functions, which include amino acid activation, RNA binding, RNA discrimination and, often, editing of incorrectly aminoacylated tRNAs (32, 56, 57). In several cases, these distinct functions are carried out by specific domains, which can function independently of the larger enzyme. “Mixing and matching” domains from distinct proteins affords the possibility of creating “artificial” synthetases, which could be engineered to have different RNA or amino acid specificities than those of the wild-type enzymes. By fusing the catalytic site of a synthetase to unrelated RNA binding domains, we have taken a first step in this direction.
In our experiments, we made a simple head-to-tail linear fusion of N terminus of CArc1p or AdScQRS to the C terminus of fragment 465N of AlaRS. We suspect that placing the nonspecific RNA binding domain on the C-terminal side of fragment 465N may have been important for the success of the experiment. In an earlier study, we showed that fragment 461N of AlaRS could be split after His249—the end of the class-defining catalytic domain—to give fragments 249N and 212C. These fragments spontaneously assembled in vivo to give a protein that executed G3:U70-specific charging of microhelixAla (58). These experiments supported the concept that, in principle, early tRNA synthetases could have developed as noncovalent assemblies of fragments that synthesize aminoacyl adenylates with those that bind RNA. These and earlier experiments (summarized in refs. 32 and 59) also demonstrated explicitly that the domain for adenylate synthesis and RNA binding elements were arranged in a linear “head-to-tail” way along the sequence for AlaRS. Thus, our fusion proteins duplicated the strategy used by the natural enzyme.
Acknowledgments
We thank Drs. Lluís Ribas de Pouplana, Shana Kelley, Brian Steer, and Tamara Hendrickson for many helpful discussions. This work was supported by National Institutes of Health Grant GM23562. J.W.C. was a National Institutes of Health postdoctoral fellow (1997–1999).
Abbreviations
- AlaRS
alanyl-tRNA synthetase
- 461N
461-aa N-terminal fragment
References
- 1.Schimmel P. Annu Rev Biochem. 1987;56:125–158. doi: 10.1146/annurev.bi.56.070187.001013. [DOI] [PubMed] [Google Scholar]
- 2.Meinnel T, Mechulam Y, Blanquet S. In: tRNA: Structure, Biosynthesis, and Function. Söll D, RajBhandary U L, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 251–292. [Google Scholar]
- 3.Arnez J G, Moras D. Trends Biochem Sci. 1997;22:211–216. doi: 10.1016/s0968-0004(97)01052-9. [DOI] [PubMed] [Google Scholar]
- 4.Nagel G M, Doolittle R F. J Mol Evol. 1995;40:487–498. doi: 10.1007/BF00166617. [DOI] [PubMed] [Google Scholar]
- 5.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Nature (London) 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 6.Moras D. Trends Biochem Sci. 1992;17:159–164. doi: 10.1016/0968-0004(92)90326-5. [DOI] [PubMed] [Google Scholar]
- 7.Webster T A, Tsai H, Kula M, Mackie G A, Schimmel P. Science. 1984;226:1315–1317. doi: 10.1126/science.6390679. [DOI] [PubMed] [Google Scholar]
- 8.Hountondji C, Dessen P, Blanquet S. Biochimie. 1986;68:1071–1078. doi: 10.1016/s0300-9084(86)80181-x. [DOI] [PubMed] [Google Scholar]
- 9.Ludmerer S W, Schimmel P. J Biol Chem. 1987;262:10801–10806. [PubMed] [Google Scholar]
- 10.Cusack S, Härtlein M, Leberman R. Nucleic Acids Res. 1991;19:3489–3498. doi: 10.1093/nar/19.13.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delarue M, Moras D. BioEssays. 1993;15:675–687. doi: 10.1002/bies.950151007. [DOI] [PubMed] [Google Scholar]
- 12.Schimmel P, Ribas de Pouplana L. Cell. 1995;81:983–986. doi: 10.1016/s0092-8674(05)80002-9. [DOI] [PubMed] [Google Scholar]
- 13.Noller H F. In: The RNA World. Gesteland R F, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 137–156. [Google Scholar]
- 14.Weiner A M, Maizels N. Proc Natl Acad Sci USA. 1987;84:7383–7387. doi: 10.1073/pnas.84.21.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maizels N, Weiner A M. In: The RNA World. Gesteland R F, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 577–602. [Google Scholar]
- 16.Kikuchi Y, Ando Y, Shiba T. Nature (London) 1986;323:824–826. doi: 10.1038/323824a0. [DOI] [PubMed] [Google Scholar]
- 17.Chapman K B, Bystrom A S, Boeke J D. Proc Natl Acad Sci USA. 1992;89:3236–3240. doi: 10.1073/pnas.89.8.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isel C, Ehresmann C, Keith G, Ehresmann B, Marquet R. J Mol Biol. 1995;247:236–250. doi: 10.1006/jmbi.1994.0136. [DOI] [PubMed] [Google Scholar]
- 19.Francklyn C, Schimmel P. Nature (London) 1989;337:478–481. doi: 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- 20.Francklyn C, Musier-Forsyth K, Schimmel P. Eur J Biochem. 1992;206:315–321. doi: 10.1111/j.1432-1033.1992.tb16929.x. [DOI] [PubMed] [Google Scholar]
- 21.Martinis S A, Schimmel P. Proc Natl Acad Sci USA. 1992;89:65–69. doi: 10.1073/pnas.89.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frugier M, Florentz C, Giegé R. Proc Natl Acad Sci USA. 1992;89:3990–3994. doi: 10.1073/pnas.89.9.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nureki O, Niimi T, Muto Y, Kanno H, Kohno T, Muramatsu T, Kawai G, Miyazawa T, Giegé R, Florentz C, Yokoyama S. In: The Translation Apparatus. Nierhaus K H, Franceschi F, Subramamian A R, Erdmann V A, Wittmann-Liebold B, editors. New York: Plenum; 1993. pp. 59–66. [Google Scholar]
- 24.Frugier M, Florentz C, Giegé R. EMBO J. 1994;13:2218–2226. doi: 10.1002/j.1460-2075.1994.tb06499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamann C S, Hou Y-M. Biochemistry. 1995;34:6527–6532. doi: 10.1021/bi00019a034. [DOI] [PubMed] [Google Scholar]
- 26.Quinn C L, Tao N, Schimmel P. Biochemistry. 1995;34:12489–12495. doi: 10.1021/bi00039a001. [DOI] [PubMed] [Google Scholar]
- 27.Saks M E, Sampson J R. EMBO J. 1996;15:2843–2849. [PMC free article] [PubMed] [Google Scholar]
- 28.Schimmel P, Giege R, Moras D, Yokoyama S. Proc Natl Acad Sci USA. 1993;90:8763–8768. doi: 10.1073/pnas.90.19.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, Lambowitz A M. Cell. 1993;75:1071–1081. doi: 10.1016/0092-8674(93)90317-j. [DOI] [PubMed] [Google Scholar]
- 30.Buechter D D, Schimmel P. Biochemistry. 1993;32:5267–5272. doi: 10.1021/bi00070a039. [DOI] [PubMed] [Google Scholar]
- 31.Park S J, Schimmel P. J Biol Chem. 1988;263:16527–16530. [PubMed] [Google Scholar]
- 32.Jasin M, Regan L, Schimmel P. Nature (London) 1983;306:441–447. doi: 10.1038/306441a0. [DOI] [PubMed] [Google Scholar]
- 33.Ho C, Jasin M, Schimmel P. Science. 1985;229:389–393. doi: 10.1126/science.3892692. [DOI] [PubMed] [Google Scholar]
- 34.Putney S D, Sauer R T, Schimmel P R. J Biol Chem. 1981;256:198–204. [PubMed] [Google Scholar]
- 35.Regan L, Bowie J, Schimmel P. Science. 1987;235:1651–1653. doi: 10.1126/science.2435005. [DOI] [PubMed] [Google Scholar]
- 36.Glasfeld E, Schimmel P. Biochemistry. 1997;36:6739–6744. doi: 10.1021/bi970151n. [DOI] [PubMed] [Google Scholar]
- 37.Wang C C, Schimmel P. J Biol Chem. 1999;274:16508–16512. doi: 10.1074/jbc.274.23.16508. [DOI] [PubMed] [Google Scholar]
- 38.Ribas de Pouplana L, Schimmel P. Biochemistry. 1997;36:15041–15048. doi: 10.1021/bi971788+. [DOI] [PubMed] [Google Scholar]
- 39.Higuchi R, Krummel B, Saiki R K. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whelihan E F, Schimmel P. EMBO J. 1997;16:2968–2974. doi: 10.1093/emboj/16.10.2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buechter D D, Schimmel P. Biochemistry. 1995;34:6014–6019. doi: 10.1021/bi00018a002. [DOI] [PubMed] [Google Scholar]
- 42.Francklyn C, Musier-Forsyth K, Schimmel P. Eur J Biochem. 1992;206:315–321. doi: 10.1111/j.1432-1033.1992.tb16929.x. [DOI] [PubMed] [Google Scholar]
- 43.Calendar R, Berg P. Biochemistry. 1966;5:1690–1695. doi: 10.1021/bi00869a034. [DOI] [PubMed] [Google Scholar]
- 44.Calendar R, Berg P. In: Procedures in Nucleic Acids Research. Cantoni R, Davies D R, editors. New York: Harper & Row; 1966. pp. 384–399. [Google Scholar]
- 45.Simos G, Segref A, Fasiolo F, Hellmuth K, Shevchenko A, Mann M, Hurt E C. EMBO J. 1996;15:5437–5448. [PMC free article] [PubMed] [Google Scholar]
- 46.Simos G, Sauer A, Fasiolo F, Hurt E C. Mol Cell. 1998;1:235–242. doi: 10.1016/s1097-2765(00)80024-6. [DOI] [PubMed] [Google Scholar]
- 47.Hou Y M, Schimmel P. Nature (London) 1988;133:140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- 48.McClain W H, Foss K. Science. 1988;240:793–796. doi: 10.1126/science.2452483. [DOI] [PubMed] [Google Scholar]
- 49.Putney S D, Royal N J, Neuman de Vegvar H, Herlihy W C, Biemann K, Schimmel P. Science. 1981;213:1497–1500. doi: 10.1126/science.7025207. [DOI] [PubMed] [Google Scholar]
- 50.Francklyn C, Schimmel P. Nature (London) 1989;337:478–481. doi: 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- 51.Buechter D D, Schimmel P. Biochemistry. 1993;32:5267–5272. doi: 10.1021/bi00070a039. [DOI] [PubMed] [Google Scholar]
- 52.Musier-Forsyth K, Usman N, Scaringe S, Doudna J, Green R, Schimmel P. Science. 1992;253:784–786. doi: 10.1126/science.1876835. [DOI] [PubMed] [Google Scholar]
- 53.Musier-Forsyth K, Shi J-P, Henderson B, Bald R, Fürste J P, Erdmann V A, Schimmel P. J Am Chem Soc. 1995;117:7253–7254. [Google Scholar]
- 54.Beuning P J, Yang F, Schimmel P, Musier-Forsyth K. Proc Natl Acad Sci USA. 1997;94:10150–10154. doi: 10.1073/pnas.94.19.10150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ribas de Pouplana L, Buechter D, Sardesai N Y, Schimmel P. EMBO J. 1998;17:5449–5457. doi: 10.1093/emboj/17.18.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waye M M Y, Winter G, Wilkinson A J, Fersht A R. EMBO J. 1983;2:1827–1829. doi: 10.1002/j.1460-2075.1983.tb01665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin L, Hale S P, Schimmel P. Nature (London) 1996;384:33–34. doi: 10.1038/384033b0. [DOI] [PubMed] [Google Scholar]
- 58.Sardesai N, Schimmel P. J Am Chem Soc. 1998;123:3269–3270. [Google Scholar]
- 59.Schimmel P, Ripmaster T. Trends Biochem Sci. 1995;20:333–334. doi: 10.1016/s0968-0004(00)89067-2. [DOI] [PubMed] [Google Scholar]