Abstract
Cell polarization in response to external cues is critical to many eukaryotic cells. During pheromone-induced mating in Saccharomyces cerevisiae, the mitogen-activated protein kinase (MAPK) Fus3 induces polarization of the actin cytoskeleton toward a landmark generated by the pheromone receptor. Here, we analyze the role of Fus3 activation and cell cycle arrest in mating morphogenesis. The MAPK scaffold Ste5 is initially recruited to the plasma membrane in random patches that polarize before shmoo emergence. Polarized localization of Ste5 is important for shmooing. In fus3 mutants, Ste5 is recruited to significantly more of the plasma membrane, whereas recruitment of Bni1 formin, Cdc24 guanine exchange factor, and Ste20 p21-activated protein kinase are inhibited. In contrast, polarized recruitment still occurs in a far1 mutant that is also defective in G1 arrest. Remarkably, loss of Cln2 or Cdc28 cyclin-dependent kinase restores polarized localization of Bni1, Ste5, and Ste20 to a fus3 mutant. These and other findings suggest Fus3 induces polarized growth in G1 phase cells by down-regulating Ste5 recruitment and by inhibiting Cln/Cdc28 kinase, which prevents basal recruitment of Ste5, Cdc42-mediated asymmetry, and mating morphogenesis.
INTRODUCTION
A major question in biology is how specialized structures are formed during polarized morphogenesis when cells differentiate or migrate. Although much is known about how cell polarity is generated by polarity establishment proteins, cytoskeleton, and vesicular transport (Leof, 2000; Pruyne and Bretscher, 2000; Iijima et al., 2002; Nelson, 2003; Pruyne et al., 2004), less is known about how it is initiated by external stimuli and coordinated with signaling events that control proliferation. For example, mitogen-activated protein kinase (MAPK) cascades act downstream of many receptors, and they also regulate morphogenesis and proliferation in a variety of contexts, including nervous system development, T cell polarization, chemotaxis, and tumorigenesis (Qi and Elion, 2005a; Stowers et al., 1995; Giniger, 2002; Huang et al., 2004; Xia and Karin, 2004; Guo et al., 2006; Pullikuth and Catling, 2007).
The mating pathway of the yeast Saccharomyces cerevisiae is an excellent example of MAPK cascade control of cell division and specialized morphogenesis in response to G protein-coupled receptor (GPCR) activation (Marsh et al., 1991). During sexual reproduction, peptide pheromones secreted from haploid “a” cells and “α” cells (a factor and α factor, respectively) bind to GPCRs on opposite cell type (Ste3 and Ste2), which activates a heterotrimeric G protein-coupled MAPK cascade. Sufficient pathway activation causes G1 phase arrest and polarized growth in the direction of highest pheromone from nearby cells (termed shmooing), with eventual fusion at the tips of growing cells. This response to the spatial gradient of pheromone is termed chemotropism (Segall, 1993).
Many of the steps in the MAPK pathway have been defined previously (Bardwell, 1995; Dohlman, 2002). The pheromone-bound GPCR dissociates a heterotrimeric G protein into Gα monomer (Gpa1) and Gβγ heterodimer (Ste4 and Ste18, respectively). The free Gβγ dimer binds multiple targets including Ste20, a p21-activated protein kinase, and Ste5, an MAPK cascade scaffold protein essential for the activation of MAPK kinase kinase (MAPKKK) Ste11 by Ste20 and transmission of the pheromone signal down through the Ste7 MAPKK to two MAPKs, Fus3 and Kss1. The Fus3 MAPK is most essential for mating and phosphorylates numerous targets including transcription factors (e.g., Ste12, Dig1, Dig2, and Tec1), an inhibitor of G1 cyclin-dependent kinase and regulator of chemotropism (Far1), a formin (Bni1), and various signaling components in the pathway (e.g., Ste5, Ste7, and Sst2).
The initial development of cell polarity is through the mating pheromone receptor and G protein engagement of proteins that normally regulate cell polarity during budding, including the essential Rho-type GTPase Cdc42 (Johnson, 1999; Bretscher and Pruyne, 2000; Pruyne et al., 2004). This event may involve actin-mediated receptor clustering that consolidates sufficient Cdc42 and associated proteins at the cell cortex to induce shmooing (Ayscough and Drubin, 1998). Orientation of the growth axis toward the pheromone source requires the Far1 scaffold, which binds the mating G protein Gβγ dimer and links it to the Cdc42/Cdc24 guanine exchange factor complex (Gulli and Peter, 2001; Chang and Peter, 2005; Park and Bi, 2007). Polarized growth requires Bni1, a scaffold protein that nucleates the assembly of actin cables independently of Arp2/Arp3 and is activated by Rho GTPases (Evangelista et al., 2003; Moseley and Goode, 2006). During the α factor response, Rho1 and Cdc42 are required for cortical localization of Bni1 at the shmoo tip (Qi and Elion, 2005b).
The Fus3 and Kss1 MAPKs provide distinct functions for chemotropism (Farley et al., 1999; Paliwal et al., 2007). Fus3 is essential for shmooing (Farley et al., 1999; Matheos et al., 2004), and may regulate actin polarization through phosphorylation of Bni1 (Matheos et al., 2004). Kss1 is required for optimal shmooing (Farley et al., 1999), and it regulates the dynamic range of the shmooing response to pheromone (Paliwal et al., 2007). Active Fus3 binds to Gpa1, the Gα subunit (Metodiev et al., 2002), which has led to the speculation that the pool of Fus3 that regulates Bni1 is bound to Gpa1 (Matheos et al., 2004). Bni1 is required for full activation of Fus3 and for polarized cortical localization of the Ste5 MAPK cascade scaffold, the Cdc24 guanine exchange factor for Cdc42 and Fus3 (Qi and Elion, 2005b). That Bni1 is poorly localized in a fus3 mutant (Matheos et al., 2004) but is also required for cortical recruitment of Ste5, which activates Fus3 (Qi and Elion, 2005b) and can recruit Fus3 to the plasma membrane (van Drogen et al., 2001), raises questions about the hierarchy of localization events required for MAPK activation and morphogenesis.
To better understand the relationship between the activation of Fus3 and induction of cell polarity during pheromone stimulation, we analyzed the localization of Ste5, Bni1, Cdc24, and Ste20 in mutants with defects in MAPK signaling. We identify novel functions for Fus3 in down-regulating unrestrained cortical recruitment of Ste5 and in promoting polarized cortical recruitment of Ste20 and Cdc24 in addition to Bni1. By contrast, Cln/Cdc28 blocks cortical recruitment of Ste5, Bni1, and Ste20 in G1 phase cells and prevents mating morphogenesis. Our findings suggest that Fus3 globally regulates Cdc42-mediated asymmetry through its role as an inhibitor of Cln/Cdc28, and reveal a general strategy for how a morphological switch in dividing eukaryotic cells can be regulated. They also provide strong support for the proposal that Cln2/Cdc28 kinase phosphorylation of Ste5 blocks its membrane recruitment (Strickfaden et al., 2007). We find that Ste5 exists in multiple pools at the plasma membrane and that polarization of Ste5 is important for morphogenesis. Under conditions of decreased Cdc28 and elevated MAPK activation, Ste5 accumulates in an internal pool that colocalizes with N-[3-triethylammoniumpropyl]-4-[p-diethylaminophenylhexatrienyl] pyridinium dibromide (FM4-64) and thus may be vacuolar and/or endocytic. These and other results raise the possibility that down-regulation of Ste5 involves targeting to the vacuole.
MATERIALS AND METHODS
Media and Materials
Yeast cells were grown in selective synthetic complete (SC) medium (containing yeast nitrogenous base supplemented with amino acids) or rich yeast extract-peptone (YEP) medium containing dextrose, raffinose, or galactose at a final concentration of 2% and 1 mM CuSO4 to induce CUP1-GFP-STE5. For some experiments (i.e., STE5-3xYFP, STE5-3xGFP, GFP-STE20, and GFP-ste20Δ334-369 strains), the SC medium contained low-fluorescence yeast nitrogen base with H3BO3 instead of H3BO4 (Sheff and Thorn, 2004) plus 20 mg/l adenine, 50 mg/l l-arginine HCl, 80 mg/l l-aspartic acid, 63 mg/l l-histidine HCl, 50 mg/l l-isoleucine, 219 mg/l l-Leucine, 50 mg/l l-lysine HCl, 20 mg/l l-methionine, 50 mg/l l-phenylalanine, 100 mg/l l-threonine, 81.7 mg/l l-tryptophan, 50 mg/l l-tyrosine, 22.4 mg/l uracil, and 140 mg/l l-valine (Sigma-Aldrich, St. Louis, MO). Antibiotics were used at the following concentrations: 100 μg/ml ampicillin (Sigma-Aldrich), 250 μg/ml Geneticin (G418; Calbiochem, San Diego, CA), and 300 μg/ml hygromycin b (Invitrogen, Carlsbad, CA). α factor was used at 5–50 nM for bar1Δ strains and at 5 μM for BAR1 strains. Standard methods were used for growth, DNA manipulations, and transformations of yeast (Sambrook et al., 1989; Amberg et al., 2005). The yeast strains are listed in Supplemental Table 1, plasmids in Supplemental Table 2, and oligonucleotides in Supplemental Table 3. Details of strain construction and fluorescent protein tagging can be found in Supplemental Material.
Live Cell Fluorescence Microscopy
Ste5-3xGFP and Ste5-3xYFP.
The cultures were grown overnight in low-fluorescence media at 30°C to mid-logarithmic phase (A600 ∼ 0.2–0.6) and normalized to A600 ∼0.4 with fresh low-fluorescence medium. One milliliter of culture was pelleted and concentrated into ∼100 μl of medium, then it was sonicated and α factor was added to 5 μM. Cells were incubated at room temperature, and then 4.5 μl of cell suspension was spotted on slides and covered with a coverslip for visualization. Controls showed that α factor induction of the 10× concentrated cells led to G1 arrest and shmoo formation with similar efficiency and kinetics as for unconcentrated cells. The cdc28-4 strains were pregrown at room temperature, pelleted, and then resuspended into fresh medium either with or without α factor. The remainder of the culture was transferred to prewarmed medium and incubated at 37°C for 3 h, and then it was pelleted and resuspended into fresh 37°C medium with or without α factor.
GFP-Ste5.
Strains harboring a CUP1-GFP-STE5 centromeric plasmid (pSKM21) were pregrown overnight in selective SC medium to early logarithmic phase (A600 ∼ 0.4), followed by addition of 1 mM CuSO4 for 1 h, and then they were pelleted and resuspended in fresh YPD medium containing 1 mM CuSO4 and treated with α factor with shaking at 30°C before processing for visualization.
Bni1-GFP, GFP-Ste20, and GFP-Cdc24.
Strains harboring BNI1-GFP (EBL334) and GFP-STE20 (EBL511) on centromeric plasmids were grown as described for GFP-STE5 except media lacked CuSO4. Strains harboring the 415MET-GFPS65T-A8-CDC24 centromeric plasmid (EBL664) were pregrown overnight to logarithmic phase in selective SC medium, and then they were diluted into selective SC medium containing 0.19 mM methionine and grown to early logarithmic phase (A600 ∼ 0.1). Finally, they were pelleted and resuspended in fresh medium containing 50 nM α factor and incubated with shaking at 30°C. Ste5-3xGFP and Ste5-3xYFP were visualized on a Nikon TE2000E motorized inverted microscope with a filter wheel setup with exciter 484/15, the dichroic, and emitter 517/30 for green fluorescent protein (GFP), and with exciter 500/20, the dichroic, and emitter 535/30 for yellow fluorescent protein (YFP). Images were captured with a Hamamatsu ORCA ER digital camera and MetaMorph 7 software (Microscopy Center, Department of Cell Biology, Harvard Medical School). GFP-Ste5, GFP-Ste20, and GFP-Cdc24 were visualized on a Zeiss Axioscope 2 microscope (Carl Zeiss, Thornwood, NY) linked to a Hamamatsu C4742-95 digital camera (Hamamatsu, Bridgewater, NJ) with filters from Chroma Technology (Brattleboro, VT) (set 41018 for GFP, set 51006 Texas Red filter for fluorescein isothiocyanate) (laboratory microscope). The percentage of cells showing a particular localization pattern was determined by tallying 100 to ∼450 cells from two transformants in at least two experiments.
Visualization of Actin Cytoskeleton
Cells were fixed with 4% formaldehyde for 45 min and then stained with rhodamine-phalloidin (final concentration 5 U/ml; Invitrogen) in the dark for 1 h. Cells were washed twice in phosphate-buffered saline, and then they were resuspended in Vectashield mounting medium (Vector Laboratories, Burlingame, CA) before visualization under a microscope.
FM 4-64 Staining
FM4-64 (Invitrogen) was dissolved in dimethyl sulfoxide to a concentration of 2.5 mM. Cells were grown at room temperature to A600 ∼0.2–0.8. One milliliter of cells was harvested and resuspended in 50 μl of YPD, and then cells were incubated with 0.5 μl of FM4-64 for 20 min. Cells were washed with 1 ml of YPD to remove excess dye, and then they were incubated in 5 ml of YPD for 90 min followed by washing with water and resuspending in low-fluorescence medium.
Yeast Whole Cell Extracts (WCE) and Immunoblot Analysis
Yeast strains were grown at 30°C in selective synthetic complete medium containing 2% dextrose to an A600 of ∼0.4–0.6, and then they were treated with α factor. Whole cell extracts were prepared as described previously (Elion et al., 1993) using modified H buffer with 250 mM NaCl. To detect Fus3 and Kss1 phosphorylation, 200 μg of total protein was separated by 10% SDS-polyacrylamide gel electrophoresis (PAGE), and then the sample was transferred onto a nitrocellulose membrane (Whatman Schleicher and Schuell, Keene, NH). The membrane was blocked with 5% skim milk in Tris-buffered saline containing 0.1% Tween 20, and then in the same buffer containing rabbit anti-phospho-p44/p42 antibody (1:1000 dilution, Cell Signaling Technology, Beverly, MA), followed by washing and incubation in the same buffer containing horseradish peroxidase-coupled goat anti-rabbit antibody (1:10,000 dilution; Bio-Rad, Hercules, CA). After the signal was visualized with enhanced chemiluminescence (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom), the membrane was stripped and reprobed with anti-Fus3 polyclonal antibody as described previously (Elion et al., 1993). To detect Ste5-Myc9, 50 μg of WCE protein was separated by 8% SDS-PAGE and transferred to Immobilon polyvinylidene difluoride membrane (Millipore, Billerica, MA). The membrane was blocked with 5% milk in PBS containing 0.1% Tween 20 (PBST), and then it was incubated with anti-Myc monoclonal antibody (mAb) 9E10 in PBST buffer containing 5% bovine serum albumin for 2 h followed by incubation with goat anti-mouse secondary antibody (1:10,000 dilution; Bio-Rad) for another 1 h. Antibodies were from the following sources: anti-Myc mAb 9E10 (ascites from Harvard University antibody facility) and anti-Tcm1 mAb (gift of J. Warner, Albert Einstein College of Medicine). Densitometry of immunoblot bands was done with Scion Image (NIH Image) software (National Institutes of Health, Bethesda, MD).
RESULTS
Fus3 Is Required for Ste5-CTM to Induce Actin Polarization and Shmoo Formation
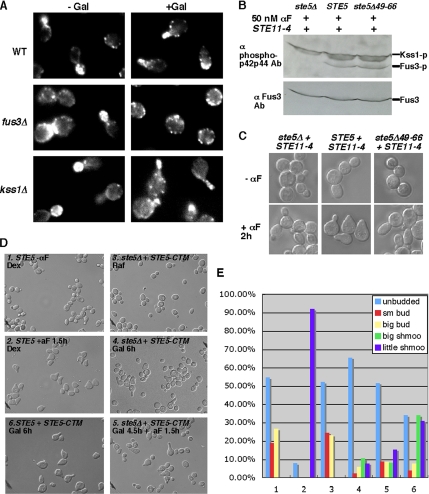
Previous data suggest that polarized cortical recruitment of Ste5 requires activation of Fus3 and Kss1 (Qi and Elion, 2005b). To determine whether Fus3 regulates the actin cytoskeleton and polarized growth independently of a role in Ste5 recruitment, we examined whether the plasma membrane-localized Ste5-CTM fusion protein could induce polarization of the actin cytoskeleton and polarized growth in a fus3 null mutant (fus3Δ). The expression of the GAL1-STE5-CTM gene in a W303a FUS3 KSS1 strain induces cell cycle arrest, actin polarization, and shmoo formation (Figure 1A) as found previously (Pryciak and Huntress, 1998). In contrast, GAL1-STE5-CTM poorly induces actin polarization and shmoo formation in a fus3Δ strain, although the cells undergo G1 arrest. This contrasts a kss1Δ strain that shmoos as efficiently as the FUS3 KSS1 strain, although the shmoos are longer and thinner (Figure 1A). Therefore, Fus3 provides essential functions for actin polarization independently of its role in Ste5 recruitment.
Figure 1.
Polarized cortical localization of Ste5 correlates with shmoo formation. (A) Ste5-CTM poorly induces actin polarization in a fus3Δ mutant. WT (EY699), fus3Δ (EY701), and kss1Δ (EY725) cells expressing Ste5-CTM were stained with phalloidin and representative fields are shown. (B) Activation of MAPKs Fus3 and Kss1 in a STE11-4 strain expressing Ste5 or Ste5Δ49-66 (Ste5ΔNLS) treated with 50 nM α factor for 1 h. Activated Fus3 and Kss1 were detected with anti-phospho p42p44 antibody using 200 μg of whole cell extract. (C) Nomarski images of representative cells from B. (D) Ste5-CTM is an inefficient inducer of shmooing. Shmoo formation in STE5-CTM was induced for 6 h in SC medium containing 2% galactose with or without 50 nM α factor addition for 90 min. Dex, 2% dextrose; Raf, 2% raffinose; Gal, 2% galactose. (E) Quantitation of cell morphology of samples shown in D. sm bud, small budding cells.
Changing the Distribution of Ste5 at the Plasma Membrane Affects Shmoo Formation
Ste5 localizes to the plasma membrane by binding the receptor-activated Gβγ dimer (Whiteway et al., 1995; Inouye et al., 1997; Feng et al., 1998) and plasma membrane lipids through a polybasic plasma membrane (PM) domain that seems to bind acidic phospholipids (Winters et al., 2005) and a pleckstrin homology (PH)-like domain involving phosphoinositide binding (Garrenton et al., 2006). To explore whether localization of Ste5 at the cell cortex has a role in shmoo formation beyond activation of Fus3 and Kss1, we examined whether a Ste5Δ49-66 mutant protein that does not accumulate at the shmoo tip (Mahanty et al., 1999) is able to induce shmoos when its defect in MAPK activation is bypassed. Ste5Δ49-66 can bind Gβ (Ste4) and the MAPK cascade kinases, but it does not accumulate at the shmoo tip of signaling competent cells (Mahanty et al., 1999). This defect is due to the loss of amino acids that weakly bind lipids (Winters et al., 2005) and a nuclear localization signal that stimulates cortical recruitment of Ste5 (Mahanty et al., 1999; Wang and Elion, 2003). When combined with a constitutively active Ste11 mutant protein, Ste11T596I (encoded by STE11-4; Stevenson et al., 1992), the Ste5Δ49-66 mutant protein efficiently activates the expression of FUS1-lacZ (Mahanty et al., 1999), suggesting it activates Fus3 and Kss1. Immunoblot analysis of whole cell extracts made from STE11-4 ste5Δ49-66 and STE11-4 STE5 cells treated with α factor for 60 min shows similar levels of active Fus3 and Kss1 (Figure 1B), but there is little shmoo formation in the STE11-4 ste5Δ49-66 strain compared with the STE11-4 STE5 strain (Figure 1C). This result implies that stable association of Ste5 with Gβγ at the plasma membrane is important for shmoo formation.
We also examined whether delocalizing the distribution of Ste5 at the plasma membrane influences shmoo formation, by comparing shmoo formation in STE5 and GAL1-STE5-CTM cells grown in medium containing 2% galactose in the absence or presence of α factor. After 90-min exposure to 50 nM α factor, 92% of the STE5 cells had shmoos (Figure 1, D and E, 2), with 100% of total cells in G1 phase. However, only 17% of the STE5-CTM cells had shmoos after 6 h of galactose induction, even though ∼90% of the cells underwent G1 phase arrest (Figure 1, D and E, 4). The inclusion of α factor to the STE5-CTM cells increased shmoo formation only slightly (Figure 1, D and E, 5), whereas expression of wild-type Ste5 with Ste5-CTM led to 64% of the cells having shmoos (Figure 1, D and E, 6). Collectively, these findings suggest that a polarized distribution of Ste5 at the cell cortex is important for shmoo formation.
Ste5-3xGFP and Ste5-3xYFP Reveal Multiple Random and Polarized Pools of Ste5
Previous work suggests the existence of cortical pools of Ste5 in dividing cells and at very early time points after α factor addition (Wang and Elion, 2003; Wang et al., 2005); however, the cortical pool was difficult to detect. To better detect Ste5 localization, Ste5-triple GFP and Ste5-triple YFP derivatives (STE5-3xGFP, STE5-3xYFP; using triple GFP and triple YFP from Wu and Pollard, 2005) were inserted in place of the chromosomal STE5 gene in S288c and W303a strain backgrounds (see Supplemental Material). The constructs were functional based on the ability of the MATa STE5-3xGFP and MATa STE5-3xYFP strains to arrest in G1 phase, undergo shmoo formation in dose–response experiments and form diploids. In logarithmically dividing cells, it is possible to more readily detect basal recruitment of Ste5 to the plasma membrane, which is visualized as occasional, randomly distributed spots of fluorescence on most cells (Figure 2A, shown is S288c background, similar results found in W303a background). The spots seem to accumulate at the cortex, but they are also found internally within the cytoplasm. A band of cortical accumulation of Ste5-3xGFP and Ste5-3xYFP is also at the junction of a low percentage of budded cells (∼5%), and it seems equivalent to the basal cortical pool detected with GFP-Ste5 in live cells (Wang et al., 2005). The addition of 5 μM α factor for ∼5 min increases the number of densely staining spots of Ste5-3xGFP and Ste5-3xYFP at the plasma membrane; these spots are randomly distributed (Ste5-3xGFP, representative shown) with a coalescence of spots into patches resembling the beginning of cell polarity in some cells (Ste5-3xYFP, representative cell shown) (Supplemental Figure 2 for tally of FUS3 cells). Longer α factor exposure (i.e., 90 min) reveals characteristic polarized shmoo tip staining (see Supplemental Figure 2). These findings reveal multiple pools of Ste5: nonpolarized cortical spots, internal spots, a larger cortical pool at the junction of some large budded cells, and the polarized pool at the emerging shmoo tip in α factor-treated cells.
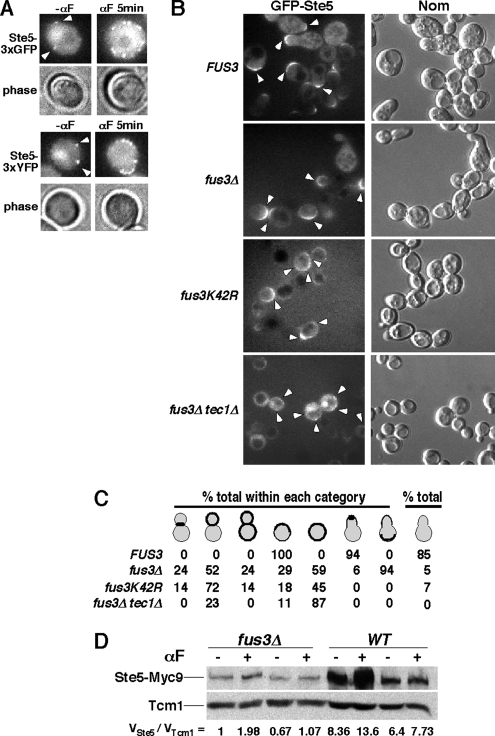
Figure 2.
Fus3 regulates Ste5 recruitment. (A) Random recruitment of Ste5-3xGFP and Ste5-3xYFP to plasma membrane before and after α factor treatment. Ste5-3xGFP (EYL4653) and Ste5-3xYFP (EYL4654) were treated with 5 μM α factor for 5 min. (B) Increased nonpolarized plasma membrane localization of GFP-Ste5 in fus3Δ, fus3K42R and tec1Δ mutants. ste5Δ (EY1775), ste5Δ fus3Δ (EY1774), ste5Δ fus3K42R (EYL4692), and ste5Δ fus3Δ tec1Δ (EYL4685) strains harboring CUP1-GFP-STE5 (pSKM21) were induced with 1 mM CuSO4 for 1 h, and then treated with 50 nM α factor for 90 min. (C) Tally of GFP-Ste5 localization in photographed cells. Thick lines and dots indicate plasma membrane recruitment; gray indicates cytoplasmic pool. Nuclear pool not indicated. (D) Immunoblot analysis of Ste5-Myc9 abundance in FUS3 and fus3Δ strains. Total Ste5 protein was monitored in whole cell extracts from cultures treated as in B. Anti-Myc mAb 9E10 was used to detect Ste5-Myc9. The amount of Ste5-Myc9 was normalized to ribosomal protein Tcm1 by densitometry. VSte5, arbitrary densitometry value of Ste5-Myc9 and VTcm1, arbitrary densitometry value of Tcm1.
Fus3 Prevents Random Distribution of Ste5 at the Cell Cortex during Shmoo Formation
To determine whether Fus3 regulates Ste5 localization, we first compared the localization of GFP-Ste5 in FUS3 ste5Δ and fus3Δ ste5Δ strains. A fus3 mutant does not arrest properly in G1 phase, leading to a mixed population of dividing cells that can have some shmoo morphology (Elion et al., 1990; Farley et al., 1999). Strikingly, more depolarized GFP-Ste5 accumulates across the plasma membrane in fus3Δ mutant cells than in FUS3 cells (Figure 2, B and C). The cortical distribution of GFP-Ste5 is much more depolarized and random in the fus3Δ cells, and it is detected in both budded and unbudded cells. This pattern contrasts the polarized localization of GFP-Ste5 in FUS3 cells that are either unbudded or shmoos (Figure 2C and Supplemental Figure 2). Greater and more depolarized cortical localization of Ste5-3xGFP also occurs in the fus3Δ mutant (see Supplemental Figure 2), and it can be detected at suboptimal α factor induction conditions (i.e., 10 and 25 nM; data not shown) and in multiple strain backgrounds (i.e., S288c and W303a). Collectively, these findings demonstrate that Fus3 inhibits depolarized cortical recruitment of Ste5.
The GFP signal from GFP-Ste5 and Ste5-3xGFP is routinely weaker in fus3Δ cells compared with FUS3 cells, suggesting there is less Ste5 protein in the absence of Fus3. We performed immunoblot analysis of whole cell extracts prepared from fus3Δ and FUS3 strains of a functional Ste5-Myc9 protein expressed under identical conditions as GFP-Ste5, because it is easier to detect than GFP-Ste5. Ste5-Myc9 levels are reduced in the fus3Δ cells compared with FUS3 cells (Figure 2D). Therefore, Fus3 positively regulates Ste5 abundance, and the apparent increase in GFP-Ste5 cortical recruitment in the fus3 strains is likely to be an underestimate due to there being less GFP-Ste5 protein.
The Catalytic Activity of Fus3 Is Required to Down-Regulate Cortical Recruitment of Ste5
To determine whether the catalytic activity of Fus3 is required for down-regulation of Ste5 recruitment, we compared GFP-Ste5 localization in FUS3 ste5Δ and fus3K42R ste5Δ strains during α factor stimulation. The fus3K42R cells display an even greater defect in GFP-Ste5 localization than the fus3Δ cells, with more obvious localization to the cell cortex that is delocalized in both budded and unbudded cells (Figure 2B). Therefore, Fus3 regulates Ste5 localization through its function as a protein kinase.
We looked at GFP-Ste5 in a fus3Δ tec1Δ double mutant to determine whether Fus3 regulates Ste5 localization indirectly through Tec1, which is phosphorylated by Fus3 and then degraded (Bao et al., 2004; Chou et al., 2004). A fus3Δ mutant has elevated levels of active Kss1 as a result of increased Tec1 levels. The fus3Δ tec1Δ double mutant displays somewhat greater depolarized cortical localization of GFP-Ste5 recruitment than the fus3Δ single mutant (Figure 2, B and C). These findings argue that Fus3 regulates cortical recruitment of Ste5 independently of Tec1.
Fus3 Is Essential for Cortical Recruitment of Ste20, Cdc24, and Bni1 during α Factor Stimulation
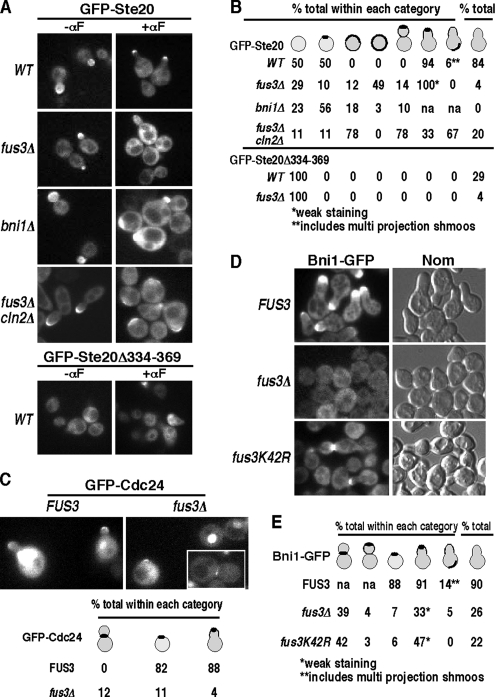
Ste20 binds to Cdc42 through a Cdc42/Rac interactive binding (CRIB) domain within residues 334-369 (Peter et al., 1996; Leberer et al., 2000; Lamson et al., 2002; Ash et al., 2003) that is not required for kinase activity but is required for Cdc42-mediated polarized localization of Ste20 during budding (Lamson et al., 2002) and shmooing (Figure 3, A and B, GFP-Ste20Δ334-369 in FUS3 strain). Bni1 does not control Cdc42-mediated asymmetry during α factor stimulation, because GFP-Ste20 is still polarized in a bni1Δ mutant (Qi and Elion, 2005b). To determine whether Fus3 regulates Cdc42-mediated asymmetry, we directly compared the localization of GFP-Ste20 in fus3Δ and bni1Δ mutants. Strikingly, there is a pronounced defect in polarized recruitment of GFP-Ste20 in unbudded fus3Δ cells treated with α factor (Figure 3, A and B; 10% polarized recruitment in fus3Δ compared with 50% in FUS3 with marginal detection at tip of occasional shmoos), together with an increase in the percentage of unbudded cells that displayed enhanced cortical recruitment around the plasma membrane. This localization pattern is similar to that of GFP-Ste5. In contrast, the cortical localization of GFP-Ste20 is still polarized in a bni1Δ mutant (Figure 3, A and B; 56% of unbudded bni1Δ cells display polarized cortical recruitment of GFP-Ste20 compared with 50% of BNI1 cells), as shown previously (Qi and Elion, 2005b). A slight defect in Ste20 localization is apparent based on a few cells that display slightly broader cortical localization of Ste20; however, this localization was still asymmetrical. These findings reveal that Fus3 regulates Cdc42-mediated asymmetry and put Fus3 upstream of Cdc42 and Ste20 in a localization pathway.
Figure 3.
Fus3 is required for cortical recruitment of Bni1-GFP, GFP-Cdc24, and for polarized recruitment of GFP-Ste20 during pheromone response. (A) Localization of GFP-Ste20 in WT, fus3Δ, bni1Δ, and fus3Δ cln2Δ strains and GFP-Ste20 (Δ334-369) in WT before and after α factor treatment. (B) Tally of cortical pattern of GFP-Ste20 and GFP-Ste20Δ334-369 localization after α factor treatment in A. (C) Fus3 is required for GFP-Cdc24 recruitment. FUS3 and fus3Δ strains expressing GFP-Cdc24 were grown overnight in medium containing 0.19 mM methionine, and then treated with α factor. Tally of cortical pattern of GFP-Cdc24 localization shown below. (D) Recruitment of Bni1-GFP in FUS3, fus3Δ, and fus3K42R cells. (E) Tally of cortical pattern of Bni1-GFP localization. For A–E, cells were treated with 50 nM α factor for 90 min. Nom, Nomarski. Strains: WT (EY957), fus3Δ (EY1095), and bni1Δ (EYL917). Plasmids: GFP-Cdc24 (EBL664), Bni1-GFP (EBL334), and GFP-Ste20 (EBL511).
We compared the localization of GFP-Ste20 in a fus3Δ strain to that of GFP-Cdc24 and Bni1-GFP. Fus3 is essential for polarized cortical recruitment of GFP-Cdc24 during α factor induction (Figure 3C; 11% of unbudded fus3Δ cells compared with 82% of unbudded FUS3 cells), but it has no obvious role in regulating the nuclear or the cortical pools during vegetative growth (data not shown). Fus3 is also essential for cortical localization of Bni1 (Figure 3, D and E; 7% of unbudded fus3Δ cells and 6% of fus3K42R unbudded cells compared with 88% of unbudded FUS3 cells). We detect a greater recruitment defect in fus3Δ cells than reported previously (Matheos et al., 2004), most likely because of lower Bni1-GFP levels (the BNI1-GFP gene has an ADH1 promoter rather than GAL1). A low level of shmoo formation and Bni1-GFP cortical recruitment occurs in both fus3 mutants, but the Bni1-GFP signal is weak compared with that in the FUS3 strain. Bni1-GFP also accumulates at the tip of small buds and at the junction of bi-lobed budded cells similar to its mitotic pattern of localization (Moseley and Goode, 2006). These findings, together with those of Matheos et al., 2004 and Qi and Elion, 2005b, place cortical recruitment of Bni1 downstream of Fus3 and cortical recruitment of Cdc24 downstream of Bni1.
GPA1 Down-Regulates Polarized Cortical Recruitment of GFP-Ste5 and Promotes Shmooing
Gpa1 (Gα) binds to and inhibits Gβγ (Ste4/Ste18), binds to Fus3 (Metodiev et al., 2002), and binds to and stimulates Vps34/Vps15 phosphatidylinositol-3-kinase at the endosome (Slessareva et al., 2006). We determined the likelihood of whether Fus3 down-regulates Ste5 cortical recruitment through Gpa1 as suggested previously for Bni1 (Matheos et al., 2004) by comparing the localization of GFP-Ste5 in gpa1Δ and fus3Δ strains. To prevent unregulated activation of the mating pathway through loss of Gpa1 repression of Gβγ (Ste4/Ste18), we made a ste5Δ gpa1Δ double mutant that conditionally expresses GFP-STE5 from the CUP1 promoter. In the absence of α factor treatment, GFP-Ste5 accumulates in a polarized pattern at the cortex of more cells in the gpa1Δ strain than in the GPA1 strain (Figure 4, A and B). In addition, 10% of gpa1Δ cells resemble shmoos. After 5 min of α factor induction, a stronger GFP-Ste5 signal is detected at the cortex of the gpa1Δ strain, with 20% of the cells resembling shmoos, and most unbudded cells already have polarized cortical localization of GFP-Ste5 (Figure 4, A and B). Longer α factor treatment results in more rapid decline in the intensity of GFP-Ste5 cortical signal in the gpa1Δ strain compared with GPA1 control. The decline in GFP-Ste5 recruitment at the shmoo tip at later time points (i.e., >2 h after α factor addition) is similar to what has been noted previously (Mahanty et al., 1999). Interestingly, fewer gpa1Δ cells exhibit obvious shmoos compared with the GPA1 cells: after 90 min of α factor treatment, 23% of gpa1Δ cells are shmoos compared with 73% of GPA1 cells (Figure 4B). These findings show that Gpa1 is required for optimal shmooing. They support the existence of an active down-regulatory mechanism for Ste5 recruitment, and they suggest that Gpa1 both inhibits and sustains the polarized pool of Ste5 at the shmoo tip. Moreover, given that gpa1Δ and fus3Δ mutations have different effects on GFP-Ste5 localization, the findings also suggest that Gpa1 and Fus3 down-regulate Ste5 by distinct mechanisms.
Figure 4.
Cortical recruitment of GFP-Ste5 and Bni1-GFP does not require GPA1 or FAR1. (A) Localization of GFP-Ste5 in GPA1 and gpa1Δ strains. GPA1 ste5Δ (EY1775) and gpa1Δ ste5Δ (EYL4640) cells harboring CUP1-GFP-STE5 (pSKM21) were grown for 1 h in medium containing 1 mM CuSO4, and then they were induced with 50 nM α factor. (B) Tally of cortical recruitment of GFP-Ste5 in unbudded and shmooing cells after α factor addition. Percentage of shmoos exhibiting GFP-Ste5 tip staining and percentage of shmoos in total cells are shown. (C) Cortical recruitment of Bni1-GFP in GPA1 and gpa1Δ cells. The ste5Δ (EY1775) and gpa1Δ ste5Δ (EYL4640) strains express GAL1p-STE5-CTM (EBL206) and BNI1-GFP (EBL334). Percentage of shmoos with Bni1-GFP tip staining and percentage of shmoos in total cells are shown below. (D) Cortical recruitment of Bni1-GFP in FAR1 and far1Δ cells. FAR1 (EY957) and far1Δ (EY1262) strains expressing Bni1-GFP (EBL334). Cells were treated with 50 nM α factor for 90 min at A600 = 0.4. Tally of Bni1-GFP localization in different categories of cells and percentage of shmoos in total cells are shown below. (E) Recruitment of GFP-Ste5 in FAR1 and far1Δ cells after α factor stimulation. Strains: ste5Δ (EY1775) and ste5Δ far1Δ (EY2019) expressing CUP1-GFP-STE5 (EBL367; pSKM21). Tally of GFP-Ste5 localization in different types of cells and percentage of shmoos in total cells are shown below. (F) Far1 is not required for shmoo formation. FAR1 (EY957) and far1Δ (EY1262) cells expressing GAL1p-STE5-CTM (EBL206) were grown for 6 h in medium containing 2% galactose. Percentage of shmoos in total cells is shown below.
Fus3 Regulates Polarized Recruitment of Bni1 Independently of Gpa1
We tested the hypothesis that Fus3 regulates Bni1 from the pool of active Fus3 that is bound to Gpa1 (Matheos et al., 2004) by examining the localization of Bni1-GFP in a gpa1Δ ste5Δ double mutant that conditionally expresses STE5-CTM from the GAL1 promoter. Bni1-GFP was efficiently localized to the cortex of emerging shmoos in the ste5Δ gpa1Δ GAL1-STE5-CTM strain after Ste5-CTM expression, with even more pronounced shmoo formation occurring in the ste5Δ gpa1Δ GAL1-STE5-CTM strain compared with the ste5Δ GAL1-STE5-CTM strain (Figure 4C). Therefore, Gpa1 does not play a critical role in localization of Bni1, and a different pool of Fus3 must regulate this localization event.
Far1 Is Required for G1 Phase Specificity and Optimal Recruitment of Ste5 and Bni1
Fus3 phosphorylates and stabilizes the Far1 protein. Far1 inhibits Cln/Cdc28 kinase, and it orients polarized growth in the direction of the highest pheromone source by binding to Gβγ (Ste4/Ste18) and the Cdc24/Bem1/Cdc42 complex (Chang, 1993; Bloom and Cross, 2007). To determine whether Fus3 regulates Ste5 and Bni1 recruitment through Far1, we looked at the localization of GFP-Ste5 and Bni1-GFP in a far1Δ strain after α factor treatment for 90 min. A far1 mutant does not arrest properly in G1 phase due to high levels of G1 cyclin-dependent kinase (Cln/Cdc28), leading to a population of cells that are dividing but manifest morphological features resembling pheromone-treated cells (Chang and Herskowitz, 1990; Chang, 1993). Bni1-GFP and GFP-Ste5 were both recruited in a polarized manner to the cortex of unbudded cells and occasional shmoos in addition to being localized at the junction between budding cells (Figure 4, D and E). The polarized localization of GFP-Ste5 contrasted the delocalized pattern in the fus3Δ mutant (Figure 2), which also has high levels of Cln/Cdc28 kinase (Cherkasova et al., 1999). Overexpression of Ste5 bypasses the G1 arrest defect of a far1Δ mutant (Leberer et al., 1993) through Fus3- and Kss1-dependent functions (Elion et al., 1991; Cherkasova et al., 1999). Overexpression of plasma membrane localized Ste5-CTM bypassed the G1 arrest defect of the far1Δ mutant, and it induced cell cycle arrest and shmoo formation in the far1Δ cells as efficiently as in FAR1 cells (Figure 4F). Therefore, Far1 is not required for polarized cortical recruitment of Ste5 and Bni1, although it is critical for G1 phase specificity.
CLN2 Inhibits Polarized Cortical Recruitment of Ste5, Bni1, and Ste20
Prior analysis of fus3 point mutants reveals a strict correlation between G1 arrest and the ability of cells to shmoo (Farley et al., 1999). We assessed the localization of Ste5, Bni1, and Ste20 in a fus3Δ cln2Δ strain, because a cln2Δ mutation restores the greatest level of G1 arrest to a fus3Δ mutant that is bar1Δ (Satterberg, 1994; Cherkasova et al., 1999; Cherkasova and Elion, 2001). Remarkably, the localization of GFP-Ste5 was more polarized in the fus3Δ cln2Δ strain with a significant increase in the percentage of shmoos (Figure 5A). Analysis of GFP-Ste5 localization in a fus3Δ cln1Δ double mutant revealed a more subtle increase in polarized localization of Ste5. These findings suggest that Cln2/Cdc28 normally prevents polarized localization of Ste5 with much less contribution by Cln1/Cdc28.
Figure 5.
Null mutations in CLN1 and CLN2 restore polarized cortical recruitment of GFP-Ste5 and Bni1-GFP in fus3Δ cells. (A) Cortical recruitment of GFP-Ste5 in FUS3, fus3Δ, fus3Δ cln1Δ, and fus3Δ cln2Δ strains after α factor stimulation. FUS3 (EY1775), fus3Δ (EY1774), fus3Δ cln1Δ (EYL4684), and fus3Δ cln2Δ (EYL4649) strains harboring CUP1p-GFP-STE5 (pSKM21) were prepared and analyzed as in Figure 2B. Tally of GFP-Ste5 localization in different categories of cells and percentage of shmoos in total cells are shown below. (B) Recruitment of Bni1-GFP in FUS3, fus3Δ, fus3Δ cln1Δ, and fus3Δ cln2Δ strains after α factor stimulation. FUS3 (EY957), fus3Δ (EY1095), fus3Δ cln1Δ (EY1094), and fus3Δ cln2Δ (EY1093) strains harboring Bni1-GFP (EBL334) were prepared and analyzed as in Figure 4D. Tally of cortical recruitment of Bni1-GFP in different categories of cells and percentage of shmoos in total cells are shown below.
The fus3Δ defect in Bni1-GFP localization was also corrected in the fus3Δ cln2Δ strain, with highly polarized localization of Bni1-GFP occurring at the tip of partially emerged shmoos (Figure 5B). Although there was nearly complete restoration of a polarized localization of Bni1-GFP at the cortex of the fus3Δ cln2Δ cells, the level of cortical recruitment of Bni1-GFP was less than in FUS3 CLN2 cells, and the fus3Δ cln2Δ cells only underwent a partial initiation of polarized growth. In contrast, a more mitotic pattern of polarized localization of Bni1-GFP occurred in the fus3Δ cln1Δ mutant, which does not arrest as efficiently in G1 phase compared with the fus3Δ cln2Δ strain (Figure 5B). The localization of GFP-Ste20 was also compared in fus3Δ and fus3Δ cln2Δ cells, and a similar result was obtained to that of GFP-Ste5; restoration of polarized localization in unbudded cells (Figure 3, A and B). Collectively, these findings argue that Cln2/Cdc28 normally prevents polarized cortical localization of Ste5, Bni1, and Ste20 in G1 phase cells. Because the level of cortical recruitment of GFP-Ste5, Bni1-GFP, and GFP-Ste20 is lower in the fus3Δ cln2Δ strain compared with the FUS3 CLN2 strain and the fus3Δ cln2Δ cells do not form complete shmoo extensions it is possible that Fus3 provides additional functions for cortical recruitment and polarized growth and/or that the remaining G1 cyclins exert inhibitory functions that decrease cortical recruitment and polarized growth.
Hyperelongation in a cdc28-4 Strain Is Blocked by a fus3Δ kss1Δ Double Mutation
To further examine whether Cdc28 prevents onset of shmooing induced by the mating pathway, we examined the morphology of a cdc28-4 strain in the absence or presence of FUS3/KSS1. cdc28-4 is a conditional allele that confers a G1 Start defect (Reed, 1980) by blocking the action of G1 cyclins. We compared the morphology of cdc28-4 and cdc28-4 fus3Δ kss1Δ strains grown at room temperature or shifted to 37°C for 3 h in the absence or presence of α factor. The cells also harbored CUP1-GFP-STE5, which was induced by the inclusion of copper in the medium. At nonpermissive temperature (37°C), the cdc28-4 cells became enlarged and elongated compared with at room temperature, and they underwent hypershmooing in the presence of α factor as shown by very long projections and the presence of cells with more than one projection (Figure 6A; weaker GFP-Ste5 fluorescence signal occurs at high temperature). By contrast, hyperelongation did not occur in the cdc28-4 fus3 kss1 strain at 37°C either in the absence or presence of α factor. The cells were round, similar to cdc28-4 cells grown at room temperature (Figure 6A). Therefore, the mating MAPKs constitutively promote onset of mating morphogenesis and Cdc28 counteracts this polarization. Loss of Cdc28 kinase causes hyperelongation that is mediated by the mating MAPKs.
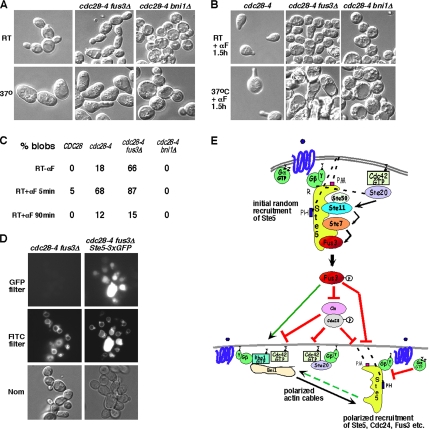
Figure 6.
Inactivation of Cdc28 stimulates cortical recruitment of Ste5. (A) The cdc28-4 mutant has an elongated morphology that is dependent on FUS3 KSS1. Comparison of cell morphology and GFP-Ste5 localization in cdc28-4 (EYL2190) and cdc28-4 fus3Δ kss1Δ (EYL2184) cells expressing CUP1p-GFP-STE5 pregrown at room temperature or 37°C for 3 h followed by 1-h induction in 500 μM CuSO4 to induce GFP-Ste5 expression, followed by further incubation at indicated temperature for 3 h without or with addition of 50 nM α factor. (B) Pattern of cortical recruitment of Ste5-3xGFP in cdc28-4, cdc28-4 fus3Δ, and cdc28-4 bni1Δ cells. Strains cdc28-4 (EYL4705), cdc28-4 fus3Δ (EYL4710), and cdc28-4 bni1Δ (EYL4711) with integrated Ste5-3xGFP were pregrown at room temperature overnight, and then they were shifted to prewarmed 37°C medium for 3 h followed by incubation at same temperature. (C) Pattern of different Ste5-3xGFP pools in cdc28-4, cdc28-4 fus3Δ, and cdc28-4 bni1Δ cells before and after α factor stimulation. Strains cdc28-4 (EYL4705), cdc28-4 fus3Δ (EYL4710), and cdc28-4 bni1Δ (EYL4711) were grown at room temperature. The different arrowheads point to speckles, blobs, and shmoo tip pools of Ste5-3xGFP.
The cdc28-4 Mutation Induces Cortical Recruitment of Ste5
During the course of this analysis, Strickfaden et al., 2007 found that Ste5 is phosphorylated by Cln2/Cdc28 complexes in vitro and that mutation of 8 potential Cdc28 phosphorylation sites (TP/SP) near the PM domain to glutamate (EE) blocks nonpolarized cortical localization of Ste5 that occurs when it is overexpressed in ste4Δ ste7Δ cells. These and other findings led to the elegant proposal that phosphorylation of Ste5 by Cln2/Cdc28 creates negative charges that interfere with binding of the polybasic PM domain to acidic phospholipids. Our prior finding of polarized localization of GFP-Ste5 in a fus3Δ cln2Δ double mutant is consistent with this model. A physiologically relevant prediction of the model of Strickfaden et al. (2007) is that loss of Cdc28 function in G1 phase through a cdc28-4 mutation should increase cortical recruitment of Ste5 to the plasma membrane during mitotic cell division (i.e., in the absence of mating pheromone). In addition, the cdc28-4 mutation should result in more polarized recruitment of Ste5 and polarized growth during α factor stimulation.
We compared cortical speckling of Ste5-3xGFP in isogenic CDC28 and cdc28-4 strains (of W303a background) grown at room temperature and after a 3-h shift to 37°C. The cdc28-4 strain grew more slowly than the CDC28 strain (doubling time of ∼3 h rather than ∼100 min), indicating that the cdc28-4 mutation causes a partial loss of Cdc28 function at permissive temperature. Remarkably, there were more cortical speckles of Ste5-3xGFP in cdc28-4 cells grown at room temperature compared with the CDC28 cells (Figure 6B). Shifting the cells to 37°C for 3 h increased slightly the ability to detect cortical speckling in the CDC28 strain; however, even more cortical speckling of Ste5-3xGFP occurred in the cdc28-4 strain (Figure 6B; note that overall intensity of the GFP signal is somewhat reduced at 37°C compared with at room temperature). The increase in cortical speckling was detected in both unbudded and budded cdc28-4 cells. These findings support the proposal that Cdc28 inhibits basal recruitment of Ste5 and also suggest that Cdc28 prevents Ste5 recruitment throughout the cell cycle.
Strikingly, even greater Ste5-3xGFP cortical speckling was detected in a cdc28-4 fus3Δ strain compared with the cdc28-4 strain at both room temperature and 37°C (Figure 6, B and C). When grown at room temperature, cdc28-4 cells are enlarged compared with CDC28 cells, as expected for reduced G1 cyclin-dependent kinase (Figure 6B). Interestingly, the cdc28-4 fus3Δ cells are smaller than the cdc28-4 cells (Figure 6, B and C), consistent with a basal repressive role for Fus3 inhibition of Cln/Cdc28 (Cherkasova et al., 1999). The simultaneous loss of Cdc28 and Fus3 after the 37°C shift also led to an increase in coalesced speckles in the cdc28-4 fus3Δ strain compared with the cdc28-4 strain. The additive effect of cdc28-4 and fus3Δ mutations indicates that Fus3 inhibits Ste5 cortical recruitment during mitotic growth independently of any role it might have in inhibition of Cdc28.
The increase in Ste5 cortical recruitment in the cdc28-4 fus3Δ strain compared with the cdc28-4 strain was also apparent after 5-min exposure with α factor, with the amount of cortical speckling following the order CDC28 FUS3 < cdc28-4 FUS3 < cdc28-4 fus3 (data not shown). Loss of Cdc28 function led to a more pronounced cortical recruitment of Ste5-3xGFP at the shmoo tip of cdc28-4 cells compared with CDC28 cells after 90 min α factor treatment, although random cortical speckles remained in the cdc28-4 fus3Δ strain (Figure 6C). Furthermore, the loss of Cdc28 function reversed the fus3Δ polarization defect: Although the fus3Δ mutation still blocked cell elongation at room temperature (Figures 6B and Figure 7A, compare cdc28-4 with cdc28-4 fus3Δ), some cell elongation and shmoo-like morphologies are detected after the 37°C temperature shift (Figures 6B and 7A). In addition, very obvious hyperelongation and shmooing occurs after 90-min exposure to α factor (Figures 6C and 7B, compare cdc28-4 and cdc28-4 fus3Δ). The cdc28-4 mutation restores more polarized growth to the fus3Δ strain than does the cln2Δ mutation (compare Figure 7, A and B, with Figure 5A). However, Ste5-3xGFP is still more broadly distributed at the shmoo tip cortex, and the shmoos are broader in the cdc28-4 fus3Δ cells compared with the cdc28-4 cells (Figures 6C and 7B), indicating that Fus3 is needed for polarization apart from its role in Cdc28 inhibition. Collectively, these results substantiate the interpretation that Fus3 and Cdc28 provide opposing morphogenesis functions during vegetative growth and shmooing. In addition, they show that Fus3 down-regulates Ste5 cortical recruitment through a function separate from inhibition of Cln/Cdc28.
Figure 7.
Hyperactive shmoo formation in cdc28-4 strains depends on Bni1. (A and B) Morphology of cdc28-4, cdc28-4 fus3Δ, and cdc28-4 bni1Δ strains at room temperature and 37°C either without (A) or with (B) α factor treatment. (C) Percentage of blobs in CDC28, cdc28-4, cdc28-4 fus3Δ, and cdc28-4 bni1Δ strains in different conditions. (D) FM4-64 staining of cdc28-4 fus3Δ strain (EYL4704) with or without integrated STE5-3xGFP grown at room temperature. Nom, Nomarski pictures of corresponding fields are shown in the bottom panel. (E) Cartoon summarizing genetic interactions. Fus3 regulates polarized recruitment of Ste5 and cell polarity proteins through inhibition of G1 cyclin dependent kinase (Cln/Cdc28), which inhibits polarized recruitment of Ste5 and cell polarity proteins in G1 phase. Fus3 has a distinct function(s) that down-regulates cortical recruitment of Ste5 and prevents random accumulation at plasma membrane. Gpa1 negatively regulates cortical recruitment of Ste5. Polarized localization of Ste5, Cdc24, and Fus3 is dependent upon polarization of the actin cytoskeleton by Bni1. Polarization of Ste5 positively stimulates polarized growth (dotted green arrow).
Ste5-3XGFP Accumulates in FM4-64-staining Structures in cdc28-4 and cdc28-4 fus3Δ Cells
The analysis of Ste5-3xGFP in cdc28-4 and cdc28-4 fus3Δ strains revealed the presence of Ste5 in two internal pools, one pool that was typically round and another pool that was always much larger in diameter and could be more irregularly shaped (Figure 6C). The round internal pool was nuclear based on DAPI staining of live cells, and it was consistent with previous work showing increased nuclear accumulation of Ste5 in a cdc28-4 strain (Mahanty et al., 1999). The second internal pool was novel and has not been noted previously. It consisted of large irregularly shaped areas of fluorescence or chains of areas of fluorescence that will be referred to as Ste5-3xGFP blobs. The blobs were most readily detected in cdc28-4 fus3Δ cells and could be detected in either rich YPD medium or minimal low fluorescence medium (Supplemental Figure 3, A and B; representative examples are shown in the 5 min α factor panels for cdc28-4 and cdc28-4 fus3Δ of Figure 6C and tallied at room temperature in Figure 7C. The GFP signal is weaker for all Ste5-3xGFP pools at 37°C). The Ste5-3xGFP blobs are not readily detected in the CDC28 strain at room temperature but are readily detected in ∼18–21% of cdc28-4 cells grown in minimal low fluorescence medium either at room temperature or after 37°C shift. By contrast, the Ste5-3xGFP blobs are detected in the majority of cdc28-4 fus3Δ cells grown at room temperature (66%; Supplemental Figure 3, A and B, and Figure 7C, GFP; and D). The addition of α factor causes a dramatic increase in the intensity of the blobs in the cdc28-4 fus3Δ cells at the 5-min time point (i.e., 87% cells show obvious blobs; Figures 6C and 7C room temperature; Supplemental Figure 3, A and B).
The presence of the Ste5-3xGFP blobs is transient during the course of α factor stimulation. Although the blobs are readily detected after 5 min α factor treatment, they disappear entirely or are significantly less prominent after 90 min α factor treatment in most cdc28-4 and cdc28-4 fus3Δ cells at room temperature (Figures 6C and 7C and Supplemental Figure 3, A and B). Thus, loss of Fus3 and addition of α factor promotes accumulation of Ste5 in the blobs, but this accumulation (or ability to detect the accumulation) is transient during α factor stimulation. Furthermore, GFP-Ste5 does not seem to accumulate in blobs in a cdc28-4 fus3Δ kss1Δ strain that lacks Kss1 (Figure 6A), suggesting that Kss1 is required for accumulation of Ste5 within the blobs.
We explored the identity of the Ste5-3xGFP blobs. The blobs were internal rather than cortical based on imaging numerous cells with different exposure times and planes of view. The blobs were unlikely to be nuclear based on staining with 4,6-diamidino-2-phenylindole (DAPI) and Hoescht 33342 (which was possible to do in live cells despite some overlap of the emission spectra of the Ste5-3xGFP with DAPI and Hoescht 33342 and poor retention of both nuclear stains). The blobs did not represent high autofluorescence of cells that were dying based on phloxine b staining or autofluorescence from internal fluorescent pools based on analysis of an isogenic strain that lacked GFP (Figure 7D, top left). To determine whether the blobs represented an endocytic or vacuolar pool of Ste5-3xGFP, we stained live cells with FM4-64, a lipophilic styryl dye that is taken up via the endocytic pathway and stains the membranes of acidic compartments including vacuole and endosomes in vacuolar sorting mutants (Vida and Emr, 1995; Rieder et al., 1996). Strikingly, there is a 100% correspondence between Ste5-3xGFP blob fluorescence being within the outlines of FM4-64 staining membranes, regardless of whether the blobs are weakly or strongly fluorescent. The localization of blobs within the confines of FM4-64 membranes is easiest to visualize in the cdc28-4 fus3Δ cells that exhibit strong blob fluorescence (Figure 7D). Therefore, loss of Cdc28 and Fus3 function leads to accumulation of Ste5-3xGFP in FM4-64 compartments that may be vacuoles or endocytic vesicles.
Bni1 Is Required for Cortical and Blob Accumulation of Ste5-3xGFP and Shmoo Formation in a cdc28-4 Strain
Recent studies would argue that the actin cytoskeleton is not important for targeting Ste5 to the plasma membrane because Ste5 associates directly through the PM and PH-like domains (Winters et al., 2005; Garrenton et al., 2006). We reexamined the role of Bni1 in random and polarized cortical recruitment of Ste5 and whether it regulates Ste5 recruitment independently of Cdc28 by looking at Ste5-3xGFP in a cdc28-4 bni1Δ strain. Loss of Bni1 reduced the level of basal cortical speckling of Ste5-3xGFP in the cdc28-4 strain both at room temperature and after a 37°C shift or 5 min α factor treatment, and it completely blocked polarized cortical localization of Ste5-3xGFP and shmoo formation after 90 min α factor treatment (Figures 6C and 7B). In addition, the internal Ste5-3xGFP blobs are completely blocked from accumulating (Figures 6C and 7C). Therefore, Bni1 regulates cortical recruitment of Ste5 speckles, accumulation of Ste5 in FM4-64 structures in addition to polarized recruitment. These findings place Bni1 downstream of Cdc28 and upstream of Ste5 in a localization pathway and suggest that Bni1 or the actin cytoskeleton is required for localization of Ste5 to the vacuole.
DISCUSSION
A summary of the genetic interactions we have found to influence cortical localization and polarity during α factor stimulation is shown in Figure 7E. We find that the MAPK Fus3 globally regulates plasma membrane localization of several key proteins required for mating MAPK activation and polarized morphogenesis (i.e., Ste5, Cdc24, Ste20, and Bni1; Figures 3 and 4). Fus3 acts early in the hierarchy of cell polarization, because it regulates Cdc42-mediated asymmetry. This interpretation is based on use of Ste20 CRIB domain as a reporter for Cdc42 polarity (Figure 3) and pleiotropic effects of the fus3Δ mutation on localization of multiple proteins that associate with Cdc42 (i.e., Ste20, Bni1, and Cdc24). By contrast, Cdc42-mediated asymmetry is still maintained in a bni1Δ mutant, which puts Bni1 downstream of both Cdc42 and Fus3 in the hierarchy of control that ultimately leads to polarized localization of Ste5, Fus3, and Cdc24 (Qi and Elion, 2005b; Figure 7E). The outcome of this regulatory device is a complete dependence of mating morphogenesis on the level of Fus3 activation, a biological logic that links pathway specificity to entry into differentiation.
Much of the control of polarity by Fus3 occurs through inhibition of Cln/Cdc28 kinase that normally drives the G1-to-S phase transition and onset of budding (Figures 6 and 7). Our analysis reveals a strong counteractive control mechanism by Cln/Cdc28 kinase that blocks the assembly of a mating morphogenesis machinery in unbudded dividing cells in the absence of significant levels of Fus3 activation (Figures 5, 6, and 7). Implicit in this model (Figure 7E) is the notion that Cln/Cdc28 serves as a master gatekeeper of onset of mating differentiation in dividing cells by blocking stable recruitment of Ste5 together with Bni1 and Ste20. This interpretation is based on the observation that polarized localization of Ste5, Ste20 and Bni1 is restored to a fus3Δ mutant by a null mutation in the CLN2 cyclin (Figures 3 and 5) and that greater suppression of the fus3Δ defect occurs with a cdc28-4 mutation compared with individual cln2Δ and cln1Δ mutations (Figures 6 and 7). Further strong support comes from the remarkable observation that loss of Cdc28 function at permissive or restrictive temperatures is sufficient to induce random cortical recruitment of Ste5 in the absence of α factor (Figure 6, B and C). That the increase in random cortical recruitment of Ste5 also occurs in budding cdc28-4 cells at nonpermissive temperature, raises the possibility that Clb/Cdc28 kinases may perform similar functions as the G1 phase Cln/Cdc28 kinases to block mating morphogenesis in S, G2, and M phases. A second possibility is that there is a reduction in the inhibitory action of Clb2/Cdc28 on SBF, which controls transcription of CLN1 and CLN2 genes, thus leading to CLN1,2 gene expression outside of the normal cell cycle window (Bloom and Cross, 2007). The restoration of α factor-induced shmooing to a cdc28-4 fus3Δ strain, coupled with the absence of shmooing in a cdc28-4 bni1Δ strain (Figures 6 and 7), argues that the loss of Cdc28 function restores polarized localization of multiple polarity proteins in addition to Ste5.
The simplest interpretation of our findings is that Cln2/Cdc28 kinase, and to lesser extent Cln1/Cdc28 kinase, directly phosphorylates Ste5, Bni1, Ste20, and possibly other targets, and in so doing, either prevents their recruitment to the plasma membrane or makes the proteins less able to form mating-specific complexes that promote MAPK activation and shmoo formation rather than budding. The prevention of cortical recruitment could occur through direct interference with the mechanism of recruitment such as has been hypothesized for Ste5 (Strickfaden et al., 2007), or by an indirect effect. For Bni1, no direct evidence of a link to Cdc28 has been established; however, Bni1 does have consensus Cdc28 phosphorylation sites. Fus3 has been thought to directly phosphorylate Bni1 from a pool associated with Gpa1 (Gα) (Matheos et al., 2004); however, our results raise the possibility that another pool of Fus3 controls Bni1 or else the control is indirectly through Cdc28 inhibition. Our new findings are still consistent with our previous speculation that mating pheromone may engage Ste4 to bind to Rho1 and recruit Bni1 (Qi and Elion, 2005b). We have analyzed the phenotype of a Bni1KK492,493EE mutated in the postulated Fus3 docking site and of Bni1S1344A mutated in the best potential MAPK phosphorylation site, and we have not found obvious defects in actin polarization, shmoo formation or quantitative mating (Qi and Elion, unpublished observations). Further work is needed to clarify the role of Fus3 in phosphorylation of Bni1. For Ste20, there is ample evidence of direct phosphorylation by Cln/Cdc28 kinase both in vitro and in vivo (Oehlen and Cross, 1998; Wu et al., 1998; Oda et al., 1999). However, the full spectra of Cdc28 phosphorylation sites in Ste20 (Oda et al., 1999; Gruhler et al., 2005) have not yet been tested for their importance in localization. In addition, Cdc28 is known to phosphorylate Cdc24 in such a way as to stimulate its role in bud emergence (McCusker et al., 2007). Therefore, loss of Cdc28 phosphorylation by Fus3 inhibition could make Cdc24 more competent for mating complexes such as with Far1 or Ste5.
There is ample evidence that Ste5 is a substrate of Cln2/Cdc28 kinase in vitro (Strickfaden et al., 2007). These findings are consistent with our previous work that basal phosphorylation of Ste5 during vegetative growth is blocked in a cdc28-4 strain at nonpermissive temperature (Flotho et al., 2004). Moreover, Cdc28-dependent phosphorylation still occurs when Ste5 is kept in the cytoplasm by a ste5Δ49-66 nuclear localization signal (NLS) mutation, suggesting that Cdc28 phosphorylates Ste5 in the cytoplasm (Flotho et al., 2004), in support of the model of Strickfaden et al., 2007. Although the elegant study of Strickfaden et al., 2007 shows that mutation of putative Cdc28 sites surrounding the PM/NLS domains blocks signal transduction, evidence to show that Cdc28 phosphorylation has an effect on Ste5 localization under physiological conditions has been lacking. Furthermore, an alternative reasonable interpretation is that the mutations nonspecifically interfere with Ste5 function. Our finding that the cdc28-4 mutation stimulates random cortical recruitment of Ste5 provides the first in vivo support for the model proposed by Strickfaden et al., 2007 and fits with the expectation of initial recruitment being dependent on PM domain stabilization of a Ste5 interaction with Gβγ that is not initially polarized during isotropic α factor exposure. Ste5 accumulates in nuclei of cdc28-4 cells (Mahanty et al., 1999; Figure 6). This nuclear localization could be due to a greater pool of free Ste5 that accumulates in the absence of binding to membranes, or from a loss of Cdc28 phosphorylation interfering with recognition of Ste5 for nuclear export or dissociation by the importin (most likely Kap95; Mahanty et al., 1999) that binds the NLS overlapping PM.
Several other lines of evidence support the notion that Cdc28 prevents onset of mating morphogenesis during cell division through inhibitory effects on Ste5 localization and other proteins such as Bni1. First, overexpression of CLN2 and CLN1 inhibits activity through the mating MAPK cascade (Oehlen and Cross, 1994) near the Ste11 MAPKKK step (Wassmann and Ammerer, 1997). Second, there is a strict correlation between the ability of a variety of fus3 mutants to undergo G1 arrest and to induce shmoo formation that is not found for other outputs such as transcriptional activation (Farley et al., 1999). Third, stable polarized localization of Ste5 is important for shmoo formation, and it can be considered a rate-limiting inducer of the process (Figure 1). Fourth, in a cdc28-4 strain, cell elongation in the absence of α factor and α factor-induced hypershmooing are both dependent upon Fus3 and Kss1 (Figure 6A). A prediction of our findings is that cells that are in stationary phase with low levels of Cdc28 kinase (Mendenhall et al., 1987) should be poised for shmooing and mating. This prediction is borne out by the observation of polarized cortical recruitment of Ste5 in unbudded fus3Δ cells entering stationary phase (Yu and Elion, unpublished data) and of intratetrad ascospore mating (Taxis et al., 2005; Knop, 2006). Thus, the control mechanism seems to be designed to protect dividing cells in conditions of nutrients to continue to divide rather than mate. Interestingly, it has been found that the S. pombe Ste11 transcription factor required for mating is only active in G1 phase when Cdc2 levels are lowest, and they preclude inhibitory phosphorylation of Ste11 (Kjaerulff et al., 2006). Our findings raise the possibility that this control mechanism could be enforced through a mating-specific control loop that inhibits Cdc2 in G1 phase.
Our results reveal multiple pools of Ste5 that include cortical speckles, nuclear, cytosolic, shmoo tip, and FM4-64 compartment. Ste5 may regulate distinct pools of associated proteins within these pools or be differentially regulated at these sites. For example, both Bni1 and Stt4 localize in cortical speckles at the cell cortex (Audhya and Emr, 2002; Yu and Elion, unpublished data). We find that the initial cortical recruitment is random in unbudded and budding cells (Figure 2). This finding is consistent with the fact that MAPK signaling is not restricted to unbudded cells (Oehlen and Cross, 1994; Wassmann and Ammerer, 1997). The coalescence of Ste5-3xGFP cortical speckles into larger areas after brief (5 min) α factor treatment in CDC28 FUS3 cells (Figure 2A) and after a 37°C shift in cdc28-4 fus3Δ cells (Figure 6B) suggests that cooperative interactions may occur either at the level of Ste5–Ste5 interactions or interactions between other signaling components that influence Ste5, such as the receptor and G protein.
Our results reveal a potent role for Fus3 in down-regulation of cortical recruitment of Ste5. The potent role of Fus3 in down-regulation of Ste5 cortical recruitment is particularly apparent in the cdc28-4 fus3Δ double mutant where it is easy to visualize a large pool of Ste5 initially accumulating at the cell cortex by 5 min of α factor treatment (Supplemental Figure 6B). Fus3-regulated polarized recruitment of Ste5 still occurs in gpa1Δ cells (Figure 4), suggesting Fus3 down-regulation of polarized cortical recruitment of Ste5 it is not strictly dependent on Gpa1. Gpa1 plays a role in down-regulating Ste5 cortical localization (Figure 4); further work is required to know whether this control involves Fus3. The down-regulation of Ste5 cortical recruitment by Fus3 occurs by a mechanism that is distinct from control of Ste5 abundance (Figure 2D; Flotho et al., 2004). It is also independent of Fus3 inhibition of Cln/Cdc28 kinase, because it still operates in a far1Δ mutant (Figure 4) that has as high levels of CLN1/CLN2 mRNA and Cln/Cdc28 kinase activity as in a fus3Δ mutant (Cherkasova et al., 1999). Fus3 control of Ste5 localization could be direct. Fus3 phosphorylates Ste5 in vitro at multiple sites (Kranz et al., 1994; Flotho et al., 2004; Kranz and Elion, unpublished data). Only threonine 287 has been demonstrated to be a bona fide site in vitro (Bhattacharyya et al., 2006). Further work is required to determine the significance of the full spectrum of phosphorylations; however, it is conceivable that Fus3 phosphorylation of Ste5 at threonine 287 mediates part of the down-regulatory recruitment event we have described. It is also conceivable that some of the sites overlap those recognized by Cln2/Cdc28 due to overlap in the recognition motifs. The Ste2 receptor is internalized by an endocytotic pathway (Jenness and Spatrick, 1986) that is important for shmoo formation (Vallier et al., 2002), but further work is required to determine whether Ste2 is internalized with Ste5 and other linked components. The apparent vacuolar localization of Ste5-3xGFP in cdc28-4 and cdc28-4 fus3Δ cells that is dependent upon Bni1 could suggest that Ste5 is down-regulated by actin-mediated endocytic targeting to the vacuole in CDC28 (wild-type) cells. An alternative view is that the loss of multiple phosphorylation events destabilizes Ste5 and leads to its accumulation in the vacuole for proteolysis. Such a control could be specific for Ste5 or part of an autophagic response normally activated under conditions of nutrient stress (Yorimitsu and Klionsky, 2005). The absence of Ste5 accumulation in FM4-64 structures in a cdc28-4 fus3Δ kss1Δ strain raises the possibility that the enhanced levels of active Kss1 in the cdc28-4 fus3Δ strain drive Ste5 into the FM4-64 structures. Although further work is needed to define the mechanism of Fus3 down-regulation of Ste5 localization, it seems likely that it is important for polarized growth, given that disruption of polarized localization of Ste5 interferes with shmoo formation (Figure 1).
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by NIGMSRO1 46962 and Milton Fund grants (to E.A.E.) and by the Department of Biological Chemistry and Molecular Pharmacology for access to the Nikon Imaging Center in November 2007. We thank Pablo Marina Losada for help with imaging software and microscopes and Annette Flotho for constructing several yeast strains. We thank Tom Pollard and Jian-Qiu Wu for triple GFP and triple YFP plasmids, Mark Longtine and Kurt Thorn for FP plasmids, and Jennifer Waters and Lara Petrak for assistance in the Department of Cell Biology Microscope Facility (Nikon Imaging Center). We are also grateful to Dan Klionsky for helpful discussion, Riki Eggert for Hoescht 33342, and Thierry Doan and David Rudner for FM4-64.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-08-0757) on February 6, 2008.
REFERENCES
- Amberg D. C., Burke D., Strathern J. N. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- Audhya A., Emr S. D. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc-1-mediated MAP kinase cascade. Dev. Cell. 2002;2:593–605. doi: 10.1016/s1534-5807(02)00168-5. [DOI] [PubMed] [Google Scholar]
- Ash J., Wu C., Larocque R., Jamal M., Stevens W., Osborne M., Thomas D. Y., Whiteway M. Genetic analysis of the interface between Cdc42p and the CRIB domain of Ste20p in Saccharomyces cerevisiae. Genetics. 2003;163:9–20. doi: 10.1093/genetics/163.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough K. R., Drubin D. G. A role for the yeast actin cytoskeleton in pheromone receptor clustering and signalling. Curr. Biol. 1998;8:927–930. doi: 10.1016/s0960-9822(07)00374-0. [DOI] [PubMed] [Google Scholar]
- Bao M. Z., Schwartz M. A., Cantin G. T., Yates J. R., 3rd, Madhani H. D. Pheromone-dependent destruction of the Tec1 transcription factor is required for MAP kinase signaling specificity in yeast. Cell. 2004;119:991–1000. doi: 10.1016/j.cell.2004.11.052. [DOI] [PubMed] [Google Scholar]
- Bardwell L. A walk-through of the yeast mating pheromone response pathway. Peptides. 2005;26:339–350. doi: 10.1016/j.peptides.2004.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya R. P., Reményl A., Good A., M. C., Bashor C. J., Fallck A. M., Lim W. A. The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science. 2006;311:822–826. doi: 10.1126/science.1120941. [DOI] [PubMed] [Google Scholar]
- Bloom J., Cross F. R. Multiple levels of cyclin specificity in cell-cycle control. Nat. Rev. Mol. Cell Biol. 2007;8:149–160. doi: 10.1038/nrm2105. [DOI] [PubMed] [Google Scholar]
- Chang F. Stop that cell cycle. Curr. Biol. 1993;3:693–695. doi: 10.1016/0960-9822(93)90071-u. [DOI] [PubMed] [Google Scholar]
- Chang F., Herskowitz I. Identification of a gene necessary for cell cycle arrest by a negative growth factor of yeast: FAR1 is an inhibitor of a G1 cyclin, CLN2. Cell. 1990;63:999–1011. doi: 10.1016/0092-8674(90)90503-7. [DOI] [PubMed] [Google Scholar]
- Chang F., Peter M. Yeast make their mark. Nat. Cell Biol. 2005;3:294–299. doi: 10.1038/ncb0403-294. [DOI] [PubMed] [Google Scholar]
- Cherkasova V., Elion E. A. far4, far5, and far6 define three genes required for efficient activation of MAPKs Fus3 and Kss1 and accumulation of glycogen. Curr. Genet. 2001;40:13–26. doi: 10.1007/s002940100217. [DOI] [PubMed] [Google Scholar]
- Cherkasova V., Lyons D. M., Elion E. A. Fus3p and Kss1p control G1 arrest in Saccharomyces cerevisiae through a balance of distinct arrest and proliferative functions that operate in parallel with Far1p. Genetics. 1999;151:989–1004. doi: 10.1093/genetics/151.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S., Huang L., Liu H. Fus3-regulated Tec1 degradation through SCFCdc4 determines MAPK signaling specificity during mating in yeast. Cell. 2004;119:981–990. doi: 10.1016/j.cell.2004.11.053. [DOI] [PubMed] [Google Scholar]
- Dohlman H. G. G proteins and pheromone signaling. Annu. Rev. Physiol. 2002;64:129–152. doi: 10.1146/annurev.physiol.64.081701.133448. [DOI] [PubMed] [Google Scholar]
- Elion E. A., Brill J. A., Fink G. R. Functional redundancy in the yeast cell cycle: FUS3 and KSS1 have both overlapping and unique functions. Cold Spring Harb. Symp. Quant. Biol. 56:41–49. doi: 10.1101/sqb.1991.056.01.007. [DOI] [PubMed] [Google Scholar]
- Elion E. A., Grisafi P. L., Fink G. R. FUS3 encodes a cdc2+/CDC28-related kinase required for the transition from mitosis into conjugation. Cell. 1990;60:649–664. doi: 10.1016/0092-8674(90)90668-5. [DOI] [PubMed] [Google Scholar]
- Elion E. A., Satterberg B., Kranz J. E. FUS3 phosphorylates multiple components of the mating signal transduction cascade: evidence for STE12 and FAR1. Mol. Biol. Cell. 1993;4:495–510. doi: 10.1091/mbc.4.5.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista Zigmond M. S., Boone C. Formins: signaling effectors for assembly and polarization of actin filaments. J. Cell Sci. 2003;116:2603–2611. doi: 10.1242/jcs.00611. [DOI] [PubMed] [Google Scholar]
- Farley F., Satterberg B., Goldsmith E. A., Elion E. A. Relative dependance of different outputs of the Saccharomyces cerevisiae pheromone response pathway on the MAP kinase Fus3. Genetics. 1999;151:1425–1444. doi: 10.1093/genetics/151.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Song L. Y., Kincaid E., Mahanty S. K., Elion E. A. Functional binding between Gbeta and the LIM domain of Ste5 is required to activate the MEKK Ste11. Curr. Biol. 1998;8:267–278. doi: 10.1016/s0960-9822(98)70108-3. [DOI] [PubMed] [Google Scholar]
- Flotho A., Simpson D. M., Qi M., Elion E. A. Localized feedback phosphorylation of Ste5p scaffold by associated MAPK cascade. J. Biol. Chem. 2004;279:47391–47401. doi: 10.1074/jbc.M405681200. [DOI] [PubMed] [Google Scholar]
- Garrenton L. S., Young S. L., Thorner J. Function of the MAPK scaffold protein, Ste5, requires a cryptic PH domain. Genes Dev. 2006;20:1946–1958. doi: 10.1101/gad.1413706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E. How do Rho family GTPases direct axon growth and guidance? A proposal relating signaling pathways to growth cone mechanics. Differentiation. 2002;70:385–396. doi: 10.1046/j.1432-0436.2002.700801.x. [DOI] [PubMed] [Google Scholar]
- Gruhler A., Olsen J. V., Mohammed S., Mortensen P., Faergeman N. J., Mann M., Jensen O. N. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell Proteomics. 2005;4:310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- Gulli M. P., Peter M. Temporal and spatial regulation of Rho-type guanine-nucleotide exchange factors: the yeast perspective. Genes Dev. 2001;15:365–379. doi: 10.1101/gad.876901. [DOI] [PubMed] [Google Scholar]
- Guo W., Pylayeva Y., Pepe A., Yoshioka T., Muller W. J., Inghirami G., Giancotti F. G. Beta 4 integrin amplifies ErbB2 signaling to promote mammary tumorigenesis. Cell. 2006;126:489–502. doi: 10.1016/j.cell.2006.05.047. [DOI] [PubMed] [Google Scholar]
- Huang C., Jacobson K., Schaller M. D. MAP kinases and cell migration. J Cell Sci. 2004;117:4619–4628. doi: 10.1242/jcs.01481. [DOI] [PubMed] [Google Scholar]
- Iijima M., Huang Y. E., Devreotes P. Temporal and spatial regulation of chemotaxis. Dev. Cell. 2002;3:469–478. doi: 10.1016/s1534-5807(02)00292-7. [DOI] [PubMed] [Google Scholar]
- Inouye C., Dhillon N., Thorner J. Ste5 RING-H2 domain: role in Ste4-promoted oligomerization for yeast pheromone signaling. Science. 1997;278:103–106. doi: 10.1126/science.278.5335.103. [DOI] [PubMed] [Google Scholar]
- Jenness D. D., Spatrick P. Down regulation of the alpha-factor pheromone receptor in S. cerevisiae. Cell. 1986;46:345–353. doi: 10.1016/0092-8674(86)90655-0. [DOI] [PubMed] [Google Scholar]
- Johnson D. I. Cdc 42, An essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 1999;63:54–105. doi: 10.1128/mmbr.63.1.54-105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjaerulff S., Andersen N. R., Borup M. T., Nielsen O. Cdk phosphorylation of the Ste11 transcription factor constrains differentiation-specific transcription to G1. Genes Dev. 2006;21:347–359. doi: 10.1101/gad.407107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M. Evolution of the hemiascomycete yeasts: on life styles and the importance of inbreeding. Bioessays. 2006;28:696–708. doi: 10.1002/bies.20435. [DOI] [PubMed] [Google Scholar]
- Kranz J. E., Satterberg B., Elion E. A. The MAP kinase Fus3 associates with and phosphorylates the upstream signaling component Ste5. Genes Dev. 1994;8:313–327. doi: 10.1101/gad.8.3.313. [DOI] [PubMed] [Google Scholar]
- Lamson R. E., Winters M. J., Pryciak P. M. Cdc42 regulation of kinase activity and signaling by the yeast p21-activated kinase Ste20. Mol. Cell Biol. 2002;22:2939–2951. doi: 10.1128/MCB.22.9.2939-2951.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E., Dignard D., Thomas D. Y., Leeuw T. A conserved Gbeta binding (GBB) sequence motif in Ste20p/PAK family protein kinases. J. Biol. Chem. 2000;381:427–431. doi: 10.1515/BC.2000.055. [DOI] [PubMed] [Google Scholar]
- Leberer E., Dignard D., Harcus D., Hougan L., Whiteway M., Thomas D. Y. Cloning of Saccharomyces cerevisiae STE5 as a suppressor of a Ste20 protein kinase mutant: structural and functional similarity of Ste5 to Far1. Mol. Gen. Genet. 1993;241:241–254. doi: 10.1007/BF00284675. [DOI] [PubMed] [Google Scholar]
- Leof E. B. Growth factor receptor signalling: location, location, location. Trends Cell Biol. 2000;10:343–348. doi: 10.1016/s0962-8924(00)01795-5. [DOI] [PubMed] [Google Scholar]
- Mahanty S. K., Wang Y., Farley F. W., Elion E. A. Nuclear shuttling of yeast scaffold Ste5 is required for its recruitment to the plasma membrane and activation of the mating MAPK cascade. Cell. 1999;98:501–512. doi: 10.1016/s0092-8674(00)81978-9. [DOI] [PubMed] [Google Scholar]
- Marsh L., Neiman A. M., Herskowitz I. Signal transduction during pheromone response in yeast. Annu. Rev. Cell. Dev. Biol. 1999;7:699–728. doi: 10.1146/annurev.cb.07.110191.003411. [DOI] [PubMed] [Google Scholar]
- Matheos D., Metodiev M., M., Muller E., Stone D., Rose M. D. Pheromone-induced polarization is dependent on the Fus3p MAPK acting through the formin Bni1p. J. Cell Biol. 2004;165:99–109. doi: 10.1083/jcb.200309089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker D., Denison C., Anderson S., Egelhofer T. A., Yates J. R., 3rd, Gygi S. P., Kellogg D. R. Cdk1 coordinates cell-surface growth with the cell cycle. Nat. Cell Biol. 2007;9:506–515. doi: 10.1038/ncb1568. [DOI] [PubMed] [Google Scholar]
- Mendenhall M. D., Jones C. A., Reed S. I. Dual regulation of the yeast CDC28–p40 protein kinase complex: cell cycle, pheromone, and nutrient limitation effects. Cell. 1987;50:927–935. doi: 10.1016/0092-8674(87)90519-8. [DOI] [PubMed] [Google Scholar]
- Metodiev M. V., Matheos D., Rose M. D., Stone D. E. Regulation of MAPK function by direct interaction with the mating-specific Galpha in yeast. Science. 2002;296:1483–1486. doi: 10.1126/science.1070540. [DOI] [PubMed] [Google Scholar]
- Moseley J. B., Goode B. L. The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol. Mol. Biol. Rev. 2006;70:605–645. doi: 10.1128/MMBR.00013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson W. J. Adaptation of core mechanisms to generate cell polarity. Nature. 2003;422:766–774. doi: 10.1038/nature01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y., Huang K., Cross F. R., Cowburn D., Chait B. T. Accurate quantitation of protein expression and site-specific phosphorylation. Proc. Natl. Acad. Sci. USA. 1999;96:6591–6596. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehlen L. J., Cross F. R. G1 cyclins CLN1 and CLN2 repress the mating factor response pathway at Start in the yeast cell cycle. Genes Dev. 1994;8:1058–1070. doi: 10.1101/gad.8.9.1058. [DOI] [PubMed] [Google Scholar]
- Oehlen L. J., Cross F. R. Potential regulation of Ste20 function by the Cln1-Cdc28 and Cln2-Cdc28 cyclin-dependent protein kinases. J. Biol. Chem. 1998;273:25089–25097. doi: 10.1074/jbc.273.39.25089. [DOI] [PubMed] [Google Scholar]
- Paliwal S., Inglesias P. A., Campbell K., Hiloti Z., Groisman A., Levchenko A. MAPK-mediated biomodal gene expression and gradient sensing in yeast. Nature. 2007;446:46–51. doi: 10.1038/nature05561. [DOI] [PubMed] [Google Scholar]
- Park H. O., Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M., Neiman A. M., Park H. O., van Lohuizen M., Herskowitz I. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 1996;15:7046–7059. [PMC free article] [PubMed] [Google Scholar]
- Pryciak P. M., Huntress F. A. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gbetagamma complex underlies activation of the yeast pheromone response pathway. Genes Dev. 1998;12:2684–2697. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D., Bretscher A. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 2000;113:365–375. doi: 10.1242/jcs.113.3.365. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Legesse-Miller A., Gao L., Dong Y., Bretscher A. Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 2004;20:559–591. doi: 10.1146/annurev.cellbio.20.010403.103108. [DOI] [PubMed] [Google Scholar]
- Pullikuth A. K., Catling A. D. Scaffold mediated regulation of MAPK signaling and cytoskeletal dynamics: a perspective. Cell. Signal. 2007;19:1621–1632. doi: 10.1016/j.cellsig.2007.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi M., Elion E. A. MAP kinase pathways. J. Cell Sci. 2005a;118:3569–3572. doi: 10.1242/jcs.02470. [DOI] [PubMed] [Google Scholar]
- Qi M., Elion E. A. Formin-induced actin cables are required for polarized recruitment of the Ste5 scaffold and high level activation of MAPK Fus3. J. Cell Sci. 2005b;118:2837–2848. doi: 10.1242/jcs.02418. [correction published in J. Cell Sci. (2007). 120, 712] [DOI] [PubMed] [Google Scholar]
- Reed S. I. The selection of S. cerevisiae mutants defective in the start event of cell division. Genetics. 1980;95:561–577. doi: 10.1093/genetics/95.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder S. E., Banta L. M., McCaffery J. M., Emr S. D. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol. Biol. Cell. 1996;7:985–999. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Segall J. E. Polarization of yeast cells in spatial gradients of alpha mating factor. Proc. Natl. Acad. Sci. USA. 1993;90:8332–83236. doi: 10.1073/pnas.90.18.8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff M. A., Thorn K. S. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- Slessareva J. E., Routt S. M., Temple B., Bankaitis V. A., Dohlman H. G. Activation of the phosphatidylinositol 3-kinase Vps34 by a G protein alpha subunit at the endosome. Cell. 2006;126:191–203. doi: 10.1016/j.cell.2006.04.045. [DOI] [PubMed] [Google Scholar]
- Stevenson B. J., Rhodes N., Errede B., Sprague G. F., Jr Constitutive mutants of the protein kinase STE11 activate the yeast pheromone response pathway in the absence of the G protein. Genes Dev. 1992;6:1293–1304. doi: 10.1101/gad.6.7.1293. [DOI] [PubMed] [Google Scholar]
- Stowers L., Yelon D., Berg L. J., Chant J. Regulation of the polarization of T cells toward antigen-presenting cells by Ras-related GTPase CDC42. Proc. Natl. Acad. Sci. USA. 1995;92:5027–5031. doi: 10.1073/pnas.92.11.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickfaden S. C., Winters M. J., Ben-Ari G., Lamson R. E., Tyers M., Pryciak P. M. A mechanism for cell-cycle regulation of MAP kinase signaling in a yeast differentiation pathway. Cell. 2007;128:519–531. doi: 10.1016/j.cell.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxis C., Keller P., Kavagiou Z., Jensen L. J., Colombelli J., Bork P., Stelzer E. H., Knop M. Spore number control and breeding in Saccharomyces cerevisiae: a key role for a self-organizing system. J. Cell Biol. 2005;171:627–640. doi: 10.1083/jcb.200507168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Klionsky D. J. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L. G., Segall J. E., Snyder M. The alpha-factor receptor C-terminus is important for mating projection formation and orientation in Saccharomyces cerevisiae. Cell Motil. Cytoskeleton. 2002;53:251–266. doi: 10.1002/cm.10073. [DOI] [PubMed] [Google Scholar]
- van Drogen F., Stucke V. M., Jorritsma G., Peter M. MAP kinase dynamics in response to pheromones in budding yeast. Nat. Cell Biol. 2001;3:1051–1059. doi: 10.1038/ncb1201-1051. [DOI] [PubMed] [Google Scholar]
- Vida T. A., Emr S. D. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Chen W., Simpson D. M., Elion E. A. Cdc24 regulates nuclear shuttling and recruitment of the Ste5 scaffold to a heterotrimeric G protein in S. cerevisiae. J. Biol. Chem. 2005;280:13084–13096. doi: 10.1074/jbc.M410461200. [DOI] [PubMed] [Google Scholar]
- Wang Y., Elion E. A. Nuclear export and plasma membrane recruitment of the Ste5 scaffold are coordinated with oligomerization and association with signal transduction components. Mol. Biol. Cell. 2003;14:2543–2558. doi: 10.1091/mbc.E02-10-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmann K., Ammerer G. Overexpression of the G1-cyclin gene CLN2 represses the mating pathway in Saccharomyces cerevisiae at the level of the MEKK Ste11. J. Biol. Chem. 1997;272:13180–13188. doi: 10.1074/jbc.272.20.13180. [DOI] [PubMed] [Google Scholar]
- Whiteway M. S., Wu C., Leeuw T., Clark K., Fourest-Lieuvin A., Thomas D. Y., Leberer E. Association of the yeast pheromone response G protein beta gamma subunits with the MAP kinase scaffold Ste5p. Science. 1995;269:1572–1575. doi: 10.1126/science.7667635. [DOI] [PubMed] [Google Scholar]
- Winters M. J., Lamson R. E., Nakanishi H., Neiman A. M., Pryciak P. M. A membrane binding domain in the ste5 scaffold synergizes with gbetagamma binding to control localization and signaling in pheromone response. Mol. Cell. 2005;20:21–32. doi: 10.1016/j.molcel.2005.08.020. [DOI] [PubMed] [Google Scholar]
- Wu C., Leeuw T., Leberer E., Thomas D. Y., Whiteway M. Cell cycle- and Cln2p-Cdc28p-dependent phosphorylation of the yeast Ste20p protein kinase. J. Biol. Chem. 1998;273:28107–28115. doi: 10.1074/jbc.273.43.28107. [DOI] [PubMed] [Google Scholar]
- Wu J. Q., Pollard T. D. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- Xia Y., Karin M. The control of cell motility and epithelial morphogenesis by Jun kinases. Trends Cell Biol. 2004;14:94–101. doi: 10.1016/j.tcb.2003.12.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.