Abstract
In eukaryotic cells, proteasomes play an essential role in intracellular proteolysis and are involved in the control of most biological processes through regulated degradation of key proteins. Analysis of 20S proteasome localization in human cell lines, using ectopic expression of its CFP-tagged α7 subunit, revealed the presence in nuclear foci of a specific and proteolytically active complex made by association of the 20S proteasome with its PA28γ regulator. Identification of these foci as the nuclear speckles (NS), which are dynamic subnuclear structures enriched in splicing factors (including the SR protein family), prompted us to analyze the role(s) of proteasome-PA28γ complexes in the NS. Here, we show that knockdown of these complexes by small interfering RNAs directed against PA28γ strongly impacts the organization of the NS. Further analysis of PA28γ-depleted cells demonstrated an alteration of intranuclear trafficking of SR proteins. Thus, our data identify proteasome-PA28γ complexes as a novel regulator of NS organization and function, acting most likely through selective proteolysis. These results constitute the first demonstration of a role of a specific proteasome complex in a defined subnuclear compartment and suggest that proteolysis plays important functions in the precise control of splicing factors trafficking within the nucleus.
INTRODUCTION
Proteasomes are large proteolytic complexes that play a central role in intracellular proteolysis and are particularly important for the tightly controlled degradation of many critical regulatory proteins (Ciechanover and Schwartz, 2004). The term proteasomes defines a family of complexes that share a common proteolytic core (the 20S proteasome) but differ in the regulators associated with the complex (Demartino and Slaughter, 1993; Rechsteiner and Hill, 2005). This diversity of proteasome complexes might reflect the necessity for the cell to degrade hundreds of different proteins at the same time, often in a highly selective and regulated manner and in certain cases only at precise locations in the cell (Pines and Lindon, 2005). However, whether the activity of each of these complexes is restricted to a particular set of substrates or to particular cellular domains is presently unclear.
The 20S proteasome is a cylindrical-shaped protein complex, comprising 28 subunits arranged in four stacked rings: two outer rings each made of seven alpha-type subunits (α1–α7) and two inner rings each made of seven beta-type subunits (β1–β7). The catalytic sites are enclosed in the inner catalytic chamber made by the β-type subunits (Groll et al., 1997). In cells, the 20S proteasome is found associated with a variety of regulators that modulate its functions. The best understood 20S regulator is the 19S complex, which binds in an ATP-dependent manner to both ends of the 20S cylinder to form the 26S proteasome that is, in particular, involved in the degradation of ubiquitylated proteins (Coux et al., 1996; Voges et al., 1999). The 20S proteasome can also associate with a variety of other regulators (DeMartino and Slaughter, 1999), including PA200 (Ortega et al., 2005) and the complexes of the PA28 family, PA28α/β and PA28γ (Rechsteiner and Hill, 2005), to form other proteolytic complexes that likely carry out specific functions. PA28α/β is a heptamer of the two closely related PA28α and PA28β subunits (Whitby et al., 2000). It can bind to both ends of 20S proteasome (Gray et al., 1994) and also can be integrated into “hybrid proteasomes” in which one molecule of 19S complex and one molecule of PA28α/β are bound at each end of 20S proteasome (Cascio et al., 2002). PA28α/β is essentially cytoplasmic and, in vitro, has been shown to stimulate the peptidase, but not the protease, activities of the 20S proteasome (Ma et al., 1992). It is thought to play an important role in the production of MHC class I antigenic peptides (Groettrup et al., 1996). PA28γ, initially called “Ki antigen,” is similar to PA28α and β (Kandil et al., 1997; Tanahashi et al., 1997), but forms only homoheptamers and is mostly nuclear. Its functions are poorly characterized, but studies in mice (Murata et al., 1999; Barton et al., 2004) and flies (Masson et al., 2003) suggest a role for this complex in cell cycle progression and apoptosis. Furthermore, PA28γ has been recently identified as a key player in the degradation of the oncogene SRC-3 (Li et al., 2006) and of the CKI p21 (Chen et al., 2007; Li et al., 2007) and was shown to act as a novel regulator of Cajal Bodies integrity under genotoxic stress (Cioce et al., 2006).
To better understand the specific functions of the various proteasome complexes, it is important to know where in the cells each type of complex is present and active. However, in most previous studies of proteasome localization, only immunofluorescence analyses of the 20S proteasome have been conducted, and it was not known whether the fluorescence corresponded to free 20S or 20S bound to the 19S complex (i.e., the 26S proteasome), or to other regulators. In general, the 20S proteasome appears distributed in both cytoplasmic and nuclear compartments (Reits et al., 1997; Wojcik and DeMartino, 2003), and most of it is found in the soluble fraction after cell lysis. A small percentage, however, seems to be associated with defined cellular structures. In the cytoplasm, the 20S proteasome is found associated with the cytoskeleton (Grossi de Sa et al., 1988), the endoplasmic reticulum (Brooks et al., 2000), and a perinuclear structure that coincides with the centrosome (Wigley et al., 1999). In the nucleus, the 20S proteasome is found in distinct nuclear regions, including PML bodies (Rockel and von Mikecz, 2002; Wojcik and DeMartino, 2003) and nuclear speckles (Rockel and von Mikecz, 2002; Rockel et al., 2005).
Nuclear speckles (NS), also called interchromatin granule clusters (IGCs), correspond to subnuclear domains enriched in components of the pre-mRNA splicing machinery such as the SR protein family (Misteli et al., 1997; Rockel and von Mikecz, 2002; Lamond and Spector, 2003; Rockel et al., 2005). They are highly dynamic domains with a constant in-out shuttling of splicing factors that undergo posttranslational modifications (particularly phosphorylation/dephosphorylation) necessary for their subsequent recruitment at transcription sites (Misteli et al., 1998). Indeed, disassembly of NS leads to dramatic reduction of accumulation of splicing factors at transcription sites and perturbs coordination between transcription and splicing (Sacco-Bubulya and Spector, 2002). The presence of proteasomes in NS, together with the fact that proteasome inhibitors promote the accumulation of SR protein SC35 in NS (Rockel and von Mikecz, 2002), suggests that, as with phosphorylation, proteolysis might be intrinsically involved in the control of the intranuclear trafficking of splicing factors and their repartition between NS and transcription sites.
In this study, we show that a specific form of proteasomes (i.e., 20S proteasome-PA28γ complexes) is present in the NS in an active form. Also, we demonstrate that a reduction of PA28γ expression by siRNA triggers a profound alteration of nuclear speckles organization and affects the recruitment of SR proteins to transcription sites.
MATERIALS AND METHODS
Antibodies and Compounds
Antibodies against green fluorescent protein (GFP) were obtained from Roche Diagnostic (Meylan, France). Antibodies against α3 (PW8115), α4 (PW8120), α6 (PW8100), α7 (PW8110), Mss1/Rpt1 (PW8825), Sug1/Rpt6 (PW9265), PA28γ (PW8190), and PA28β (PW8280) were purchased from Affiniti Research Products (Nottingham, UK), and antibodies against SC35 (S4045), SF2/ASF (32–4500), and HA (12CA5) were purchased from Sigma (L'isle d'Abeau, France), Zymed (South San Francisco, CA), and Roche Diagnostic, respectively. Alexa- and HRP-conjugated secondary antibodies were purchased respectively from Interchim (Montluçon, France) and Bio-Rad SA (Hercules, CA). Suc-Leu-Leu-Val-Tyr-7amido-4-methylcoumarin (suc-LLVY-AMC, Bachem Biochimie SARL, Voisins Le Bretonneux, France), MG132 (Boston Biochem, Cambridge, MA), DRB (Sigma), and actinomycin D (Sigma) were dissolved in DMSO. DQ-Ovalbumin (DQ-OVA) was purchased from Calbiochem (La Jolla, CA).
Plasmid Constructs and Small Interfering RNA
Human cDNA (768 base pairs) encoding the full-length α7 (PSMA3 or C8, Accession no. P25788), with or without the final stop codon, was amplified from a human fibroblast cDNA library by PCR and inserted into pcDNA3 or pML1-ECFP vectors and verified by sequencing. The cDNA coding for α7-ECFP and enhanced cyan fluorescent protein (ECFP) were then inserted into pTRE2 vector (Clontech, Saint-Germain-en-Laye, France). The pEGFP-hSC35, pcDNA3-Tat, and pEGFP-SF2/ASF constructs were generated in the laboratory of J. Tazi and E. Bertrand, respectively. To construct mammalian expression vectors for bimolecular fluorescence complementation (BiFC) analyses, sequences encoding amino acids 1–154 of enhanced yellow fluorescent protein (EYFP; N-terminal fragment [YN]) and amino acids 155–239 of EYFP (C-terminal fragment of YFP [YC]) were cloned into pcDNA3. Full-length α7 cDNA (without the final stop) was inserted in pcDNA3-YN and full-length PA28γ cDNA in pcDNA3-YC to create α7-NY and CY-PA28γ fusion proteins, respectively. Small interfering RNA (siRNA) duplexes against human PA28γ were purchased from Ambion (Austin, TX; PSME3, ID: 138669; siRNA 1) or MWG-Biotech (Ebersberg, Germany) (siRNA 2; Cioce et al., 2006). Scrambled duplexes (Silencer negative control 1) were purchased from Ambion. A PA28γ construct resistant to the siRNA 2 was made by mutation of six nucleotides in PA28γ cDNA subcloned in pcDNA3-3HA vector.
Cell Culture and Transfection
HeLa cells (ATCC, Manassas, VA), U2OS-tTA (Theis-Febvre et al., 2003), 2E11 cell line (Boireau et al., 2007; du Chéné et al., 2007), and MelJuso cell line expressing constitutively the LMP2-GFP fusion protein (Reits et al., 1997) were cultured as described in the respective articles. U2OS-tTA cells expressing stably α7-CFP or CFP were established as described (Theis-Febvre et al., 2003). Plasmid transfections in cells were carried out using the JetPEI reagent (Qbiogene, Carlsbad, CA). Twenty-five nanomoles per milliliter of each siRNA were transfected into cells with the oligofectamine reagent (Invitrogen, Carlsbad, CA), following the manufacturer's instructions.
Immunoprecipitation and Immunoblot Analysis
Cells were lysed for 15 min at 4°C in lysis buffer (50 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 0.5 mM EDTA, 1 mM ATP, 1 mM DTT, 10% glycerol, 0.5% IGEPAL CA630). This treatment does not solubilize NS components (not shown), which can be later solubilized using a buffer containing 0.3% Triton X-100. Lysates were clarified by centrifugation for 10 min at 10,000 × g, and the protein concentration of the supernatant was determined using BSA as a standard (Bradford reagent assay, Pierce, Rockford, IL). Total lysate (200 μg) was precleared for 30 min, and immunoprecipitations were performed using 3 μl of indicated antibodies and protein A or G magnetic beads (Dynal, Lake Success, NY) in lysis buffer containing 100 mM NaCl. After several washes, bead pellets were boiled in SDS sample buffer, separated by SDS-PAGE and transferred to nitrocellulose membranes. Immunoblotting was performed as described previously (Baldin et al., 1993). Visualization was performed by enhanced chemiluminescence (Perkin Elmer-Cetus, Norwalk, CT).
Proteasome Activity Assay
Proteasome activity was measured on beads of immunopurified proteasome by incubation at 37°C for 20 min in 100 μl of activity buffer (20 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 1 mM ATP, 1 mM DTT, 10% glycerol) containing 100 μM suc-LLVY-AMC. The fluorescence of the free AMC generated by proteasome activity (λex = 380 nm, λ em = 440 nm) was measured on a spectrofluorimeter (Perkin Elmer-Cetus). Proteasome activity in living cells was analyzed in U2OS cells as described (Rockel et al., 2005).
Indirect Immunofluorescence
Cells were transfected or not with expression vectors encoding the mentioned cDNAs. At indicated times after the transfection and/or induction, cells were fixed using 3.7% formaldehyde and then permeabilized with 0.25% Triton X-100 and cold methanol. Proteasome subunits were detected by incubating the cells with the first antibody, followed by a second incubation with Alexa-594– or 488–conjugated goat anti-mouse (or anti-rabbit) antibody (1:1000; Molecular Probes, Eugene, OR). In all cases, DNA was stained with DAPI dye (Sigma). In situ hybridization was performed as described (Verheggen et al., 2002). Microscopic examinations were performed with a 63×/1.32 NA or 100×/1.4 NA oil immersion objective lens under a Leica DMRA microscope and a 12-bit Micromax YHS 1300 camera (Deerfield, IL). Images were acquired as TIF files using the MetaMorph 6.2 imaging software (Universal Imaging, West Chester, PA).
Fluorescence Recovery after Photobleaching
Fluorescence recovery after photobleaching (FRAP) analyses were performed at 37°C on HeLa cells transiently expressing α7-GFP with a laser scanning microscope (LSM Meta 510; Zeiss, Jena, Germany). The argon laser spectral line (wavelength of 488 nm) was set to an intensity not greater than 0.3% of its total power (458, 477, 488, 514, 30 mW) for image collection. A 40×/1.2 NA water immersion objective was used. After 12 prebleach scans (1 scan/s), a region of interest (ROI) was photobleached using 15 iterations at 3 mW laser power. Twelve-bit images were collected before, immediately after, and at defined intervals after bleaching (once every 0.8 s). Our total experiment time was set to 130 s, which was sufficient for complete recovery from photobleaching. The mobile fraction (R) was determined using the formula: R = (F∞ − F0)/(Fi − F0). τ1/2 was directly read off of the graph as the time when fluorescence had recovered by 50%.
RT-PCR
Total RNA was extracted using the Mini RNA isolation II kit (Zymo Research, Orange, CA). One microgram of RNA was reverse-transcribed using the first-strand cDNA synthesis kit (GE Healthcare, Waukesha, WI) according to the supplier's specifications. cDNAs were amplified with the Taq polymerase (Invitrogen) using specific primers corresponding to S26 and artificial gene mRNA (100 and 193 nucleotides, respectively).
RESULTS
Ectopically Expressed Proteasome α7 Subunit Accumulates into Nuclear Foci
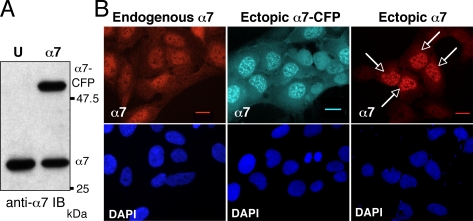
To study the subcellular localization of the 20S proteasome in living cells and to avoid overproduction of the protein by transient expression, we established stable U2OS-tTA cell lines expressing 20S α-subunits in fusion with CFP, under an inducible promotor (tet-off). In the tested cell lines, exogenous α-CFPs were expressed at a level similar to that of endogenous α proteins (Figure 1A; α7-CFP as an example). Surprisingly, in addition to the relatively even cytoplasmic and nuclear distribution of α7 detected by indirect immunofluorescence in parental U2OS-tTA cells, distinct nuclear foci were apparent in α7-CFP expressing cells (Figure 1B; compare panel endogenous α7 to panel ectopic α7-CFP), which contrasted with the images obtained with cell lines expressing other tagged subunits such as α3-CFP (not shown) or LMP2 (Reits et al., 1997). Importantly, despite the apparent brightness of the α7-CFP containing foci, quantification of the fluorescence using the Metamorph software showed that α7-CFP containing foci represent only 7.7% of the total cellular α7-CFP and 22% of the nuclear α7-CFP.
Figure 1.
Ectopic α7-CFP and α7 accumulate into nuclear foci. (A) Expression of the α7-CFP protein in cells. Total cellular extracts (30 μg) of human osteosarcoma U2OS-tTA (U) or U2OS-tTA cells stably transfected with vectors conditionally expressing α7-CFP (α7) cultured in the absence of tetracycline for 48 h were separated by electrophoresis and analyzed by immunoblotting using anti-α7 antibodies. (B) The cellular distribution of α7 in U2OS-tTA cells either untransfected (endogenous α7), stably transfected with pTRE2-α7-CFP (ectopic α7-CFP), or transiently transfected with pcDNA3-α7 (ectopic α7) was analyzed by indirect immunofluorescence using anti-α7 antibodies (red) or by CFP fluorescence (cyan) on fixed cells. Arrows indicate transiently transfected cells. Observation with a 63× objective. Bar, 10 μm.
Although this peculiar distribution could reflect the presence of aggregates accumulating upon expression of the CFP-tagged subunit, several lines of evidence indicated that this was not the case. First, we observed that this effect was independent of the presence of the CFP moiety on the subunit: expression of unmodified α7 protein also gave rise to accumulation of the subunit into similar nuclear foci (Figure 1B, right panel). Second, upon prolonged induction of the α7-CFP protein, no defect on cell cycle progression could be detected (data not shown), as would be expected if proteasome function were altered. Third, we observed that the distribution of α7-CFP dramatically changed during mitosis: α7-CFP diffused throughout the cell during metaphase and reformed foci in telophase (Figure S1). In addition, gel filtration analysis of extracts from U2OS-α7-CFP cells revealed the presence of α7-CFP protein only in fractions containing proteasomes (data not shown). Altogether, these experiments indicate that α7-CFP–containing nuclear foci are not aggregates. Furthermore, FRAP (White and Stelzer, 1999) analysis of the mobility of α7-GFP showed that, after laser photobleaching of α7-GFP nuclear foci, the mobile fraction of α7-GFP corresponded to 77% (±2.9%) of the protein present in the foci and that the half time (τ1/2) of complete α7-GFP recovery in the bleached area was 5.75 s (±1.43). This recovery rate is considerably slower than that of GFP alone (τ1/2 = 0.3 s, see Kruhlak et al., 2000), but equivalent to that of many other mobile nuclear proteins, including transcription factors and RNA splicing components such as SR proteins (τ1/2 = 3–6 s; Phair and Misteli, 2000), showing that α7-CFP localization into nuclear foci is in fact dynamic.
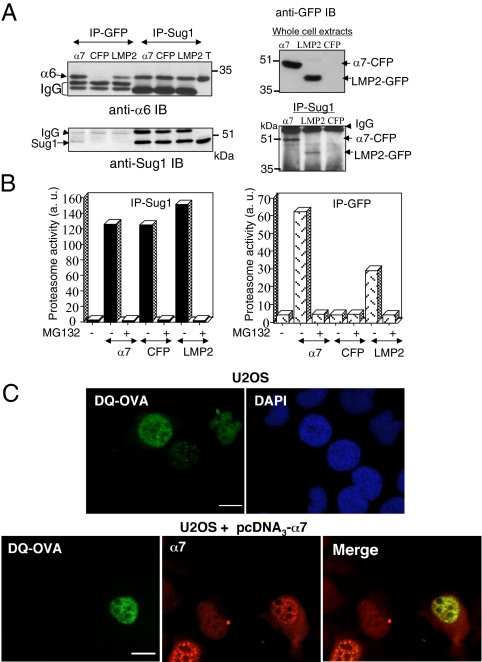
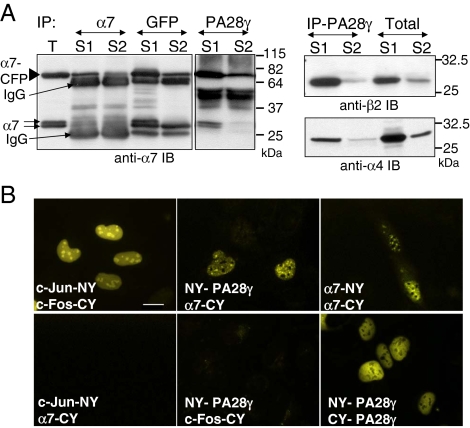
Biochemical experiments indicated that α7-CFP was readily incorporated into proteasomal complexes (Figure 2). Immunoprecipitation on whole cell extracts demonstrated the presence of other 20S proteasome subunits (α6 subunit) as well as 19S complex subunits (Sug1 protein, an ATPase of this complex) in anti-GFP immunoprecipitates and showed that α7-CFP is also present in anti-Sug1 immunoprecipitates (Figure 2A). The immunoprecipitated CFP-containing complexes were proteolytically active when tested against the proteasome substrate suc-LLVY-AMC (Figure 2B, right panel). Morever, using an in situ proteasome-dependent proteolysis assay (Rockel et al., 2005), based on the microinjection of a fluorogenic substrate (DQ-OVA) into the cell nucleus, we demonstrated that α7-CFP complexes were active in the nuclear foci. After microinjection of DQ-OVA in parental U2OS cell nuclei, we confirmed that its degradation (monitored by the appearance of fluorescence) occurs throughout the nucleoplasm but also in discrete foci, as previously published (Rockel et al., 2005; Figure 2C, top panel). However, in U2OS cells expressing ectopic α7, the fluorescent foci resulting from DQ-OVA degradation colocalized completely, in number and shape, with α7 positive nuclear foci (Figure 2C, bottom panel). Based on these observations, it can be concluded that the proteasomes complexes containing α7-CFP protein are active in protein degradation.
Figure 2.
α7-CFP is incorporated into active proteasome complexes that accumulate in nuclear foci. (A) Total cell lysate from human osteosarcoma U2OS-tTA cells stably transfected with vectors conditionally expressing α7-CFP (α7) or CFP (CFP) cultured in the absence of tetracycline for 48 h and from MelJuso cells stably expressing LMP2-GFP (LMP2) were subjected to immunoprecipitation (IP) with anti-GFP or anti-Sug1 antibodies. Immunoprecipitated proteins as well as 30 μg of U2OS-tTA total extract (T) were separated by electrophoresis and analyzed by immunoblotting using the indicated antibodies. (B) Proteins immunoprecipitated with either anti-Sug1 or anti-GFP antibodies from total cell extracts were tested for proteasomal activity using the peptide suc-LLVY-AMC as a substrate, in the presence (+) or not (−) of 50 μM MG132 (first bar: no IP in the assay). (C) Proteasomes are active in nuclear foci. U2OS-tTA cells (top panel) or U2OS-tTA cells transiently expressing α7 (bottom panel) were microinjected (24 h after transfection) with the fluorogenic substrate protein DQ-OVA (0.5 mg/ml). The substrate was injected into the nuclei. Cells were incubated 10 min at 37°C and then fixed, and the fluorescent DQ-OVA degradation products were visualized by the appearance of green fluorescence. α7 was detected by indirect immunofluorescence with anti-α7 (red; 63× objective). Bar, 10 μm. U2OS-tTA-α7-CFP cells were not used in this experiment because of the overlap of the fluorescence spectrums of CFP and of DQ-OVA degradation products.
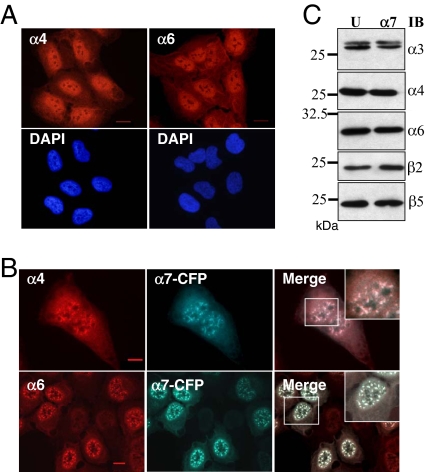
Finally, we found that whole 20S proteasomes were present in α7-CFP–containing nuclear foci: indirect immunofluorescence showed that in contrast to the diffuse nuclear localization observed in U2OS parental cells (Figure 3A), other tested 20S subunits, namely α4 and α6, localized into the same nuclear foci upon α7 expression (Figure 3B), without any significant variations of their cellular level (Figure 3C). Altogether, these data show that ectopic expression of α7 at moderate level entails recruitment to nuclear foci of genuine, active proteasome complexes and not accumulation of free α7.
Figure 3.
Ectopic expression of α7-CFP promotes recruitment of whole 20S proteasomes into the nuclear foci. Parental U2OS-tTA cells (A) and U2OS-tTA-α7-CFP cells induced for 48 h for α7-CFP expression (B) were subjected to indirect immunofluorescence using antibodies directed against the α4 and α6 subunits of the 20S proteasome (red). α7-CFP fusion protein was detected by CFP fluorescence. Wide-field overlay images and enlargements of marked area are shown. All observations were done with a 63× objective. Bar, 10 μm. (C) Expression of α7-CFP does not alter expression level of other 20S proteasome subunits. Thirty micrograms of total protein extract from U2OS-tTA (U) and induced U2OS-tTA-α7-CFP (α7) cells were separated by electrophoresis and analyzed by immunoblot (IB) using the indicated antibodies.
Identification of the Nuclear Foci as the Nuclear Speckles
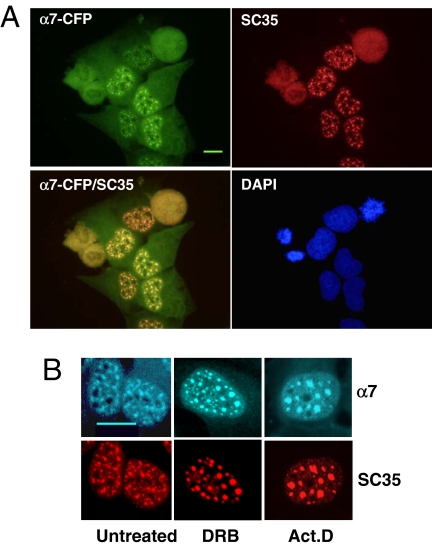
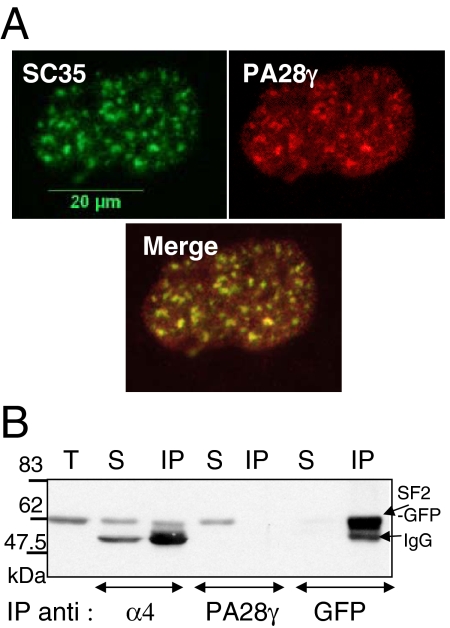
A critical point was to define the nature of the nuclear foci evidenced by expression of α7-CFP. Because previous reports described that 20S proteasomes localized to subnuclear domains such as PML bodies (Rockel and von Mikecz, 2002; Wojcik and DeMartino, 2003) or NS (Chen et al., 2002; Rockel and von Mikecz, 2002), we analyzed by indirect immunofluorescence the potential colocalization of α7-CFP with markers of those two nuclear subdomains, using antibodies raised against PML and SR proteins. Although the nuclear foci were clearly distinct from the PML bodies (data not shown), we observed a complete colocalization of α7-CFP with SC35 protein (Figure 4A), a member of the SR protein family that is concentrated in NS (Misteli et al., 1997; Lamond and Spector, 2003). To verify that the localization of α7-CFP in NS was not a coincidence, we treated the cells with transcriptional inhibitors (actinomycin D and DRB) that cause NS to enlarge and round up (Spector, 1993; for a review, see Lamond and Spector, 2003). In these cases, we observed similar morphological changes for the foci labeled by α7-CFP and SC35 (Figure 4B), indicating that proteasome complexes containing α7-CFP and NS are tightly connected.
Figure 4.
The nuclear foci enriched in 20S proteasome-α7-CFP complexes correspond to the NS. (A) U2OS-tTA-α7-CFP were fixed 48 h after α7-CFP induction, permeabilized, and stained with a mAb directed against the phosphorylated NS protein SC35 (red) and with DAPI dye (blue). α7-CFP fluorescence is in green for easier codetection (α7-CFP/SC35 image). Wide-field of individual detection and overlay images are presented (63× objective). Bar, 10 μm. (B) Forty-eight-hour–induced U2OS-tTA-α7-CFP cells, untreated or treated with DRB (100 μM) or actinomycin D (10 μg/ml) for 2 h before fixation, were stained with anti-SC35 (red); α7-CFP expression was directly visualized by CFP fluorescence (cyan; 100× objective). Bar, 10 μm.
20S Proteasome-PA28γ Complexes, But Not 26S Proteasome Complexes, Are Recruited to NS upon α7 Expression
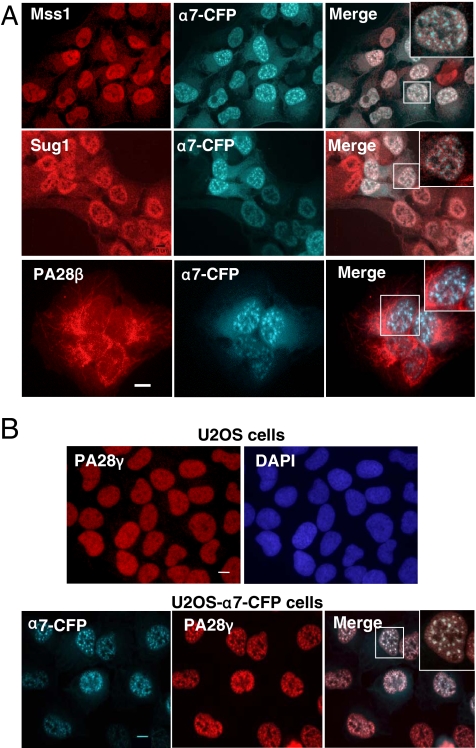
To determine to which regulator(s) the 20S proteasome was associated with in the NS, we performed indirect immunofluorescence analyses using specific antibodies raised against components of different regulatory complexes. Although colocalization of α7-CFP and two ATPases (Mss1 and Sug1) of the 19S regulator of the 20S proteasome could be detected in both the cytoplasm and the nucleus of the cells, these ATPases did not accumulate together with α7-CFP into the NS, unlike the α4 subunit of the 20S proteasome (Figure 5A and Figure S2, A and B). These results indicate that 26S proteasome is not the proteasomal complex recruited to the NS upon α7 ectopic expression. We thus tested for the presence of other regulators of the 20S proteasome. Strikingly, in U2OS cells expressing α7-CFP, PA28γ accumulated with α7 (and thus with 20S proteasomes) in these foci (Figure 5B and Figure S2, A and B), whereas, in contrast, the related regulator PA28α/β did not (Figure 5A, bottom panel). We also noted that, during mitosis, when the foci reversibly redistribute throughout the cell, the localization of PA28γ strictly followed that of α7-CFP–containing 20S proteasome (Figure S1).
Figure 5.
PA28γ, but not other 20S proteasome regulatory complexes, is recruited together with 20S proteasome into the nuclear foci. (A) U2OS-tTA-α7-CFP cells induced for 48 h were analyzed by indirect immunofluorescence using antibodies directed against the Mss1/Rpt1 and Sug1/Rpt6 ATPase subunits of the 19S complex (red) or PA28β. α7-CFP fusion protein was detected by CFP fluorescence. (B) Subcellular localization of PA28γ (indirect immunofluorescence, red) in parental U2OS-tTA cells and in induced U2OS-tTA-α7-CFP cells. α7-CFP was detected by direct fluorescence. Wide field of individual areas of detection, overlay images, and enlargements of marked area are presented. All observations were done with a 63× objective. Bar, 10 μm.
Together, these experiments show that ectopic expression of α7 (-CFP) entails accumulation into the NS of 20S proteasome-PA28γ complexes, which are active in protein degradation (Figure 2C). This accumulation is specific for this type of proteasome complex, because the 19S regulatory complex of the 20S proteasome, visualized here by following its Sug1 and Mss1 subunits, is not recruited in the NS, even though α7-CFP–containing 20S proteasomes can associate to the 19S complex elsewhere in the cells, as shown by coimmunoprecipitation experiments (Figure 2, A and B).
We confirmed the interaction between α7-CFP–containing 20S proteasome and PA28γ in cell extracts by coimmunoprecipitation from mildly solubilized (S1 fraction, no extraction of α7-CFP nuclear foci) or triton-solubilized cell extracts (S2 fraction, obtained after solubilization of α7-CFP nuclear foci) using antibodies directed against either α7, GFP, or PA28γ (Figure 6A). Even though the majority of 20S proteasome-PA28γ complexes was present in the S1 fraction, the complexes were also present in the S2 fraction enriched in NS (Figure 6A) and were active when tested for degradation of the proteasomal peptide substrate Suc-LLVY-AMC (data not shown). Notably, the distribution of the complexes between the S1 and S2 fractions is in agreement with the low percentage of α7-CFP present in the nuclear foci, estimated by fluorescence quantification (see above). We then used the BiFC technique to evaluate directly, in cellulo, the ability of α7 to interact with PA28γ in the NS. This technique is based on the fact that the YFP chromophore can be reconstituted in cells when two chimeric proteins, each harboring half of the chromophore, interact (Kerppola, 2006). When α7 fused to the C-terminal half of YFP (α7-CY) was coexpressed in cells with PA28γ fused to the N-terminal half of YFP (NY-PA28γ), the fluorescence could be reconstituted throughout the nuclei, with a marked accumulation in nuclear foci, which most likely correspond to NS (Figure 6B). Interaction was also observed in the nucleus between molecules of PA28γ, as expected because PA28γ forms heptamers and curiously between molecules of α7 as well (Figure 6B; see Discussion). Together, these results confirm the in cellulo interaction between 20S proteasome and its PA28γ regulator and strongly suggest that 20S proteasome-PA28γ complexes are indeed present in the NS.
Figure 6.
α7 and PA28γ are physically associated in total cell extracts, in a nuclear foci-enriched fraction and in cellulo. (A) After lysis of induced U2OS-tTA-α7-CFP cells, the cell extract was clarified by centrifugation, and the supernatant was collected (S1). The insoluble material was then resuspended in lysis buffer containing 0.3% Triton X-100 (15 min at 4°C) and clarified by centrifugation, yielding the S2 supernatant. Proteins immunoprecipitated from the S1 and S2 supernatants, using the indicated antibodies, were then separated by SDS-PAGE and analyzed by immunoblotting using antibodies directed against the proteasome subunits α7 (left panel), β2 (right top panel) or α4 (right bottom panel). T: 30 μg of total proteins. (B) The interaction between α7-CFP and PA28γ was visualized in cellulo by BiFC. Proteins indicated in each panel were coexpressed in HeLa cells. Twenty-four hours after transfection of expression vectors, cells were incubated at 32°C for a further 24 h to promote chromophore maturation and then fixed to allow YFP fluorescence monitoring. c-Fos (118–210) and c-Jun (257–318) constructs (Hu et al., 2002) were used as positive (Jun-NY/Fos-CY) or negative (Jun-NY/α7-CY and NY-PA28γ/Fos-CY) BiFC controls (63× objective). Bar, 10 μm.
PA28γ Is Present in the Nuclear Speckles without Expression of α7-CFP
To exclude the possibility that the localization of 20S-PA28γ complexes in the NS was only due to α7 ectopic expression, we examined the localization of PA28γ in parental U2OS cells. Because in fixed cells no clear specific intranuclear localization of PA28γ could be detected, soluble PA28γ was removed by simultaneous fixation and permeabilization of the cells. Under these conditions, we clearly visualized the colocalization of the remaining endogenous PA28γ with SC35 protein in the NS (Figure 7A). Therefore, 20S-PA28γ complexes are present in the NS without α7 ectopic expression, although their NS localization is strongly enhanced by it. Curiously, we were unable to detect the endogenous 20S proteasome in the NS, even when different conditions of permeabilization and various antibodies were used, although these complexes have been seen previously by others in the NS (Rockel and von Mikecz, 2002; Rockel et al., 2005). However, we could visualize a physical interaction between 20S proteasome and SR proteins in coimmunoprecipitation experiments using extracts from cells expressing α7 and SF2/ASF-GFP proteins (Figure 7B). By contrast, SF2/ASF was not detected in anti-PA28γ immunoprecipitates, possibly because the PA28γ epitope was masked by the interaction with SF2/ASF-GFP.
Figure 7.
Presence of PA28γ in the NS without α7-CFP expression and association of 20S proteasome with SF2/ASF protein. (A) Parental U2OS-tTA cells were permeabilized and fixed simultaneously to remove most of soluble PA28γ complexes. Cells were then stained with anti-SC35 (green) and anti-PA28γ (red), and an overlay image is shown (100× objective). (B) U2OS-tTA cells, cotransfected with pcDNA3-α7 and pEGFP-SF2/ASF vectors, were lysed 24 h after transfection in lysis buffer containing 0.3% Triton X-100. Two hundred micrograms of total cell extract were subjected to immunoprecipitation using either anti-α4, anti-PA28γ or anti-GFP antibodies. Immunoprecipitated proteins were then separated by SDS-PAGE (10%) and visualized by immunoblotting using anti-GFP antibody. T, 30 μg of total cells extract; S, supernatant of the IP (1/10); IP, immunoprecipitation.
Roles of 20S Proteasome-PA28γ Complexes in the Structural Integrity of the NS
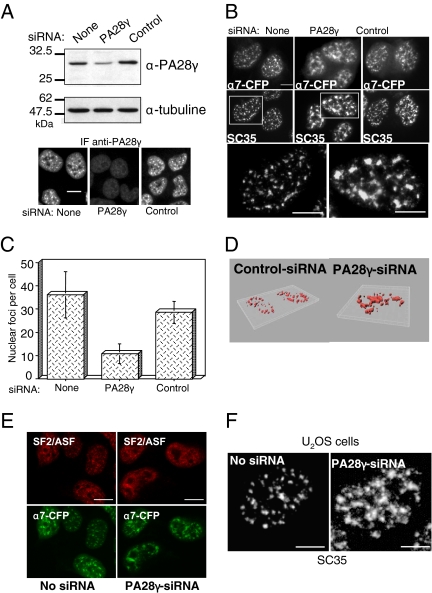
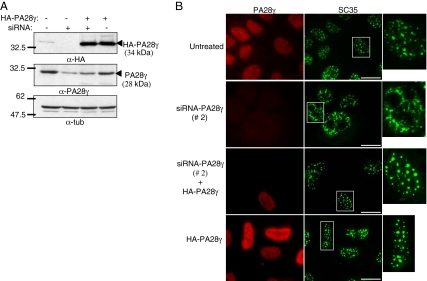
The results described above demonstrate for the first time that 20S proteasome-PA28γ complexes are present in the NS and suggest a specific role of these complexes in this subnuclear domain. To gain insight into this role, we investigated the effect of the absence of 20S-PA28γ complexes, using siRNA duplexes directed against PA28γ. Because the complete absence of PA28γ appeared to be toxic to our cells, we could only analyze a partial knockdown of PA28γ (decreased ∼70% from its normal level; Figure 8A). Very interestingly, we observed that in cells treated with PA28γ-siRNA, but not in control cells, α7-CFP nuclear foci were fewer and appeared significantly larger and less dot-like compared with untreated cells (Figure 8, B, top panels, and C). Furthermore, when the distribution of either endogenous SC35 (Figure 8, B and D) and SF2/ASF (Figure 8E), or exogenous SF2/ASF-GFP (see Figure 11A), was analyzed by immunofluorescence, the same structural alteration of the NS was observed, showing that not only the distribution of the proteasome, but that of the entire NS population was affected by PA28γ knockdown. Importantly, ectopic expression of α7 was not required for these effects, because the same alteration of the NS was observed in parental U2OS cells (Figure 8F) upon PA28γ knockdown. Moreover, the same results were obtained using a second siRNA duplex (siRNA 2; Figure 9, A and B), directed against another PA28γ mRNA region and already validated in a previous study (Cioce et al., 2006). Importantly, the effect of this siRNA was reversed by coexpression of a construct resistant to the siRNA (Figure 9B). Therefore, alteration of the NS is clearly due to PA28γ depletion, showing that endogenous 20S-PA28γ complexes are necessary for typical NS organization in U2OS cells.
Figure 8.
Proteasome-PA28γ complexes are required for the integrity of the NS. (A) U2OS-tTA-α7-CFP cells were induced for α7-CFP expression and concomitantly transfected with control-siRNA or PA28γ-siRNA (siRNA 1) duplexes. Cells were recovered 48 h after transfection, and the expression level of PA28γ was analyzed either by immunoblotting or by indirect immunofluorescence using an anti-PA28γ antibody. Bar, 10 μm. (B) NS were visualized by the fluorescence of α7-CFP and by indirect immunofluorescence using anti-SC35 antibodies in induced U2OS-tTA-α7-CFP cells untransfected or transfected with PA28γ- or control-siRNA duplexes. Enlargements of the marked areas are presented. Bar, 10 μm. (C) Quantification of the number of α7-CFP–labeled NS in cells treated with PA28γ- or control-siRNA duplexes. Quantification was performed visually, by counting on the pictures the number of compact fluorescent foci in each cell. The values correspond to the means of five independent experiments (n = 100 cells, ±SD). (D) 3D reconstruction. SC35 localization was detected by indirect immunofluorescence in induced U2OS-tTA-α7-CFP cells treated with control- or PA28γ (1)-siRNA. Fixed cells were observed with a Leica DMRA microscope equipped with a 63× PL APO (NA = 1.32) oil immersion objective and a N2.1 (Leica) filter set. Stacks of images were acquired using a piezo stepper (Physik Instruments, Waldbronn, Germany), Metamorph 7.1 (Molecular Devices, Menlo Park, CA) and a Micromax 1300YHS CCD camera (Princeton Research Instruments, Princeton, NJ). Stacks were further deconvolved using a MLE algorithm and the Huygens 2.9 software (Scientific Volume Imaging, Hilversrum, The Netherlands) and analyzed in 3D using Imaris 5.3 (Bitplane, Zurich, Switzerland). (E) NS were visualized by the fluorescence of α7-CFP and by indirect immunofluorescence using anti-SF2/ASF antibodies in induced U2OS-tTA-α7-CFP cells untransfected or transfected with PA28γ- or control-siRNA duplexes. Bar, 10 μm. (F) In parental U2OS-tTA cells, untreated or treated with PA28γ-siRNA duplexes, NS were visualized by indirect immunofluorescence using anti-SC35 antibodies. Bar, 10 μm.
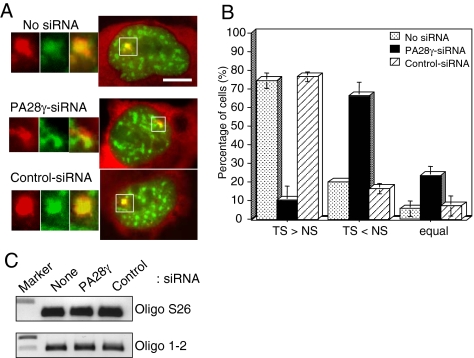
Figure 11.
PA28γ-siRNA reduces SF2/ASF accumulation at transcription sites. 2E11 cells were cotransfected with pcDNA3-Tat, pEGFP-SF2/ASF, and control- or PA28γ-siRNA (1) duplexes. After 48 h, cells were fixed and subjected to in situ hybridization using a MS2-Cy3 probe. (A) Right, merged images of the intracellular distribution of SF2/ASF-GFP (green) and MS2 TS and mRNA (red) under the indicated conditions. Left, magnification of the TS (white square): MS2 detection (red), SF2/ASF-GFP (green), merge (100× objective). Bar, 10 μm. (B) Quantification of the level of SF2/ASF-GFP recruited at the TS. The relative levels of SF2/ASF-GFP at the TS and at the NS was estimated by quantification of the green fluorescence using the Metamorph software, in cells expressing similar levels of transcripts (estimated by quantification of MS2-Cy3 at TS) and SF2/ASF-GFP in the NS. The values correspond to the means of four independent experiments (n = 40 cells, ±SD). See Figure S3 for illustration of the quantification procedure. (C) PA28γ-siRNA does not affect the level of transcription. Relative mRNA levels for the ribosomal S26 subunit (control) and for the artificial gene in 2E11 cells untreated or treated with the indicated siRNA were analyzed by RT-PCR and agarose gel electrophoresis.
Figure 9.
Specificity of the effect of the siRNA directed against PA28γ. U2OS-tTA cells were treated or not with PA28γ-siRNA duplexes #2, PA28γ-siRNA duplexes #2, and pcDNA3-3HA-PA28γ (allowing the expression of a PA28γ construct refractory to siRNA #2), and pcDNA3-3HA-PA28γ alone, as indicated. (A) Expression levels of endogenous or exogenous PA28γ were analyzed by immunoblotting using anti-PA28γ or -HA antibodies. (B) Analysis of SC35 and PA28γ localization by indirect immunofluorescence as described in Figure 8. Bar, 10 μm.
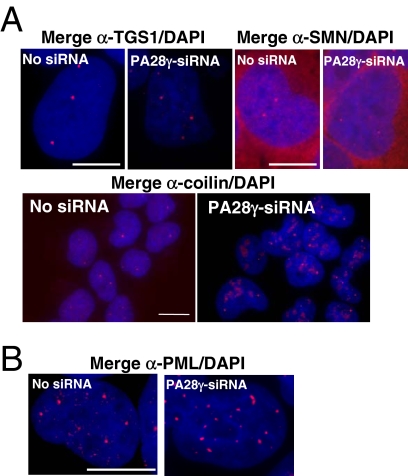
To test whether the effect of PA28γ knockdown was restricted to the NS, we analyzed by indirect immunofluorescence the organization of other known nuclear domains such as PML bodies and Cajal bodies (CB; Figure 10). In cells treated with PA28γ-siRNA, no changes were observed in PML bodies (Figure 10B) or in the number and appearance of CBs detected by anti-TGS1 (trimethylguanosine synthase) or anti-SMN antibodies (Figure 10A, top panels). However, we noticed a redistribution of coilin (an autoantigen marker of CBs). In cells knocked down for PA28γ, coilin was still present in the CBs, but appeared additionally in other nuclear regions (Figure 10A, bottom panel). Most likely, the partial delocalization of coilin was due to the fact that PA28γ plays a role in its nuclear distribution (Cioce et al., 2006). Altogether, our results suggest that PA28γ knockdown does not alter the whole nuclear organization but impacts mostly NS integrity, even though it is clear that PA28γ must have functions outside the NS.
Figure 10.
Effect of PA28γ siRNA on subnuclear domains. Localization of proteins marker of Cajal (A) and PML bodies (B) was analyzed in induced U2OS-tTA-α7-CFP cells treated with control- or PA28γ-siRNA, as described in Figure 8. Cells were stained with antibodies raised against three components of CB: TGS1, SMN, and coilin (A), or with antibodies raised against the protein PML (B). Bar, 10 μm.
NS are known to be highly dynamic structures in which splicing factors undergo posttranslational modifications (particularly phosphorylations) that are required for the SR proteins to leave the NS and be subsequently recruited at transcription sites (Misteli et al., 1998). We thus tested whether knockdown of PA28γ altered the capacity of SR proteins to be recruited at a specific transcription site. For this purpose, we used the 2E11 (also called pU2OS_Exo1) cell line, that harbors an integrated inducible HIV-1 reporter gene containing a splice site for SF2/ASF. An additional RNA tag (MS2) was inserted to facilitate its visualization by in situ hybridization (Boireau et al., 2007; du Chéné et al., 2007). The ability of SF2/ASF to be recruited to nascent pre-mRNAs was estimated by measuring the relative level of the protein at the transcription site (TS, visualized as a nuclear dot by in situ hybridization with a MS2-Cy3 probe) compared with its level at a neighboring NS (Figure 11A and Figure S3). To eliminate the variation of GFP level, due to transfection efficiency, only foci of cells presenting a similar level of SF2/ASF-GFP in NS were scored. Results were summarized in the histogram shown in Figure 11B. In untreated cells or with control-siRNA treatment, the level of SF2/ASF-GFP at the TS was generally higher than in the NS, reflecting the normal recruitment of SF2/ASF-GFP to TS. In contrast, with PA28γ-siRNA treatment, the level of SF2/ASF-GFP at the TS was most of the time lower than at the NS without affecting the transcription level of the artificial gene (Figure 11C). These results show that under PA28γ-siRNA treatment, the recruitment of SR proteins to the active TS is largely reduced, although not abolished.
Together, these experiments demonstrate that 20S proteasme-PA28γ complexes play an important role in NS organization and dynamics and mediate a process involved in the regulation of accumulation and/or targeting of splicing factors to transcription sites.
DISCUSSION
We show here that active 20S proteasome-PA28γ complexes are present in the NS and that reduction of PA28γ levels, using siRNA, dramatically alters NS organization and nuclear trafficking of splicing factors. Our data thus demonstrate a new role of 20S-PA28γ complexes in nuclear organization and provide novel important information for our understanding of both the specific functions and localization of the various forms of proteasomes and the regulation of NS structure and dynamics.
Our discovery originates from the serendipitous observation that ectopic expression of the proteasomal α7 subunit triggers accumulation of 20S proteasome-PA28γ complexes in the NS. This observation raises the intriguing and puzzling problem of how this effect is mediated. One possibility was that overexpression of α7 led to an increase of the cellular concentration of 20S proteasome, but we found no evidence to support this supposition. One alternative hypothesis is that α7 ectopic expression entails incorporation of more than two α7 subunits into individual 20S proteasome complexes. Indeed, some plasticity in proteasome assembly has been documented in yeast, in which α3 could be replaced by α4 (Velichutina et al., 2004). In this respect, it is remarkable that α7 seems to possess a greater propensity to interact to PA28γ than other subunits. When we used the proteasome subunit α3, no interaction with PA28γ could be detected by BiFC, contrary to the result obtained for α7, suggesting that 1) PA28γ does not interact identically with all α subunits of the proteasome and 2) within the 20S proteasome, the α7 subunit possesses certain sequence and/or structural features that makes it a favored partner for PA28γ. Thus, a proteasome with more than one α7 subunit per α-ring could either have a higher affinity for PA28γ or impose a particular conformation to the complex formed with PA28γ, two mechanisms that in turn could impact on the steady-state nuclear distribution of the 20S-PA28γ complex.
In any case, even if the exact reasons for accumulation of 20S-PA28γ in NS upon α7 ectopic expression remain to be elucidated, several arguments show that the presence of 20S-PA28γ complexes in the NS is physiological, albeit not easily detectable normally. In particular, the presence of active 20S proteasome in the NS, as previously documented by others (Rockel et al., 2005), together with our results demonstrating that 1) PA28γ can be detected in the NS without ectopic α7 expression, 2) knockdown of endogenous PA28γ alters NS morphology, and 3) 20S proteasomes can interact with PA28γ in the NS (as illustrated by colocalization, affinity purification, and in vivo interaction; BiFC), demonstrate that these complexes constitute an important active form of the proteasome in the NS.
Regarding the function(s) of 20S-PA28γ complexes in the NS, it is most likely that these complexes mediate the local degradation of specific proteins, even though they certainly have other substrates in other nuclear regions. We hypothesized that substrates of 20S-PA28γ complexes in the NS could be the SR proteins themselves because it has been reported that the intrinsic NS component SC35 is stabilized by proteasome inhibitors (Rockel and von Mikecz, 2002). However, in our cells, expression of α7-CFP or knockdown of PA28γ did not alter the half-life of SC35 (data not shown). Other attractive substrates for 20S-PA28γ complexes are kinases or phosphatases present in the NS (Colwill et al., 1996; Misteli and Spector, 1996; Wang et al., 1998; Ko et al., 2001). Indeed, these enzymes control the phosphorylation status of SR proteins and thereby their recruitment at transcription sites (Misteli et al., 1998). Thus, alteration of the expression level of one or several of these enzymes could explain the alteration of both the morphology of the NS and the recruitment of SR proteins to transcription sites seen upon PA28γ knockdown. Obviously, to understand the role(s) of 20S-PA28γ complexes in the NS, an important development for the future will be to identify their nuclear substrates, particularly those localized in the NS.
Nevertheless, even without this information, our observations provide additional demonstration that, within the family of proteasome complexes, 20S proteasome-PA28γ complexes have specific functions in the cell nucleus. Until recently, the functions of PA28γ have remained quite elusive. Genetic manipulations in mice and Drosophila have suggested that it might be involved in cell cycle progression and have anti-apoptotic functions (Rechsteiner and Hill, 2005). However, its exact role in these processes is not understood. Biochemical analyses have shown that PA28γ can activate peptide degradation by the 20S proteasome in vitro (Realini et al., 1997), but it has been demonstrated only recently that PA28γ is indeed involved in the degradation of proteins, namely the HCV (hepatitis C virus) core protein (Moriishi et al., 2003), the SRC-3 oncogene, and the CKI p21 (Li et al., 2006, 2007; Chen et al., 2007). PA28γ has also been identified as a novel regulator of Cajal bodies integrity (Cioce et al., 2006), but it was unclear in this case whether 20S proteasome was required for this process. We now additionally show that 20S-PA28γ complexes are involved in the control of nuclear trafficking of splicing factors, because reduction of PA28γ levels by siRNA strongly perturbs NS morphology and impairs recruitment of SR proteins (SF2/ASF) at transcription sites. The physiological importance of this role is presently unclear, because knockdown of PA28γ did not appear to affect transcription—as evaluated by RT-PCR of a specific gene (Figure 11C) or globally by 5-fluorouracil (5-FU) incorporation (Figure S4A), nor to impair globally the specificity of the splicing machinery, because no variations were detected on various endogenous and transfected genes subjected to alternative splicing (Figure S4, B and C). Our results are similar to previously described data (Caceres et al., 1997; Misteli et al., 1998) showing that an SF2/ASF mutant (Δ-RS) was able to perform alternative splicing in vivo, in a manner very similar to the wild type, in absence of its recruitment to transcription sites, and suggesting that recruitment to the TS does not always reflect the functional properties of splicing factors. However, one possibility in our case is that the low level of SF2/ASF still present at TS is in fact sufficient for normal splicing.
Another question arising from our data are how 20S-PA28γ complexes are targeted to the NS. Our FRAP analyses demonstrated that α7-GFP (and therefore the proteasome) mobility is high in the NS, indicating that the complexes shuttle into and out of the NS. Currently, the mechanism of the targeting of the complex to the NS is unclear, because neither proteasome subunits nor PA28γ display signature motifs of NS proteins, such as RS domains, for example. Possibly, protein(s) binding to 20S proteasome and/or PA28γ could act as adaptor(s) facilitating the docking of the complex to the NS. Interestingly, because we detected a physical interaction between the 20S proteasome and SF2/ASF, SR proteins themselves could be responsible for targeting proteasomes to the NS.
In conclusion, the data presented here demonstrate that 20S proteasome associated with its PA28γ regulator plays a critical role in the NS because it appears necessary for their structural organization and for intranuclear trafficking of splicing factors. Our data also suggest that the specificity of proteasome-dependent proteolysis, which is often considered as being regulated at the level of the ubiquitylation, can be controlled by complex mechanisms operating at the level of the proteasome and its intracellular localization. Further analysis of the role of 20S proteasome-PA28γ complexes in the NS should provide new information on the mechanisms involved in the control of nuclear organization. Our results highlight a novel layer of complexity in the regulation of NS architecture and function, as well as SR protein trafficking, and suggest that intracellular proteolysis is intrinsically involved in the regulation of nuclear plasticity.
Supplementary Material
ACKNOWLEDGMENTS
We thank E. Reits (University of Amsterdam, The Netherlands) for MelJuso cells, U. Seifert (Charité University, Berlin, Germany) for PA28γ cDNA, and R. Bordonné (IGMM-CNRS, Montpellier, France) for antibodies (TGS1 and SMN). We also thank our colleagues for their help, suggestions, and criticisms and the cell imaging facility (Montpellier RIO Imaging) for the quality of its service. This work was supported by a grant from the Association pour la Recherche sur le Cancer to O.C. (ARC no. 3243), an ACI-BCMS from the French Research Ministry (O.C.), the European contract no. QLG1-CT-2001-02026 (O.C.), and a grant to J.T. from the Agence Nationale de la Recherche (ANR-05-BLAN-0261-01). C.D. was a recipient of a fellowship from the Fondation de la Recherche Médicale (FRM). E.B. and M.P. laboratories are “Equipes labelisées de la Ligue Nationale contre le Cancer.”
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-07-0637) on February 6, 2008.
REFERENCES
- Baldin V., Lukas J., Marcote M. J., Pagano M., Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993;7:812–821. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- Barton L. F., Runnels H. A., Schell T. D., Cho Y., Gibbons R., Tevethia S. S., Deepe G. S., Jr, Monaco J. J. Immune defects in 28-kDa proteasome activator gamma-deficient mice. J. Immunol. 2004;172:3948–3954. doi: 10.4049/jimmunol.172.6.3948. [DOI] [PubMed] [Google Scholar]
- Boireau S., et al. The transcriptional cycle of HIV-1 in real-time and live cells. J. Cell Biol. 2007;179:291–304. doi: 10.1083/jcb.200706018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks P., Murray R. Z., Mason G. G., Hendil K. B., Rivett A. J. Association of immunoproteasomes with the endoplasmic reticulum. Biochem. J. 2000;352:611–615. [PMC free article] [PubMed] [Google Scholar]
- Caceres J. F., Misteli T., Screaton G. R., Spector D. L., Krainer A. R. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascio P., Call M., Petre B. M., Walz T., Goldberg A. L. Properties of the hybrid form of the 26S proteasome containing both 19S and PA28 complexes. EMBO J. 2002;21:2636–2645. doi: 10.1093/emboj/21.11.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Rockel T., Steinweger G., Hemmerich P., Risch J., Von Mikecz A. Subcellular recruitment of fibrillarin to nucleoplasmic proteasomes: implications for processing of a nucleolar autoantigen. Mol. Biol. Cell. 2002;13:3576–3587. doi: 10.1091/mbc.02-05-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Barton L. F., Chi Y., Clurman B. E., Roberts J. M. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Mol. Cell. 2007;26:843–852. doi: 10.1016/j.molcel.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., Schwartz A. L. The ubiquitin system: pathogenesis of human diseases and drug targeting. Biochim. Biophys. Acta. 2004;1695:3–17. doi: 10.1016/j.bbamcr.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Cioce M., Boulon S., Matera A. G., Lamond A. I. UV-induced fragmentation of Cajal bodies. J. Cell Biol. 2006;175:401–413. doi: 10.1083/jcb.200604099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwill K., Pawson T., Andrews B., Prasad J., Manley J. L., Bell J. C., Duncan P. I. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 1996;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- Coux O., Tanaka K., Goldberg A. L. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- Demartino G. N., Slaughter C. A. Regulatory proteins of the proteasome. Enzyme Protein. 1993;47:314–324. doi: 10.1159/000468689. [DOI] [PubMed] [Google Scholar]
- DeMartino G. N., Slaughter C. A. The proteasome, a novel protease regulated by multiple mechanisms. J. Biol. Chem. 1999;274:22123–22126. doi: 10.1074/jbc.274.32.22123. [DOI] [PubMed] [Google Scholar]
- du Chéné I., et al. Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J. 2007;26:424–435. doi: 10.1038/sj.emboj.7601517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray C. W., Slaughter C. A., Demartino G. N. PA28 activator protein forms regulatory caps on proteasome stacked rings. J. Mol. Biol. 1994;236:7–15. doi: 10.1006/jmbi.1994.1113. [DOI] [PubMed] [Google Scholar]
- Groettrup M., Soza A., Eggers M., Kuehn L., Dick T. P., Schild H., Rammensee H. G., Koszinowski U. H., Kloetzel P. M. A role for the proteasome regulator PA28-alpha in antigen presentation. Nature. 1996;381:166–168. doi: 10.1038/381166a0. [DOI] [PubMed] [Google Scholar]
- Groll M., Ditzel L., Lowe J., Stock D., Bochtler M., Bartunik H. D., Huber R. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Grossi de Sa M. F., Martins de Sa C., Harper F., Olink-Coux M., Huesca M., Scherrer K. The association of prosomes with some of the intermediate filament networks of the animal cell. J. Cell Biol. 1988;107:1517–1530. doi: 10.1083/jcb.107.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C. D., Chinenov Y., Kerppola T. K. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- Kandil E., Kohda K., Ishibashi T., Tanaka K., Kasahara M. Pa28 subunits of the mouse proteasome—primary structures and chromosomal localization of the genes. Immunogenetics. 1997;46:337–344. doi: 10.1007/s002510050281. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K. Visualization of molecular interactions by fluorescence complementation. Nat. Rev. Mol. Cell Biol. 2006;7:449–456. doi: 10.1038/nrm1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko T. K., Kelly E., Pines J. CrkRS: a novel conserved Cdc2-related protein kinase that colocalises with SC35 speckles. J. Cell Sci. 2001;114:2591–2603. doi: 10.1242/jcs.114.14.2591. [DOI] [PubMed] [Google Scholar]
- Kruhlak M. J., Lever M. A., Fischle W., Verdin E., Bazett-Jones D. P., Hendzel M. J. Reduced mobility of the alternate splicing factor (ASF) through the nucleoplasm and steady state speckle compartments. J. Cell Biol. 2000;150:41–51. doi: 10.1083/jcb.150.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond A. I., Spector D. L. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Li X., Amazit L., Long W., Lonard D. M., Monaco J. J., O'Malley B W. Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGgamma-proteasome pathway. Mol. Cell. 2007;26:831–842. doi: 10.1016/j.molcel.2007.05.028. [DOI] [PubMed] [Google Scholar]
- Li X., Lonard D. M., Jung S. Y., Malovannaya A., Feng Q., Qin J., Tsai S. Y., Tsai M. J., O'Malley B W. The SRC-3/AIB1 coactivator is degraded in a ubiquitin- and ATP-independent manner by the REGgamma proteasome. Cell. 2006;124:381–392. doi: 10.1016/j.cell.2005.11.037. [DOI] [PubMed] [Google Scholar]
- Ma C. P., Slaughter C. A., DeMartino G. N. Identification, purification, and characterization of a protein activator (PA28) of the 20 S proteasome (macropain) J. Biol. Chem. 1992;267:10515–10523. [PubMed] [Google Scholar]
- Masson P., Lundgren J., Young P. Drosophila proteasome regulator REGgamma: transcriptional activation by DNA replication-related factor DREF and evidence for a role in cell cycle progression. J. Mol. Biol. 2003;327:1001–1012. doi: 10.1016/s0022-2836(03)00188-8. [DOI] [PubMed] [Google Scholar]
- Misteli T., Caceres J. F., Clement J. Q., Krainer A. R., Wilkinson M. F., Spector D. L. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J. Cell Biol. 1998;143:297–307. doi: 10.1083/jcb.143.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T., Caceres J. F., Spector D. L. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- Misteli T., Spector D. L. Serine/threonine phosphatase 1 modulates the subnuclear distribution of pre-mRNA splicing factors. Mol. Biol. Cell. 1996;7:1559–1572. doi: 10.1091/mbc.7.10.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriishi K., et al. Proteasome activator PA28gamma-dependent nuclear retention and degradation of hepatitis C virus core protein. J. Virol. 2003;77:10237–10249. doi: 10.1128/JVI.77.19.10237-10249.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata S., Kawahara H., Tohma S., Yamamoto K., Kasahara M., Nabeshima Y., Tanaka K., Chiba T. Growth retardation in mice lacking the proteasome activator PA28gamma. J. Biol. Chem. 1999;274:38211–38215. doi: 10.1074/jbc.274.53.38211. [DOI] [PubMed] [Google Scholar]
- Ortega J., Bernard Heymann J., Kajava A. V., Ustrell V., Rechsteiner M., Steven A. C. The axial channel of the 20S proteasome opens upon binding of the PA200 activator. J. Mol. Biol. 2005;346:1221–1227. doi: 10.1016/j.jmb.2004.12.049. [DOI] [PubMed] [Google Scholar]
- Phair R. D., Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- Pines J., Lindon C. Proteolysis: anytime, any place, anywhere? Nat. Cell Biol. 2005;7:731–735. doi: 10.1038/ncb0805-731. [DOI] [PubMed] [Google Scholar]
- Realini C., Jensen C. C., Zhang Z., Johnston S. C., Knowlton J. R., Hill C. P., Rechsteiner M. Characterization of recombinant REGalpha, REGbeta, and REGgamma proteasome activators. J. Biol. Chem. 1997;272:25483–25492. doi: 10.1074/jbc.272.41.25483. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M., Hill C. P. Mobilizing the proteolytic machine: cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 2005;15:27–33. doi: 10.1016/j.tcb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Reits E.A.J., Benham A. M., Plougastel B., Neefjes J., Trowsdale J. Dynamics of proteasome distribution in living cells. EMBO J. 1997;16:6087–6094. doi: 10.1093/emboj/16.20.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockel T. D., Stuhlmann D., von Mikecz A. Proteasomes degrade proteins in focal subdomains of the human cell nucleus. J. Cell Sci. 2005;118:5231–5242. doi: 10.1242/jcs.02642. [DOI] [PubMed] [Google Scholar]
- Rockel T. D., von Mikecz A. Proteasome-dependent processing of nuclear proteins is correlated with their subnuclear localization. J. Struct. Biol. 2002;140:189–199. doi: 10.1016/s1047-8477(02)00527-0. [DOI] [PubMed] [Google Scholar]
- Sacco-Bubulya P., Spector D. L. Disassembly of interchromatin granule clusters alters the coordination of transcription and pre-mRNA splicing. J. Cell Biol. 2002;156:425–436. doi: 10.1083/jcb.200107017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D. L. Nuclear organization of pre-mRNA processing. Curr. Opin. Cell Biol. 1993;5:442–447. doi: 10.1016/0955-0674(93)90009-f. [DOI] [PubMed] [Google Scholar]
- Tanahashi N., et al. Molecular properties of the proteasome activator PA28 family proteins and gamma-interferon regulation. Genes Cells. 1997;2:195–211. doi: 10.1046/j.1365-2443.1997.d01-308.x. [DOI] [PubMed] [Google Scholar]
- Theis-Febvre N., Filhol O., Froment C., Cazales M., Cochet C., Monsarrat B., Ducommun B., Baldin V. Protein kinase CK2 regulates CDC25B phosphatase activity. Oncogene. 2003;22:220–232. doi: 10.1038/sj.onc.1206107. [DOI] [PubMed] [Google Scholar]
- Velichutina I., Connerly P. L., Arendt C. S., Li X., Hochstrasser M. Plasticity in eucaryotic 20S proteasome ring assembly revealed by a subunit deletion in yeast. EMBO J. 2004;23:500–510. doi: 10.1038/sj.emboj.7600059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheggen C., Lafontaine D. L., Samarsky D., Mouaikel J., Blanchard J. M., Bordonne R., Bertrand E. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J. 2002;21:2736–2745. doi: 10.1093/emboj/21.11.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges D., Zwickl P., Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- Wang H. Y., Lin W., Dyck J. A., Yeakley J. M., Songyang Z., Cantley L. C., Fu X. D. SRPK2, a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J. Cell Biol. 1998;140:737–750. doi: 10.1083/jcb.140.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby F. G., Masters E. I., Kramer L., Knowlton J. R., Yao Y., Wang C. C., Hill C. P. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature. 2000;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- White J., Stelzer E. Photobleaching GFP reveals protein dynamics inside live cells. Trends Cell Biol. 1999;9:61–65. doi: 10.1016/s0962-8924(98)01433-0. [DOI] [PubMed] [Google Scholar]
- Wigley W. C., Fabunmi R. P., Lee M. G., Marino C. R., Muallem S., DeMartino G. N., Thomas P. J. Dynamic association of proteasomal machinery with the centrosome. J. Cell Biol. 1999;145:481–490. doi: 10.1083/jcb.145.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik C., DeMartino G. N. Intracellular localization of proteasomes. Int. J. Biochem. Cell Biol. 2003;35:579–589. doi: 10.1016/s1357-2725(02)00380-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.