Abstract
Endosomal transport is critical for cellular processes ranging from receptor down-regulation and retroviral budding to the immune response. A full understanding of endosome sorting requires a comprehensive picture of the multiprotein complexes that orchestrate vesicle formation and fusion. Here, we use unsupervised, large-scale phenotypic analysis and a novel computational approach for the global identification of endosomal transport factors. This technique effectively identifies components of known and novel protein assemblies. We report the characterization of a previously undescribed endosome sorting complex that contains two well-conserved proteins with four predicted membrane-spanning domains. Vps55p and Vps68p form a complex that acts with or downstream of ESCRT function to regulate endosomal trafficking. Loss of Vps68p disrupts recycling to the TGN as well as onward trafficking to the vacuole without preventing the formation of lumenal vesicles within the MVB. Our results suggest the Vps55/68 complex mediates a novel, conserved step in the endosomal maturation process.
INTRODUCTION
Endosomal protein sorting is a highly conserved cellular process that is required for receptor down-regulation, viral replication, and development (Katzmann et al., 2002; Morita and Sundquist, 2004). Much progress has been made in recent years in identifying distinct multiprotein complexes that regulate the fusion of primary endocytic vesicles, the maturation of an early endosome into a multivesicular body (MVB), and the recycling of proteins and lipids to other organelles (Gruenberg and Stenmark, 2004; Bonifacino and Rojas, 2006). The ESCRT-I, -II, and -III complexes (endosomal-sorting complex required for transport), which act in sequence to regulate the formation of internal vesicles at the MVB, play a central role in endosome biogenesis (Katzmann et al., 2002; Hurley and Emr, 2006). Other transport complexes regulate the targeting and fusion of endosomes or endosome-derived vesicles with other organelles (Bowers and Stevens, 2005; Bonifacino and Rojas, 2006).
Many components of the endosomal-sorting machinery were first identified in yeast genetic screens for mutants that are defective in protein transport to the yeast vacuole, which requires functional endosomal sorting and recycling (Bowers and Stevens, 2005). Most of these components have a conserved role in endosome biogenesis in higher cells. Mammalian homologues have been identified for most if not all yeast ESCRT subunits (von Schwedler et al., 2003; Hurley and Emr, 2006), and at least some of the machinery that regulates recycling from the MVB is also well conserved. For example, the five-subunit retromer complex that mediates endosome-to-trans-Golgi network (TGN) transport in yeast is associated with tubular regions of the endosome in mammalian cells and mediates retrograde transport (Arighi et al., 2004; Seaman, 2004). In the past 20 years, classical genetic screens have identified more than 50 vacuole protein-sorting (VPS) genes (Bowers and Stevens, 2005). Surprisingly, a recent screen of the yeast genome-wide deletion set identified more than 350 genes with mild to severe vacuolar protein-sorting defects (Bonangelino et al., 2002). This represents a sevenfold increase in the number of candidate VPS genes and motivates the development of new strategies to prioritize genes for further study.
Exploiting the full potential of global phenotypic screens requires quantitative, objective strategies for collecting and interpreting data and defining gene function (Carpenter and Sabatini, 2004; Quenneville and Conibear, 2006). Yeast knockout screens that record fitness defects under conditions of chemical, environmental, or genetic perturbation have successfully identified the target pathways of bioactive compounds and increased our understanding of genetic redundancy (Tong et al., 2004; Parsons et al., 2006; St. Onge et al., 2007). Although growth rate is easily measured, assays that record biologically relevant attributes for a given pathway are expected to be more effective for systematically identifying the underlying molecular machinery.
Here, we use a sensitive biochemical assay for the genome-scale phenotypic analysis of yeast knockout mutants to define the relative contribution of each gene to the process of endosomal transport. Using a novel computational approach to identify statistically significant clusters, we show that new and known transport complexes can be predicted in an unbiased manner based on objective phenotypic criteria. As a result, we have identified and characterized a new endosome sorting complex. Our data indicate that the Vps55/68 complex acts with or downstream of the ESCRT machinery to regulate a novel step in endosome biogenesis.
MATERIALS AND METHODS
Plasmid and Strain Construction
The VPS68 gene was amplified from yeast genomic DNA using primers that added flanking HindIII and SacI sites and cloned into pCR Blunt, creating pMS2. The HindIII-SacI insert from pMS2 was subcloned into the same sites in pRS316, creating pMS4. Quick-change mutagenesis was used to create a BamHI site immediately before the stop codon of VPS68 in pMS4, resulting in pMS7. To epitope-tag VPS68, pMS7 was digested with BamHI and a 127-bp BglII fragment encoding a triple-hemagglutinin (HA) epitope tag was inserted, creating pMS8. This plasmid was found to complement the carboxypeptidase Y (CPY)-sorting defects of a vps68 mutant.
A 1.6-kb PCR fragment encoding VPS55 was amplified from genomic DNA using primers that added a 5′ XbaI site, digested with XbaI and HindIII, and cloned into pRS315. Quickchange mutagenesis was used to create a BamHI site immediately before the stop codon, creating pLC158. Epitope tagging was accomplished by the ligation of either double-stranded oligonucleotide linkers (encoding triple AU1 or myc tags) or a DNA fragment (encoding a triple-HA tag) into the BamHI site of pLC158 to create pBW2, pBW5, or pBW8, respectively; each plasmid complemented the CPY-sorting defect of vps55Δ strains. Sequences encoding Vps55-HA were PCR-amplified from pBW8 and cotransformed with linearized pRS316 into a vps55Δ strain followed by plasmid rescue, creating pCS23. pCS7 was made by cotransforming linearized pRS316 with a PCR fragment containing GFP-Snc1 sequences from pGS (Lewis et al., 2000) into yeast, followed by plasmid rescue. A BamHI/XhoI fragment from pSRG22 encoding Vps21 tagged with a single c-myc epitope was cloned into the BamHI/SalI site of YEp351 (Guthrie and Fink, 1991) creating pSRG102. pHY5 encodes VPS27 under control of the GAL1 promoter (Nothwehr et al., 2000). Plasmids for the expression of green fluorescent protein (GFP)-tagged Ste3, Sna3, and ALP have been previously described: pJLU34 (Urbanowski and Piper, 2001); pGFP-Sna3 (Reggiori and Pelham, 2001); and pRS426-GFP-ALP (Cowles et al., 1997).
The yeast strain BY4741 and its gene deletion derivatives BY4741-6279 BY4741-6842, and BY4741-5588 from the Saccharomyces Deletion Project (Winzeler et al., 1999) were obtained through Open Biosystems (Huntsville, AL). Other yeast strains used in this work are described in Table 1. BWY4 was made by crossing BY4741-6842 and BY4742-16279, selecting vps55Δ::kanR vps68Δ::kanR double mutant segregants, and confirming complementation of CPY secretion phenotypes by plasmid-borne wild-type VPS55 and VPS68 genes. To replace VPS68 or VPS55 coding sequences with the kanR marker, strains were transformed with a PCR product derived from BY4741-6279 and BY4741-6842 genomic DNA.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| SF838-9D | MATα ura3-52 leu2-3,112 his4-519 ade6 gal2 pep4-3 | Rothman and Stevens (1986) |
| RPY10 | MATα ura3-52 leu2-3,112 his4-519 ade6 gal2 | Piper et al. (1995) |
| SNY128 | MATα ura3-1 leu2-3,112 his3-11,15 trp1-1 ade2-1 can1-100 pep4-ΔH3 pho8Δ::ADE2 VPS10::HA vps27Δ::LEU2 | Nothwehr et al. (2000) |
| NDY1181 | MATα ura3 leu2 his4 bar1-1 GAL1-STE3Δ365::HA | Chen and Davis (2000) |
| BLY1 | MATα ura3-52 leu2-3,112 his4-519 ade6 gal2 pho8Δ::X | Bowers et al. (2000) |
| BWY4 | MATalys2 MET15 vps55Δ::KanR vps68Δ::kanR | This study |
| MSY1 | SF838-9D vps68Δ::kanR | This study |
| MSY2 | RPY10 vps68Δ::kanR | This study |
| KEBY37 | BLY1 vps27Δ::LEU2 | Bowers et al. (2000) |
| MSY3 | BLY1 vps68Δ::kanR | This study |
| MSY11 | KEBY37 vps68Δ::kanR | This study |
| LCY434 | RPY10 vps55Δ::KanR | This study |
| BWY10 | BY4741 vps68Δ::kanR SEC7::3GFP | This study |
| BWY12 | SNY128 vps68Δ::kanR | This study |
| BWY13 | LCY294 vps68Δ::kanR | This study |
| BWY14 | NDY1181 vps68Δ::kanR | This study |
| LCY294 | SF838-9D VPS10::HA | Conibear and Stevens (2000) |
Screening of Yeast Knockout Sets
Manipulation and screening of yeast knockout collections was performed essentially as described (Burston et al., 2008). Colony arrays on 384-array stock plates were pinned four times to create 1536 arrays suitable for large-scale assays, using a Virtek automated colony arrayer (Bio-Rad, Hercules, CA). For immunoblot analysis, colony arrays were replicated in parallel on YPD plates and on YPD plates overlaid with a nitrocellulose filter. After incubation at 30°C for 18 h, CPY bound to filters was detected by immunoblotting as described (Conibear and Stevens, 2002). Blots were developed with enhanced chemiluminescence (ECL) and exposed to film. Digital images of CPY secretion and colony growth were acquired using an Epson 2400 flat-bed scanner (Long Beach, CA). Image densitometry was accomplished using Quantity One software (Ver 4.2.1, Bio-Rad) or the spot-finding program GridGrinder (gridgrinder.sourceforge.net). Raw densitometry values were subjected to background subtraction, normalized, and filtered to eliminate absent or very slow-growing strains. A median endosomal-sorting score was then calculated for each open reading frame (ORF) based on replicate data points from each of the three yeast knockout collections. Genome-wide localization data (Huh et al., 2003) was simplified as follows to reduce the number of categories: vacuole = vacuole/vacuolar membrane; nucleus = nucleus/nucleolus; bud = bud/budneck/cell periphery; endoplasmic reticulum (ER) = ER/nuclear periphery; and Golgi = Golgi/Golgi-to-vacuole/ER-to-Golgi/Golgi-to-ER.
Phenotypic Testing of Yeast Miniarrays
The top 279 CPY-secreting mutants were rearrayed in 384-array format along with 93 randomly chosen knockouts and 8 controls. For growth inhibition assays, miniarrays were replicated four times to parallel YPD plates and YPD plates containing inhibitors (2.5 μg/ml amphotericin, 12 mM caffeine, 50 mM CaCl2, 10 μg/ml nystatin, 0.05% SDS, 50 μg/ml calcofluor white, and 10–20 μg/ml benomyl). For secretion assays, miniarrays were pinned to YPD or SC plates covered with nitrocellulose filters, which were probed with antibodies to pro-alpha factor or CPY as described (Conibear and Stevens, 2002; Burston et al., 2008). ECL luminescence or colony fluorescence on calcofluor white–containing plates was captured with a FluorS Max Multi-imager (Bio-Rad; Lam et al., 2006; Burston et al., 2008). The APNE assay was as described (Wolf and Fink, 1975). Each experiment was carried out in triplicate on miniarrays from three different knockout sets (MATa, MATα, and homozygous diploid).
Module Isolation Method of Scoring Cluster Significance
The raw phenotypic dataset was subjected to background subtraction, normalization, and filtering, and a median phenotypic score was calculated for each gene deletion based on replicate data points from each of the three yeast knockout collections. Pairwise Euclidean distances were computed and converted into ranks. Single-linkage agglomerative hierarchical clustering was conducted on the ranked distances, resulting in the sequential formation of 278 (=279 − 1) clusters, each of which is associated with a level reflecting the (ranked) distance at which the merge occurred. For each cluster, survival time was computed as the difference between the level at which the cluster was first formed (“birth”) and the level at which it was merged into a larger cluster (“death”; Ling, 1973). Each observed survival time is converted into a p value by computing the upper tail probability from the associated negative hypergeometric distribution. For computational reasons, we used an approximation known to work well in our context: namely, we substituted the geometric distribution for the negative hypergeometric. For a cluster of size c that is born at level or time b, the approximate null distribution of the survival time is given by a geometric distribution with success probability p:
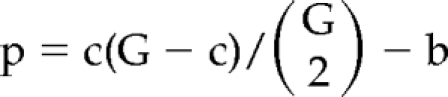 |
where G is 279 is the number of genes. Isolation scores were computed as −log(p value). This index of cluster significance arises from a graph process approach to the ranked pairwise distances and is driven by the concept of module isolation (Miso). For a full treatment of the Miso methodology (see Brumm, Conibear, Wasserman, and Bryan, unpublished data).
To provide strong control of the family-wise error rate, we computed Bonferroni adjusted p values. In summary tables (Supplementary Information), the set of clusters produced by p value threshholding was grouped into families, which are defined as groups of clusters in which each member either contains or is contained by another member. Within each cluster family, the latest-forming cluster was used to describe results for that family in all tables. The “wild-type” cluster of mutants exhibiting an essentially wild-type phenotypic profile was defined as the first cluster to hold more than 50 genes. Clusters containing this wild-type cluster and clusters contained within it were removed from the output.
The Miso method for computing cluster significance has been implemented in R code (R Code Development Team, 2005) and as a Cytoscape plug-in (Shannon et al., 2003), both available at www.stat.ubc.ca/∼jenny/webSupp/schluterGlobDisc/.
Coprecipitation Experiments and Western Blotting
Immunoprecipitation of CPY, Vps10p, ALP, and Ste3p was performed under denaturing conditions from radiolabeled extracts as described (Conibear and Stevens, 2000, 2002) using appropriate polyclonal antisera. Anti-Ste3p antiserum was a gift from G. Sprague (University of Oregon, Eugene, OR). For coprecipitation experiments, log phase cells were converted to spheroplasts and stored at −70°C (Conibear and Stevens, 2000). Spheroplasts from 10 OD600 cells were resuspended in 1 ml lysis buffer (1.2 M sorbitol, 25 mM KPO4, pH 7.5, 50 mM NaCl, 1% CHAPSO, 1% PMSF) and incubated with 2 μl of rabbit anti-HA antiserum for 1 h at 4°C, after which 15 μl of protein G Sepharose (Amersham, Piscataway, NJ) was added for 1 h at 4°C. The pellets were washed three times in wash buffer (50 mM KPO4, pH 7.5, 50 mM NaCl, 1% CHAPSO) and subjected to SDS-PAGE. Coprecipitated proteins were detected by Western blotting with monoclonal antibodies to HA (HA11, Covance Research Products, Madison, WI) or AU1 (Bio/Can Scientific, Mississauga, ON, Canada) followed by HRP-labeled anti-mouse secondary antibody (Sigma). Blots were developed with ECL and either exposed to x-ray film (X-OMAT LS, Eastman Kodak, Rochester, NY), or the luminescent images were captured with a Fluor S Max Multi-imager (Bio-Rad).
Fluorescence Microscopy
Indirect immunofluorescence microscopy, fluorescent microscopy of yeast cells expressing GFP-tagged proteins, and staining with vital dyes were carried out as described (Conibear and Stevens, 2000, 2002). Uptake of 40 μM FM4-64 and/or 10 μM NBD-PC (Avanti Polar Lipids, Alabaster, AL) by live cells was performed at 30°C for 15 min, after which cells were resuspended in YPD and incubated for 30 min at 30°C. Cells were subsequently washed twice in PBS before visualization on concanavalin A–coated slides. Cells were viewed using a 100× oil-immersion objective on a Zeiss Axioplan2 fluorescence microscope (Thornwood, NY), and images were captured with a CoolSnap camera using MetaMorph software (Universal Imaging, West Chester, PA) and adjusted using Adobe Photoshop (San Jose, CA).
Cell Surface Recycling Assay
Preparative and analytical protease shaving were performed as previously described (Chen and Davis, 2000). GAL1-driven expression of Ste3Δ365p was induced by addition of 2% galactose to log phase cultures and terminated by the addition of 3% glucose. Pheromone treatment was carried out for 45 min by adding 0.5 vol of a-factor–containing supernatant. Mock treatment was carried out using a similarly prepared supernatant lacking a-factor. This was followed directly by preparative shaving, in which 6 × 108 cells were digested with 2000 U of Pronase (Calbiochem-Novabiochem, La Jolla, CA) in 9 ml of 10 mM sodium azide and 10 mM sodium fluoride. Cells were returned to culture in YPDS medium (YPD, 1 M sorbitol). To assess the amount of surface-exposed receptor at each time point, aliquots containing 5 × 107 cells were removed and subjected to a second protease-shaving treatment using 10 mg/ml Pronase or 10 mg/ml bovine serum albumin as a control. Ste3p was detected by SDS-PAGE and Western blotting with anti-HA antibodies.
Endosome-recycling Assay
The late endosome-recycling assay was carried out as previously described (Nothwehr et al., 2000). Strains carrying a GAL1-VPS27 plasmid were grown overnight and allowed to double twice the next day in selective medium containing 2% raffinose, and 0, 45, 60, 75, and 90 min after 2% galactose was added to induce expression of VPS27, cells were fixed, spheroplasted, and costained with antibodies against the HA epitope and Vph1p. The distribution of Vph1p and Vps10p was determined in at least 100 cells for each time point.
RESULTS
Genome-Scale Quantitative Analysis of Endosomal Protein Sorting
For the genome-scale identification of genes that contribute to endosome function, we adapted an existing biochemical assay that measures the receptor-mediated sorting of newly synthesized CPY (Conibear and Stevens, 2002). Defects in transport between the late endosome and the Golgi prevent the recycling of the CPY receptor and block CPY sorting to the vacuole, resulting in its secretion from the cell (Bowers and Stevens, 2005). We assessed CPY secretion in three independent yeast knockout sets in replicate using a modified immunoblot technique (Figure 1A) and calculated a median value for each knockout strain. We used this value, which reflects each gene's relative contribution to endosomal sorting, to derive a “sorting index” for each gene represented in the knockout collection (Table S1).
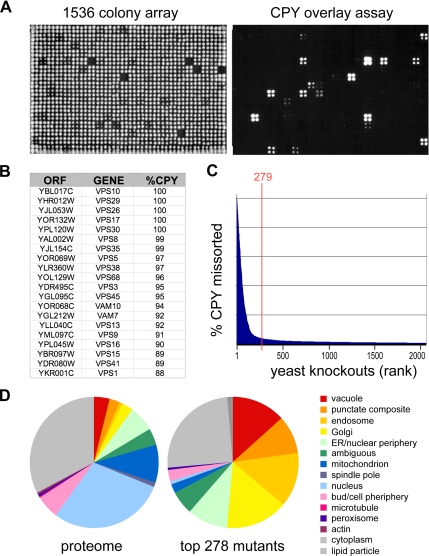
Figure 1.
Genome-scale identification of protein-sorting mutants. (A) Yeast knockout strains were plated as 1536 arrays on solid agar (left) or on agar covered with a nitrocellulose filter. Secreted CPY retained on the filter was visualized by Western blotting (right). (B) Top 20 CPY-secreting mutants. Genome-wide screens for CPY missorting were carried out in duplicate on three independent deletion collections, and a median endosomal-sorting index was computed based on densitometry of digital images. (C) Ranked secretion values for the 2000 top CPY-secreting mutants. The top 279 strains were chosen for further analysis. (D) Pie chart showing the subcellular localization of proteins corresponding to the 279 mutants based on a genome-wide GFP fusion study (Huh et al., 2003). Vacuole, endosome, “punctate composite,” Golgi, and ER localizations were enriched in the mutant set compared with the proteome.
Many of the highest-scoring genes corresponded to VPS, PEP, or VAM genes previously implicated in CPY sorting (Bonangelino et al., 2002; Bowers and Stevens, 2005; Figure 1B). All of the VPS genes identified by classical genetic approaches were found in the top 100 hits, with the exception of three (VPS11, VPS33, VPS53) that were missed because of strain construction errors in two of the three knockout collections. We chose a set of 279 mutants that have an appreciable CPY secretion phenotype for further analysis (Figure 1C). Comparison with localization data from a large-scale GFP fusion study (Huh et al., 2003) indicates that this set is enriched for genes whose products localize to the Golgi, endosome, vacuole, and ER, organelles previously implicated in the intracellular transport of CPY (Figure 1D).
Identification of Functionally Related Genes by Phenotypic Miniarray Profiling
The top 279 mutants represent a comprehensive set of genes involved in Golgi/late endosome transport. This set is expected to include most components of endosomal transport pathways and complexes, with the exception of those with essential or redundant functions. Because genes that act together can often be identified based on shared mutant phenotypes (Schuldiner et al., 2005; Parsons et al., 2006; Quenneville and Conibear, 2006), we reasoned that quantitative phenotypic profiling might resolve these genes into functional groupings that represent individual complexes or transport steps. We rearrayed the selected 279 mutants and 93 randomly chosen controls to create a “miniarray” that reduces experimental variation by allowing phenotypes to be evaluated on a single plate. Three different miniarrays, containing strains from each of the three deletion sets, were subjected to 13 diverse phenotypic assays. To identify mutants defective in the Golgi/endosome recycling of the pro-alpha factor–processing enzyme Kex2p, the secretion of unprocessed pro-alpha factor was measured by immunoblot assay. A fluorescence assay based on the binding of Calcofluor white to cell-surface chitin (Lam et al., 2006) was used to estimate the cell surface activity of the chitin synthase Chs3p, which recycles between the cell surface, Golgi, and chitosomes/early endosomes. CPY secretion was reevaluated on rich and minimal nutrient conditions. In addition, the colorimetric APNE assay was used to measure the amount of active CPY present in the vacuole (Wolf and Fink, 1975). Finally, we assessed the sensitivity of mutant strains to a variety of chemical inhibitors (benomyl, SDS, amphotericin, nystatin, CaCl2, calcofluor white, minimal medium, and caffeine; see Materials and Methods for details) based on colony size.
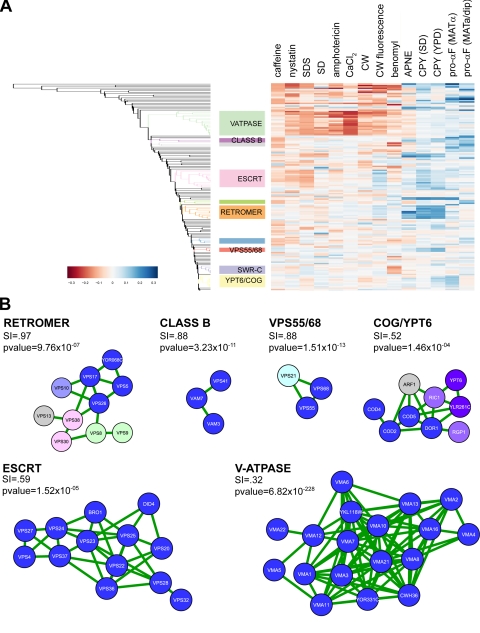
Replicate testing of the three miniarrays under experimental and control conditions and quantitation of the resulting images created a database of ∼300,000 raw measurements. Hierarchical clustering was performed after subjecting this raw phenotypic dataset to background subtraction, normalization and filtering, and calculating median values for each gene deletion (Figure 2A and Table S2). Many gene clusters were enriched for subunits of protein complexes implicated in endosome/vacuolar transport, including components of the vacuolar H+-ATPase (V-ATPase), ESCRT, and class B Vps complexes, whereas other clusters contained genes of unknown function. Inspection of the heatmap in Figure 2A shows that protein-sorting and chemical sensitivity phenotypes make distinct contributions to the functional groupings (e.g., retromer vs. V-ATPase).
Figure 2.
Unbiased prediction of protein complexes based on phenotypic profiling. Miniarrays containing the top CPY-secreting strains were tested for phenotypes including CPY sorting, pro-alpha factor processing, sensitivity to chemical inhibitors, and chitin content. A graph theoretic approach was used to compute p values that identify significant clusters. (A) Enlargement of region of heat map enriched for significant clusters. Clusters corresponding to known protein complexes are indicated. (B) Clusters of three or more genes that have a significance score (p) ≤ 0.05 and a sorting index (SI) >13% are illustrated. The protein complex represented by the greatest number of genes in a given cluster is indicated; known subunits of that complex are shown in blue. Subunits of additional complexes in the same cluster are also color-coded. The relative contribution of each complex to the process of endosomal sorting was estimated by calculating the median sorting index for all elements of that cluster.
Unbiased Discovery of Protein Complexes via a Cluster Significance Index
Prior knowledge of gene function can be used to guide the selection of functionally related gene clusters, but does not contribute to the discovery of new complexes or pathways for which no information is available (Conibear, 2005; Quenneville and Conibear, 2006). To exploit the phenotypic profiling approach in settings in which it is either impossible or undesirable to incorporate external information, we developed an objective, unsupervised method suitable for identifying biologically coherent gene groups in genome-wide experimental data. We derived a cluster-specific index of significance (or “p value”) using a novel graph theoretic approach that allows the computation of p values through a simple formula based on established probability theory (Ling, 1973). This method, which we have named Miso (for module isolation), is available in R code and as a plug-in for the genomics software Cytoscape (Shannon et al., 2003) and will be fully elaborated in a dedicated report (Brumm, Conibear, Wasserman, and Bryan, unpublished data).
We computed this cluster significance index for the miniarray data. Gene clusters with Bonferroni-adjusted p values <0.05 were treated as statistically significant. The hierarchical nature of the algorithm induces interrelationships among the clusters. After resolving the clusters into nonredundant groupings, we identified a total of 21 significant gene clusters (Table 2). Of the 10 clusters with 3 or more components, 8 correspond to known protein complexes, providing proof of the utility of our significance index. To evaluate the relative importance of each cluster, we computed an endosome-sorting score based on the median sorting index of its components. Taken together, these analyses provide an unbiased prediction of the protein complexes and pathways that collectively constitute the endosomal transport machinery, along with an indication of their relative contribution to the process.
Table 2.
Significant nonredundant gene groupings identified by the algorithm
| Bonferroni p value | Sorting index | Cluster ID | Cluster size | Complexa |
|---|---|---|---|---|
| 6.82E-228 | 32.4 | 231 | 18 | V-ATPASE (18) |
| 1.10E-84 | 51.9 | 272 | 2 | PI3 Kinase (2) |
| 3.07E-32 | 3.0 | 252 | 2 | |
| 1.46E-24 | 7.3 | 266 | 2 | |
| 4.16E-23 | 81.0 | 202 | 2 | CLASS D VPS (2) |
| 2.48E-19 | 76.9 | 160 | 2 | Overlapping ORFs (2) |
| 1.51E-13 | 87.6 | 121 | 3 | VPS55/68 (2) |
| 3.23E-11 | 88.2 | 247 | 3 | CLASS B VPS (3) |
| 9.64E-11 | 29.7 | 220 | 2 | Overlapping ORFs (2) |
| 1.33E-10 | 20.1 | 222 | 2 | |
| 1.92E-10 | 63.8 | 150 | 2 | GARP (2) |
| 3.95E-10 | 25.3 | 169 | 4 | INO80 (2) |
| 5.00E-09 | 6.6 | 163 | 2 | |
| 7.16E-09 | 17.1 | 249 | 3 | RSC (2) |
| 9.76E-09 | 97.0 | 211 | 10 | Retromer (4) |
| 2.70E-07 | 99.3 | 200 | 2 | Retromer (2) |
| 5.56E-07 | 13.5 | 154 | 6 | SWR-C (6) |
| 1.52E-05 | 59.0 | 233 | 13 | ESCRT (13) |
| 4.34E-05 | 10.2 | 86 | 2 | |
| 1.46E-04 | 51.9 | 164 | 9 | COG/YPT6 (4/4) |
| 1.05E-03 | 8.3 | 213 | 3 |
The list of ORFs included in each cluster is in Table S3.
a The name of a protein complex is shown when at least two genes in a cluster encode physically interacting proteins (number of subunits present in the cluster is indicated in parentheses). When more than one complex is represented, the one with the greatest number of included subunits is shown.
Identification of New and Known Complexes
The six clusters containing three or more genes that have the highest sorting index are shown in Figure 2. Each of these top-scoring clusters consisted primarily of genes encoding components of a particular protein complex or sorting pathway (Table S3). The largest cluster corresponded precisely to the V-ATPase, an ATP-dependent protein pump responsible for endosome/vacuole acidification (Graham et al., 2003). This cluster, which contains 18 genes that encode all 13 nonredundant V-ATPase subunits, 2 overlapping ORFs, and 3 dedicated V-ATPase assembly factors, lacks only VPH1 and STV1, which encode two isoforms of the 100-kDa stator subunit with partially overlapping functions. The second-largest cluster contains 13 genes corresponding to all known components of ESCRT-I, -II, and -III complexes apart from the newly described ESCRT-I subunit Mvb12 (Chu et al., 2006; Curtiss et al., 2007; Kostelansky et al., 2007; Oestreich et al., 2007) and also includes the ESCRT-associated factors Vps27p, Vps4p, and Bro1p (Bowers and Stevens, 2005). The fact that all three ESCRT subcomplexes were found in a single cluster is consistent with their highly interdependent function in MVB formation.
A significant score (p = 9.76 × 10−9) was obtained for a larger retromer-containing cluster that included the CPY receptor Vps10p, two subunits of the phosophatidylinositol (PI) lipid kinase complex that produces the MVB pool of PI3P (Burda et al., 2002) and three other genes with late endosome functions. Because the retromer complex binds a sorting signal in the Vps10p cytosolic domain (Nothwehr et al., 2000) and contains functionally important PX domains that bind PI3P at the MVB (Burda et al., 2002; Seaman, 2005), this cluster may correspond to a pathway relevant for retromer function. The remaining two PI3 kinase subunits, Vps15p and Vps34p, formed a distinct cluster with p = 1.10 × 10−84, which could reflect their role in creating additional cellular pools of PI3P (Slessareva et al., 2006).
Other endosomal transport complexes identified by the algorithm mediate fusion with the endosome (class D Vps) or vacuole (class B Vps), whereas the GARP and COG tethering complexes, the Rab GTPase Ypt6p and its heteromeric exchange factor Rgp1/Ric1 are implicated in the fusion of endosome-derived transport vesicles at the Golgi (Bonifacino and Rojas, 2006; Figure 2B). Surprisingly, not all complexes with highly significant p values function primarily in endosomal transport. One cluster (p = 5.56 × 10−7) contains six subunits of SWR-C, a chromatin-remodeling complex that prevents the spread of silent chromatin (Krogan et al., 2003; Kobor et al., 2004; Mizuguchi et al., 2004). Loss of SWR-C function could cause the aberrant silencing of endosomal trafficking genes. However, CPY secretion in htz1 and swr1 mutants was SIR2-independent (not shown), suggesting SWR-C regulates endosome transport by a distinct mechanism. Other groups included subunits of the RSC and INO80 chromatin-remodeling complexes (Table 2). Each of the clusters corresponding to chromatin-remodeling complexes had a relatively low endosomal-sorting index, consistent with a minor or indirect role in protein traffic.
Discovery of the New Endosomal-sorting Complex Vps55/68
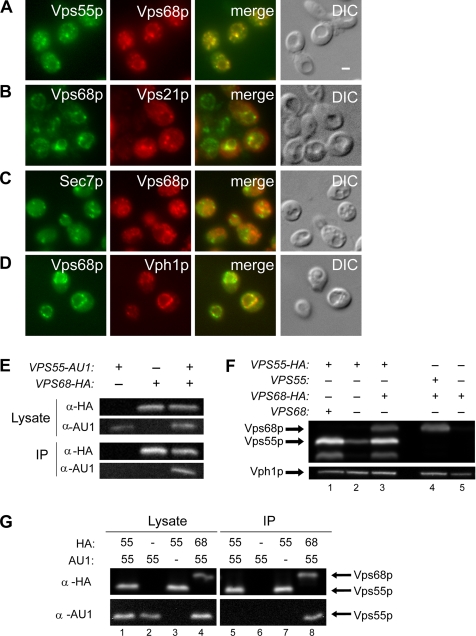
Most groupings identified by the algorithm corresponded to well-characterized complexes with established roles in endosomal transport, providing immediate validation of the predictions. Interestingly, one high-scoring cluster (p value = 1.51 × 10−13; sorting index = 87.6) contained two relatively uncharacterized genes, VPS55 and VPS68, and the Rab GTPase VPS21 (Figure 2B). Both VPS55 and VPS68 are predicted to encode small four-transmembrane domain proteins that are unrelated to each other, but have orthologues in higher organisms, including humans (Belgareh-Touze et al., 2002; data not shown). VPS68 was originally identified in a screen of the yeast deletion collection for mutants defective in CPY sorting (Bonangelino et al., 2002) and was localized to the vacuolar membrane in a genome-wide study of GFP fusion proteins (Huh et al., 2003). In contrast, Vps55p was reported to act at the late endosome and colocalize with the endosomal marker Snf7p (Belgareh-Touze et al., 2002; Huh et al., 2003). By double-label immunofluorescence microscopy of cells coexpressing two differently tagged forms of Vps68p and Vps55p, we found these proteins colocalized at the vacuolar limiting membrane and adjacent punctate structures that contain Vps21p but are devoid of the late Golgi marker Sec7p, indicating they are endosomes (Figure 3, A–D). Because these data are consistent with a common site of action, we tested the hypothesis that Vps55p and Vps68p interact physically and functionally.
Figure 3.
(A–D) Vps55p and Vps68p colocalize at endosomes and the vacuole-limiting membrane. The following strains were fixed and processed for double-label fluorescence microscopy. (A) Vps68-HA and Vps55-myc were coexpressed from plasmids in vps55 vps68 (BWY4) mutants and labeled with anti-myc mAb and anti-HA rabbit antiserum. (B) myc-tagged Vps21p was coexpressed with Vps68-HA from plasmids in vps68 cells (BY4741-6279) and double-labeled with anti-HA mAb and anti-myc rabbit antiserum. (C) Plasmid-expressed Vps68-HA was colocalized with Sec7-GFP in BWY10 cells labeled with rabbit anti-HA antiserum. (D) Plasmid-borne Vps68-HA was expressed in vps68 (BY4741-6279) cells double-labeled with an antiserum to endogenous Vph1 and anti-HA mAb. (E) Vps55p and Vps68p form a complex. Proteins were precipitated with anti-HA antiserum from extracts prepared from vps55 (BY4741-6842) cells expressing AU1-tagged Vps55, vps68 (BY4741-6279) cells expressing HA-tagged Vps68p, or vps55 vps68 (BWY4) cells expressing both tagged proteins, as indicated, and resolved by SDS-PAGE. Lysates and immunoprecipitates (IP) were analyzed by Western blotting with anti-HA and anti-AU1 mAbs. (F) Vps55p and Vps68p stabilize each other. vps55 (BY4741-6842; lane 1), vps68 (BY4741-6279; lane 4), or vps55 vps68 (BWY4; lanes 2, 3, and 5) strains containing plasmids for the expression of HA-tagged Vps55p and/or HA-tagged Vps68p were analyzed by Western blotting with anti-HA mAbs. Presence or absence of endogenous, untagged Vps68p or Vps55p is indicated. (G) The Vps55/68 complex contains a single copy of Vps55p. Detergent extracts were prepared from vps55 (BY4741-6842) mutant cells containing plasmids for the expression of AU1- and/or HA-tagged Vps55p (lanes 1–3 and 5–7), as indicated, and from vps55 vps68 (BWY4) expressing both AU1-tagged Vps55p and HA-tagged Vps68p (lanes 4 and 8). Proteins were immunoprecipitated with anti-HA antiserum and protein G-Sepharose, resolved by SDS-PAGE, and analyzed by Western blotting with anti-HA and anti-AU1 mAbs.
To determine if these proteins form a complex, we expressed differently tagged forms of the two proteins in the same cell. Immunoprecipitation of Vps68p with anti-HA antibodies led to the coprecipitation of Vps55p from CHAPSO-solubilized cell lysates (Figure 3E). Coprecipitation was specific, because no coenrichment of the vacuolar membrane protein Vph1p or the endosomal protein Nhx1p was found (data not shown). In addition, neither Vps55p nor Vps68p copurified with Vps21-myc expressed from a high-copy plasmid (not shown).
Proteins that are constitutively associated in a complex often depend on each other for their stability. We found that steady-state levels of Vps55-HA and Vps68-HA were greatly reduced in the absence of their interacting partners (Figure 3F). Although Vps55-HA appeared to be expressed at higher levels than Vps68-HA (Figure 3F, lane3), suggesting the Vps55/68 complex may contain multiple copies of Vps55p, no interaction was observed between coexpressed AU1- and HA-tagged versions of Vps55p under conditions where copurification of Vps55p and Vps68p was readily detected (Figure 3G). Furthermore, AU1-tagged Vps68p did not coprecipitate with Vps68-HA (not shown). Taken together, these results suggest that Vps55p and Vps68p are each present at single copy in a stable protein complex.
Vps68p Is Required for CPY Sorting But Not for Stability of the CPY Receptor
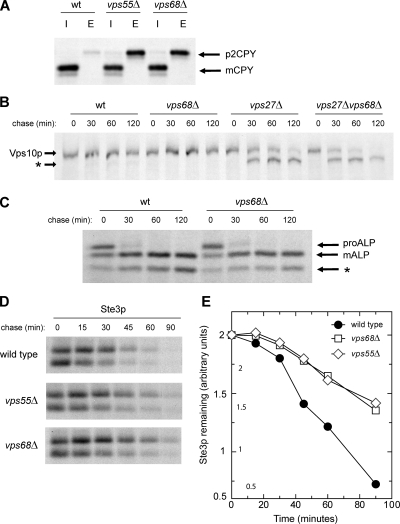
We analyzed vacuolar protein sorting in vps68 and vps55 mutants to determine the role of Vps68p in protein trafficking and test its functional relationship to Vps55p. By pulse-chase analysis of metabolically labeled cells (Figure 4A), vps68 and vps55 mutants both secreted ∼50% of newly synthesized CPY into the extracellular medium in a fully glycosylated, Golgi-modified p2 form (Stevens et al., 1982). CPY is recognized at the late Golgi by its receptor, Vps10p, and is delivered to the late endosome (Cereghino et al., 1995; Cooper and Stevens, 1996). Mutations that block Vps10p recycling back to the Golgi often result in its transport to the vacuole where it is degraded; however, the stability of Vps10p was not affected by mutation of either VPS68 or VPS55 (Figure 4B, and data not shown), and Vps10p was not missorted to the vacuolar membrane in vps68 mutants (Figure S1A).
Figure 4.
Cargo sorting in vps68 and vps55 mutants. (A) Secretion of newly synthesized CPY. Strains were labeled with [35S]methionine for 10 min and chased for 60 min in the presence of 50 μg/ml unlabeled cysteine and methionine. CPY was immunoprecipitated from intracellular (I) and extracellular (E) fractions, analyzed by SDS-PAGE, and visualized by fluorography. Arrows indicate the position of Golgi-modified (p2) CPY and that of mature, vacuolar (m) CPY. (B) VPS68 acts downstream of the class E gene VPS27. Pulse-chase labeling and immunoprecipitation of Vps10p was carried out as described above using the following strains: wild-type (BLY1), vps27 (KEBY37), vps68 (MSY3), and vps27 vps68 (MSY11). Proteolytic cleavage of Vps10p in vps27 and vps27 vps68 double mutants generates a faster-migrating cleavage product (*). (C) Kinetics of ALP maturation are unchanged in vps68 mutants. ALP was immunoprecipitated from wild-type (RPY10) or vps68 cells (MSY2) that had been radiolabeled for 10 min with [35S]methionine and chased for the times indicated. PEP4-dependent cleavage converts precursor (pro) forms of ALP into the mature (m) form. Asterisk (*) indicates a commonly observed degradation product. (D and E) Degradation of Ste3p is slowed in vps68 mutants. Ste3p was immunoprecipitated from wild-type (RPY10), vps68 (MSY2), or vps55 (LCY434) cultures that had been radiolabeled with [35S]methionine for 10 min and chased with unlabeled cysteine and methionine for the times indicated. The total amount of Ste3 remaining at each time point was quantitated by densitometry. Representative images for wild-type, vps55, and vps68 strains are shown in D, and semilog plots are shown in E. Wild type, •; vps68, □; vps55, ◇.
Defects in membrane fusion at the late endosome or vacuole stabilize Vps10p by causing it to accumulate in transport intermediates. An epistasis experiment was used to determine if VPS68 acts upstream or downstream of fusion with the MVB (Figure 4B). Mutations that prevent the fusion of Golgi-derived vesicles with the MVB block the delivery of Vps10p to the aberrant protease-active endosomal compartment that accumulates in class E mutants such as vps27 (Bryant et al., 1998). As previously reported, Vps10p was rapidly cleaved in vps27 mutants, with a half time of ∼30 min. In vps68 vps27 double mutants, Vps10p cleavage was not blocked, demonstrating that VPS68 is not required for fusion with the MVB.
Vps10p can also be stabilized by loss of fusion with the vacuole-limiting membrane (Darsow et al., 1997). Defects in vacuole fusion block the PEP4-dependent maturation of newly synthesized ALP, which follows the AP-3 pathway to the vacuole and does not transit the MVB (Cowles et al., 1997; Piper et al., 1997). However, ALP was transported to the vacuole with wild-type kinetics in vps68 mutants, indicating that Vps68p is not part of the general vacuole fusion machinery (Figure 4C). Furthermore, the efficient maturation of ALP and the intracellular fraction of CPY in vps68 cells (Figure 4A) demonstrate that PEP4-dependent vacuole protease activity is normal in these mutants.
vps68 Mutants Accumulate Endocytic and Biosynthetic Cargo at the MVB
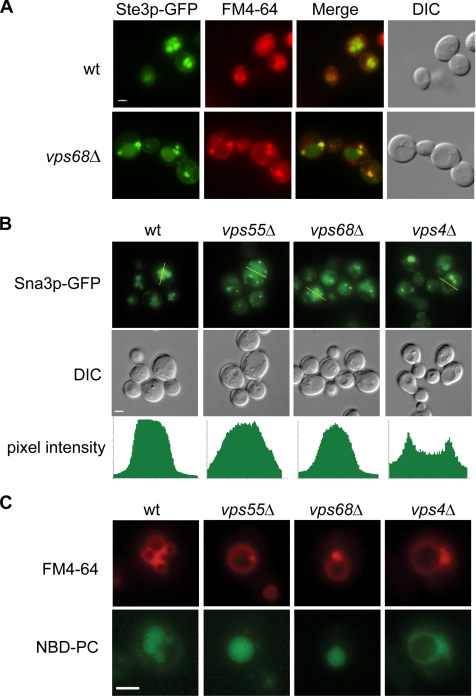
In the absence of ligand, the pheromone receptor Ste3p is constitutively endocytosed and enters the MVB pathway, resulting in its degradation in the vacuole (Chen and Davis, 2002). Ste3p down-regulation was significantly delayed in vps68 mutants, with a half time of 55 min compared with 30 min in wild-type cells. This rate of degradation was strikingly similar to that of vps55 mutants (Figure 4, D and E). To determine where the defect in Ste3p trafficking occurred, the localization of Ste3p was examined by fluorescence microscopy (Figure 5A). In wild-type cells, GFP-tagged Ste3p accumulated in the vacuole lumen, with faint labeling of the cell surface. In contrast, vps68 cells showed an accumulation of Ste3-GFP in endosomal compartments adjacent to the vacuole that also labeled with the endocytic dye FM4-64 (Figure 5A and Figure S1B). The delayed turnover and endosomal retention of vps55 and vps68 mutants is consistent with previous results showing that vps55 mutants are defective in a postendocytic step in down-regulation of the uracil transporter Fur4p (Belgareh-Touze et al., 2002).
Figure 5.
Endosomal accumulation of Ste3p and Sna3p in vps68 mutants. (A) Wild-type (wt; SF838-9D) and vps68 (MSY1) strains containing plasmids for the expression of Ste3-GFP and labeled with FM4-64 were viewed by double-label fluorescence microscopy of live cells. (B) Sna3-GFP was expressed from a plasmid in wild-type (SF838-9D), vps55 (BY4741-6842), vps68 (MSY1), and vps4 (BY4741-5588) strains. Lines (top panels) indicate the location of pixel intensity measurements (bottom panel). Note that Sna3-GFP is evenly distributed throughout the vacuolar lumen in wild-type, vps55, and vps68 strains, whereas pixel intensity is greatest at the vacuolar-limiting membrane in vps4 mutants. (C) Intracellular sorting of fluorescent lipids NBD-PC and FM4-64. NBD-PC is sorted to the vacuole lumen in wild-type, vps55, and vps68 mutants, but not in vps4 mutants. Strains are as described in A. Bar, 2 μm.
Ste3p relies on a ubiquitin tag for its sorting into lumenal vesicles at the MVB (Chen and Davis, 2002), whereas the role of ubiquitin in sorting the membrane protein Sna3p, which follows the biosynthetic pathway from the Golgi to the MVB, is less clear (Reggiori and Pelham, 2001; Stawiecka-Mirota et al., 2007). By fluorescence microscopy, Sna3-GFP, like Ste3-GFP, accumulated in internal structures in vps68 and vps55 single and double mutants (Figure 5B and Figure S1B). VPS68 is therefore required for the efficient transport of both endocytic and biosynthetic cargos at the MVB. In contrast, VPS68 was not required for the correct localization of other proteins that do not transit the MVB, including GFP-ALP and GFP-Snc1p (Lewis et al., 2000; Figure S1C).
The accumulation of proteins in aberrant endosomes adjacent to the vacuole is characteristic of ESCRT mutants that are defective in the formation of intralumenal MVB vesicles. However, in vps68 mutants the endosomal structures are less pronounced, and some Ste3p and Sna3p can clearly be seen in the vacuolar lumen in vps68 and vps55 vps68 mutants, suggesting that the budding of vesicles into the MVB lumen is not blocked (Figure 5, A and B, and Figure S1B). As a more direct test of lumenal vesicle formation, we examined the localization of the fluorescent lipid NBD-PC, which is sorted to the vacuole lumen via the MVB pathway (Hanson et al., 2002). NBD-PC clearly labeled the lumen of the vacuole of vps55 and vps68 strains, whereas FM4-64 was found at the vacuole-limiting membrane (Figure 5C). In contrast, both NBD-PC and FM4-64 colocalized at the limiting membrane in vps4 mutants. Therefore, the endosomal transport defects seen in vps68 mutants are distinct from those of class E vps mutants.
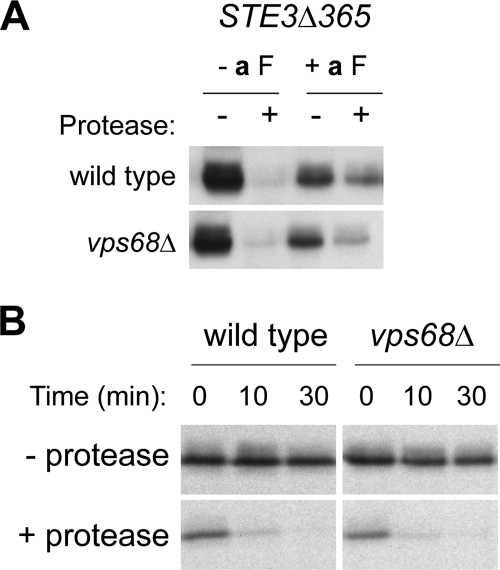
VPS68 Is Not Required for Efficient Recycling of Ste3Δ365p to the Cell Surface
Trafficking routes that lead out of the endosome include a recycling pathway to the cell surface, a retrograde transport pathway to the Golgi, and a delivery pathway to the vacuole (Conibear and Tam, 2006). Changes in the relative rates of any of these trafficking steps could cause cargo proteins to accumulate in endosomal compartments. Rapid recycling to the cell surface has been demonstrated for a truncated form of Ste3p that is subject to ligand-induced, but not constitutive, endocytosis (Ste3Δ365p; Chen and Davis, 2000). To determine if vps68 mutants exhibit a recycling defect that could account for the endosomal accumulation of Ste3p, strains induced to express GAL1-driven Ste3Δ365-HA for 2 h were grown in the presence of a-factor, the peptide ligand for Ste3p, to allow uptake and transport to endosomes. Before a-factor addition, Ste3Δ365p was present at the surface of wild-type and vps68 mutant cells where it was largely protease-sensitive (Figure 6A). After a 45-min incubation with a-factor, a significant fraction of the Ste3Δ365 was protected from protease in an internal pool consistent with a partial redistribution to endosomes. After a-factor withdrawal, Ste3Δ365p continues to recycle to the cell surface but is no longer internalized. Because protease treatment of intact spheroplasts digests only surface-exposed receptor, the rate at which the intracellular pool of Ste3Δ365p acquires protease sensitivity provides a measure of recycling (Chen and Davis, 2000). In wild-type cells (Figure 6B), most of the receptor was surface-accessible after 10 min in medium lacking a-factor. The rate at which Ste3Δ365p regained protease sensitivity was not reduced in vps68 strains, indicating that recycling back to the cell surface is not blocked in these mutants.
Figure 6.
Loss of VPS68 does not slow the endosome-to-cell surface recycling of Ste3Δ365p. (A) Ligand-dependent internalization of Ste3Δ365p. Wild type (NDY1181) and vps68 (BWY14) mutant cells were exposed to a-factor to induce endocytosis, and treated with protease to cleave surface-exposed Ste3Δ365 protein. Ste3Δ365p was detected in cell extracts by Western blotting with anti-HA mAb. (B) Cell surface recycling of internalized Ste3Δ365. Wild-type and vps68 strains expressing Ste3Δ365 were treated as described in A and incubated in fresh medium lacking a-factor. Aliquots removed at the times indicated were subjected to a second protease treatment. The time course with which the internal pool of Ste3Δ365 regains protease sensitivity provides a measure of cell surface recycling.
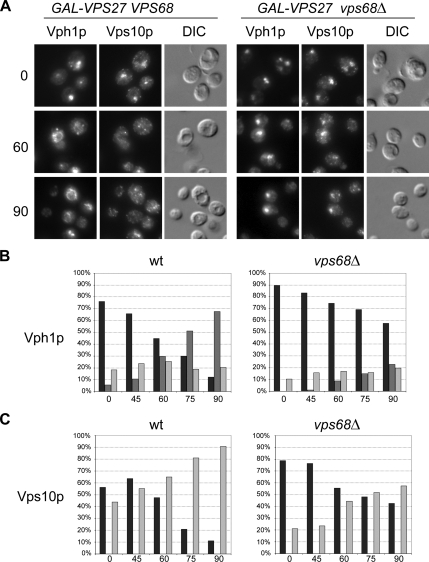
vps68 Mutants Are Defective in Two Trafficking Pathways Leaving the Endosome
Taken together, the localization, pulse-chase immunoprecipitation and epistasis results suggest that vps68 mutants have defects in exit from the late endosome. Although no alteration was found in the rate of transport to the cell surface, two other transport pathways lead out of the MVB. We used an assay that measures exit from the late endosome to distinguish Golgi recycling from vacuolar delivery (Bryant and Stevens, 1997; Nothwehr et al., 2000). In strains lacking the class E gene VPS27, resident TGN and newly synthesized vacuolar proteins that transit the MVB are trapped in the aberrant form of the MVB termed the “class E” compartment. When expression of VPS27 is restored, trafficking out of this compartment resumes, and TGN proteins such as Vps10p and vacuolar proteins such as Vph1p regain their wild-type cellular distribution. In agreement with previous observations, Vps10p and Vph1p colocalized in punctate structures next to the vacuole that are characteristic of class E mutants when VPS27 was not expressed (Figure 7A). Ninety minutes after adding galactose to restore GAL1-driven VPS27 expression in wild-type cells, class E compartments were rarely seen and Vph1p had redistributed to the vacuolar membrane in 70% of cells, whereas Vps10p had regained its punctate localization in Golgi and endosomes in 90% of cells. In contrast, 90 min after restoring VPS27 expression to vps68 mutants, class E-like structures that contained both Vps10p and Vph1p were still visible in a large proportion (>40%) of the cells (Figure 7, B and C). Because recycling from the late endosome to the Golgi as well as late endosome-to-vacuole transport are significantly delayed in vps68 mutants, Vps68p appears to act parallel to or downstream of the ESCRT complex to regulate two separate pathways from the late endosome.
Figure 7.
VPS68 is required for two transport pathways out of the late endosome. Wild-type (SNY128) and vps68 (BWY12) strains containing the vps27Δ mutation and carrying pHY5 for the GAL1-driven expression of VPS27 were grown to log phase in raffinose medium. Aliquots removed 0, 45, 60, 75, and 90 min after the addition of galactose were processed for the localization of integrated, HA-tagged Vps10p and endogenous Vph1p by double-label immunofluorescence microscopy. (A) Representative images of cells fixed 0, 60, and 90 min after galactose addition. (B and C) Localization of Vph1p (B) and Vps10p (C) was scored for 100–200 cells at each time point. Dark gray bars, vacuole; black bars, class E compartment; light gray bars, dispersed/ambiguous. Bar, 2 μm.
DISCUSSION
A goal of genome-scale studies is to identify all components of a given process and classify them using objective high throughput techniques. By identifying endosomal-trafficking factors using a biochemical, genome-wide assay and assigning them to complexes and pathways by phenotypic miniarray profiling, we find that large-scale phenotypic data can be used to describe a cellular process in molecular detail.
A high-density genetic interaction map of a subset of genes implicated in ER function was previously used to identify protein complexes in the early secretory pathway (Schuldiner et al., 2005). This approach, referred to as an E-MAP (epistatic miniarray profile), has recently been extended to other processes (Collins et al., 2007; St. Onge et al., 2007). Selecting genes already linked to a specific process or organelle enriches for genetic interactions and functional relationships but makes it difficult to identify genes previously unconnected to the process of interest (Conibear, 2005). The two-part method we describe here has several advantages. The initial genome-wide phenotypic screening provides an unbiased approach for the identification of genes that contribute most to a given process and ensures that all nonessential subunits of relevant complexes are present in the set. The subsequent generation of a phenotypic miniarray profile (P-MAP) does not require the construction of an exhaustive set of double mutants and is therefore less labor intensive than E-MAP techniques and more readily applied to other systems (e.g., RNA interference in human cells). In principle, a P-MAP could be constructed using available phenotypic data from large-scale drug sensitivity studies (Parsons et al., 2006). However, we find biochemical readouts of phenotypes relevant to trafficking processes—such as CPY or pro-alpha factor secretion, or cell surface chitin synthesis—lead to the identification of a greater number of complexes, with more significant p values, than growth-based assays (unpublished observations).
The identification of the SWR-C chromatin-remodeling complex in a study of vesicle transport demonstrates that systematic phenotypic analysis can provide information about pathways distantly related to the one under study. Because we have used an unsupervised statistical approach to find the most relevant complexes, without reference to gene ontology or other large-scale datasets, we expect this method could be widely applied to uncharacterized cellular processes to map out relevant complexes and pathways and define their relative contribution to the phenotype of interest.
The Vps55/68 Complex Is Required for Two Transport Steps out of the Late Endosome
In addition to describing known endosomal-sorting complexes, our unbiased predictive process identified a new membrane-associated complex, Vps55/68, which is required for a late stage of endosomal transport. Recent systematic proteomics studies have identified many yeast endosomal transport complexes but did not find all of the complexes predicted by our study, including Vps55/68 (Gavin et al., 2006; Krogan et al., 2006). Phenotypic methods may be more effective at identifying proteins that associate via hydrophobic, transient, or weak interactions or those linked by enzyme-substrate relationships, such as PI3K production and retromer function. Phenotypic profiling and physical interaction analysis are therefore complementary approaches whose integration is expected to yield high confidence predictions when applied to other processes.
Our epistasis analysis indicates the Vps55/68 complex acts with or downstream of the ESCRT machinery to regulate a novel step in late endosome sorting and/or maturation. Loss of ESCRT subunits blocks the formation of inward-budding vesicles at the MVB and causes multiple cargoes to accumulate in an aberrant “class E” endosomal compartment next to the vacuole. It is not clear how defects in lumenal vesicle formation resulting from loss of the ESCRT machinery delay transport to the vacuolar and slow recycling to the Golgi (Piper et al., 1995). Preventing inward budding of vesicles may alter the protein and lipid composition of the endosome limiting membrane, and thus indirectly inhibit membrane budding and fusion processes. We found that mutation of either VPS68 or VPS55 similarly slows the transit of biosynthetic and endocytic cargo proteins through the MVB, but does not block the formation of intralumenal vesicles or cause the accumulation of an aberrant protease-active compartment. A subset of class E vps mutants (including did2/fti1/vps46, vps60, and vta1) that affect ESCRT function by regulating the AAA ATPase Vps4p exhibit mild CPY-sorting defects (<20%) and partial impairment of lumenal MVB vesicle formation (Azmi et al., 2006; Lottridge et al., 2006). Our results do not preclude a similar role for the Vps55/68 complex in regulating ESCRT proteins and increasing the efficiency of lumenal vesicle formation; however, the strong CPY-sorting defects of vps55 and vps68 mutants suggest they have a different function. Taken together, our data support the model that VPS55 and VPS68 regulate endosome maturation at a step downstream of the ESCRT machinery.
As endosomes mature, they lose the ability to fuse with primary endocytic vesicles and acquire the ability to fuse with the vacuole/lysosome. The maturation process may effect this change in fusogenic potential by the recruitment or loss of specific components of the fusion machinery. Interestingly, other small four-transmembrane domain proteins, including Yip1p and Sys1p, are required for the recruitment of small GTPases to specific compartments (Yang et al., 1998; Behnia et al., 2004; Setty et al., 2004). Although we were unable to detect a physical association between the Vps55/68 complex and the endosomal Rab GTPase Vps21p, the presence of these proteins in the same phenotypic cluster suggests the Vps55/68 complex may interact transiently with Vps21p to fulfill a specific step in endosome maturation. The compartment-specific recruitment of additional trafficking factors is a plausible model for Vps55/68 function worthy of future investigation.
Vps55p and Vps68p are each represented by two different isoforms in humans. Both SMP1 (small membrane protein 1) and c21orf4 encode predicted homologues of Vps68p, whereas OB-RGRP and LEPROTL1 are homologous to Vps55p (Bailleul et al., 1997). OBRGRP has previously been shown to complement the CPY-trafficking defects of a vps55 mutant when expressed in yeast, indicating functional conservation (Belgareh-Touze et al., 2002). The presence of two sets of Vps55p- and Vps68p-related proteins raises the interesting possibility that they may form distinct, functionally divergent complexes in higher cells. Further characterization of the role of the Vps55/68 complex in endosomal trafficking may yield insight into the processes of endosomal maturation in yeast and mammalian cells.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Rob Piper (University of Iowa), Matthew Seaman (University of Cambridge), Scott Emr (Cornell University), and Hugh Pelham (MRC Laboratory of Molecular Biology, Cambridge, UK) for providing plasmids, to George Sprague for antibodies and to Amy Roth and Nick Davis (both of Wayne State University) for advice and reagents. We thank Rick White for creating the Cytoscape plug-in and members of the Conibear lab for helpful discussions. This work was supported by funding from the Canadian Institutes of Health Research (CIHR; E.C.), the National Sciences and Engineering Research Council of Canada (J.B.), and National Institutes of Health Grant GM32448 (T.H.S.). Additional support was provided by Michael Smith Foundation for Health Research Career Awards (E.C. and J.B.) and a CIHR New Investigator Award (E.C.).
Abbreviations used:
- CPY
carboxypeptidase Y
- ALP
alkaline phosphatase
- MVB
multivesicular body
- ESCRT
endosomal-sorting complex required for transport.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-07-0659) on January 23, 2008.
REFERENCES
- Arighi C. N., Hartnell L. M., Aguilar R. C., Haft C. R., Bonifacino J. S. Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J. Cell Biol. 2004;165:123–133. doi: 10.1083/jcb.200312055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi I., Davies B., Dimaano C., Payne J., Eckert D., Babst M., Katzmann D. J. Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J. Cell Biol. 2006;172:705–717. doi: 10.1083/jcb.200508166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailleul B., Akerblom I., Strosberg A. D. The leptin receptor promoter controls expression of a second distinct protein. Nucleic Acids Res. 1997;25:2752–2758. doi: 10.1093/nar/25.14.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnia R., Panic B., Whyte J. R., Munro S. Targeting of the Arf-like GTPase Arl3p to the Golgi requires N-terminal acetylation and the membrane protein Sys1p. Nat. Cell Biol. 2004;6:405–413. doi: 10.1038/ncb1120. [DOI] [PubMed] [Google Scholar]
- Belgareh-Touze N., Avaro S., Rouille Y., Hoflack B., Haguenauer-Tsapis R. Yeast Vps55p, a functional homolog of human obesity receptor gene-related protein, is involved in late endosome to vacuole trafficking. Mol. Biol. Cell. 2002;13:1694–1708. doi: 10.1091/mbc.01-12-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino C. J., Chavez E. M., Bonifacino J. S. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:2486–2501. doi: 10.1091/mbc.02-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J. S., Rojas R. Retrograde transport from endosomes to the trans-Golgi network. Nat. Rev. Mol. Cell Biol. 2006;7:568–579. doi: 10.1038/nrm1985. [DOI] [PubMed] [Google Scholar]
- Bowers K., Levi B. P., Patel F. I., Stevens T. H. The sodium/proton exchanger Nhx1p is required for endosomal protein trafficking in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2000;11:4277–4294. doi: 10.1091/mbc.11.12.4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers K., Stevens T. H. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2005;1744:438–454. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Bryant N. J., Piper R. C., Gerrard S. R., Stevens T. H. Traffic into the prevacuolar/endosomal compartment of Saccharomyces cerevisiae: a VPS45-dependent intracellular route and a VPS45-independent, endocytic route. Eur. J. Cell Biol. 1998;76:43–52. doi: 10.1016/S0171-9335(98)80016-2. [DOI] [PubMed] [Google Scholar]
- Bryant N. J., Stevens T. H. Two separate signals act independently to localize a yeast late Golgi membrane protein through a combination of retrieval and retention. J. Cell Biol. 1997;136:287–297. doi: 10.1083/jcb.136.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda P., Padilla S. M., Sarkar S., Emr S. D. Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase. J. Cell Sci. 2002;115:3889–3900. doi: 10.1242/jcs.00090. [DOI] [PubMed] [Google Scholar]
- Burston H. E., Davey M., Conibear E. Genome-wide analysis of membrane transport using yeast knockout arrays. In: Vancura A., editor. Methods in Molecular Biology, vol. Membrane Trafficking. Totowa, NY: Humana Press; 2008. (in press) [DOI] [PubMed] [Google Scholar]
- Carpenter A. E., Sabatini D. M. Systematic genome-wide screens of gene function. Nat. Rev. 2004;5:11–22. doi: 10.1038/nrg1248. [DOI] [PubMed] [Google Scholar]
- Cereghino J. L., Marcusson E. G., Emr S. D. The cytoplasmic tail domain of the vacuolar protein sorting receptor Vps10p and a subset of VPS gene products regulate receptor stability, function, and localization. Mol. Biol. Cell. 1995;6:1089–1102. doi: 10.1091/mbc.6.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Davis N. G. Recycling of the yeast a-factor receptor. J. Cell Biol. 2000;151:731–738. doi: 10.1083/jcb.151.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Davis N. G. Ubiquitin-independent entry into the yeast recycling pathway. Traffic. 2002;3:110–123. doi: 10.1034/j.1600-0854.2002.030204.x. [DOI] [PubMed] [Google Scholar]
- Chu T., Sun J., Saksena S., Emr S. D. New component of ESCRT-I regulates endosomal sorting complex assembly. J. Cell Biol. 2006;175:815–823. doi: 10.1083/jcb.200608053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S. R., et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Conibear E. An E-MAP of the ER. Cell. 2005;123:366–368. doi: 10.1016/j.cell.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Conibear E., Stevens T. H. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol. Biol. Cell. 2000;11:305–323. doi: 10.1091/mbc.11.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E., Stevens T. H. Studying yeast vacuoles. Methods Enzymol. 2002;351:408–432. doi: 10.1016/s0076-6879(02)51861-9. [DOI] [PubMed] [Google Scholar]
- Conibear E., Tam Y. Y. Protein Trafficking: Mechanisms and Regulation. Austin, TX: Landes Bioscience; 2006. The endocytic pathway. [Google Scholar]
- Cooper A. A., Stevens T. H. Vps10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J. Cell Biol. 1996;133:529–541. doi: 10.1083/jcb.133.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles C. R., Snyder W. B., Burd C. G., Emr S. D. Novel Golgi to vacuole delivery pathway in yeast: identification of a sorting determinant and required transport component. EMBO J. 1997;16:2769–2782. doi: 10.1093/emboj/16.10.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtiss M., Jones C., Babst M. Efficient cargo sorting by ESCRT-I and the subsequent release of ESCRT-I from multivesicular bodies requires the subunit Mvb12. Mol. Biol. Cell. 2007;18:636–645. doi: 10.1091/mbc.E06-07-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsow T., Rieder S. E., Emr S. D. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J. Cell Biol. 1997;138:517–529. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A. C., et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Graham L. A., Flannery A. R., Stevens T. H. Structure and assembly of the yeast V-ATPase. J. Bioenerg. Biomembr. 2003;35:301–312. doi: 10.1023/a:1025772730586. [DOI] [PubMed] [Google Scholar]
- Gruenberg J., Stenmark H. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Fink G. Methods in Enzymology. Vol. 194. San Diego, CA: Academic Press; 1991. Guide to yeast genetics and molecular biology; p. 933. [PubMed] [Google Scholar]
- Hanson P. K., Grant A. M., Nichols J. W. NBD-labeled phosphatidylcholine enters the yeast vacuole via the pre-vacuolar compartment. J. Cell Sci. 2002;115:2725–2733. doi: 10.1242/jcs.115.13.2725. [DOI] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Hurley J. H., Emr S. D. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann D. J., Odorizzi G., Emr S. D. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell. Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- Kobor M. S., Venkatasubrahmanyam S., Meneghini M. D., Gin J. W., Jennings J. L., Link A. J., Madhani H. D., Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostelansky M. S., Schluter C., Tam Y. Y., Lee S., Ghirlando R., Beach B., Conibear E., Hurley J. H. Molecular architecture and functional model of the complete yeast ESCRT-I heterotetramer. Cell. 2007;129:485–498. doi: 10.1016/j.cell.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N. J., et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Krogan N. J., et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- Lam K. K., Davey M., Sun B., Roth A. F., Davis N. G., Conibear E. Palmitoylation by the DHHC protein Pfa4 regulates the ER exit of Chs3. J. Cell Biol. 2006;174:19–25. doi: 10.1083/jcb.200602049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M. J., Nichols B. J., Prescianotto-Baschong C., Riezman H., Pelham H. R. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol. Biol. Cell. 2000;11:23–38. doi: 10.1091/mbc.11.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling R. F. A probability theory of cluster analysis. J. Am. Stat. Assoc. 1973;68:159–164. [Google Scholar]
- Lottridge J. M., Flannery A. R., Vincelli J. L., Stevens T. H. Vta1p and Vps46p regulate the membrane association and ATPase activity of Vps4p at the yeast multivesicular body. Proc. Natl. Acad. Sci. USA. 2006;103:6202–6207. doi: 10.1073/pnas.0601712103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G., Shen X., Landry J., Wu W. H., Sen S., Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Morita E., Sundquist W. I. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- Nothwehr S. F., Ha S. A., Bruinsma P. Sorting of yeast membrane proteins into an endosome-to-Golgi pathway involves direct interaction of their cytosolic domains with Vps35p. J. Cell Biol. 2000;151:297–310. doi: 10.1083/jcb.151.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich A. J., Davies B. A., Payne J. A., Katzmann D. J. Mvb12 is a novel member of ESCRT-I involved in cargo selection by the multivesicular body pathway. Mol. Biol. Cell. 2007;18:646–657. doi: 10.1091/mbc.E06-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons A. B., et al. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell. 2006;126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- Piper R. C., Bryant N. J., Stevens T. H. The membrane protein alkaline phosphatase is delivered to the vacuole by a route that is distinct from the VPS-dependent pathway. J. Cell Biol. 1997;138:531–545. doi: 10.1083/jcb.138.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper R. C., Cooper A. A., Yang H., Stevens T. H. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J. Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville N. R., Conibear E. Toward the systems biology of vesicle transport. Traffic. 2006;7:761–768. doi: 10.1111/j.1600-0854.2006.00436.x. [DOI] [PubMed] [Google Scholar]
- Reggiori F., Pelham H. R. Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J. 2001;20:5176–5186. doi: 10.1093/emboj/20.18.5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman J. H., Stevens T. H. Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell. 1986;47:1041–1051. doi: 10.1016/0092-8674(86)90819-6. [DOI] [PubMed] [Google Scholar]
- Schuldiner M., et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Seaman M. N. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J. Cell Biol. 2004;165:111–122. doi: 10.1083/jcb.200312034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M. N. Recycle your receptors with retromer. Trends Cell Biol. 2005;15:68–75. doi: 10.1016/j.tcb.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Setty S. R., Strochlic T. I., Tong A. H., Boone C., Burd C. G. Golgi targeting of ARF-like GTPase Arl3p requires its Nalpha-acetylation and the integral membrane protein Sys1p. Nat. Cell Biol. 2004;6:414–419. doi: 10.1038/ncb1121. [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slessareva J. E., Routt S. M., Temple B., Bankaitis V. A., Dohlman H. G. Activation of the phosphatidylinositol 3-kinase Vps34 by a G protein alpha subunit at the endosome. Cell. 2006;126:191–203. doi: 10.1016/j.cell.2006.04.045. [DOI] [PubMed] [Google Scholar]
- Stawiecka-Mirota M., Pokrzywa W., Morvan J., Zoladek T., Haguenauer-Tsapis R., Urban-Grimal D., Morsomme P. Targeting of Sna3p to the endosomal pathway depends on its interaction with Rsp5p and multivesicular body sorting on its ubiquitylation. Traffic. 2007;8:1280–1296. doi: 10.1111/j.1600-0854.2007.00610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Onge R. P., Mani R., Oh J., Proctor M., Fung E., Davis R. W., Nislow C., Roth F. P., Giaever G. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat. Genet. 2007;39:199–206. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens T., Esmon B., Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 1982;30:439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- R Code Development Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2005. [Google Scholar]
- Tong A. H., et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Urbanowski J. L., Piper R. C. Ubiquitin sorts proteins into the intralumenal degradative compartment of the late-endosome/vacuole. Traffic. 2001;2:622–630. doi: 10.1034/j.1600-0854.2001.20905.x. [DOI] [PubMed] [Google Scholar]
- von Schwedler U. K., et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- Winzeler E. A., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Wolf D. H., Fink G. R. Proteinase C (carboxypeptidase Y) mutant of yeast. J. Bacteriol. 1975;123:1150–1156. doi: 10.1128/jb.123.3.1150-1156.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Matern H. T., Gallwitz D. Specific binding to a novel and essential Golgi membrane protein (Yip1p) functionally links the transport GTPases Ypt1p and Ypt31p. EMBO J. 1998;17:4954–4963. doi: 10.1093/emboj/17.17.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.