Abstract
Misfolded or improperly assembled proteins in the endoplasmic reticulum (ER) are exported into the cytosol and degraded via the ubiquitin–proteasome pathway, a process termed ER-associated degradation (ERAD). Saccharomyces cerevisiae Hrd1p/Der3p is an ER membrane-spanning ubiquitin ligase that participates in ERAD of the cystic fibrosis transmembrane conductance regulator (CFTR) when CFTR is exogenously expressed in yeast cells. Two mammalian orthologues of yeast Hrd1p/Der3p, gp78 and HRD1, have been reported. Here, we demonstrate that gp78, but not HRD1, participates in ERAD of the CFTR mutant CFTRΔF508, by specifically promoting ubiquitylation of CFTRΔF508. Domain swapping experiments and deletion analysis revealed that gp78 binds to CFTRΔF508 through its ubiquitin binding region, the so-called coupling of ubiquitin to ER degradation (CUE) domain. Gp78 polyubiquitylated in vitro an N-terminal ubiquitin-glutathione-S-transferase (GST)-fusion protein, but not GST alone. This suggests that gp78 recognizes the ubiquitin that is already conjugated to CFTRΔF508 and catalyzes further polyubiquitylation of CFTRΔF508 in a manner similar to that of a multiubiquitin chain assembly factor (E4). Furthermore, we revealed by small interfering RNA methods that the ubiquitin ligase RMA1 functioned as an E3 enzyme upstream of gp78. Our data demonstrates that gp78 cooperates with RMA1 with E4-like activity in the ERAD of CFTRΔF508.
INTRODUCTION
In the eukaryotic cell secretory pathway, a significant proportion of proteins that enter the endoplasmic reticulum (ER) are specifically extracted from the ER and targeted to the cytosol, where they are degraded by the ubiquitin–proteasome system in a process known as ER-associated degradation (ERAD) (Meusser et al., 2005). Whereas misfolded, improperly assembled, and metabolically regulated endogenous proteins are targeted by the ERAD machinery as an ER quality control system, in some cases the machinery is usurped by exogenous factors, such as viruses and bacterial toxins (Meusser et al., 2005). In ERAD, protein ubiquitylation plays a role in both protein extraction from the ER and proteasomal protein degradation. This crucial protein modification is mediated by a set of three enzymes: a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3) (Hershko et al., 2000). In some cases, a multiubiquitin chain assembly factor (E4) is also involved (Koegl et al., 1999; Hoppe, 2005). Although the precise mechanism has not been clarified, the E4 protein acts to promote multiubiquitylation of mono- or oligoubiquitylated substrates. Ufd2p, an E4 enzyme, is implicated in yeast ERAD, but it is not known whether E4 enzymes play a role in ERAD in mammals (Hoppe, 2005; Richly et al., 2005).
Cystic fibrosis transmembrane conductance regulator (CFTR) is a cell surface chloride ion channel. A large portion of CFTR is degraded by the ERAD machinery due to inefficient folding (Kopito, 1999). Several mutations in CFTR are associated with the severe genetic disorder cystic fibrosis (CF), but the majority of CF patients carry the ΔF508 mutation (Kopito, 1999). The ubiquitin ligases CHIP, Fbs1, and RMA1/RNF5 have been implicated in the clearance of CFTR in mammalian cells (Meacham et al., 2001; Yoshida et al., 2002; Younger et al., 2006). Genetic analysis has suggested that another ubiquitin ligase, Hrd1p/Der3p, is also involved in ERAD of CFTR (Gnann et al., 2004).
Saccharomyces cerevisiae Hrd1p is an integral membrane ubiquitin ligase that contains a RING finger domain facing the cytosol, which functions predominantly with the E2 enzyme Ubc7p in the ERAD pathway (Hampton et al., 1996; Bordallo et al., 1998; Bays et al., 2001; Deak and Wolf, 2001). Although there is no CFTR orthologue in yeast, Hrd1p is involved in the ERAD of human CFTR when it is exogenously expressed in yeast cells (Gnann et al., 2004).
There are two mammalian proteins that are structurally and functionally related to yeast Hrd1p, gp78/autocrine motility factor receptor and HRD1/Synoviolin (Fang et al., 2001; Amano et al., 2003; Kikkert et al., 2004). Gp78 was originally identified as a 78-kDa cell surface receptor for a tumor cell autocrine motility factor that promotes tumor metastasis (Nabi and Raz, 1987). Subsequently, it was shown that gp78 localizes not only to the cell surface but also to the ER (Wang et al., 1997). Similar to Hrd1p, gp78 is a RING finger-dependent ubiquitin ligase, an integral ER membrane protein, and it is involved in ERAD, in cooperation with Ube2g2/UBC7, the mammalian counterpart of yeast Ubc7p (Fang et al., 2001; Liang et al., 2003; Song et al., 2005; Lee et al., 2006). Recently, Chen et al. (2006) demonstrated that both the E2 binding site and the coupling of ubiquitin to ER degradation (CUE) domain of gp78 are essential for its ubiquitin ligase activity. Although recent biochemical and structural studies have helped define the ubiquitin binding properties of CUE domains (Ponting, 2000; Kang et al., 2003; Prag et al., 2003), the role of the gp78 CUE domain in ubiquitin ligation remains unclear. HRD1 is also a RING finger-dependent ubiquitin ligase, located in the ER, and it is involved in ERAD, in cooperation with Ube2g2 (Kaneko et al., 2002; Nadav et al., 2003; Kikkert et al., 2004).
Although Hrd1p is involved in the ERAD of exogenously expressed human CFTR in yeast cells, it is not known whether its mammalian counterparts are involved in the ERAD of CFTR in mammalian cells. Here, we demonstrate that gp78, but not HRD1, is involved in the ERAD of a mutant form of CFTR in mammalian cells, and that this activity is dependent on the CUE domain of gp78. We also show that the gp78 CUE domain confers an E4-like ubiquitin ligase activity to gp78, which catalyzes multiubiquitin chain assembly on CFTRΔF508 after initial ubiquitylation by RMA1 in vivo.
MATERIALS AND METHODS
Plasmids, Antibodies, and Small Interfering RNA (siRNA) Duplexes
The open reading frame (ORF) of mouse gp78 (BC040338) was cloned as follows: a 5′ 1038-base pair fragment was amplified from a mouse embryonic day 15.5 cDNA library (Invitrogen, Carlsbad, CA) by polymerase chain reaction (PCR), and inserted into pT7Blue (Clonetech, Mountain View, CA). A 3′ 1381-base pair fragment that included the 3′ untranslated region was isolated from a mouse liver cDNA library (Marathon cDNA library; Clonetech) by rapid amplification of cDNA ends, and inserted into pT7Blue. The full-length ORF of gp78 was generated by fusing the two fragments together at the SalI site in the region of overlap of the two fragments. The full-length gp78 ORF was inserted into pcDNA3.1 (−) (Invitrogen), then it was subcloned into pcDNA3.1/Myc-His(+)A (Invitrogen) to generate gp78-Myc. The cDNA for mouse HRD1 (NM_028769) was amplified from a mouse embryonic day 17 cDNA library (MATCHMAKER cDNA library; Clonetech), and then it was inserted into pcDNA3.1/Myc-His(+)A to generate HRD1-Myc. The single point mutations in the RING domains of gp78 and HRD1, C337/374S (for Rfm gp78-Myc) and C329S (for Rfm HRD1-Myc), respectively, were generated by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The point mutations in the CUE domain of gp78, MFP463/464/465AAR, were generated by using the same kit.
To generate wild-type and Rfm GGH-Myc, a 5′ 1479-base pair fragment of wild-type or Rfm gp78 with a BamHI site at its 3′ end, and a 3′ 438-base pair fragment of HRD1 carrying a BamHI site at its 5′ end were amplified by PCR, fused at the BamHI site, and introduced into pcDNA3.1/Myc-His(+)A. Glycine and serine residues corresponding to the additional BamHI site were subsequently inserted into the joint of the chimeric protein. GHH-Myc and Rfm GHH-Myc were obtained by fusing a 5′ 927-base pair fragment of gp78 and a 3′ 1047-base pair fragment of wild-type or Rfm HRD1. HHG-Myc and Rfm HHG-Myc were generated by fusing a 5′ 1338-base pair fragment of wild-type or Rfm HRD1 with a 3′ 438-base pair fragment of gp78. HGG-Myc and Rfm HGG-Myc were generated by fusing a 5′ 789-base pair fragment of mHRD1 and a 3′ 993-base pair fragment of wild-type or Rfm gp78. Glycine and serine residues corresponding to the insertional BamHI site were incorporated into all chimeric proteins. GXG-Myc was obtained by fusing a 5′ 927-base pair fragment and a 3′ 438-base pair fragment of gp78, which carried the glycine and serine residues of the BamHI site. The CUE domain deletion of gp78-Myc was generated by fusing a 5′ 1323-base pair fragment and a 3′ 438-base pair fragment of gp78, which carried the glycine and serine residues of the BamHI site. The CUE domain insertion of HRD1-Myc was generated by fusing a 5′ 1194 base pair fragment of HRD1 carrying a HindIII site at its 3′ end with a 3′ 648 base pair fragment of GGH-Myc carrying a HindIII site at its 5′ end. Lysine and leucine residues of the HindIII site were incorporated into the CUE domain insertional mutant HRD1-Myc, in addition to the glycine and serine residues of the insertion site of GGH-Myc.
To generate GST-gp78, a 3′ 993-base pair fragment of gp78 was subcloned into pGEX5X-1 (GE Healthcare Bio-Sciences, Little Chalfont, Buckinghamshire, United Kingdom). The same region, but lacking the CUE domain, was subcloned into the same vector to generate GST-ΔCUE. To generate GST-HRD1, a 1047-base pair fragment of HRD1 was subcloned into pGEX5X-1.
The expression vector for hemagglutinin (HA)-ubiquitin was described previously (Sato et al., 2006). The expression vector for C-terminally Myc-tagged Derlin1, -2, and -3 were described previously (Oda et al., 2006). The expression vector for Flag-RMA1 was described in Younger et al. (2006). The expression vector for CFTRΔF508-GFP was a kind gift from Dr. Ron R. Kopito (Stanford University, Stanford, CA).
Mouse monoclonal anti-valosin containing protein (VCP) antibodies were purchased from Affinity BioReagents (Golden, CO). Monoclonal anti-green fluorescent protein (GFP) antibody and polyclonal anti-Myc antibodies used for immunoblot analysis were purchased from Roche Diagnostics (Indianapolis, IN) and Santa Cruz Biotechnology (Santa Cruz, CA), respectively. Monoclonal anti-Myc antibody clone 9E10 and monoclonal anti-Flag antibody clone M2 was purchased from Santa Cruz Biotechnology. Polyclonal anti-CHIP antibody was purchased from Calbiochem (San Diego, CA), and monoclonal anti-actin antibody was from Chemicon International (Temecula, CA).
siRNA duplexes targeting CHIP (HSS145537 and HSS145538) and gp78 (HSS100447 and HSS100449) were purchased from Invitrogen, and those for RMA1 were from Dharmacon RNA Technologies (Lafayette, CO) and QIAGEN (Valencia, CA) (SI03157014). The negative control siRNA duplexes were purchased from Dharmacon RNA Technologies.
Cell Culture and Transfection
Human embryonic kidney (HEK)293 cells were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum and antibiotics (100 U/ml penicillin G and 100 U/ml streptomycin). Plasmids and siRNA duplexes were transfected into cells by using FuGENE 6 transfection reagent (Roche Diagnostics) or Lipofectamine RNAiMAX (Invitrogen), respectively, according to the manufacturer's instructions.
In Vivo Ubiquitylation Assay
Cells were lysed in buffer containing 50 mM Tris-HCl, pH 8.0, 1% NP-40, and 150 mM NaCl, and then they were centrifuged at 13,000 × g. The supernatants were incubated with anti-GFP antibody (mouse monoclonal; Roche Diagnostics), and immune complexes were captured using protein G-Sepharose (GE Healthcare Bio-Sciences). Immunoprecipitates were subjected to immunoblot analysis by using an anti HA-antibody (rabbit polyclonal; Santa Cruz Biotechnology). Immunoreactive proteins were detected using Can Get Signal (Toyobo Engineering, Osaka, Japan).
Metabolic Labeling and Immunoprecipitation
Cells were incubated for 30 min in methionine- and cysteine-free DMEM supplemented with 2 mM glutamine and 10% dialyzed fetal bovine serum. Cells were then pulsed with 4.1 Mbq/3.5-cm dish of EXPRESS labeling mixture (PerkinElmer Life and Analytical Sciences, Boston, MA), and chased with fresh complete medium. Cells were lysed in buffer containing 50 mM Tris-HCl, pH 8.0, 1% NP-40, and 150 mM NaCl, and supernatants were incubated with anti-GFP antibody (rabbit polyclonal or mouse monoclonal, MBL International [Woburn, MA], or Roche, respectively). Immune complexes were captured using protein A-Sepharose (GE Healthcare Bio-Sciences). Beads were washed twice in the same buffer used to lyse the cells, and then they were boiled in Laemmli sample buffer. Immunoprecipitates were separated by SDS-polyacrylamide gel electrophoresis (PAGE), and radioactivity of specific bands was quantified using a Storm PhosphorImager (GE Healthcare Bio-Sciences).
In Vitro Ubiquitin Binding Assay
GST-fusion proteins expressed in Escherichia coli were captured on glutathione (GSH)-Sepharose beads. Equal amounts of GST fusion proteins, as measured by Coomassie Blue staining, and oligoubiquitin mixture were incubated in buffer containing 50 mM Tris-HCl, pH 8.0, and 150 mM NaCl at 4°C for 1 h. GST-fusion proteins conjugated to GSH-Sepharose were collected by centrifugation, and coprecipitating ubiquitin was analyzed by immunoblot using anti-ubiquitin antibody (mouse monoclonal; Laboratories, South San Francisco, CA).
Protein Preparation for In Vitro Ubiquitylation Assay
Bovine ubiquitin was purchased from Sigma-Aldrich (St. Louis, MO). The ORF of human Ube2g2 was amplified by reverse transcription-PCR. cDNAs corresponding to Ube2g2 and ubiquitin-GST fusion protein (ub-GST) were cloned into pT7–7. The ub-GST could only be a substrate but not a source of ubiquitylation reaction in in vitro assay system, because GST was fused to the C terminus of ubiquitin and thereby prevents ubiquitin activation by E1 enzyme. His6-E1 baculovirus and bacterial expression were performed as described previously (Iwai et al., 1999). pGEX-2T was used for expression and purification of GST from bacteria (GE Healthcare Bio-Sciences). Ubiquitin ligases (gp78-Myc, gp78ΔCUE-Myc, HRD1-Myc, HRD1+CUE-Myc, and Flag-RMA1) were obtained from respective ubiquitin ligase-overexpressing HEK293 cells. The ubiquitin ligase-overexpressing cells were lysed in buffer containing 50 mM Tris-HCl, pH 8.0, 1% NP-40, and 150 mM NaCl, and then they were centrifuged at 13,000 × g. Membrane-spanning ubiquitin ligases were solubilized into supernatants with 1% NP-40. The supernatants were incubated with 10 μg of anti-Myc antibody (mouse monoclonal clone 9E10; Santa Cruz Biotechnology) or 10 μg of anti-Flag antibody (mouse monoclonal clone M2; Santa Cruz Biotechnology), and immune complexes were captured using 10 μl of protein G-Sepharose (GE Healthcare Bio-Sciences).
In Vitro Ubiquitylation Assay
One microgram of GST or ub-GST was incubated for 2 h at 37°C in a reaction volume of 20 μl of Tris-HCl, pH 7.5, containing 5 μg of ubiquitin, 100 ng of E1, 200 ng of Ubc7, 5 mM MgCl2, 2 mM dithiothreitol, 0.5 mM ATP, 10 mM creatine phosphate, 1 μg of creatine phosphokinase, and ubiquitin ligase immune complexes captured on 10 μl of protein G-Sepharose (described above). Reactions were terminated by the addition of 20 μl of Laemmli sample buffer, and proteins were separated by 10% SDS PAGE and visualized by immunoblot using anti-GST antibody (mouse monoclonal; Santa Cruz Biotechnology).
RESULTS
Gp78 Is Involved in ERAD of CFTRΔF508
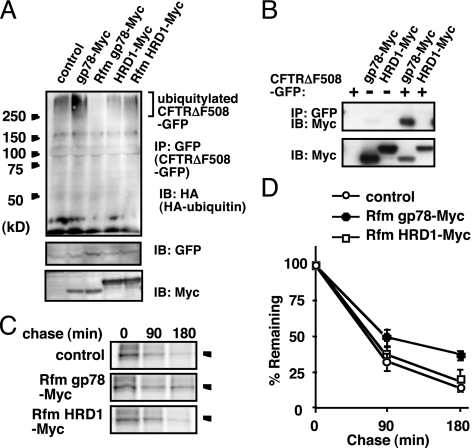
To evaluate whether gp78 and HRD1 were involved in the clearance of CFTR in mammalian cells, we examined the effect of gp78 and HRD1 expression on polyubiquitylation of the most common mutant form of CFTR, CFTRΔF508. Expression vectors for a CFTRΔF508 fused with GFP at the C terminus (CFTRΔF508-GFP) and an ubiquitin fused with HA at the N terminus (HA-ubiquitin) were transiently transfected into HEK293 cells. Ubiquitylated CFTRΔF508-GFP was detected by immunoprecipitation with anti-GFP antibody followed by immunoblot using anti-HA antibody. As shown in Figure 1A, polyubiquitylation of CFTRΔF508-GFP was enhanced by coexpression of a gp78 fused with Myc epitope at the C terminus (gp78-Myc), but it was not affected by coexpression of HRD1-Myc. Expression of a RING finger mutant of gp78 (Rfm gp78-Myc), but not HRD1 (Rfm HRD1-myc), exerted a dominant-negative effect that suppressed polyubiquitylation of CFTRΔF508-GFP (Figure 1A). Similar results were obtained when the immunoprecipitated proteins were analyzed under denaturing conditions, which indicated that the ubiquitylation we observed was not due to nonspecific ubiquitylated proteins that coimmunoprecipitated with CFTRΔF508-GFP (Supplemental Figure 1). Thus, gp78 specifically promoted polyubiquitylation of CFTRΔF508.
Figure 1.
Gp78 but not HRD1 is involved in the ERAD of CFTRΔF508. (A) Gp78 promotes ubiquitylation of CFTRΔF508. HEK293 cells transiently expressing CFTRΔF508-GFP and HA-ubiquitin, with or without (control lane) the indicated ubiquitin ligases, were lysed and subjected to immunoprecipitation by using anti-GFP antibody, followed by immunoblot using anti-HA antibody. (B) Gp78 physically interacts with CFTRΔF508. HEK293 cells transiently expressing CFTRΔF508-GFP and the indicated ubiquitin ligases were lysed and subjected to immunoprecipitation by using anti-GFP antibody, and immune complexes were analyzed by immunoblot using anti-Myc antibody. (C) Rfm gp78 delays ERAD of CFTRΔF508. HEK293 cells transiently expressing CFTRΔF508-GFP, together with Rfm gp78-Myc or Rfm HRD1-Myc, were pulse-labeled for 30 min with [35S]methionine, and after the indicated chase period, CFTRΔF508-GFP was immunoprecipitated and analyzed by autoradiography. (D) Quantitation of the data presented in C. Radioactivity of CFTRΔF508-GFP at each time point was normalized to the value at chase time point 0 h. The data represents the averages and SD of three independent experiments. IP, immunoprecipitation; IB, immunoblot.
Next, we examined whether gp78 physically interacted with CFTRΔF508 in HEK293 cells. Immunoprecipitation with anti-GFP antibody followed by immunoblot using anti-Myc antibody revealed that gp78-Myc, but not HRD1-Myc, physically interacted with CFTRΔF508-GFP (Figure 1B). Similar binding was also observed for Rfm gp78-Myc (data not shown). Notably, p97/VCP, which cooperates with gp78 in ERAD (Zhong et al., 2004; Ballar et al., 2006), coimmunoprecipitated with CFTRΔF508 when gp78-Myc was expressed in cells (Supplemental Figure 2). These results indicated that gp78 associates with CFTRΔF508 and that it recruits p97 to CFTRΔF508 complexes.
Ubiquitylation of CFTRΔF508 is followed by proteasomal degradation. We examined the effect of gp78 on the degradation of CFTRΔF508 by using a pulse-chase experiment. Coexpression of Rfm gp78-Myc, but not Rfm HRD1-Myc, exerted a dominant-negative effect that delayed the degradation of CFTRΔF508-GFP (Figure 1, C and D). This was consistent with the previous observation that gp78, but not HRD1, physically associated with CFTRΔF508, and that Rfm gp78, but not Rfm HRD1 markedly inhibited the ubiquitylation of CFTRΔF508. Thus, similar to yeast Hrd1p, gp78 seemed to be involved in the ERAD of CFTRΔF508 in mammalian cells. Coexpression of wild-type gp78-Myc had no effect on the degradation of CFTRΔF508-GFP (Supplemental Figure 3), even though it enhanced the ubiquitylation of CFTRΔF508-GFP. Overexpression of gp78 has been shown to accelerate the degradation of other gp78 substrates (Fang et al., 2003; Song et al., 2005; Chen et al., 2006), which suggests that additional factor(s) are required for the degradation of CFTRΔF508.
The CUE Domain of gp78 Is Required for CFTRΔF508 Binding
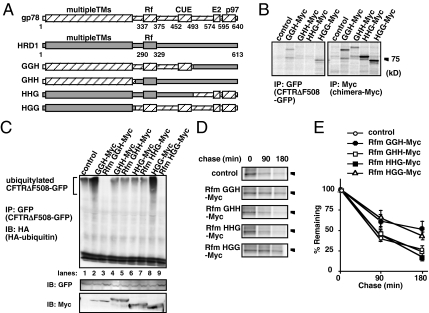
To identify the structural basis of the different functions of gp78 and HRD1 in the ERAD of CFTRΔF508, we generated a set of chimeric proteins, each of which contained the N-terminal multiple transmembrane region, the central region containing the RING finger domain, and the C-terminal region of either gp78 or HRD1 (Figure 2A). First, we examined the binding of each chimera to CFTRΔF508-GFP. The two chimeras that contained the central region of gp78, GGH-Myc and HGG-Myc, coimmunoprecipitated with CFTRΔF508-GFP (Figure 2B, left), whereas the two chimeras that contained the central region of HRD1, GHH-Myc and HHG-Myc, did not. These results indicated that the binding of gp78 to CFTRΔF508 is mediated by its central region. Similar binding was also observed for Rfm chimeric proteins, which contained mutations in their RING finger domains (data not shown).
Figure 2.
The central region of gp78 is important for the ERAD of CFTRΔF508. (A) Schematic representation of chimeric gp78 and HRD1 proteins. The multiple-transmembrane region, RING finger domain, E2 binding region, and p97 binding region are indicated as TM, Rf, E2, and p97, respectively. Hatched and gray regions show gp78 and HRD1 sequences, respectively. (B) Chimeras containing the central region of gp78 physically associate with CFTRΔF508. HEK293 cells transiently expressing CFTRΔF508-GFP and the indicated chimeric proteins were labeled for 1 h with [35S]methionine, and cell lysates were subjected to immunoprecipitation using anti-GFP antibody (left) or anti-Myc antibody (right). Twofold more cell lysate was used for immunoprecipitation with anti-GFP antibody. Immune complexes were analyzed by SDS-PAGE followed by autoradiogram. (C) Chimeras containing the central region of gp78 promote ubiquitylation of CFTRΔF508. Expression vectors for CFTRΔF508-GFP and HA-ubiquitin were transfected into HEK293 cells, together with expression vectors for the indicated chimeric proteins. Ubiquitylated CFTRΔF508-GFP was detected by immunoprecipitation with anti-GFP antibody followed by immunoblot using anti-HA antibody. (D) Rfm chimeras containing the central region pf gp78 delay the degradation of CFTRΔF508. HEK293 cells were pulse-labeled for 30 min with [35S]methionine, and chased for the indicated periods, and then CFTRΔF508-GFP was immunoprecipitated and processed for autoradiography. (E) The radioactivity of CFTRΔF508-GFP at each time point in (D) was normalized to the value at chase period 0 h. The data represents the averages and SD of three independent experiments.
We next examined the ability of the chimeric proteins to induce polyubiquitylation of CFTRΔF508-GFP. Polyubiquitylation of CFTRΔF508-GFP was enhanced by coexpression of GGH-Myc and HGG-Myc (Figure 2C, lanes 2 and 8), and it was reduced by coexpression of the corresponding Rfm chimeras by dominant-negative effect (lanes 3 and 9). Neither wild-type nor Rfm chimeras that contained the central region of HRD1 had any effect on polyubiquitylation. These results indicated that the binding of the central region of gp78 to CFTRΔF508-GFP strongly correlates with specific polyubiquitylation of CFTRΔF508-GFP.
We also investigated the effect of the chimeric proteins on the degradation of CFTRΔF508-GFP by using a pulse-chase experiment. Coexpression of Rfm chimeras containing the central region of gp78 delayed the degradation of CFTRΔF508-GFP, whereas chimeras containing the central region of HRD1 did not (Figure 2, D and E). These results suggested that the central region of gp78 is responsible for specific recognition of CFTRΔF508, and subsequent ubiquitylation and degradation.
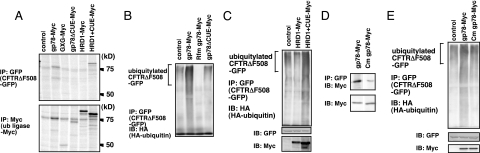
The most apparent difference between the central regions of gp78 and HRD1 is the presence of a CUE domain in gp78. We generated a CUE domain-deletion mutant of gp78-Myc (gp78ΔCUE-Myc) and a CUE domain-insertional mutant of HRD1-Myc (HRD1+CUE-Myc), and we expressed these proteins in cells. Coimmunoprecipitation experiments demonstrated that deletion of the CUE domain of gp78 resulted in loss of affinity for CFTRΔF508-GFP and that insertion of the CUE domain into HRD1 conferred CFTRΔF508-GFP binding activity to HRD1 (Figure 3A). These results indicated that the CUE domain of gp78 is both necessary and sufficient for its association with CFTRΔF508.
Figure 3.
The CUE domain of gp78 is essential for recognition of CFTRΔF508. (A) HRD1+CUE but not gp78ΔCUE physically associates with CFTRΔF508. HEK293 cells expressing CFTRΔF508-GFP with or without (control lane) gp78-Myc, GXG-Myc, gp78ΔCUE-Myc, HRD1-Myc, or HRD1+CUE-Myc were labeled for 1 h with [35S]methionine, and cell lysates were subjected to immunoprecipitation using anti-GFP antibody (top) or anti-Myc antibody (bottom). Twofold more cell lysate was used for immunoprecipitation with anti-GFP antibody. Immune complexes were analyzed by autoradiogram. (B) Gp78ΔCUE does not affect CFTRΔF508 ubiquitylation. Expression vectors for CFTRΔF508-GFP and HA-ubiquitin were transfected into HEK293 cells together with expression vectors for the indicated variants of gp78. Ubiquitylated CFTRΔF508-GFP was detected by immunoprecipitation with anti-GFP antibody followed by immunoblot using anti-HA antibody. (C) HRD1+CUE acquired CFTRΔF508-GFP-ubiquitylating activity. Expression vectors for CFTRΔF508-GFP and HA-ubiquitin were transfected into HEK293 cells together with expression vectors for the indicated variants of HRD1. Ubiquitylated CFTRΔF508-GFP was detected by immunoprecipitation with anti-GFP antibody followed by immunoblot using anti-HA antibody. (D) Point mutations in the CUE domain of gp78 (Cm gp78-Myc) nearly abolish its association with CFTRΔF508. HEK293 cells expressing CFTRΔF508-GFP, together with gp78-Myc or Cm gp78-Myc, were lysed and subjected to immunoprecipitation using anti-GFP antibody, and immune complexes were analyzed by immunoblot using anti-Myc antibody. (E) Cm gp78-Myc does not have CFTRΔF508 ubiquitylation activity. Expression vectors for CFTRΔF508-GFP and HA-ubiquitin were transfected into HEK293 cells together with expression vectors for gp78-Myc or Cm gp78-Myc. Ubiquitylated CFTRΔF508-GFP was detected by immunoprecipitation with anti-GFP antibody followed by immunoblot using anti-HA antibody.
The importance of the CUE domain in the polyubiquitylation of CFTRΔF508 was next examined. As shown in Figure 3B, coexpression of gp78ΔCUE-Myc had no apparent effect on polyubiquitylation. This result was consistent with the loss of affinity of gp78ΔCUE for CFTRΔF508-GFP. Conversely, an insertion of the CUE domain into HRD1 conferred the ubiquitylating activity for CFTRΔF508 on HRD1 (Figure 3C). This was also consistent with the result that HRD1+CUE acquired the affinity for CFTRΔF508-GFP (Figure 3A). Together, these results indicated that the CUE domain of gp78 is prerequisite and sufficient for CFTRΔF508 recognition and subsequent ubiquitylation.
The association of gp78 with CFTRΔF508 was mediated by its intrinsic CUE domain, which also has ubiquitin binding activity (Supplemental Figure 4; Zhong et al., 2004; Chen et al., 2006). To determine whether the ubiquitin binding region of the CUE domain overlapped the CFTRΔF508 binding region, we generated a gp78 mutant in which the ubiquitin binding site in the CUE domain, MFP (amino acids 463-465), was mutated to AAR. The association of this mutant (Cm gp78-Myc) with CFTRΔF508-GFP was nearly undetectable (Figure 3D), and the ubiquitylation activity of Cm gp78 for CFTRΔF508-GFP was also abolished (Figure 3E). These results indicated that the ubiquitin binding site of gp78 is also the site of association of gp78 with CFTRΔF508.
Gp78 Has E4-like Ubiquitin Ligase Activity
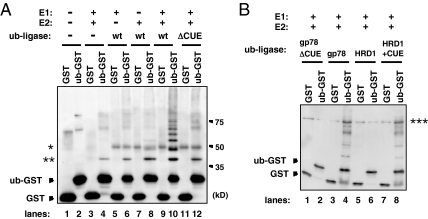
Through its intrinsic CUE domain, gp78 associated with both ubiquitin and CFTRΔF508. We were interested in whether gp78 recognized ubiquitin that was already attached to CFTRΔF508, and we catalyzed further ubiquitylation in a manner similar to that of the so-called E4 enzymes. We examined the in vitro ubiquitin ligase activity of gp78 toward GST alone, or an N-terminal ub-GST, which mimics monoubiquitylated GST. For in vitro ubiquitylation analysis, we used gp78, gp78ΔCUE, HRD1, and HRD1+CUE enzymes, which were overexpressed in HEK293 cells and immunopurified with anti-Myc antibody as described in Materials and Methods. gp78 ubiquitylated ub-GST, but not GST alone, after 2-h incubation at 37°C, which indicated that gp78 selectively ubiquitylates an already ubiquitin-conjugated substrate (Figure 4A, lanes 9 and 10). Notably, the CUE domain deletion mutant of gp78 had very low ubiquitylation activity toward GST and ub-GST (Figure 4A, lanes 11 and 12). We also observed that gp78 preferentially binds to oligoubiquitin rather than ubiquitin monomers (Supplemental Figure 4). These results indicated that gp78 possesses an E4-like ubiquitin ligase activity in vitro. As HRD1 inserted with the CUE domain acquired gp78-like CFTRΔF508-associating and -ubiquitylating activity in vivo, we tested whether HRD1+CUE acquired an E4-like ubiquitin ligase activity in vitro. As shown in Figure 4B, HRD1+CUE specifically ubiquitylated ub-GST, indicating the CUE domain conferred the E4-like activity on HRD1. Again, HRD1+CUE did not ubiquitylate the GST, as was shown for gp78 (Figure 4B, lanes 3 and 7). Although we could not rule out the effect of proteins that coimmunoprecipitated with ubiquitin ligases, these results indicate that gp78 specifically ubiquitylated ubiquitin-conjugated substrates in the CUE domain-dependent manner.
Figure 4.
Gp78 and CUE domain inserted HRD1 have an E4-like ubiquitin ligase activity in vitro. (A) Gp78 has an E4-like ubiquitin ligase activity in vitro. GST or ub-GST was incubated with ubiquitin, E1, E2 (Ube2g2), and ubiquitin ligase (wild-type gp78 or gp78ΔCUE), as indicated, and then it was analyzed by immunoblot using anti-GST antibody. (B) HRD1+CUE has an E4-like ubiquitin ligase activity in vitro. GST or ub-GST was incubated with ubiquitin, E1, E2 (Ube2g2), and ubiquitin ligase (wild-type gp78, gp78ΔCUE, wild-type HRD1, and HRD1+CUE), and then it was analyzed by immunoblot using anti-GST antibody. *, nonspecific contamination in the E3 immunoprecipitates; **, nonspecific monoubiquitylated ub-GST by Ube2g2; and ***, unidentified signal.
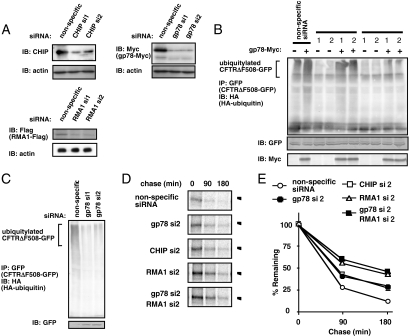
If gp78 serves as an E4-like enzyme for multiubiquitylation of CFTRΔF508 in vivo, then some other E3 ubiquitin ligase would have to mediate the initial ubiquitylation of intact CFTRΔF508. It has been reported that the ubiquitin ligases CHIP and RMA1 are involved in an early step of CFTR clearance, before CFTR is dislocated from the ER (Meacham et al., 2001; Younger et al., 2006). To examine whether CHIP or RMA1 served as an upstream E3 enzyme for the E4-like activity of gp78, we generated CHIP and RMA1 knockdown cells by using siRNA duplexes (Figure 5A). Although gp78 promoted the ubiquitylation of CFTRΔF508 in CHIP knockdown cells, it was unable to promote ubiquitylation in RMA1 knockdown cells (Figure 5B), suggesting that RMA1 is specifically required as an upstream E3 enzyme for the subsequent E4-like activity of gp78. We tested this putative E3/E4 cooperation in in vitro ubiquitylation assay by the incubation of GST or ub-GST with RMA1 and gp78 (Supplemental Figure 5). No apparent cooperative ubiquitylation for GST was observed, suggesting that RMA1 had substrate specificity and did not exert its E3 activity on GST. Ubiquitylation of CFTRΔF508 was also impaired in gp78 knockdown cells, which confirmed the importance of endogenous gp78 (Figure 5C). Knockdown of gp78 also caused a delay in the degradation of CFTRΔF508, and this delay was more pronounced when RMA1 was knocked down (Figure 5, D and E). Simultaneous knockdown of gp78 and RMA1 did not additively delay the degradation of CFTRΔF508 (Figure 5, D and E). These data suggested that RMA1 functions upstream of gp78. Knockdown of CHIP also caused a delay in the degradation of CFTRΔF508 (Figure 5, D and E), comparable with that caused by gp78 knockdown. This result was confirmed with other siRNA duplexes (Supplemental Figure 6). These results suggested that the degradation of CFTRΔF508 is mediated by at least two independent pathways—one pathway mediated by CHIP, and one pathway mediated by RMA1-gp78 (Figure 5, D and E).
Figure 5.
RMA1 functions upstream of gp78. (A) Knockdown efficiency was examined by immunoblot analysis for CHIP, gp78, and RMA1, 48 h after siRNA duplexes were transfected into HEK293 cells. Two independent sequences were adopted for knockdown of each gene. (B) RMA1 knockdown specifically inhibits gp78 activity. HEK293 cells were cotransfected with siRNA duplexes targeting CHIP or RMA1, and expression vectors for CFTRΔF508-GFP, HA-ubiquitin, and gp78-Myc. CFTRΔF508-GFP was immunoprecipitated using anti-GFP antibody, followed by immunoblot using anti-HA antibody. (C) Endogenous gp78 is involved in the ubiquitylation of CFTRΔF508. HEK293 cells were cotransfected with siRNA duplexes targeting gp78, and expression vectors for CFTRΔF508-GFP and HA-ubiquitin. The ubiquitylation of CFTRΔF508-GFP was examined 48 h after transfection. (D) Knockdown of gp78, CHIP, and RMA1 delay the degradation of CFTRΔF508. An expression vector for CFTRΔF508-GFP and siRNA duplexes were transfected into HEK293 cells. Cells were pulse-labeled for 30 min with [35S]methionine, and then they were chased for indicated periods of time. CFTRΔF508-GFP was immunoprecipitated and processed for autoradiography. (E) The radioactivity of CFTRΔF508-GFP at each time point in D was normalized to the value at chase period 0 h.
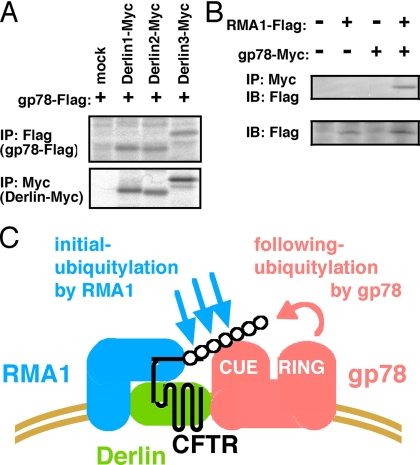
Previous work showed that RMA1 physically associates and functionally cooperates with Derlin-1, a putative channel protein for protein retrotranslocation during ERAD (Younger et al., 2006). We tested physical interaction of gp78 with Derlin and RMA1 by immunoprecipitation with specific antibodies. Gp78 associates with all members of mammalian Derlin family proteins, Derlin1, -2, and -3 (Figure 6A); and with RMA1 (Figure 6B). Together, RMA1 and gp78 could associate each other via Derlin(s) and may function successively in Derlin-containing complexes to polyubiquitylate CFTRΔF508 as E3- and E4-like enzymes, respectively (Figure 6C).
Figure 6.
Gp78 interacts with Derlins and RMA1. (A) Gp78 associates with Delin proteins. HEK293 cells transiently expressing gp78-Flag and the Myc-tagged Derlin family proteins were labeled for 1 h with [35S]methionine, and cell lysates were subjected to immunoprecipitation using anti-Flag antibody (top) or anti-Myc antibody (bottom). Immune complexes were analyzed by SDS-PAGE followed by autoradiogram. (B) Gp78 associates with RMA1. HEK293 cells transiently expressing gp78-Myc and Flag-RMA1 were lysed and subjected to immunoprecipitation using anti-Myc antibody, and immune complexes were analyzed by immunoblot using anti-Flag antibody. (C) Model of sequential ubiquitylation of CFTRΔF508 by RMA1 and gp78-containing complexes.
DISCUSSION
Hrd1p has been implicated in the ERAD of exogenously expressed human CFTR in yeast cells (Gnann et al., 2004). In the current study, we examined whether the mammalian orthologues of yeast Hrd1p also participated in the ERAD of CFTR in mammalian cells. We demonstrated that gp78, one of the two mammalian Hrd1p orthologues, is involved in the ERAD of CFTRΔF508, the most common mutant form of CFTR, in mammalian cells. Analysis of overexpression of ectopic gp78 and knockdown of endogenous gp78 showed that gp78 plays a role in the ERAD of CFTRΔF508 in HEK293 cells (Figure 5).
We also demonstrated that the CUE domain of gp78 is functionally significant in the ERAD of CFTRΔF508. Previous studies have precisely mapped the specific sites of interaction between conserved amino acid side chains of the CUE domain and side chains of ubiquitin (Kang et al., 2003; Prag et al., 2003). The amino acids identified in these previous studies were conserved in the CUE domain of gp78, which suggested that the association of gp78 with CFTRΔF508 (Figure 3D) occurs through a ubiquitin moiety. This mechanism of binding to substrates via an intervening ubiquitin molecule is used by the so-called multiubiquitin chain elongation factor E4. E4 likely recognizes a ubiquitin moiety already attached to a mono- or oligoubiquitylated substrate, and it catalyzes further ubiquitin elongation (Koegl et al., 1999; Hoppe, 2005; Richly et al., 2005).
Ufd2, originally identified E4, cannot elongate ubiquitin chain to ub-protein A without Ufd4, a cooperative E3 (Koegl et al., 1999). This would be different from the case of gp78 reported here, which can ubiquitylate ub-GST without cooperative E3 in vitro. The precise enzymatic mechanism of Ufd2 remains unclear. Recently, Tu et al. (2007) described the crystal structure of full-length Ufd2, and they showed its own E3 activity. Based on the crystal structure and functional analysis, they proposed alternative models of how Ufd2 (E4) and Ufd4 (E3) can cooperate to polyubiquitylate substrate. The first model is a “sequential model” in which substrate is first ubiquitylated by E3 followed by the elongation of ubiquitin chain by E4. In this model, E4 can catalyze the ubiquitylation reaction without E3. The other model is a “cooperative model” in which E3 and E4 cooperatively catalyze the polyubiquitylation reaction. If the enzymatic mechanism of E4-like activity of gp78 is sequential, gp78 will ubiquitylate ub-GST without cooperative E3. Considering the in vitro ubiquitylation analysis reported here, we propose that gp78 may sequentially ubiquitylate CFTRΔF508 as an E4-like enzyme after the initial ubiquitylation by E3.
It has been reported that various E4 enzymes require their specific E3 enzymes (e.g., the E4 enzyme Ufd2 cooperates with the E3 enzyme Ufd4 and the E4 enzyme p300 cooperates with the E3 enzyme Mdm2) (Koegl et al., 1999; Grossman et al., 2003; Hoppe, 2005). In the current study, we showed that RNA interference-mediated knockdown of RMA1 strongly reduced gp78-mediated ubiquitylation of CFTRΔF508, whereas knockdown of CHIP had no apparent effect. Both RMA1 and CHIP have been reported to possess E3 activity and to ubiquitylate CFTRΔF508. Our results indicate that the E4-like enzyme gp78 sequentially cooperates with RMA1 in the ubiquitylation of CFTRΔF508. In addition, the interaction between gp78 and RMA1 putatively mediated via Derlin(s) suggest a direct transfer of CFTRΔF508 from E3 to E4 in the same complex (Figure 6C). RMA1 seems to act cotranslationally and CHIP posttranslationally to select CFTR for ERAD (Younger et al., 2006). It takes 10 min to synthesize CFTR; so, it is possible that pools of CFTR that are initially ubiquitylated by RMA1 may not be immediately retrotranslocated and degraded. Such stalled forms of ubiquitylated CFTR might become substrates of deubiquitinating enzymes. Therefore, the binding of polyubiquitylated CFTR by the CUE domain of gp78 might ward off the effects of deubiquitylating enzymes and serve to maintain the length or extend of ubiquitin chains attached to CFTR during the time gap between its initial ubiquitylation and retrotranslocation from the ER membrane. CHIP seems to act posttranslationally, so the ubiquitin chains attached to CFTR would be recognized immediately and there would be no need for an E4 enzyme. However, we should point out that the study was done with CFTRΔF508 transiently overexpressed in HEK293 cells; thus, the study still leaves open the exact extent to which the E4-like activity of gp78 is relevant to the physiological and clinical situation.
In vitro study shown in Figure 4 indicated that gp78 specifically ubiquitylated ubiquitin-fused GST, but not intact GST. However, it is not clear which lysine residue(s) in Ub-GST was ubiquitylated by gp78. E4 enzyme was originally defined as a ubiquitin ligase that elongates the ubiquitin chain on the mono- or oligoubiquitins already attached on the substrates. If gp78 catalyzed extensive ubiquitylation on the ubiquitin moiety of Ub-GST, gp78 should be classified as the E4 enzyme; however, if gp78 ubiquitylated GST moiety of Ub-GST, it will not be a typical E4 enzyme itself, but is a specific class of ubiquitin ligase, which needs the ubiquitin moiety on the substrate for its ligase activity. Although we cannot conclude which is the case in the present study, we can attribute this gp78 activity as “E4-like activity,” because it needs initial ubiquitylation by other ubiquitin ligase in the ERAD of CFTRΔF508.
Previously, it was reported that mutations in the CUE domain of gp78 that impaired its ability to undergo self-ubiquitylation and to ubiquitylate the substrate CD3-δ did not affect the physical association of gp78 with CD3-δ (Chen et al., 2006). This observation suggests that a ubiquitin moiety is not needed for the association of gp78 and CD3-δ and that gp78 has a substrate recognition domain for CD3-δ that is independent of the ubiquitin binding domain. Thus, Gp78 may function as an E3 ubiquitin ligase in the ERAD of CD3-δ, and as an E4-like enzyme in the ERAD of CFTRΔF508.
Two models of substrate recognition by gp78 are possible. In the first, gp78 directly recognizes a substrate through a specific recognition domain, which is distinct from the CUE domain. If a substrate has an affinity for this putative substrate recognition domain of gp78, gp78 associates with the intact substrate and catalyzes its ubiquitylation. Alternatively, if a substrate does not have a high enough affinity for the substrate recognition domain, gp78 cooperates with RMA1 to polyubiquitylate the substrate. Another possibility is that cofactors of gp78 influence the enzymatic activity of gp78. Recently, it was proposed that several factors, including HRD1, gp78, EDEM, Derlin, VIMP, p97/VCP, PNGase, and proteasome constitute a functional complex, which dispatches substrates effectively for succeeding steps of ERAD, including dislocation from the ER, ubiquitylation, deglycosylation, and degradation (Lilley and Ploegh, 2004, 2005; Ye et al., 2004, 2005; Li et al., 2006; Oda et al., 2006; Carvalho et al., 2006). The enzymatic specificity of gp78 may be regulated by the combination of ERAD factors present in gp78-containing complexes. Thus, gp78 may function as an E4-like enzyme when it is a component of RMA1-containing complexes. Likewise, RMA1 may catalyze oligoubiquitylation when it is present in gp78-containing complexes, and full ubiquitylation in complexes of a different molecular composition. Future studies will address the molecular determinants of gp78 enzymatic activity and the different pathways of ERAD in mammalian cells.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Ciechanover and Y. Yoshida for invaluable discussions, K. Mori for kind help, and R. Kopito for helpful discussions and providing plasmids. Portions of this work were supported by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to N.H. and K.N.).
Abbreviations used:
- CFTR
cystic fibrosis transmembrane conductance regulator
- CUE
coupling of ubiquitin to ER degradation
- ER
endoplasmic reticulum
- ERAD
endoplasmic reticulum-associated degradation
- GFP
green fluorescent protein
- HA
hemagglutinin
- siRNA
small interfering RNA.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-06-0601) on January 23, 2008.
REFERENCES
- Amano T., et al. Synoviolin/Hrd1, an E3 ubiquitin ligase, as a novel pathogenic factor for arthropathy. Genes Dev. 2003;17:2436–2449. doi: 10.1101/gad.1096603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballar P., Shen Y., Yang H., Fang S. The role of a novel p97/valosin-containing protein (VCP)-interacting motif of GP78 in endoplasmic reticulum-associated degradation. J. Biol. Chem. 2006;281:35359–35368. doi: 10.1074/jbc.M603355200. [DOI] [PubMed] [Google Scholar]
- Bays N. W., Gardner R. G., Seelig L. P., Joazeiro C. A., Hampton R. Y. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat. Cell Biol. 2001;3:24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- Bordallo J., Plemper R. K., Finger A., Wolf D. H. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol. Biol. Cell. 1998;9:209–222. doi: 10.1091/mbc.9.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P., Goder V., Rapoport T. A. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Chen B., Mariano J., Tsai Y. C., Chan A. H., Cohen M., Weissman A. M. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc. Natl. Acad. Sci. USA. 2006;103:341–346. doi: 10.1073/pnas.0506618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak P. M., Wolf D. H. Membrane topology and function of Der3/Hrd1p as a ubiquitin-protein ligase (E3) involved in endoplasmic reticulum degradation. J. Biol. Chem. 2001;276:10663–10669. doi: 10.1074/jbc.M008608200. [DOI] [PubMed] [Google Scholar]
- Fang S., Ferrone M., Yang C., Jensen J. P., Tiwari S., Weissman A. M. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2001;98:14422–14427. doi: 10.1073/pnas.251401598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S., Lorick K. L., Jensen J. P., Weissman A. M. RING finger ubiquitin protein ligases: implications for tumorigenesis, metastasis and for molecular targets in cancer. Semin. Cancer Biol. 2003;13:5–14. doi: 10.1016/s1044-579x(02)00095-0. [DOI] [PubMed] [Google Scholar]
- Gnann A., Riordan J. R., Wolf D. H. Cystic fibrosis transmembrane conductance regulator degradation depends on the lectins Htm1p/EDEM and the Cdc48 protein complex in yeast. Mol. Biol. Cell. 2004;15:4125–4135. doi: 10.1091/mbc.E04-01-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman S. R., Deato M. E., Brignone C., Chan H. M., Kung A. L., Tagami H., Nakatani Y., Livingston D. M. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science. 2003;300:342–344. doi: 10.1126/science.1080386. [DOI] [PubMed] [Google Scholar]
- Hampton R. Y., Gardner R. G., Rine J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell. 1996;7:2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A., Varshavsky A. Basic Medical Research Award. The ubiquitin system. Nat. Med. 2000;6:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- Hoppe T. Multiubiquitylation by E4 enzymes: ‘one size’ doesn't fit all. Trends Biochem. Sci. 2005;30:183–187. doi: 10.1016/j.tibs.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Iwai K., Yamanaka K., Kamura T., Minato N., Conaway R. C., Conaway J. W., Klausner R. D., Pause A. Identification of the von Hippel-lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl. Acad. Sci. USA. 1999;96:12436–12441. doi: 10.1073/pnas.96.22.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M., Ishiguro M., Niinuma Y., Uesugi M., Nomura Y. Human HRD1 protects against ER stress-induced apoptosis through ER-associated degradation. FEBS Lett. 2002;532:147–152. doi: 10.1016/s0014-5793(02)03660-8. [DOI] [PubMed] [Google Scholar]
- Kang R. S., Daniels C. M., Francis S. A., Shih S. C., Salerno W. J., Hicke L., Radhakrishnan I. Solution structure of a CUE-ubiquitin complex reveals a conserved mode of ubiquitin binding. Cell. 2003;113:621–630. doi: 10.1016/s0092-8674(03)00362-3. [DOI] [PubMed] [Google Scholar]
- Kikkert M., Doolman R., Dai M., Avner R., Hassink G., van Voorden S., Thanedar S., Roitelman J., Chau V., Wiertz E. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J. Biol. Chem. 2004;279:3525–3534. doi: 10.1074/jbc.M307453200. [DOI] [PubMed] [Google Scholar]
- Koegl M., Hoppe T., Schlenker S., Ulrich H. D., Mayer T. U., Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- Kopito R. R. Biosynthesis and degradation of CFTR. Physiol. Rev. 1999;79:S167–S173. doi: 10.1152/physrev.1999.79.1.S167. [DOI] [PubMed] [Google Scholar]
- Lee J. N., Song B., Debose-Boyd R. A., Ye J. Sterol-regulated degradation of Insig-1 mediated by the membrane-bound ubiquitin ligase, gp78. J. Biol. Chem. 2006;281:39308–39315. doi: 10.1074/jbc.M608999200. [DOI] [PubMed] [Google Scholar]
- Li G., Zhao G., Zhou X., Schindelin H., Lennarz W. J. The AAA ATPase p97 links peptide N-glycanase to the endoplasmic reticulum-associated E3 ligase autocrine motility factor receptor. Proc. Natl. Acad. Sci. USA. 2006;103:8348–8353. doi: 10.1073/pnas.0602747103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J. S., Kim T., Fang S., Yamaguchi J., Weissman A. M., Fisher E. A., Ginsberg H. N. Overexpression of the tumor autocrine motility factor receptor Gp78, a ubiquitin protein ligase, results in increased ubiquitinylation and decreased secretion of apolipoprotein B100 in HepG2 cells. J. Biol. Chem. 2003;278:23984–23988. doi: 10.1074/jbc.M302683200. [DOI] [PubMed] [Google Scholar]
- Lilley B. N., Ploegh H. L. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429:834–840. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- Lilley B. N., Ploegh H. L. Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. USA. 2005;102:14296–14301. doi: 10.1073/pnas.0505014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham G. C., Patterson C., Zhang W., Younger J. M., Cyr D. M. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- Meusser B., Hirsch C., Jarosch E., Sommer T. ERAD: the long road to destruction. Nat. Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- Nabi I. R., Raz A. Cell shape modulation alters glycosylation of a metastatic melanoma cell-surface antigen. Int. J. Cancer. 1987;40:396–402. doi: 10.1002/ijc.2910400319. [DOI] [PubMed] [Google Scholar]
- Nadav E., Shmueli A., Barr H., Gonen H., Ciechanover A., Reiss Y. A novel mammalian endoplasmic reticulum ubiquitin ligase homologous to the yeast Hrd1. Biochem. Biophys. Res. Commun. 2003;303:91–97. doi: 10.1016/s0006-291x(03)00279-1. [DOI] [PubMed] [Google Scholar]
- Oda Y., Okada T., Yoshida H., Kaufman R. J., Nagata K., Mori K. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J. Cell Biol. 2006;172:383–393. doi: 10.1083/jcb.200507057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C. P. Proteins of the endoplasmic-reticulum-associated degradation pathway: domain detection and function prediction. Biochem. J. 2000;351:527–535. [PMC free article] [PubMed] [Google Scholar]
- Prag G., Misra S., Jones E. A., Ghirlando R., Davies B. A., Horazdovsky B. F., Hurley J. H. Mechanism of ubiquitin recognition by the CUE domain of Vps9p. Cell. 2003;113:609–620. doi: 10.1016/s0092-8674(03)00364-7. [DOI] [PubMed] [Google Scholar]
- Richly H., Rape M., Braun S., Rumpf S., Hoege C., Jentsch S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Sato S., Chiba T., Sakata E., Kato K., Mizuno Y., Hattori N., Tanaka K. 14-3–3eta is a novel regulator of parkin ubiquitin ligase. EMBO J. 2006;25:211–221. doi: 10.1038/sj.emboj.7600774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B. L., Sever N., DeBose-Boyd R. A. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol. Cell. 2005;19:829–840. doi: 10.1016/j.molcel.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Tu D., Li W., Ye Y., Brunger A. T. Inaugual Article; Structure and function of the yeast U-box-containing ubiquitin ligase Ufd2p. Proc. Natl. Acad. Sci. USA. 2007;104:15599–15606. doi: 10.1073/pnas.0701369104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. J., Benlimame N., Nabi I. The AMF-R tubule is a smooth ilimaquinone-sensitive subdomain of the endoplasmic reticulum. J. Cell Sci. 1997;110:3043–3053. doi: 10.1242/jcs.110.24.3043. [DOI] [PubMed] [Google Scholar]
- Ye Y., Shibata Y., Kikkert M., van Voorden S., Wiertz E., Rapoport T. A. Inaugural article: recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. USA. 2005;102:14132–14138. doi: 10.1073/pnas.0505006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Shibata Y., Yun C., Ron D., Rapoport T. A. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- Yoshida Y., et al. E3 ubiquitin ligase that recognizes sugar chains. Nature. 2002;418:438–442. doi: 10.1038/nature00890. [DOI] [PubMed] [Google Scholar]
- Younger J. M., Chen L., Ren H. Y., Rosser M. F., Turnbull E. L., Fan C. Y., Patterson C., Cyr D. M. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- Zhong X., Shen Y., Ballar P., Apostolou A., Agami R., Fang S. AAA ATPase p97/valosin-containing protein interacts with gp78, a ubiquitin ligase for endoplasmic reticulum-associated degradation. J. Biol. Chem. 2004;279:45676–45684. doi: 10.1074/jbc.M409034200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.