Abstract
Small nucleolar RNAs (snoRNAs) associate with specific proteins forming small nucleolar ribonucleoprotein (snoRNP) particles, which are essential for ribosome biogenesis. The snoRNAs are transcribed, processed, and assembled in snoRNPs in the nucleoplasm. Mature particles are then transported to the nucleolus. In yeast, 3′-end maturation of snoRNAs involves the activity of Rnt1p endonuclease and cleavage factor IA (CFIA). We report that after inhibition of CFIA components Rna14p and Rna15p, the snoRNP proteins Nop1p, Nop58p, and Gar1p delocalize from the nucleolus and accumulate in discrete nucleoplasmic foci. The U14 snoRNA, but not U3 snoRNA, similarly redistributes from the nucleolus to the nucleoplasmic foci. Simultaneous depletion of either Rna14p or Rna15p and the nuclear exosome component Rrp6p induces accumulation of poly(A)+ RNA at the snoRNP-containing foci. We propose that the foci detected after CFIA inactivation correspond to quality control centers in the nucleoplasm.
INTRODUCTION
Small nucleolar RNAs (snoRNAs) are an abundant group of nonprotein coding RNAs in eukaryotic cells (Kiss, 2002). The snoRNAs associate with specific proteins forming small nucleolar ribonucleoprotein (snoRNP) particles, the majority of which participates in the biogenesis of ribosomes in the nucleolus (for recent reviews, see Bachellerie et al., 2002; Filipowicz and Pogacic, 2002; Kiss, 2002; Tran et al., 2004). The known snoRNAs fall into two major classes named box C/D and box H/ACA, which are characterized by distinctive sequence elements and associated proteins.
The snoRNAs are transcribed as precursors that undergo posttranscriptional processing to generate the mature functional forms. Although most vertebrate snoRNAs are processed from the intronic regions of mRNA precursors (Maxwell and Fournier, 1995, Bachellerie et al., 2002), only a few yeast snoRNAs are intron encoded. The majority of yeast snoRNAs are encoded in independent monocistronic or polycistronic transcripts that are transcribed by RNA polymerase II. Intronic snoRNAs can be produced either by splicing-dependent or by splicing-independent processing, whereas the maturation of independently transcribed units involves both endonucleolytic cleavage and exonucleolytic trimming (Filipowicz and Pogacic, 2002).
In yeast, the U14 and U3 box C/D snoRNAs constitute paradigms for the study of snoRNA biosynthesis. U14 is cotranscribed with snR190, another box C/D snoRNA (Li et al., 1990). Cleavage of the dicistronic precursor by the endonuclease Rnt1 separates snR190 from U14 (Chanfreau et al., 1998). The pre-U3 transcript, by contrast, is produced from a single gene and contains the 7-methyl guanosine (m7G) cap structure, which is characteristic of mRNAs, and a short 3′ extension. During maturation, the m7G cap on the pre-U3 is hypermethylated to 2,2,7-trimethylguanosine (TMG) and the 3′ end is cleaved by Rnt1p, followed by removal of the 3′-terminal extension (Kufel et al., 2000). The yeast pre-U3 also contains an intron that is removed by the pre-mRNA splicing machinery, but the splicing reaction is independent of 3′ maturation.
Mature 3′ ends of many independently transcribed snoRNAs are generated by the nuclear exosome in conjunction with specific processing factors. These include RNA binding proteins Nrd1 and Nab3, the RNA helicase Sen1 (Steinmetz et al., 2001), the PAF complex (Sheldon et al., 2005), and proteins that also participate in pre-mRNA 3′-end formation. Components shared by the mRNA and the snoRNA 3′-end formation machinery include Rna14, Rna15, Pti1, Ref2, Ssu72, and Swd2 (Morlando et al., 2002; Dheur et al., 2003; Nedea et al., 2003). Yeast Rna14p and Rna15p (CstF77 and CstF64 in humans) are essential subunits of cleavage factor 1A (CF IA) involved in both cleavage and polyadenylation of mRNA precursors (Minvielle-Sebastia et al., 1994), and in polyadenylation-independent 3′-end processing of snoRNAs (Fatica et al., 2000; Morlando et al., 2002). Pti1 and Ref2 are both required to prevent snoRNA transcriptional readthrough into downstream genes (Dheur et al., 2003). Pti1p is a homologue of the mammalian cleavage-stimulation factor subunit CstF-64 that binds directly Rna14p (Skaar and Greenleaf, 2002), and Ref2 interacts with the snoRNP-specific protein Nop1p (Morlando et al., 2004). According to a current model, nonpolyadenylated 3′ ends of snoRNAs provide entry sites for the exosome to perform the final trimming of 3′-extended precursors (Allmang et al., 1999a; van Hoof et al., 2000).
The biogenesis of snoRNPs involves complex trafficking pathways in the nucleus. Processing of snoRNAs and initial assembly of core proteins takes place cotranscriptionally, whereas complete snoRNP maturation is thought to occur during transit through the nucleoplasm and the nucleolar body in yeast or Cajal bodies in higher eukaryotes (for a recent review, see Matera et al., 2007). Subsequently, particles are routed to where they function and although other destinations exist, most snoRNPs localize to sites of ribosome synthesis within the nucleolus.
In the present study, we analyzed snoRNP localization in yeast strains carrying the cleavage factor IA (CFIA) mutant alleles rna14-1 and rna15-2. Our results reveal that in the absence of Rna14p and Rna15p, snoRNPs fail to localize in the nucleolus, being sequestered within discrete domains in the nucleoplasm. Because these domains accumulate polyadenylated RNA species when the nuclear exosome is inactivated, we propose that they correspond to RNA quality control centers in the nucleus.
MATERIALS AND METHODS
Strains and Plasmids
The strains of Saccharomyces cerevisiae used in this study are listed in Table 1. All strains were routinely grown to mid-log phase in yeast extract-peptone-dextrose (YPD) medium containing either 2% glucose or 2% galactose. Plasmids were introduced by the standard polyethylene glycol-lithium procedure, as described previously (Torchet et al., 2002). Transformed strains were grown in minimal YNB medium containing 0.67% of yeast nitrogen base supplemented with the appropriate amino acids and 2% glucose. The following expression plasmids were used in this study: Nop1p-green fluorescent protein (GFP) (Verheggen et al., 2001), Tgs1p-GFP (Mouaikel et al., 2002), Nup49p-GFP (Belgareh and Doye, 1997), and Gar1p-GFP (Trumtel et al., 2000).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| D271 | MATa ade2-1 his3 leu2 trp1 ura3 | Venema and Tollervey (1996) |
| YCBA63 | MATα as D271 but HIS3-GAL::protA-RRP41 | Torchet et al. (2002) |
| YCA12 | MATa ade2-1 his3Δ200 leu2-3,112 trp1-1 ura3-1 can1-100 Kl TRP1::rrp6 | Allmang, Petfalski et al. (1999) |
| rna14.1 | MATa ade2-1 his3-11 leu2-3,112 trp1-1 ura3-1 rna14.1 | Minvielle-Sebastia et al. (1994) |
| rna14.2 | MATa ade2-1 his3-11 leu2-3,112 trp1-1 ura3-1 rna15.2 | Amrani, Dufour et al. (1996) |
| YCBA121 | MATa ade2 his3 leu2 trp1 ura3 rna14.1 HIS3-GAL::protA-RRP41 | Torchet et al. (2002) |
| YCBA128 | MATα ade2 his3 leu2 trp1 ura3 rna15.2 HIS3-GAL::protA-RRP41 | Torchet et al. (2002) |
| YCBA132 | as rna14.1 but Kl TRP1::rrp6 | Torchet et al. (2002) |
| YCBA134 | as rna15.2 but Kl TRP1::rrp6 | Torchet et al. (2002) |
RNA Extraction and Northern Blotting
Yeast RNA extraction, Northern blotting, and deadenylation analysis were performed as described previously (Mitchell et al., 2003; LaCava et al., 2005).
Immunofluorescence and Fluorescence In Situ Hybridization
Immunofluorescence microscopy using antibodies directed against Nop1p (Aris and Blobel, 1988) and Nop58p (34B12; Encor BioTechnology, Alachua, FL) (Wu et al., 1998) was performed according to Verheggen et al. (2001). Yeast cells were processed for in situ hybridization as described previously (Verheggen et al., 2002), except that a treatment with 0.1% sodium borohydride for 10 min was introduced after rehydration, before hybridization. Hybridization stringency conditions were 2× SSC, 10% formamide for poly(A)+RNA and U3 snoRNA; and 2× SSC, 50% formamide for actin mRNA and U14 snoRNA. As a control, cells were treated with 1.5 U of RNase A (Roche Diagnostics, Mannheim, Germany) in 2× SSC at 37°C for 60 min, before the in situ hybridization with each probe. When needed, cells were first hybridized with the oligo(dT) probe, fixed in 4% formaldehyde/2× SSC for 5 min, washed two times for 10 min in 2× SSC, and then hybridized with another probe. Samples were mounted in 90% glycerol, 1 mg/ml p-phenylendiamine in phosphate-buffered saline (PBS).
Poly(A)+RNA was detected using a cyanine (Cy)3-labeled oligo(dT)50 probe (MWG-Biotech AG, Ebersberg, Germany). ACT1 mRNA was detected with a mixture of oligonucleotides (MWG-Biotech AG) with the following sequences: 1) 5′-CRTTCTGGAGGAGCRATGATCTTGACRTTCATGGAAGRTGGAGCCAAAGRG-3′, 2) 5′-CRTGGGAACATGGTGGTACCACCGGACATAACGATGTTACCGTATAATTRC-3′; 3) 5′-CRGAAGAGTACARGGACAAAACGGCRTGGATGGAARCGTAGAAGGCTGRA-3′; and 4) 5′-ARCCATACCGACCRTGATACCTTGGRGTCTTGGTCTRCCGACGATAGARG-3′.
These probes were synthesized with modified nucleotides to incorporate primary amines (amino-allyl timine is represented by R), and then they were conjugated with Cy3. The U14 probe was transcribed in vitro with digoxigenin-labeled nucleotides. The U3 (Verheggen et al., 2001) and ITS2 (Dez et al., 2006) probes were described previously.
After hybridization with digoxigenin-conjugated probes, cells were treated with 4% formaldehyde/2× SSC for 5 min and washed, first in 2× SSC and then in PBS/0.1% Triton X-100. Subsequently, cells were incubated for 10 min in PBS containing 1% skimmed milk and 1% RNAse-free bovine serum albumin. Cells were then incubated for 1 h with anti-digoxigenin antibody conjugated to fluorescein isothiocyanate (FITC) (Jackson ImmunoResearch Laboratories) followed by anti-FITC antibody conjugated to Alexa-488 (Invitrogen, Paisley, United Kingdom).
Fluorescence Microscopy
Images were acquired with a Zeiss LSM 510 META confocal microscope (Carl Zeiss, Jena, Germany) using either the PlanNeofluar 100×/1.3 or the PlanApochromat 63×/1.4 objectives. Green fluorescence was detected using the 488-nm laser line of an Ar laser (25-mW nominal output, with acoustical optical transmission filter (AOTF) transmission set to 2% and the detector gain set to 1000×) and an LP 505 filter. Red fluorescence was detected using a 543-nm HeNe laser (1-mW, AOTF transmission 82% and the detector gain to 1000×) and an LP 560 filter. In some strains, accumulation of signal led to saturation of the images. To properly visualize the fluorescent staining, the detector gain was reduced to 840–860×. 4,6-Diamidino-2-phenylindole (DAPI) staining was detected using the 405 laser line of a 405-30 diode laser and a BP420-480 filter. In all cases, the amplifier gain was set to 1×.
Fluorescence Recovery after Photobleaching (FRAP) Analysis
To prepare yeast cells for FRAP experiments, 1 ml of each cell culture grown to mid-log phase was harvested and resuspended in 100 μl of medium; 5 μl of this cell suspension was added to a coverslip coated with 40% gelatin in medium. Live cells were imaged at 23°C, 30°C, or 37°C maintained by a heating/cooling frame (LaCon, Staig, Germany) in conjunction with an objective heater (PeCon, Erbach, Germany). Each FRAP analysis started with three image scans, followed by a single bleach pulse of ∼70 ms on a defined region. A series of 97 single-section images were then collected at 155-ms intervals, with the first image acquired 2 ms after the end of bleaching. Image size was 512 × 100 pixels, and the pixel width was 14.3 nm. For imaging, the laser power was attenuated to 1% of the bleach intensity. The pinhole was set to 1 airy unit. All fluorescence values were obtained by using the Physiology package of the LSM Image Browser (Carl Zeiss). For each time series, normalization was done by measuring the fluorescence intensity in the bleached, unbleached, and background areas. The fluorescence intensity measured in the bleached area during recovery was divided by the initial value of the fluorescence in that same area:
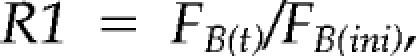 |
where FB(t) is the fluorescence in the bleached area at time t, and FB(ini) is the initial fluorescence in that same area. Each time an image is acquired, some fluorescence is lost due to the laser scan; to compensate for this loss, each R1 value was multiplied by the following factor (A):
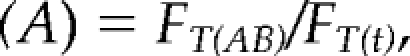 |
where FT(AB) is the total nucleolar fluorescence immediately after bleach (bleached and nonbleached areas), and FT(t) is the fluorescence in the bleach and nonbleached area at time t. Because a global loss of fluorescence was also observed after the bleach we had to compensate by multiplying the R1 value by factor (B):
where FB(final) is the fluorescence in the bleached area in the last image acquired and FT(final) is the total nucleolar fluorescence (bleached and nonbleached areas) in the last image acquired. Finally, to estimate whether a fraction of the molecules in the bleached area was immobilized, we calculated the factor (C), which yields values lower than 1 in the presence of an immobile fraction:
where FB(ini) and FB(final) represent the fluorescence in the bleached area in the first and last image acquired, respectively, and FNB(ini) and FNB(final) are the fluorescence values in the nonbleached area in the first and last image acquired, respectively. The final formula, which is the product of R1, (A), (B), and (C), represents the normalized fluorescence recovery in the bleached area over time:
RESULTS
Inactivation of Rna14p and Rna15p Delocalizes snoRNP Proteins and U14 snoRNA from the Nucleolus
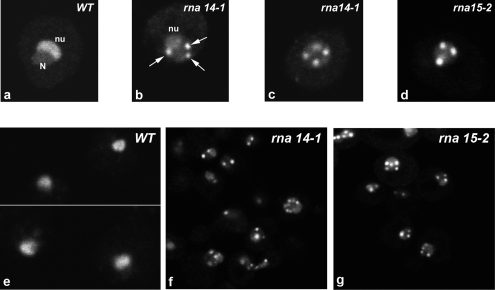
Previous studies have revealed that components of CFIA including Rna14p and Rna15p are required for 3′-end formation of small nucleolar RNAs (Fatica et al., 2000; Morlando et al., 2002). To investigate whether CFIA inactivation affects snoRNP localization, we analyzed the subcellular distribution of the box C/D snoRNP protein Nop1p in yeast strains carrying the temperature-sensitive (ts) lethal alleles rna14-1 and rna15-2 (Minvielle-Sebastia et al., 1991; Minvielle-Sebastia et al., 1994). We visualized Nop1p by fusing it to GFP (Figure 1, a–d) or by indirect immunofluorescence microscopy by using a specific antibody to detect the endogenous Nop1p (Figure 1, e–g). In the wild-type strain, Nop1p seemed uniformly distributed throughout the crescent-shaped region that is typical of the yeast nucleolus (Figure 1, a and e). However, in the rna14-1 and rna15-2 strains incubated at the nonpermissive temperature (37°C) for 1 h, Nop1p accumulated in two to six nuclear foci (Figure 1, b, c, d, f, and g). Fainter staining over a crescent-shaped region was still seen in some cells, suggesting that a fraction of the Nop1p population was still associated with the nucleolus (Figure 1b). Redistribution of Nop1p was observed as early as 30 min after shift to the nonpermissive temperature.
Figure 1.
Nop1p delocalizes from the nucleolus in rna14-1 and rna15-2 mutants. Wild-type (a) and mutant (b–d) cells expressing a Nop1p-GFP fusion protein were grown in glucose medium at 23°C, and then they were shifted to 37°C for 1 h. N, nucleoplasm; nu, nucleolus; arrows point to nuclear foci. Wild-type (e), rna14-1 (f) and rna15-2 (g) strains incubated at 37°C for 1 h were immunolabeled with anti-Nop1p antibody.
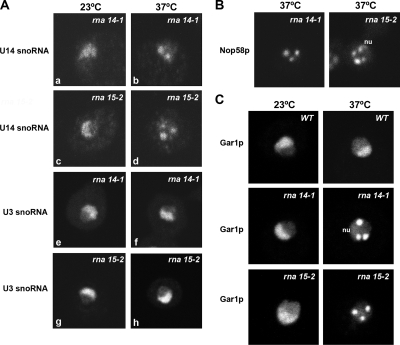
To determine whether the presence of Nop1p in nuclear foci correlated with a redistribution of snoRNAs, fluorescence in situ hybridization was performed with probes specific for the box C/D snoRNAs U3 and U14. As described previously (Verheggen et al., 2001), the U3 and U14 snoRNAs were homogeneously distributed throughout the nucleolus in wild-type cells (Supplemental Figure 1), and the same localization was observed in rna14-1 and rna15-2 strains incubated at 23°C (Figure 2A, a, c, e, and g). After transfer to 37°C for 1 h, the U14 snoRNA became concentrated in foci in both the rna14-1 and rna15-2 strains (Figure 2A, b and d). In contrast, the U3 snoRNA probe continued to decorate the typical crescent-shaped nucleolar region (Figure 2A, f and h). Thus, inhibition of CFIA activity leads to delocalization of U14 but not U3 snoRNAs from the nucleolus.
Figure 2.
Box C/D and box H/ACA snoRNP proteins, and U14 snoRNA, but not U3 snoRNA, redistribute from the nucleolus to nucleoplasmic foci in rna14-1 and rna15-2 mutants. The strains indicated were grown in glucose medium at 23°C (23°C panels) and then shifted to 37°C for 1 h (37°C panels). (A) Endogenous U14 and U3 snoRNAs were detected by in situ hybridization. (B) Endogenous Nop58p was detected by immunofluorescence. (C) Wild-type and mutant cells expressing a Gar1p-GFP fusion protein. Note that a fraction of Nop58p and Gar1p persists localized in the nucleolus (nu) at the nonpermissive temperature.
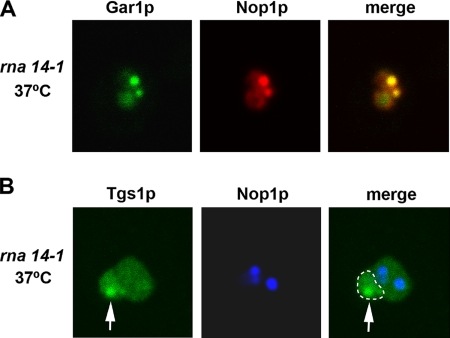
Next, we analyzed the distribution of snoRNA-associated proteins Nop58p and Gar1p in the rna14-1 and rna15-2 strains. Like Nop1p, Nop58p is a core component of box C/D snoRNPs, whereas Gar1p associates specifically with H/ACA snoRNPs (Filipowicz and Pogacic, 2002). In contrast to Nop1p and Nop58p that assemble cotranscriptionally with box C/D snoRNAs, Gar1p binds posttranscriptionally and represents a marker for mature fully functional H/ACA RNPs (Matera et al., 2007). We observed that 1 h after shift to the nonpermissive temperature, both Nop58p (Figure 2B) and Gar1p (Figure 2C) redistributed from the nucleolus to nucleoplasmic foci. Similarly to Nop1p, a fraction of Nop58p and Gar1p persisted associated with the nucleolus (Figure 2, B and C, nu). Moreover, Gar1p and Nop1p precisely colocalize in nucleoplasmic foci (Figure 3A), suggesting that inactivation of CFIA leads to sequestration of the functionally mature snoRNPs in common discrete nuclear domains.
Figure 3.
Box C/D and box H/ACA snoRNP proteins colocalize in nucleoplasmic foci. Mutant (rna14-1) cells expressing either Gar1p-GFP (A) or Tgs1-GFP (B) fusion proteins were grown in glucose medium at 23°C, and then they are shifted to 37°C for 1 h. The cells were further immunolabeled with anti-Nop1p antibody. Arrows point to the nucleolar body, and the contour of the nucleolus is outlined (white dashed line).
It is well known that the nucleolus is a dynamic compartment and that nucleolar components, including snoRNPs segregate when ribosome synthesis is inhibited (Shav-Tal et al., 2005). To address whether snoRNP redistribution observed in the rna14-1 and rna15-2 strains is a consequence of nucleolar segregation, a plasmid expressing a Tgs1-GFP fusion was introduced into the rna14-1 and rna15-2 strains and the corresponding wild-type. Tgs1p is a methyltransferase that hypermethylates 5′-cap structures of monocistronic snoRNAs, including U3 (Mouaikel et al., 2002). As described previously, Tgs1-GFP is detected throughout the nucleolus with higher concentration in a focus termed the nucleolar body (Verheggen et al., 2002). A similar distribution was observed in the wild–type and in the mutant strains grown at 23°C (data not shown), and this distribution was unaltered after transfer to 37°C (Figure 3B, an arrow points to the nucleolar body). Taken together with the normal distribution of U3 snoRNA (Figure 2A, f and h), we conclude that the overall architecture of the nucleolus is unaffected by the rna14-1 and rna15-2 mutations. Labeling of cells expressing Tgs1p-GFP with an anti-Nop1p antibody clearly shows that Nop1p is predominantly detected in foci that are excluded from the nucleolus (Figure 3B).
Simultaneous Inactivation of Rna14p or Rna15p and Depletion of Rrp6p Leads to Accumulation of Poly(A)+RNA in Nuclear Foci
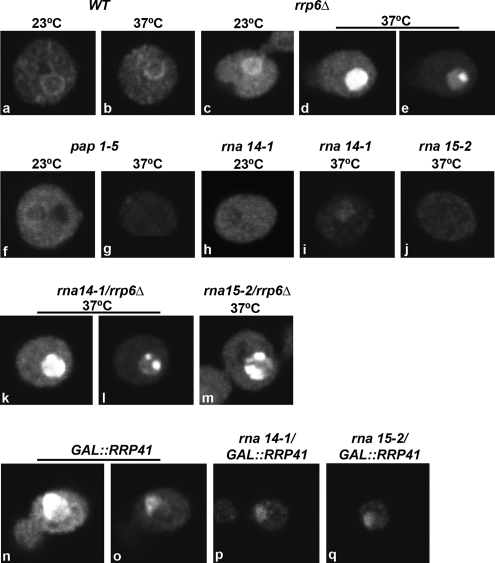
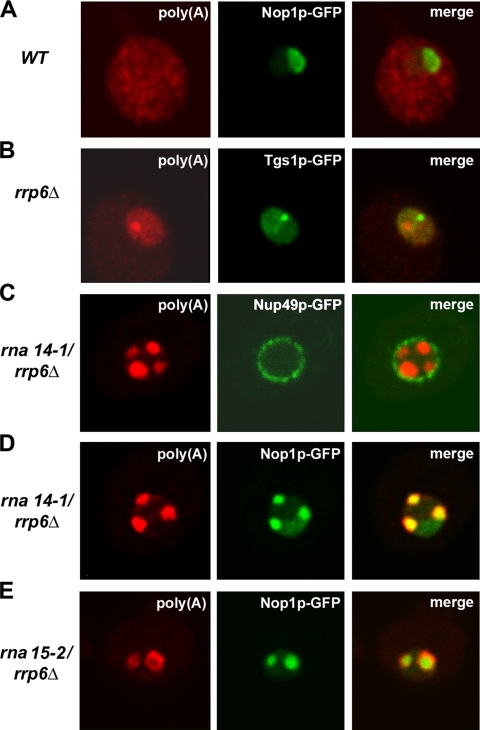
Having established that CFIA inactivation induces a rapid delocalization of snoRNPs from the nucleolus into discrete domains in the nucleoplasm, we were interested in determining the functional significance of such redistribution. We speculated that the foci may contain newly synthesized snoRNAs with aberrant 3′ ends that fail to be targeted to the nucleolus. Because a substantial fraction of aberrant transcripts are rapidly degraded in the nucleus by the combined action of the exosome and a noncanonical poly(A) polymerase activity (for review, see Houseley et al., 2006), we analyzed the effect of exosome depletion on the rna14-1 and rna15-2 strains. We performed fluorescence in situ hybridization with a labeled oligo(dT) probe to visualize polyadenylated RNAs in the cell. In wild-type cells, poly(A)+ RNA was predominantly detected throughout the cytoplasm (Figure 4, a and b; also see Figure 5A). In contrast, strains carrying the temperature-sensitive lethal pap1-5 mutation (Minvielle-Sebastia et al., 1994) showed a drastic reduction in the poly(A) signal at the nonpermissive temperature (Figure 4, f and g). Because Pap1p is the major poly(A) polymerase responsible for the synthesis of poly(A) tails of mRNAs in yeast (Patel and Butler, 1992), it is most likely that the poly(A)+ RNA detected in wild-type cells predominantly corresponds to mRNA. Total poly(A)+ RNA staining was also greatly reduced in the rna14-1 and rna15-2 strains incubated at 37°C for 1 h (Figure 4, h–j), consistent with previous reports (Minvielle-Sebastia et al., 1991; Torchet et al., 2002). By contrast, in strains lacking only Rrp6p (Figure 4, c–e), a strong accumulation of polyadenylated RNA was observed in the nucleus, as reported previously (Allmang et al., 1999a; van Hoof et al., 2000; Mitchell et al., 2003; Fang et al., 2004; Kuai et al., 2004; Houseley and Tollervey, 2006; Carneiro et al., 2007). To allow the direct comparison of fluorescence intensities, images from the rrp6-Δ mutant incubated at 23°C (Figure 4C) and 37°C (Figure 4D) were acquired using the same microscope settings. The abundant accumulation of nuclear poly(A)+ RNA in the rrp6-Δ strain saturates the image. In this strain, following an ∼20% reduction of the detector gain, unsaturated images were produced in which poly(A)+ RNA occurred nonhomogeneously distributed, with higher concentration in a discrete domain (Figure 4e). Double-labeling experiments show that in the rrp6-Δ strain poly(A)+ RNA seems concentrated in a single focus that localizes to the nucleolus but is distinct from the “nucleolar body” containing Tgs1p-GFP (Figure 5B; also see Carneiro et al., 2007).
Figure 4.
Detection of polyadenylated RNA in combined Rna14p, Rna15p, and exosome mutant strains. Poly(A)+ RNA was detected by FISH using a Cy3-labeled oligo(dT)50 probe. The indicated strains were grown at 23°C, and then they were incubated for 1 h at 37°C (a–m). The GAL::RRP41 (n and o), rna14-1/GAL::RRP41 (p), and rna15-2/GAL::RRP41 (q) strains were grown in YPD-2% galactose at 23°C, transferred to glucose medium at 23°C for 20 h, and then shifted to 37°C for 1 h. For comparison of fluorescence signals, all images were acquired with the same settings. d and e, k and l, and n and o show different exposures of the same cell.
Figure 5.
Subnuclear distribution of polyadenylated RNA. The indicated strains were grown at 23°C and incubated for 1 h at 37°C. Poly(A)+ RNA was detected by FISH using a Cy3-labeled oligo(dT)50 probe. (A) wild-type cells expressing Nop1p-GFP. (B) rrp6Δ cells expressing Tgs1p-GFP (C) rna14-1/rrp6Δ cells expressing Nup49p-GFP. (D) rna14-1/rrp6Δ cells expressing Nop1p-GFP. (E) rna15-2/rrp6Δ cells expressing Nop1p-GFP.
The rna14-1/rrp6Δ double mutant showed intense accumulation of poly(A)+ RNA in the nucleus (Figure 4k). After reducing the detector gain, the poly(A)+ staining seemed concentrated in nucleoplasmic foci (Figure 4l). A similar distribution was observed in the rna15-2/rrp6Δ double mutant (Figure 4m), although generally with lower staining intensity.
To determine whether the observed accumulation of polyadenylated RNA was a general result of inactivation of the exosome, we next analyzed strains that combine the temperature-sensitive rna14-1 or rna15-2 alleles with a GAL-regulated construct that allowed us to deplete of the core exosome exonuclease Rrp41p. After transfer to glucose medium the GAL::rrp41 strain accumulated nuclear poly(A)+ RNA, with staining predominantly in the nucleolus and without clear concentration in foci (Figure 4, n and o). A different phenotype was observed, however, at the nonpermissive temperature in the rna14-1 or rna15-2 strains that were also depleted of Rrp41p, with faint staining throughout the nucleus (Figure 4, p and q). Thus, when the rna14-1 and rna15-2 strains are incubated in the absence of Rrp41p, there is no accumulation of polyadenylated RNA species in the nucleus. We conclude that Rrp6p is specifically involved in the stabilization of polyadenylated RNAs formed in the rna14-1 and rna15-2 mutants.
We assessed whether poly(A)+ RNA accumulates at the Nop1p-foci in strains lacking both CFIA and Rrp6p activities by double-labeling. Analysis of rna14-1/rrp6Δ and rna15-2/rrp6Δ double-mutant cells expressing a GFP-tagged nucleoporin (Nup49p-GFP) under nonpermissive conditions, showed that the nucleoplasmic foci enriched in poly(A)+ RNA were frequently located in close proximity to the nuclear envelope (shown for rna14-1/rrp6Δ in Figure 5C). As shown in Figure 5, D and E, Nop1p colocalized with the poly(A) labeling in the rna14-1/rrp6Δ and rna15-2/rrp6Δ double mutants. The poly(A) labeling was often juxtaposed to the Nop1p staining, suggesting that polyadenylated RNAs accumulated at and around the Nop1p-domains.
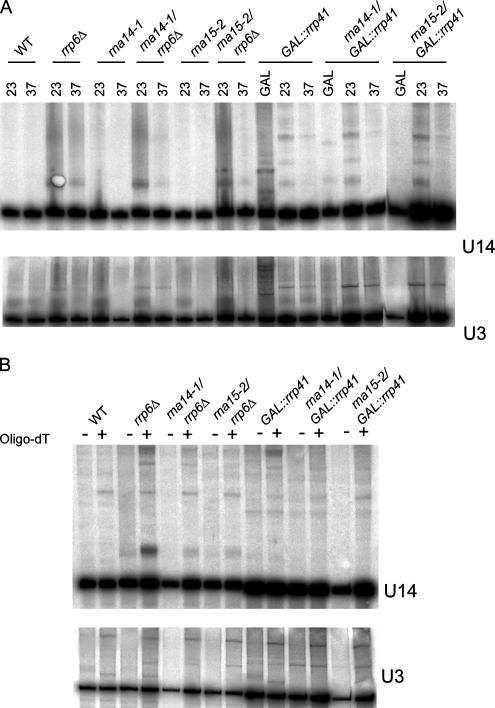
Based on the observation that U14 snoRNA redistributes from the nucleolus to nucleoplasmic foci in rna14-1 and rna15-2 strains (Figure 2A), we hypothesized that CFIA inactivation prevents normal 3′-end processing of newly synthesized U14 and that the resulting aberrant U14 RNAs are polyadenylated for subsequent degradation by the nuclear exosome. Northern blot analysis confirmed that in a rrp6Δ mutant, but not in a strain mutant for the core exosome component Rrp41p, U14 snoRNA (but not U3) retained a discrete 3′-extension (Figure 6A), as described previously (Allmang et al., 1999a; Mitchell et al., 2003; van Hoof et al., 2000). After treatment with RNase H and oligo(dT), a discrete band indicates that the rrp6Δ mutant accumulates polyadenylated forms of U14 but not U3 (Figure 6B). Surprisingly, the rna14-1 and rna15-2 mutants did not have any obvious effects (Figure 6, A and B), suggesting that Rna14p and Rna15p are not necessary for 3′-end processing of U14.
Figure 6.
Rna14p and Rna15p are not necessary for 3′-end processing of U14. Total RNA was extracted from the indicated strains. Cells were grown at 23°C, and then they were incubated for 1 h at 37°C, as indicated. Strains with GAL-regulated alleles were grown in galactose medium at 23°C, transferred to glucose medium at 23°C for 20 h, and then shifted to 37°C for 1 h. (A) Northern hybridization with probes for U14 and U3 snoRNAs. The altered gel mobility in the rrp6Δ strain represents a failure in the 3′ trimming of the U14 snoRNA, as described previously (Allmang et al., 1999; van Hoof et al., 2000; Mitchell et al., 2003). (B) Total RNA recovered from cells incubated for 1 h at 37°C was deadenylated by treatment with RNase H in the presence (+lanes) or absence (−lanes) of oligo(dT). Note that there is almost no 3′-end extended and polyadenylated U14 in the rna14-1/rrp6Δ and rna15-2/rrp6Δ double mutants.
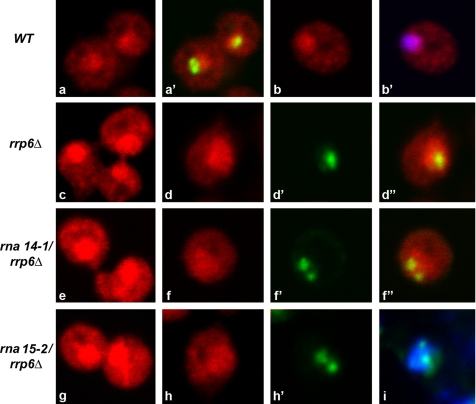
We also analyzed the subcellular distribution of pre-rRNA and actin mRNA in the double mutants. Fluorescence in situ hybridization (FISH) was performed with a probe directed against the internal spacer 2 (ITS2) region of the pre-rRNA, which is retained in the 27S component of pre-60S ribosomes (Dez et al., 2006). In wild-type cells, this probe labeled the nucleolus (stained green in a and a′, Figure 7) and the nucleoplasm (stained blue in b and b′, Figure 7). This is consistent with the view that pre-rRNA is transcribed and initially processed in the nucleolus, but late maturation steps take place in the nucleoplasm, before export to the cytoplasm (for reviews, see de la Cruz et al., 2003; Fromont-Racine et al., 2003). In the rrp6Δ cells analyzed 1 h after transfer to 37°C, a substantial increase in pre-rRNA staining was observed throughout the nucleus (Figure 7, c and d) in agreement with Northern blotting data in Dez et al. (2006). No significant labeling difference was apparent in the rna14-1/rrp6Δ (Figure 7, e and f) and rna15-2/rrp6Δ (Figure 7, g–i) double mutants relative to the cells lacking only Rrp6p (Figure 7, note that the images depicted in a, c, e, and g were acquired with the same settings). After reducing the detector gain (Figure 7, d, f, and h), the staining seems homogeneously distributed, with no concentration at the foci that contain Nop1p. These results are consistent with the observation that inactivation of Rna 14p or Rna15p had little effect on the level of 18S rRNA (Torchet et al., 2002).
Figure 7.
Detection of pre-rRNA in cells depleted of Rrp6p and Rna14p or Rna15p. The indicated strains were grown at 23°C and incubated for 1 h at 37°C. Pre-rRNA was detected by FISH with a probe directed against ITS2 (red staining). Cells were double-labeled with anti-Nop1p antibody (green staining) or DAPI (blue staining).
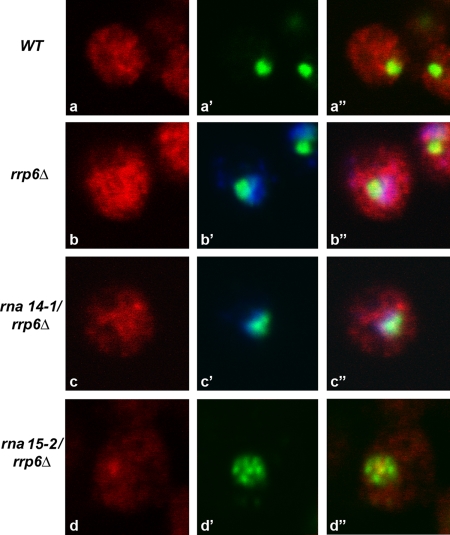
To assess the subcellular distribution of an mRNA, we performed FISH with Cy3-labeled probes complementary to exonic sequences of the ACT1 (actin) mRNA (Figure 8). In wild-type cells, the staining was detected predominantly in the cytoplasm (Figure 8, a–a″). After transfer to 37°C for 1 h, the total amount of actin mRNA staining was increased in the rrp6-Δ strain. Substantial ACT1 mRNA labeling was detected in the nucleoplasm (Figure 8, b–b″) in agreement with a role for Rrp6p in the degradation of aberrant pre-mRNAs in the nucleus. In contrast, very faint fluorescent labeling was detected in rna14-1 and rna15-2 strains at 37°C (data not shown), consistent with the previously described rapid loss of mRNAs detected when these strains were incubated at 37°C (Torchet et al., 2002). The loss of the mRNA is not due to transcription inhibition, but rather to degradation of 3′-extended forms by the core exosome (Torchet et al., 2002). The absence of Rrp6p from the rna14-1 strain (Figure 8, c–c″) but not from the rna15-2 (Figure 8, d–d″) mutant increased the level of actin mRNA labeling in the nucleus, as described previously (Torchet et al., 2002). In neither case was the actin mRNA staining concentrated at the foci that contain Nop1p.
Figure 8.
Detection of actin mRNA in cells depleted of Rrp6p and Rna14p or Rna15p. The indicated strains were grown at 23°C, and then they were incubated for 1 h at 37°C. Actin mRNA was detected by FISH (red staining). Cells were double labeled with anti-Nop1p antibody (green staining) or DAPI (blue staining).
Depletion of Rrp6p Alters Nop1p-GFP Dynamics at the rna 14- and rna 15-induced Nuclear Foci
In summary, we have shown that CFIA inactivation leads to the accumulation of snoRNP proteins in discrete nucleoplasmic domains that concentrate polyadenylated RNA species stabilized in the absence of Rrp6p. To try to identify the defect that prevents snoRNPs from accumulating in the nucleolus, we compared the mobility of Nop1p (yeast fibrillarin) in control and in rna14-1 and rna15-2 strains. Previous studies performed with mammalian cells demonstrated that fibrillarin is not stationary in the nucleolus, but constantly exchanges between the nucleolus and the nucleoplasm on a scale of seconds (Phair and Misteli, 2000; Snaar et al., 2000; Chen and Huang, 2001). According to several lines of recent evidence, assembly of macromolecular complexes in the nucleus seems to occur in a stepwise process by largely stochastic collisions of subunits (Misteli, 2007). Once assembled, proteins are transiently immobilized with different resident times that reflect their specific functional interactions (Misteli, 2007). We therefore reasoned that defects in snoRNP assembly caused by CFIA inactivation could alter the mobility of Nop1p.
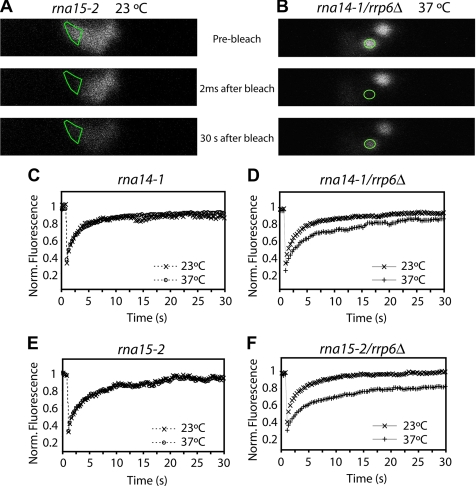
FRAP was performed in rna14-1 and rna15-2 single mutants, and rna14-1/rrp6Δ and rna15-2/rrp6Δ double mutants expressing Nop1-GFP (Figure 9). In rna14-1, rna15-2, rna14-1/rrp6Δ, and rna15-2/rrp6Δ strains at the permissive temperature (23°C), Nop1-GFP was homogeneously distributed throughout the typical crescent-shaped nucleolus. In these cells, the GFP fluorescence was irreversibly bleached by a short, high-intensity laser pulse in a region corresponding to approximately one third of the nucleolus (Figure 9A). In all cases, the fluorescent signal from the bleached area fully recovered within 15 s after bleaching (Figure 9, C and E), indicating rapid and constant exchange of Nop1-GFP similarly to what is observed in mammalian cells. One hour after transfer to 37°C, Nop1-GFP had accumulated in the nucleoplasmic foci in each of the single and double mutant strains. One focus was photobleached and the fluorescence recovery was monitored (Figure 9B). In the rna14-1 and rna15-2 single mutants the Nop1-GFP signal was rapidly recovered in the bleached nucleoplasmic focus, with kinetics similar to the recovery of the nucleolar Nop1-GFP signal seen at 23°C (Figure 9, C and E). Thus, Nop1p remains highly mobile in the foci. Recovery of Nop1-GFP fluorescence was slower in the rna14-1 and rna15-2 strains that also lacked Rrp6p when incubated at 37°C (Figure 9, D and F), suggesting that absence of Rrp6p induces a longer immobilization of Nop1p in nucleoplasmic foci. Given the significant size of the microscope point spread function compared with the dimensions of the yeast nucleus, the subnuclear volumes that were effectively photobleached were always significantly larger than the defined region of interest. For this reason we, were unable to bleach a similar circular region in the nucleolus and in the foci. Consequently, the comparison of fluorescence recovery kinetics in the different strains is only qualitative.
Figure 9.
FRAP analysis of Nop1-GFP in rna14-1 and rna15-2 single mutants and in rna14-1/rrp6Δ and rna15-2/rrp6Δ double mutants. Live cells were imaged at 23°C or 37°C, as indicated, using the 488-nm line of an Ar laser. Shown are images taken before, immediately after bleaching and after recovery for 30 s for rna15-2 at 23°C (A) and rna14-1/rrp6Δ at 37°C (B). The bleached region is outlined. The fluorescence intensity in the bleached region was measured and expressed as the normalized fluorescence (R). The recovery curves correspond to a pool of three different experiments, with at least 10 different cells analyzed per experiment. Error bars represent standard deviations. Fluorescence recovery curves are shown for the different strains at 23 and 37°C (C–F).
DISCUSSION
This study shows that after disruption of cleavage factor CFIA, snoRNPs accumulate in discrete nucleoplasmic regions or foci, failing to be properly localized to the nucleolus. The foci concentrate polyadenylated RNA species that become stabilized after inactivation of the nuclear exosome component Rrp6p, suggesting that they play a quality control function in the nucleoplasm. These domains, which become visible only when abundant aberrant products accumulate in the nucleus, may be related to the “surveillance centers” recently identified in the nucleolus (Dez et al., 2006).
Like the precursors to mRNA and snRNA, nascent snoRNA transcripts are endonucleolytically cleaved to produce free 3′ ends, which can then be processed by 3′ exonucleases to generate the mature RNAs. In yeast U3 snoRNA, this process is catalyzed by the endoribonuclease Rnt1p, the orthologue of bacterial RNase III (Kufel et al., 2000). However, other pre-snoRNAs (including U14) do not contain the hairpin structures that are preferentially recognized by Rnt1p, and these species may undergo cotranscriptional cleavage by CFIA (Fatica et al., 2000; Morlando et al., 2002). It was reported previously that mature U14 continued to accumulate after disruption of the RNT1 gene (Chanfreau et al., 1998), suggesting the involvement of an additional cleavage activity. However, our results show that mature U14 RNA is produced in strains carrying ts-lethal alleles of the CFIA subunits Rna14p and Rna15p, in good agreement with a recent study indicating that Rna15p is not necessary for 3′ end processing of U14 (Vasiljeva and Buratowski, 2006). Possibly, Rnt1p and CFIA may play redundant roles in 3′-end formation of U14 snoRNA, as suggested for U2 and U5 snRNAs (Elela et al., 1996; Chanfreau et al., 1997). Alternatively, a distinct not yet identified cleavage activity is involved in U14 3′-end formation.
Within 30 min to 1 h after inhibition of the CFIA cleavage activity, we observed a redistribution of snoRNP proteins Nop1p, Nop58p, and Gar1p, which no longer localized to the nucleolus but accumulated in a few discrete domains in the nucleoplasm. Similar mislocalization was observed for U14 snoRNA, but not for U3 snoRNA. Based on previous studies showing that in rna14-1 and rna15-2 strains several box C/D and H/AC snoRNAs are not properly 3′ end cleaved (Fatica et al., 2000; Morlando et al., 2002), we propose that the inhibition of either Rna14p or Rna15p leads to accumulation of readthrough snoRNA transcripts (other than U14) that are targeted for degradation at discrete surveillance centers in the nucleoplasm. Because CFIA-dependent snoRNAs assemble with core snoRNP proteins cotranscriptionally (Morlando et al., 2004; Ballarino et al., 2005), accumulated aberrant RNAs may form a sink in the nucleoplasm that traps any Nop1p-associated RNA in transit to the nucleolus. This could be the reason why correctly processed U14 is targeted to the foci. In contrast, U3 is not bound by Nop1p, Nop56p, and Nop58p before it reaches the final maturation stages at the nucleolar body (Kufel et al., 2000; Kufel et al., 2003). Assuming that trafficking of U3 snoRNA to the nucleolus occurs independently of Nop1p binding, U3 can escape sequestration in the foci. This could also explain U3 snoRNA persistence in the nucleolus when most Nop1p delocalized to the foci.
The precise mechanism underlying the observed CFIA-dependent accumulation of snoRNPs in nucleoplasmic foci remains unknown. Interestingly, our photobleaching results show that the exchange rate of Nop1p is similar in the nucleolus and in the foci, indicating that Nop1p does not fail to localize to the nucleolus because it is irreversibly immobilized in the foci. Rather, the data suggest that the foci provide novel binding sites for Nop1p that outcompete the normal binding sites in the nucleolus.
Accumulation of polyadenylated RNA in several discrete nucleoplasmic dots was observed within 30 min after inactivation of the essential nuclear mRNA export factor Mex67p (Segref et al., 1997). More recently, individual mRNA species were detected concentrated in a single intranuclear focus in strains carrying mutations in several nuclear mRNA export factors (Jensen et al., 2001). Because each focus is thought to represent the site of transcription, it is likely that the multiple dots that accumulate poly(A)+RNA in Mex67p mutants correspond to the transcription sites of several genes. According to the model that a system exists in the yeast nucleus to monitor the quality of 3′-end formation and inhibit the release of aberrant RNA from the transcription site (Hilleren et al., 2001), one possibility is that the Nop1p-containing nucleoplasmic foci observed in the rna14-1 and rna15-2 strains at nonpermissive temperature result from retention of abnormally processed snoRNPs at the site of transcription. However, we find this unlikely for a number of reasons. First, CFIA is required for proper processing of several snoRNAs, whereas the number of foci is, in many cells, limited to two or three. Second, Nop1p and Gar1p bind to distinct snoRNAs and yet colocalize in the same foci (Figure 3A). These results could, however, be reconciled with the cotranscriptional retention hypothesis, were snoRNA genes in the yeast nucleus to be clustered in a few transcription factories. A third observation that is incompatible with the model is the detection of U14 snoRNA in more than two foci (Figure 2A). We therefore favor the interpretation that snoRNPs synthesized in the absence of CFIA activity are released from the transcription site, but then they are trapped at specific domains in the nucleoplasm.
Although accurate decay kinetics for endogenous snoRNPs are not available, some studies suggest that U3 and U14 snoRNAs are rapidly degraded in the nucleus with a half-live of ∼1 h, whereas others (e.g., snR190) are stable over several hours (Li et al., 1990; Yadava et al., 2004). Thus, it is conceivable that the foci formed during incubation for 1 h at the nonpermissive temperature accumulate both newly synthesized RNAs and pre-existing mature snoRNPs that exited the nucleolus. Indeed, snoRNPs are constantly exchanging between the nucleolus and the nucleoplasm, as revealed by photobleaching microscopy in mammalian (Phair and Misteli, 2000; Snaar et al., 2000; Chen and Huang, 2001) and yeast cells (Figure 9).
After inactivation of Rna14p or Rna15p, actin mRNA labeling was barely visible by FISH, as expected because transfer of the rna14-1 or rna15-2 strain to 37°C for 2 h resulted in loss of ACT1 and other mRNAs tested by Northern blot analysis (Torchet et al., 2002). However, accumulation of a specific set of mRNAs in nuclear dots has been described in rna14-1 and rna15-2 cells shifted to 37°C for 30 min (Brodsky and Silver, 2000). Although it remains to be established whether these dots coincide with the Nop1p-containing foci, this observation argues that inactivation of Rna14p or Rna15p results in retention of both snoRNPs and mRNAs at discrete subnuclear domains.
The model that Nop1p-containing foci may correspond to nucleoplasmic surveillance centers is supported by the finding that they become enriched in polyadenylated RNA species after depletion of the nuclear exosome component Rrp6p. Several lines of evidence indicate that polyadenylation targets nonfunctional transcripts for degradation by the exosome. Yeast strains lacking Rrp6 accumulate polyadenylated forms of rRNA, snRNAs, snoRNAs, and other transcripts of unknown function, which are associated with promoter regions or result from intergenic transcription (for review, see Houseley et al., 2006; Struhl, 2007). A challenge for the future will be to determine whether the proposed nucleoplasmic surveillance centers do exist under physiological conditions. A prediction would be that most, if not all RNAs targeted for Rrp6-dependent degradation in the nucleoplasm are prone to self-organization, leading to the formation of discrete compartments.
Supplementary Material
ACKNOWLEDGMENTS
We thank Edouard Bertrand (CNRS, Montpellier, France) for gift of plasmids and probes and for helpful advice on yeast immunofluorescence and in situ hybridization procedures. We are also grateful to Ed Hurt (University of Heidelberg, Heidelberg, Germany) for kindly providing Nup49-GFP and Pierre-Emmanuel Gleizes (CNRS, Toulouse, France) for Gar1p-GFP. T.C. was the recipient of a fellowship from Fundação para a Ciência e Tecnologia, Portugal. This work was supported by the European Commission “QLG2-CT-2001-01554.” D.T. and L.M. were supported by the Wellcome Trust.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/10.1091/mbc.E07-10-1015) on January 30, 2008.
REFERENCES
- Allmang C., Kufel J., Chanfreau G., Mitchell P., Petfalski E., Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999a;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C., Petfalski E., Podtelejnikov A., Mann M., Tollervey D., Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′-> 5′ exonucleases. Genes Dev. 1999b;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani N., Dufour M. E., Bonneaud N., Lacroute F. Mutations in STS1 suppress the defect in 3′ mRNA processing caused by the rna15-2 mutation in Saccharomyces cerevisiae. Mol. Gen. Genet. 1996;252:552–562. doi: 10.1007/BF02172401. [DOI] [PubMed] [Google Scholar]
- Aris J. P., Blobel G. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J. Cell Biol. 1988;107:17–31. doi: 10.1083/jcb.107.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachellerie J. P., Cavaille J., Huttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- Ballarino M., Morlando M., Pagano F., Fatica A., Bozzoni I. The cotranscriptional assembly of snoRNPs controls the biosynthesis of H/ACA snoRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 2005;25:5396–5403. doi: 10.1128/MCB.25.13.5396-5403.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh N., Doye V. Dynamics of nuclear pore distribution in nucleoporin mutant yeast cells. J. Cell Biol. 1997;136:747–759. doi: 10.1083/jcb.136.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky A. S., Silver P. A. Pre-mRNA processing factors are required for nuclear export. RNA. 2000;6:1737–1749. doi: 10.1017/s1355838200001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro T., Carvalho C., Braga J., Rino J., Milligan L., Tollervey D., Carmo-Fonseca M. Depletion of the yeast nuclear exosome subunit Rrp6 results in accumulation of polyadenylated RNAs in a discrete domain within the nucleolus. Mol. Cell. Biol. 2007;27:4157–4165. doi: 10.1128/MCB.00120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G., Elela S. A., Ares M., Jr, Guthrie C. Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev. 1997;11:2741–2751. doi: 10.1101/gad.11.20.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G., Rotondo G., Legrain P., Jacquier A. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 1998;17:3726–3737. doi: 10.1093/emboj/17.13.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Huang S. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell Biol. 2001;153:169–176. doi: 10.1083/jcb.153.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J., Kressler D., Linder P. Ribosomal subunit assembly. In: Olson M.O.J., editor. In the Nucleolus. New York: Kluwer Academic/Plenum; 2003. pp. 262–290. [Google Scholar]
- Dez C., Houseley J., Tollervey D. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J. 2006;25:1534–1546. doi: 10.1038/sj.emboj.7601035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheur S., Vo le T. A., Voisinet-Hakil F., Minet M., Schmitter J. M., Lacroute F., Wyers F., Minvielle-Sebastia L. Pti1p and Ref2p found in association with the mRNA 3′ end formation complex direct snoRNA maturation. EMBO J. 2003;22:2831–2840. doi: 10.1093/emboj/cdg253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elela S. A., Igel H., Ares M., Jr RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–124. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- Fang F., Hoskins J., Butler J. S. 5-fluorouracil enhances exosome-dependent accumulation of polyadenylated rRNAs. Mol. Cell. Biol. 2004;24:10766–10776. doi: 10.1128/MCB.24.24.10766-10776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A., Morlando M., Bozzoni I. Yeast snoRNA accumulation relies on a cleavage-dependent/polyadenylation-independent 3′-processing apparatus. EMBO J. 2000;19:6218–6229. doi: 10.1093/emboj/19.22.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W., Pogacic V. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:319–327. doi: 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine M., Senger B., Saveanu C., Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- Hilleren P., McCarthy T., Rosbash M., Parker R., Jensen T. H. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature. 2001;413:538–542. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- Houseley J., LaCava J., Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Houseley J., Tollervey D. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep. 2006;7:205–211. doi: 10.1038/sj.embor.7400612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen T. H., Patricio K., McCarthy T., Rosbash M. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell. 2001;7:887–898. doi: 10.1016/s1097-2765(01)00232-5. [DOI] [PubMed] [Google Scholar]
- Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- Kuai L., Fang F., Butler J. S., Sherman F. Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2004;101:8581–8586. doi: 10.1073/pnas.0402888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel J., Allmang C., Chanfreau G., Petfalski E., Lafontaine D. L., Tollervey D. Precursors to the U3 small nucleolar RNA lack small nucleolar RNP proteins but are stabilized by La binding. Mol. Cell. Biol. 2000;20:5415–5424. doi: 10.1128/mcb.20.15.5415-5424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel J., Allmang C., Petfalski E., Beggs J., Tollervey D. Lsm Proteins are required for normal processing and stability of ribosomal RNAs. J. Biol. Chem. 2003;278:2147–2156. doi: 10.1074/jbc.M208856200. [DOI] [PubMed] [Google Scholar]
- LaCava J., Houseley J., Saveanu C., Petfalski E., Thompson E., Jacquier A., Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Li H. D., Zagorski J., Fournier M. J. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990;10:1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A. G., Terns R. M., Terns M. P. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- Maxwell E. S., Fournier M. J. The small nucleolar RNAs. Annu. Rev. Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Preker P. J., Keller W. RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3′-end processing factor. Science. 1994;266:1702–1705. doi: 10.1126/science.7992054. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Winsor B., Bonneaud N., Lacroute F. Mutations in the yeast RNA14 and RNA15 genes result in an abnormal mRNA decay rate; sequence analysis reveals an RNA-binding domain in the RNA15 protein. Mol. Cell. Biol. 1991;11:3075–3087. doi: 10.1128/mcb.11.6.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Mitchell P., Petfalski E., Houalla R., Podtelejnikov A., Mann M., Tollervey D. Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol. Cell. Biol. 2003;23:6982–6992. doi: 10.1128/MCB.23.19.6982-6992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlando M., Ballarino M., Greco P., Caffarelli E., Dichtl B., Bozzoni I. Coupling between snoRNP assembly and 3′ processing controls box C/D snoRNA biosynthesis in yeast. EMBO J. 2004;23:2392–2401. doi: 10.1038/sj.emboj.7600254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlando M., Greco P., Dichtl B., Fatica A., Keller W., Bozzoni I. Functional analysis of yeast snoRNA and snRNA 3′-end formation mediated by uncoupling of cleavage and polyadenylation. Mol. Cell. Biol. 2002;22:1379–1389. doi: 10.1128/mcb.22.5.1379-1389.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouaikel J., Verheggen C., Bertrand E., Tazi J., Bordonne R. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell. 2002;9:891–901. doi: 10.1016/s1097-2765(02)00484-7. [DOI] [PubMed] [Google Scholar]
- Nedea E., He X., Kim M., Pootoolal J., Zhong G., Canadien V., Hughes T., Buratowski S., Moore C. L., Greenblatt J. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J. Biol. Chem. 2003;278:33000–33010. doi: 10.1074/jbc.M304454200. [DOI] [PubMed] [Google Scholar]
- Patel D., Butler J. S. Conditional defect in mRNA 3′ end processing caused by a mutation in the gene for poly(A) polymerase. Mol. Cell. Biol. 1992;12:3297–3304. doi: 10.1128/mcb.12.7.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair R. D., Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- Segref A., Sharma K., Doye V., Hellwig A., Huber J., Luhrmann R., Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y., Blechman J., Darzacq X., Montagna C., Dye B. T., Patton J. G., Singer R. H., Zipori D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol. Biol. Cell. 2005;16:2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon K. E., Mauger D. M., Arndt K. M. A Requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol. Cell. 2005;20:225–236. doi: 10.1016/j.molcel.2005.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar D. A., Greenleaf A. L. The RNA polymerase II CTD kinase CTDK-I affects pre-mRNA 3′ cleavage/polyadenylation through the processing component Pti1p. Mol. Cell. 2002;10:1429–1439. doi: 10.1016/s1097-2765(02)00731-1. [DOI] [PubMed] [Google Scholar]
- Snaar S., Wiesmeijer K., Jochemsen A. G., Tanke H. J., Dirks R. W. Mutational analysis of fibrillarin and its mobility in living human cells. J. Cell Biol. 2000;151:653–662. doi: 10.1083/jcb.151.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz E. J., Conrad N. K., Brow D. A., Corden J. L. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature. 2001;413:327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- Torchet C., Bousquet-Antonelli C., Milligan L., Thompson E., Kufel J., Tollervey D. Processing of 3′-extended read-through transcripts by the exosome can generate functional mRNAs. Mol. Cell. 2002;9:1285–1296. doi: 10.1016/s1097-2765(02)00544-0. [DOI] [PubMed] [Google Scholar]
- Tran E., Brown J., Maxwell E. S. Evolutionary origins of the RNA-guided nucleotide-modification complexes: from the primitive translation apparatus? Trends Biochem. Sci. 2004;29:343–350. doi: 10.1016/j.tibs.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Trumtel S., Leger-Silvestre I., Gleizes P. E., Teulieres F., Gas N. Assembly and functional organization of the nucleolus: ultrastructural analysis of Saccharomyces cerevisiae mutants. Mol. Biol. Cell. 2000;11:2175–2189. doi: 10.1091/mbc.11.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A., Lennertz P., Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 2000;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L., Buratowski S. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol Cell. 2006;21:239–248. doi: 10.1016/j.molcel.2005.11.028. [DOI] [PubMed] [Google Scholar]
- Venema J., Tollervey D. RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J. 1996;15:5701–5714. [PMC free article] [PubMed] [Google Scholar]
- Verheggen C., Lafontaine D. L., Samarsky D., Mouaikel J., Blanchard J. M., Bordonne R., Bertrand E. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J. 2002;21:2736–2745. doi: 10.1093/emboj/21.11.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheggen C., Mouaikel J., Thiry M., Blanchard J. M., Tollervey D., Bordonne R., Lafontaine D. L., Bertrand E. Box C/D small nucleolar RNA trafficking involves small nucleolar RNP proteins, nucleolar factors and a novel nuclear domain. EMBO J. 2001;20:5480–5490. doi: 10.1093/emboj/20.19.5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P., Brockenbrough J. S., Metcalfe A. C., Chen S., Aris J. P. Nop5p is a small nucleolar ribonucleoprotein component required for pre-18 S rRNA processing in yeast. J. Biol. Chem. 1998;273:16453–16463. doi: 10.1074/jbc.273.26.16453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadava R. S., Mahen E. M., Fedor M. J. Kinetic analysis of ribozyme-substrate complex formation in yeast. RNA. 2004;10:863–879. doi: 10.1261/rna.5234204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.