Abstract
Primary cilia are sensory organelles present on most mammalian cells. The functions of cilia are defined by the signaling proteins localized to the ciliary membrane. Certain G protein–coupled receptors (GPCRs), including somatostatin receptor 3 (Sstr3) and serotonin receptor 6 (Htr6), localize to cilia. As Sstr3 and Htr6 are the only somatostatin and serotonin receptor subtypes that localize to cilia, we hypothesized they contain ciliary localization sequences. To test this hypothesis we expressed chimeric receptors containing fragments of Sstr3 and Htr6 in the nonciliary receptors Sstr5 and Htr7, respectively, in ciliated cells. We found the third intracellular loop of Sstr3 or Htr6 is sufficient for ciliary localization. Comparison of these loops revealed a loose consensus sequence. To determine whether this consensus sequence predicts ciliary localization of other GPCRs, we compared it with the third intracellular loop of all human GPCRs. We identified the consensus sequence in melanin-concentrating hormone receptor 1 (Mchr1) and confirmed Mchr1 localizes to primary cilia in vitro and in vivo. Thus, we have identified a putative GPCR ciliary localization sequence and used this sequence to identify a novel ciliary GPCR. As Mchr1 mediates feeding behavior and metabolism, our results implicate ciliary signaling in the regulation of body weight.
INTRODUCTION
Primary cilia are appendages that project from almost all human cell types (Wheatley et al., 1996). It is generally accepted that primary cilia serve important specialized signaling functions (Pazour and Witman, 2003; Marshall and Nonaka, 2006; Singla and Reiter, 2006). In the eye, photoreceptors, which are modified primary cilia, sense and respond to light. In the nose, specialized olfactory cilia detect odors and initiate signaling cascades in olfactory neurons. In the kidney, it is proposed that bending of cilia on epithelial cells by fluid flow triggers an increase in intracellular calcium mediated by an ion channel located on the cilium (Praetorius and Spring, 2001; Nauli et al., 2003). In each case, the function of the cilium is defined by the signaling proteins that are enriched in the ciliary membrane (i.e., light receptors, odorant receptors, and mechanoreceptors). Importantly, disruption of the signaling mediated by these receptors can cause disease and altered development (Davenport and Yoder, 2005; Hildebrandt and Otto, 2005; Pan et al., 2005; Bisgrove and Yost, 2006). Yet, the specific signaling proteins that localize to the vast majority of cilia in the mammalian body are unknown. Thus, the functions of primary cilia on most cell types in the body are unknown.
Neuronal primary cilia are abundant throughout the rodent brain (Bishop et al., 2007). The functional importance of these cilia is suggested by the fact that several human ciliary disorders, including Bardet-Biedl syndrome (BBS), Joubert syndrome (JS), and Meckel syndrome (MKS), have prominent functional and structural CNS phenotypes (Badano et al., 2006). Although the specific functions of neuronal cilia are unknown, certain G protein–coupled receptors (GPCRs), including somatostatin receptor 3 (Sstr3; Handel et al., 1999; Schulz et al., 2000) and serotonin receptor 6 (Htr6; Hamon et al., 1999; Brailov et al., 2000), localize to the ciliary membrane on neurons. This observation suggests that cilia may sense levels of neuromodulators in the local environment of neurons. Given that the minority of the total ciliated neurons are positive for Sstr3 (Berbari et al., 2007) and the distribution of Htr6 ciliary localization is restricted to a few regions of the brain (Hamon et al., 1999; Brailov et al., 2000), it is likely that additional GPCRs localize to neuronal cilia.
Although the ciliary membrane is continuous with the plasma membrane, only certain GPCRs localize to cilia. This is clearly demonstrated by the somatostatin receptors. There are five somatostatin receptor genes present in humans and rodents (Sstr1-5; Patel, 1999). In addition, the carboxy (C)-tail of Sstr2 can undergo alternative splicing to yield Sstr2a and Sstr2b. They are all members of the GPCR superfamily of cell-surface receptors that couple to heterotrimeric G proteins and regulate numerous downstream effectors. There are ∼950 GPCRs in the human genome, with 500 of those coding for odorant or taste receptors and the remaining 450 coding for receptors with endogenous ligands (Takeda et al., 2002). All GPCRs share a common molecular topology of seven transmembrane-spanning domains, three intracellular loops, three extracellular loops, an amino (N)-terminus outside the cell, and a C-terminus inside the cell (Dong et al., 2007). The six somatostatin receptor subtypes are expressed in the rat CNS and show overlapping regional distributions (Schulz et al., 2000). Yet, Sstr3 is selectively targeted to the ciliary membrane (Handel et al., 1999; Schulz et al., 2000). Similarly, of the 13 mammalian G protein–coupled serotonin receptor subtypes only Htr6 has been shown to localize to cilia (Hamon et al., 1999; Brailov et al., 2000). These observations suggest that Sstr3 and Htr6 contain unique sequences that mediate their localization to cilia.
Very little is known about the underlying mechanisms of sorting and trafficking of GPCRs to the cilium. One GPCR ciliary localization sequence that has been described is an adjacent hydrophobic and basic residue motif immediately C-terminal to the seventh transmembrane segment. In Caenorhabditis elegans, this sequence is required for ciliary localization of olfactory GPCRs (Dwyer et al., 2001). In mammalian cells, this motif is required for the ciliary localization of the GPCR Smoothened (Corbit et al., 2005). Although this motif is present in Sstr3 and Htr6, it is also present and conserved in all of the somatostatin and serotonin receptor subtypes. Yet, only Sstr3 and Htr6 localize to cilia. Thus, this motif may be necessary but it is not sufficient to specify ciliary localization.
We hypothesized that ciliary GPCRs contain unique sequences that mediate localization to cilia. Here, we report that the third intracellular loop of Sstr3 and Htr6 is sufficient to localize nonciliary GPCRs to cilia, suggesting that Sstr3 and Htr6 are targeted to cilia through similar mechanisms. Comparison of the sequences within these loops reveals a putative consensus sequence. This consensus sequence is present in the third intracellular loop of numerous known ciliary GPCRs and also identifies a number of candidate ciliary GPCRs. We subsequently show that one of these candidates, melanin-concentrating hormone receptor 1 (Mchr1), localizes to cilia in vitro and in vivo. These findings identify a new role for cilia in MCH signaling and, because Mchr1 is involved in the regulation of energy homeostasis, implicate ciliary signaling in the regulation of body weight.
MATERIALS AND METHODS
Animals and Tissue Processing
The mice used in this study were on a 129:BL6 background. All procedures were approved by the Institutional Animal Care and Use Committee at The Ohio State University. Animals were anesthetized by a 0.1 ml/10 g intraperitoneal injection of 2.5% tribromoethanol (Sigma-Aldrich, St. Louis, MO), killed by cardiac puncture, and perfused with phosphate-buffered saline (PBS) followed by a mixture of 2% paraformaldehyde and HistoChoice (Amresco, Solon, OH). The brains were then further fixed in 2% PFA/HistoChoice for 16–24 h at 4°C followed by cryoprotection in 30% sucrose in PBS for 16–24 h. Cryoprotected brains were embedded in Optimal Cutting Temperature compound (VWR, West Chester, PA), and sectioned in a cryostat at a thickness of 30 μm. Labelings were performed in three different animals.
Plasmid Construction
The coding sequences of the somatostatin receptors were amplified from mouse genomic DNA. The unspliced version of Sstr2 that was amplified corresponds to Sstr2a. The coding sequences of serotonin receptors 6 and 7 and melanin-concentrating hormone receptor 1 were amplified from reverse-transcribed mouse whole brain RNA using the Superscript First-Strand Synthesis RT-PCR Kit (Invitrogen, Carlsbad, CA). All coding sequences were cloned into a TA cloning vector (pSTBlue-1; Novagen, San Diego, CA). Receptor open reading frames were utilized as templates for overlap extension PCR to generate chimeric receptors, as previously described (Horton et al., 1990). Briefly, oligonucleotide primers (36mers; IDT Technologies, Coralville, IA) corresponding to the nucleotide sequence of the junction regions were generated with 5′ nucleotide sequence from one receptor and 3′ sequence from the other. The junctions were designed at conserved residues, the coding frames were maintained, and no sequences were inserted or deleted. Primers corresponding to the N-terminal and C-terminal regions of both receptors were designed with 5′ restriction sites for directional cloning. The initial PCR generated fragments of the two receptors to be combined with complementary overhanging sequence. The products from the first reaction were gel-purified and combined as template for the fusion PCR. All amplifications were performed with HiFi Platinum Taq Polymerase (Invitrogen). The final PCR products were cloned into the pEGFP-N vector (Clontech, Mountain View, CA) using the added restriction enzyme sites. All DNA constructs were sequence verified. Primer sequences are available upon request. Chimeric receptor sequences were mutated using the QuikChange Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA).
Cell Culture and Transient Transfections
IMCD-3 cells (ATCC, Manassas, VA) were maintained in DMEM:F12 media supplemented with 10% FBS, 1.2 g/l of sodium bicarbonate, and 0.5 mM sodium pyruvate (Invitrogen). Cells (n = 5 × 106) were electroporated with 10 μg DNA and plated at high density on glass coverslips. Cells were harvested 24 h after transfection for immunocytochemistry.
Immunofluorescence Procedures
Cells were fixed in 4% paraformaldehyde and permeabilized with 0.3% Triton X-100 in PBS with 2% goat serum, 0.02% sodium azide, and 10 mg/ml bovine serum albumin (BSA). The IMCD cells were then labeled with anti-acetylated α-tubulin (T-6793; Sigma-Aldrich). Brain sections were permeabilized with 0.3% Triton X-100 in PBS with 2% donkey serum, 0.02% sodium azide, and 10 mg/ml BSA and simultaneously labeled with anti-adenylyl cyclase III (ACIII) rabbit polyclonal antibody (sc-588; Santa Cruz Biotechnology, Santa Cruz, CA), used at 1:500, and anti-melanin–concentrating hormone receptor 1 goat polyclonal antibody (sc-5534; Santa Cruz Biotechnology), used at 1:250. All incubations and washes were carried out in PBS with 2% serum, 0.02% sodium azide, and 10 mg/ml BSA. Primary antibody incubations were carried out for 16–24 h at 4°C, and secondary antibody incubations were carried out for 1 h at room temperature. Secondary antibodies included the following: Alexa Fluor 546–conjugated goat anti-mouse IgG (Invitrogen), Alexa Fluor 488–conjugated donkey anti-goat IgG (Invitrogen), or Cy3-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA). Nuclei were visualized by DRAQ5 (Axxora, San Diego, CA). Slides were mounted using Immu-Mount (Thermo Scientific, Pittsburgh, PA).
Data Analysis
All samples were imaged on a Zeiss 510 META laser scanning confocal microscope (Thornwood, NY) at the Ohio State University Central Microscopy Imaging Facility. Multiple consecutive focal planes (Z-stack), spaced at 0.5–1-μm intervals, were captured. For all collected images, the brightness and contrast of each channel was adjusted using the Zeiss LSM Image Browser program. For quantitative analysis, coverslips were centered on the microscope objective, and 3–5 consecutive fields were imaged. The number of ciliated transfected cells and the number of transfected cells expressing cilia localization in each image were counted by individuals blinded to the experimental conditions. The results are expressed as the percent of transfected cells showing ciliary localization. A total of ∼100 cells were counted from three distinct transfections (n = 3) for each construct. Data are expressed as mean ± SEM.
Prediction of Human GPCR Third Intracellular Loop Sequences
Human GPCR protein sequences were downloaded from the GPCRDB information system (http://www.gpcr.org/7tm/; Horn et al., 2003). The protein sequences were input into the transmembrane hidden Markov model (TMHMM) program (version 2.0) to generate predictions of transmembrane, intracellular, and extracellular sequences (Krogh et al., 2001). A Perl program was then applied to the TMHMM output to extract the predicted third intracellular loop sequence for all human GPCRDB proteins. The resulting database was searched using the ambiguous regular expression “AX[S/A]XQ” to identify GPCRs predicted to contain this motif within their third intracellular loop.
RESULTS
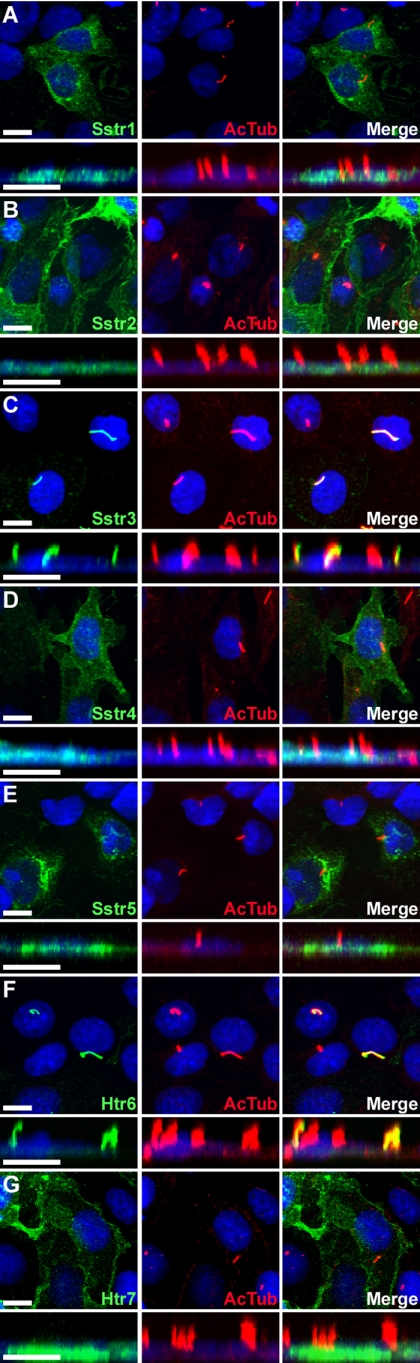
To determine whether heterologous expression could be used to identify GPCR ciliary localization sequences, we generated constructs encoding mouse somatostatin receptors one through five (Sstr1–5) fused at the C-terminus to enhanced green fluorescent protein (EGFP). These constructs were then expressed in inner medullary collecting duct (IMCD) cells, which are derived from mouse kidney and develop cilia in culture. Visualization of the subcellular localization of the fluorescently labeled receptors 24–48 h after transfection showed that of the somatostatin receptor subtypes, only Sstr3 selectively localized to cilia (Figure 1, A–E). The remaining somatostatin receptor subtypes failed to localize to cilia, as indicated by a lack of colocalization with the ciliary marker acetylated α-tubulin, and instead localized to the plasma membrane or within intracellular compartments. Similarly, transfection of IMCD cells with Htr6 and the closely related Htr7 fused at the C-terminus to EGFP revealed that Htr6 preferentially localized to cilia, whereas Htr7 localized primarily to the plasma membrane (Figure 1, F and G). Interestingly, cilia on cells expressing Sstr3 or Htr6 consistently appeared longer than cilia on untransfected cells or on cells expressing nonciliary GPCRs (Figure 1, A–G). The underlying mechanism for this difference is unknown. Overall, these results suggest that Sstr3 and Htr6 contain sequences that specify ciliary localization, and IMCD cells possess the necessary machinery to traffic specific receptors to cilia.
Figure 1.
Somatostatin receptor subtype 3 and serotonin receptor subtype 6 selectively localize to cilia when heterologously expressed in IMCD cells. (A–G) Representative confocal microscopy images of transiently transfected IMCD cells expressing somatostatin receptors 1 through 5 (Sstr1–5) and serotonin receptors 6 (Htr6) and 7 (Htr7) fused at the C-terminus to EGFP. Left, EGFP fluorescence (green) shows expression of the receptors; middle, acetylated α-tubulin (red) marks the cilia; right, merged images. Below each panel is a confocal image in the xz plane to show cilia projecting vertically from the apical surface of cells. Note that only Sstr3-EGFP (C) and Htr6-EGFP (F) show localization that overlaps with acetylated α-tubulin ciliary labeling. Sstr2 corresponds to Sstr2a. Nuclei are labeled with the DNA stain DRAQ5 (blue). All scale bars, 10 μm.
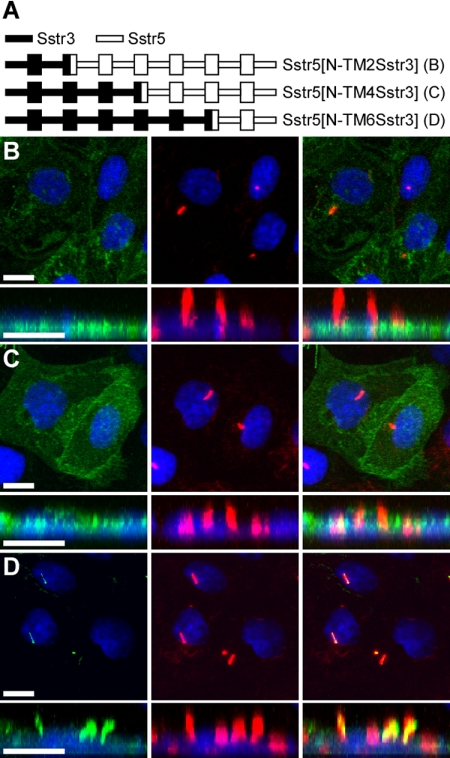
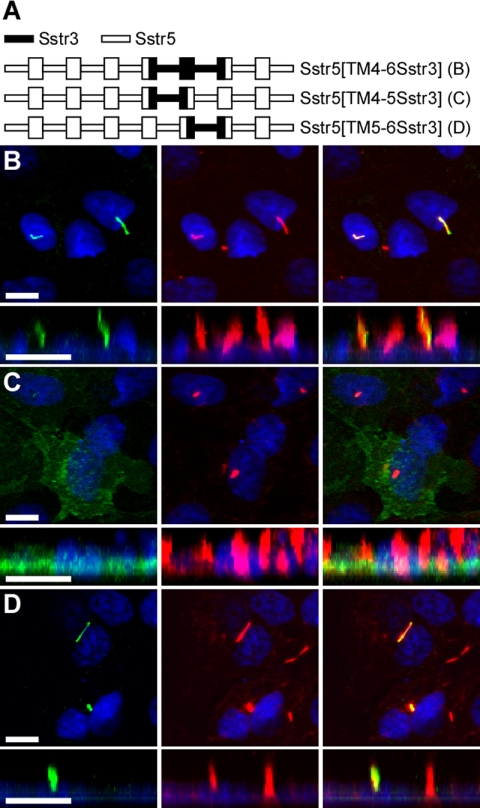
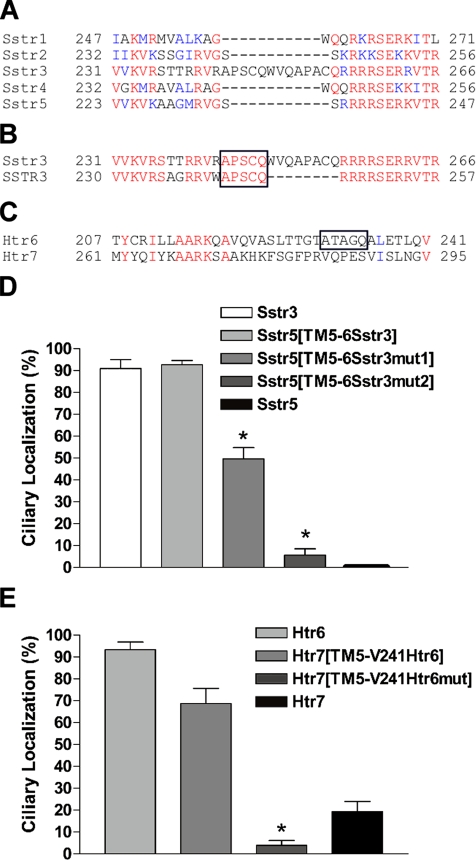
To identify the region of Sstr3 containing ciliary localization sequences, we utilized a fusion PCR approach to construct EGFP-fused chimeric receptors. Chimeras were generated containing segments of Sstr3 and the structurally and pharmacologically similar Sstr5. To minimize the likelihood of protein misfolding, the chimeric receptor fusion sites were engineered at conserved residues located within the transmembrane domains. Initially, we generated chimeric receptors of Sstr5 in which the sequence from the N-terminus to the second, fourth, or sixth transmembrane (TM) domains was substituted with the corresponding sequence from Sstr3 (Figure 2A). Expression of these chimeric receptors in IMCD cells revealed that only chimeric receptor Sstr5[N-TM6Sstr3] selectively localized to cilia (Figure 2, B–D), suggesting that sequences between the TM4 and TM6 domains in Sstr3 mediate ciliary localization. To test this hypothesis, we generated a chimeric receptor of Sstr5 in which the sequence between the TM4 and TM6 domains was replaced with the corresponding sequence from Sstr3 (Figure 3A). This chimera also localized to cilia (Figure 3B), suggesting that ciliary localization sequences are located within the third extracellular (e3) loop or the third intracellular (i3) loop of Sstr3. To distinguish between these two possibilities, we generated chimeric receptors of Sstr5 containing only the e3 or i3 loop of Sstr3 (Figure 3A). Notably, chimeric receptor Sstr5[TM4-5Sstr3] did not localize to cilia (Figure 3C), but chimeric receptor Sstr5[TM5-6Sstr3] did localize to cilia (Figure 3D), indicating that sequences within the i3 loop of Sstr3 are sufficient to localize Sstr5 to cilia. Immunoblotting of proteins isolated from cells transiently transfected with chimeric receptors Sstr5[TM4-5Sstr3] and Sstr5[TM5-6Sstr3] revealed similar expression patterns (Supplementary Figure 1), indicating that absence of cilia localization was not due to the lack of receptor expression or stability.
Figure 2.
Sequences between the fourth and sixth transmembrane domains of Sstr3 are important for ciliary localization. (A) Schematic of chimeric receptors containing portions of Sstr3 (indicated by black lines) and Sstr5 (indicated by white lines) fused at the C-terminus to EGFP. Transmembrane domains (TM) are depicted as boxes. (B–D) Representative images of transiently transfected IMCD cells expressing the indicated chimeric receptors. Left, EGFP fluorescence (green); middle, acetylated α-tubulin (red); right, merged images. Chimeric receptors Sstr5[N-TM2Sstr3] (B) and Sstr5[N-TM4Sstr3] (C) do not localize to cilia. Chimeric receptor Sstr5[N-TM6Sstr3] does localize to cilia, suggesting that ciliary localization of Sstr3 is mediated by sequences between TM4 and TM6. Nuclei are labeled with DRAQ5 (blue). All scale bars, 10 μm.
Figure 3.
The third intracellular (i3) loop of Sstr3 is sufficient to localize Sstr5 to cilia. (A) Schematic of chimeric receptors containing portions of Sstr3 (black lines) and Sstr5 (white lines) fused at the C-terminus to EGFP. TM domains are depicted as boxes. (B–D) Representative images of transiently transfected IMCD cells expressing the indicated chimeric receptors. Left, EGFP fluorescence (green); middle, acetylated α-tubulin (red); right, merged images. Chimeric receptors Sstr5[TM4-6Sstr3] (B) and Sstr5[TM5-6Sstr3] (D) localize to cilia, whereas chimeric receptor Sstr5[TM4-5Sstr3] (C) does not localize to cilia, suggesting that the i3 loop of Sstr3 contains ciliary localization sequences. Nuclei are labeled with DRAQ5 (blue). All scale bars, 10 μm.
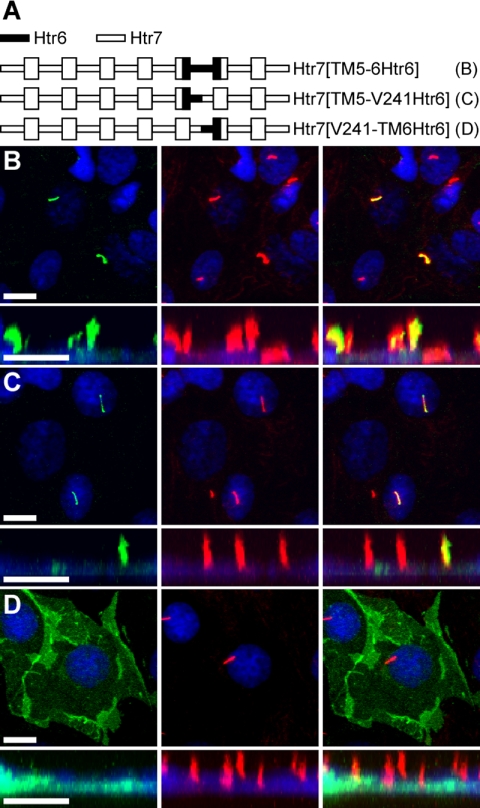
To determine whether the i3 loop of Htr6 also mediates ciliary localization, we generated a chimeric receptor in which the i3 loop of Htr7 had been substituted with the i3 loop of Htr6 (Figure 4A). Notably, this chimeric receptor selectively localized to cilia (Figure 4B). This result indicates that Sstr3 and Htr6 contain ciliary localization sequences within the same domain and suggest that they may be targeted to cilia through similar mechanisms. Because the predicted i3 loop of Htr6 (63 residues) is significantly larger than the i3 loop of Sstr3 (36 residues), we further narrowed the region of Htr6 sufficient to traffic Htr7 to the cilium by generating chimeric receptors containing only the N- or C-portion of the i3 loop (Figure 4A). V241 in Htr6, which corresponds to V295 in Htr7, was used as the fusion site. Chimeric receptor Htr7[TM5-V241Htr6] selectively localized to cilia (Figure 4C) but chimeric receptor Htr7[V241-TM6Htr6] did not (Figure 4D), indicating that sequences within the N-portion of the i3 loop of Htr6 are sufficient to localize Htr7 to cilia.
Figure 4.
The amino portion of the i3 loop of Htr6 is sufficient to localize Htr7 to cilia. (A) Schematic of chimeric receptors containing portions of Htr6 (black lines) and Htr7 (white lines) fused at the C-terminus to EGFP. TM domains are depicted as boxes. (B–D) Representative images of transiently transfected IMCD cells expressing the indicated chimeric receptors. Left, EGFP fluorescence (green); middle, acetylated α-tubulin (red); right, merged images. Chimeric receptors Htr7[TM5-6Htr6] (B) and Htr7[TM5-V241Htr6] (C) selectively localize to cilia, whereas chimeric receptor Htr7 [V241-TM6Htr6] (D) does not, suggesting that the N-portion of the i3 loop of Htr6 contains ciliary localization sequences. Nuclei are labeled with DRAQ5 (blue). All scale bars, 10 μm.
We hypothesized that ciliary localization sequences would be unique to the receptor subtype that localizes to cilia, would be conserved in species in which the receptor is ciliary, and would be similar between Sstr3 and Htr6. Comparison of the i3 loop sequences of the mouse somatostatin receptors reveals similarity between Sstr3 and the other subtypes, except for several residues in the N-portion and an insertion of 11 amino acids that is unique to Sstr3 (Figure 5A). To further narrow the sequences of interest we compared the i3 loop sequences of Sstr3 across species. Comparison of the mouse and human sequences reveals that the unique residues in the N-portion of the loop are not conserved, but 5 of the 11 amino acids inserted in mouse Sstr3 (APSCQ) are completely conserved in human SSTR3 (Figure 5B). Given that human SSTR3 also localizes to cilia when expressed in IMCD cells (data not shown), this suggested that residues within “APSCQ” might confer ciliary localization.
Figure 5.
Comparative genomics and mutational analysis identifies unique residues in Sstr3 and Htr6 that are important for ciliary localization of GPCRs. (A) Alignment of the predicted i3 loop sequences of the mouse somatostatin receptors reveals an insertion of 11 unique residues in Sstr3. The corresponding positions of the residues are indicated. Red signifies identical residues and blue signifies conserved residues. (B) Alignment of the predicted i3 loop sequence of somatostatin receptor subtype 3 from mouse (Sstr3) and human (SSTR3). Five of the unique 11 residues (APSCQ; boxed) are completely conserved between mouse and human. (C) Alignment of the predicted i3 loop sequences of mouse serotonin receptors 6 and 7 shows the presence of a unique sequence in Htr6 (ATAGQ; boxed) with modest similarity to the Sstr3 sequence. (D) Percentage of transiently transfected IMCD cells that show ciliary localization when expressing Sstr3, Sstr5, Sstr5[TM5-6Sstr3], Sstr5[TM5-6Sstr3mut1], or Sstr5[TM5-6Sstr3mut2]. Sstr3 localizes to cilia in ∼91% of transfected cells. Sstr5 never localizes to cilia. Chimeric receptor Sstr5[TM5-6Sstr3] localizes to cilia in ∼93% of transfected cells. Chimeric receptor Sstr5[TM5-6Sstr3mut1], in which the A and Q in the conserved consensus sequence have been mutated to F, localizes to cilia in ∼50% of transfected cells. Chimeric receptor Sstr5[TM5–6Sstr3mut2], in which the A and Q in the second consensus sequence have also been mutated to F, localizes to cilia in ∼6% of transfected cells. Values are expressed as mean ± SEM *Significantly different from Sstr3 and Sstr5[TM5–6Sstr3] percentages. (E) Percentage of transiently transfected IMCD cells that show ciliary localization when expressing Htr6, Htr7, Htr7[TM5-V241Htr6], or Htr7[TM5-V241Htr6mut]. Htr6 localizes to cilia in ∼93% of transfected cells. Htr7 localizes to cilia in ∼20% of transfected cells. Chimeric receptor Htr7[TM5-V241Htr6] localizes to cilia in ∼70% of transfected cells. Chimeric receptor Htr7[TM5-V241Htr6mut], in which the A and Q have been mutated to F, localizes to cilia in ∼4% of transfected cells. Values are expressed as mean ± SEM. *Significantly different from Htr6 and Htr7[TM5-V241Htr6] percentages.
We then examined the N-portion of the i3 loop of Htr6 and identified a sequence (ATAGQ) with modest similarity that is not present in Htr7 (Figure 5C). Because the A and Q at the ends of the sequence are identical between Sstr3 and Htr6, we performed site-directed mutagenesis on these residues to verify they are important for ciliary localization. The A and Q residues in chimeric receptor Sstr5[TM5-6Sstr3] were mutated to phenylalanine, and ciliary localization was then quantified in IMCD cells expressing Sstr3, Sstr5, Sstr5[TM5-6Sstr3], or Sstr5[TM5-6Sstr3mut]. Sstr3 localized to cilia in ∼91% of transfected cells, whereas Sstr5 never localized to cilia (Figure 5D). Chimeric receptor Sstr5[TM5-6Sstr3] showed a similar percentage of cilia localization (∼93%) as Sstr3 (Figure 5D and Supplementary Figure 2A). The mutated version of this chimera (Sstr5[TM5-6Sstr3mut1]) showed a dramatic reduction in the percentage of ciliary localization but still localized to cilia in ∼50% of cells (Figure 5D). However, the i3 loop of mouse Sstr3 contains a second ciliary localization consensus sequence (APACQ; Figure 5B). To test whether the second site was contributing to ciliary localization, we mutated the A and Q residues to F in Sstr5[TM5-6Sstr3mut1] and quantified ciliary localization. Notably, when all four residues were mutated in this chimera (Sstr5[TM5-6Sstr3mut2]) ciliary localization was reduced to ∼6% of cells (Figure 5D and Supplementary Figure 2B). Immunoblotting of proteins isolated from transiently transfected cells expressing chimera Sstr5[TM5-6Sstr3mut2] confirmed that the mutant chimeric receptor was expressed (Supplementary Figure 1).
Similarly, the A and Q residues in chimeric receptor Htr7 [TM5-V241Htr6] were mutated to phenylalanine and ciliary localization was then quantified in IMCD cells expressing Htr6, Htr7, Htr7[TM5-V241Htr6], or Htr7[TM5-V241Htr6mut]. Htr6 localized to cilia in ∼93% of transfected cells, whereas Htr7 localized to cilia in ∼20% of transfected cells (Figure 5E). Of note, ciliary localization of Htr7 was almost exclusively seen in cells that were expressing the receptor at a very high level. Chimeric receptor Htr7[TM5-V241Htr6] localized to cilia in ∼70% of transfected cells (Figure 5E and Supplementary Figure 2C). Remarkably, the mutated version of this chimera (Htr7[TM5-V241Htr6mut]) almost never localized to cilia (∼4%; Figure 5E and Supplementary Figure 2D). Together, these results indicate that the A and Q residues in the i3 loop of Sstr3 and Htr6 are important for ciliary localization.
To test whether these sequences could be used to predict novel ciliary GPCRs, we formulated a consensus sequence based on Sstr3 and Htr6. Because serine and alanine belong to a “strong” Gonnet Pam250 matrix conservation group (Gonnet et al., 1992), we formulated the following loose consensus sequence (AX[S/A]XQ), where position 1 is an A, position 5 is a Q, position 3 is an S or A, and positions 2 and 4 are any amino acid. As the ciliary localization sequences were identified within the i3 loop of both Sstr3 and Htr6, we searched for the consensus sequence in a database containing the sequences of the predicted i3 loop of all human GPCRs. The consensus sequence is present in the i3 loop of 11 additional GPCRs (Table 1). Remarkably, these 11 GPCRs include 3 odorant receptors, 3 cone opsins, and rhodopsin, all of which are ciliary proteins. The remaining four GPCRs (α-2A adrenergic receptor, chemokine orphan receptor 1, melanin-concentrating hormone receptor 1, and muscarinic acetylcholine receptor M5) have not been shown to localize to cilia and were classified as candidate ciliary GPCRs. Because melanin-concentrating hormone receptor 1 (Mchr1) is involved in the regulation of feeding behavior and energy balance (Pissios et al., 2006) and multiple human ciliary disorders are associated with obesity (Badano et al., 2006), Mchr1 was considered a particularly good candidate ciliary GPCR.
Table 1.
G protein–coupled receptors identified by the ciliary localization consensus sequence
| G protein–coupled receptor | i3 domain consensus sequence | Localization |
|---|---|---|
| Olfactory receptor 52N1 | ADARQ | Ciliary |
| Olfactory receptor 52N4 | ADARQ | Ciliary |
| Olfactory receptor 6V1 | ASSCQ | Ciliary |
| Opsin 1 (short-wave-sensitive) | AVAAQ | Ciliary |
| Opsin 1 (medium-wave-sensitive 2) | AVAKQ | Ciliary |
| Opsin 1 (long-wave-sensitive) | AVAKQ | Ciliary |
| Rhodopsin | AAAQQ | Ciliary |
| α-2A adrenergic receptor | ARASQ | ? |
| Melanin-concentrating hormone receptor 1 | APASQ | ? |
| Muscarinic acetylcholine receptor M5 | AKAEQ | ? |
| Chemokine orphan receptor 1 | ASSDQ | ? |
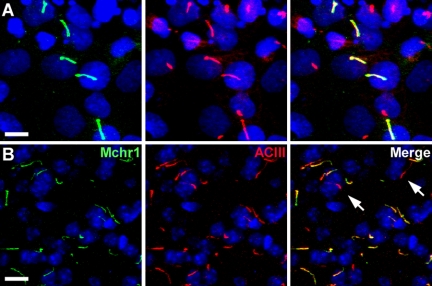
To test whether Mchr1 localizes to cilia, we cloned the coding sequence of Mchr1 from mouse cDNA, fused it to EGFP, expressed it in IMCD cells, and assessed subcellular localization. Notably, Mchr1 localized to cilia when expressed in IMCD cells (Figure 6A). We then tested whether Mchr1 localizes to cilia in tissue by labeling mouse brain sections with an antibody to Mchr1. We detected ciliary localization of Mchr1 in several brain regions (Figure 6B), including the hippocampus, nucleus accumbens, olfactory bulb, and hypothalamus. Ciliary localization was confirmed by colabeling with ACIII, which is a marker of neuronal cilia (Berbari et al., 2007; Bishop et al., 2007). These results indicate, for the first time, that Mchr1 localizes to cilia in vitro and in vivo.
Figure 6.
Melanin-concentrating hormone receptor 1 (Mchr1) localizes to cilia in vitro and in vivo. (A) Representative image of transiently transfected IMCD cells expressing Mchr1 fused at the C-terminus to EGFP. Left, EGFP fluorescence (green); middle, acetylated α-tubulin (red); right, merged image. (B) Representative image of the nucleus accumbens from an adult mouse colabeled with antibodies to Mchr1 (green; left), ACIII (red; middle), and merged (right). The majority of cilia are positive for both Mchr1 and ACIII. Cilia that are positive for ACIII but negative for Mchr1 are indicated with arrows. Nuclei are labeled with DRAQ5 (blue). All scale bars, 10 μm.
DISCUSSION
It is presumed that ciliary membrane proteins are synthesized in the endoplasmic reticulum, trafficked through the Golgi, and then transported in post-Golgi vesicles to the base of the cilium (Bloodgood, 2000). As access to the cilium is restricted and only certain proteins localize to the ciliary membrane (Bloodgood, 2000), it is thought that ciliary proteins contain targeting signals that direct them to the cilial compartment (Rosenbaum and Witman, 2002). Few mammalian ciliary localization sequences have been described (Deretic et al., 1998; Corbit et al., 2005; Geng et al., 2006; Hu et al., 2006; Jenkins et al., 2006), and only one sequence has been shown to be sufficient for ciliary targeting in mammalian cells. The N-terminal 15 amino acids of the ciliary cation channel polycystin-2 are sufficient to localize heterologous proteins to cilia (Geng et al., 2006). Within this domain is a conserved motif, RVXP, that is required for ciliary localization (Geng et al., 2006). This motif is also found at the C-terminus of olfactory cyclic nucleotide–gated channel 1b and is required for trafficking of the channel into cilia (Jenkins et al., 2006). This motif is not found in Sstr3 or Htr6, but is found in the i3 loop of Htr7, which does not localize to cilia, suggesting this motif is not functional in Htr7 or is not involved in the targeting of GPCRs to cilia.
In this work, we show that sequences within the i3 loop of Sstr3 and Htr6 are sufficient to localize nonciliary GPCRs to cilia on IMCD cells. Notably, we see the same subcellular localization results when we express the constructs in cultured primary hippocampal neurons (Berbari and Mykytyn, unpublished results). This suggests that the trafficking machinery for ciliary receptors is conserved between neurons and IMCD cells and supports the utility of this system for identifying the mechanisms of ciliary protein sorting and trafficking. Interestingly, replacing the i3 loop in Sstr3 and Htr6 with the i3 loop from Sstr5 and Htr7, respectively, does not prevent ciliary localization (data not shown). Thus, although the i3 loops are sufficient to localize nonciliary GPCRs to cilia, they are not necessary for ciliary localization of Sstr3 and Htr6. This is an intriguing result that suggests there are additional ciliary localization sequences within Sstr3 and Htr6.
The i3 loops of GPCRs, including somatostatin and serotonin receptors, are normally associated with G protein coupling and desensitization (Oksenberg et al., 1995; Gelber et al., 1999; Hipkin et al., 2000). This is the first time the i3 loop has been implicated in GPCR ciliary localization. Most known GPCR trafficking motifs are located within the C-terminus (Dong et al., 2007). However, the i3 loop of mouse vasopressin V2 receptor contains two RXR ER retention motifs that are thought to be masked under normal conditions but can be unmasked in mutant receptors and block trafficking to the plasma membrane (Hermosilla and Schulein, 2001). An RXR motif is present in the i3 loop of Sstr3 and Sstr5, but neither Htr6 nor Htr7 contain an RXR motif in the i3 loop. Thus, it is unlikely that ER retention is a mechanism for affecting ciliary localization in our system. The i3 loop of many GPCRs also contains a loose consensus sequence (BBXXB), where B is a basic residue and X is a nonbasic residue, at the junction of the i3 loop and the TM6 domain that is important for GPCR structure and activity (Kjelsberg et al., 1992; Ren et al., 1993; Egan et al., 1998). This consensus sequence is present in the C-terminal end of the i3 loop of Sstr3, Sstr5, Htr6, and Htr7 and is required for Htr6 activity (Kohen et al., 2001). However, it is likely unrelated to ciliary localization given the ciliary localization domain in Htr6 maps to the N-portion of the i3 loop.
Comparison of the sequences within the i3 loops of Sstr3 and Htr6 reveals a consensus sequence (AX[S/A]XQ) that may comprise a ciliary localization sequence. In support of this idea, the consensus sequence is also present in the i3 loop of Mchr1, which we have shown localizes to cilia. Further, mutating the A and Q residues in chimeric receptor Htr7[TM5-V241Htr6] decreases ciliary localization from ∼70% of cells to ∼4%. Mutating the A and Q residues in both consensus sequences in chimeric receptor Sstr5[TM5-6Sstr3] decreases ciliary localization from ∼93% of cells to ∼6%. It is interesting that Htr7 localizes to cilia in ∼20% of cells, whereas Sstr5 never localizes to cilia. This may be due to the fact that Sstr5 appears to be associated primarily with intracellular structures (Figure 1E), as opposed to Htr7 that appears to localize mainly to the plasma membrane (Figure 1G). It is likely that high levels of heterologously expressed proteins can overcome restrictions on ciliary localization when they are on the plasma membrane. Indeed, cotransfecting Htr7 with an empty expression vector (pcDNA3.1) at a ratio of 1:4, to lower the level of Htr7 expression, reduces ciliary localization of Htr7 to ∼6% of cells (data not shown).
Comparison of our consensus ciliary localization sequence against the i3 loop sequence of all human GPCRs identified known ciliary GPCRs and four candidate ciliary GPCRs. We have confirmed that one of these candidates, Mchr1, localizes to cilia. The consensus sequences of the remaining three candidates are present in the human protein but are not completely conserved in lower organisms. An exciting possibility is ciliary localization of some GPCRs may have evolved only in higher organisms. The development of ciliary localization would potentially be a mechanism for organisms to create additional complexity and more specialized functions in existing signaling pathways. Another possibility is our consensus sequence is too stringent. In support of this possibility, the ciliary localization sequence in human HTR6 contains a G rather than an A at position 1, suggesting there is flexibility to the consensus sequence. We also hypothesized that localization of the sequence within the i3 loop was a requirement for ciliary localization and limited our search to the i3 loop. The amount of flexibility in the consensus sequence and its location within receptors could potentially increase the number of candidate ciliary GPCRs dramatically. It will be necessary to experimentally determine what residues are required and permissive and whether the consensus sequence can be present in other domains in order to accurately predict all ciliary GPCRs.
Although the ciliary localization sequence we identified is common to Sstr3, Htr6, and Mchr1, it is not present in Smo, despite the fact that Smo localizes to cilia in vitro and in vivo (Corbit et al., 2005). One possibility is that Smo is targeted to the cilium through a different mechanism. Indeed, substitution of the hydrophobic-basic residues in the C-terminal tail of Smo with alanines prevents ciliary localization (Corbit et al., 2005), but mutation of this motif in Sstr3 or Htr6 does not prevent ciliary localization (Berbari and Mykytyn, unpublished results). Perhaps there are divergent mechanisms of ciliary trafficking between the different GPCR families. Sstr3, Htr6, and Mchr1 are members of the Rhodopsin family of GPCRs, whereas Smo is a member of the Frizzled/Smoothened family of GPCRs. Several studies have found that defects in ciliary protein transport can be associated with mislocalization of some membrane proteins but not others (Marszalek et al., 2000; Dwyer et al., 2001; Qin et al., 2005; McEwen et al., 2007), suggesting there are multiple pathways for targeting membrane proteins to the cilium.
An important outcome from these studies is the demonstration that Mchr1 localizes to neuronal cilia in regions of the mouse brain, including the hypothalamus. A role for MCH and its receptor in the regulation of feeding and metabolism is well established. Specifically, injection of MCH into the brains of mice induces a rapid increase in feeding behavior (Qu et al., 1996), whereas injection of Mchr1 antagonists reduces feeding behavior (Borowsky et al., 2002). Further, transgenic mice overexpressing MCH (Ludwig et al., 2001) are obese, and mice lacking expression of either MCH (Shimada et al., 1998) or Mchr1 (Chen et al., 2002) are lean. The fact that Mchr1 localizes to cilia in regions of the brain known to regulate feeding behavior suggests that localization of Mchr1 to cilia may be important for signaling through the receptor and proper regulation of these processes. Interestingly, mice lacking cilia in the brain or specifically on pro-opiomelanocortin–expressing cells in the hypothalamus are hyperphagic and become obese (Davenport et al., 2007). Further, obesity is a hallmark of some human ciliary disorders. The role of cilia in this phenotype in particular has been a long-standing mystery. The identification of Mchr1 as a ciliary GPCR provides, for the first time, a potential molecular mechanism to link defects in cilia with obesity.
Supplementary Material
ACKNOWLEDGMENTS
We thank W. Sadée, D. Lautenbach, L. Wallace, E. Fink, and M. Vest for technical assistance. This work was supported in part by research grant 5-FY05–39 from the March of Dimes Birth Defects Foundation (K.M.)
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/10.1091/mbc.E07-09-0942) on February 6, 2007.
REFERENCES
- Badano J. L., Mitsuma N., Beales P. L., Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- Berbari N. F., Bishop G. A., Askwith C. C., Lewis J. S., Mykytyn K. Hippocampal neurons possess primary cilia in culture. J. Neurosci. Res. 2007;85:1095–1100. doi: 10.1002/jnr.21209. [DOI] [PubMed] [Google Scholar]
- Bisgrove B. W., Yost H. J. The roles of cilia in developmental disorders and disease. Development. 2006;133:4131–4143. doi: 10.1242/dev.02595. [DOI] [PubMed] [Google Scholar]
- Bishop G. A., Berbari N. F., Lewis J. S., Mykytyn K. Type III adenylyl cyclase localizes to primary cilia throughout the adult mouse brain. J. Comp. Neurol. 2007;505:562–571. doi: 10.1002/cne.21510. [DOI] [PubMed] [Google Scholar]
- Bloodgood R. A. Protein targeting to flagella of trypanosomatid protozoa. Cell Biol. Int. 2000;24:857–862. doi: 10.1006/cbir.2000.0598. [DOI] [PubMed] [Google Scholar]
- Borowsky B., et al. Antidepressant, anxiolytic and anorectic effects of a melanin-concentrating hormone-1 receptor antagonist. Nat. Med. 2002;8:825–830. doi: 10.1038/nm741. [DOI] [PubMed] [Google Scholar]
- Brailov I., Bancila M., Brisorgueil M. J., Miquel M. C., Hamon M., Verge D. Localization of 5-HT(6) receptors at the plasma membrane of neuronal cilia in the rat brain. Brain Res. 2000;872:271–275. doi: 10.1016/s0006-8993(00)02519-1. [DOI] [PubMed] [Google Scholar]
- Chen Y., et al. Targeted disruption of the melanin-concentrating hormone receptor-1 results in hyperphagia and resistance to diet-induced obesity. Endocrinology. 2002;143:2469–2477. doi: 10.1210/endo.143.7.8903. [DOI] [PubMed] [Google Scholar]
- Corbit K. C., Aanstad P., Singla V., Norman A. R., Stainier D. Y., Reiter J. F. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- Davenport J. R., Watts A. J., Roper V. C., Croyle M. J., van Groen T., Wyss J. M., Nagy T. R., Kesterson R. A., Yoder B. K. Disruption of intraflagellar transport in adult mice leads to obesity and slow-onset cystic kidney disease. Curr. Biol. 2007;17:1586–1594. doi: 10.1016/j.cub.2007.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport J. R., Yoder B. K. An incredible decade for the primary cilium: a look at a once-forgotten organelle. Am. J. Physiol. Renal. Physiol. 2005;289:F1159–F1169. doi: 10.1152/ajprenal.00118.2005. [DOI] [PubMed] [Google Scholar]
- Deretic D., Schmerl S., Hargrave P. A., Arendt A., McDowell J. H. Regulation of sorting and post-Golgi trafficking of rhodopsin by its C-terminal sequence QVS(A)PA. Proc. Natl. Acad. Sci. USA. 1998;95:10620–10625. doi: 10.1073/pnas.95.18.10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C., Filipeanu C. M., Duvernay M. T., Wu G. Regulation of G protein-coupled receptor export trafficking. Biochim. Biophys. Acta. 2007;1768:853–870. doi: 10.1016/j.bbamem.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer N. D., Adler C. E., Crump J. G., L'Etoile N. D., Bargmann C. I. Polarized dendritic transport and the AP-1 mu1 clathrin adaptor UNC-101 localize odorant receptors to olfactory cilia. Neuron. 2001;31:277–287. doi: 10.1016/s0896-6273(01)00361-0. [DOI] [PubMed] [Google Scholar]
- Egan C. T., Herrick-Davis K., Teitler M. Creation of a constitutively activated state of the 5-hydroxytryptamine2A receptor by site-directed mutagenesis: inverse agonist activity of antipsychotic drugs. J. Pharmacol. Exp. Ther. 1998;286:85–90. [PubMed] [Google Scholar]
- Gelber E. I., Kroeze W. K., Willins D. L., Gray J. A., Sinar C. A., Hyde E. G., Gurevich V., Benovic J., Roth B. L. Structure and function of the third intracellular loop of the 5-hydroxytryptamine2A receptor: the third intracellular loop is alpha-helical and binds purified arrestins. J. Neurochem. 1999;72:2206–2214. doi: 10.1046/j.1471-4159.1999.0722206.x. [DOI] [PubMed] [Google Scholar]
- Geng L., Okuhara D., Yu Z., Tian X., Cai Y., Shibazaki S., Somlo S. Polycystin-2 traffics to cilia independently of polycystin-1 by using an N-terminal RVxP motif. J. Cell Sci. 2006;119:1383–1395. doi: 10.1242/jcs.02818. [DOI] [PubMed] [Google Scholar]
- Gonnet G. H., Cohen M. A., Benner S. A. Exhaustive matching of the entire protein sequence database. Science. 1992;256:1443–1445. doi: 10.1126/science.1604319. [DOI] [PubMed] [Google Scholar]
- Hamon M., Doucet E., Lefevre K., Miquel M. C., Lanfumey L., Insausti R., Frechilla D., Del Rio J., Verge D. Antibodies and antisense oligonucleotide for probing the distribution and putative functions of central 5-HT6 receptors. Neuropsychopharmacology. 1999;21:68S–76S. doi: 10.1016/S0893-133X(99)00044-5. [DOI] [PubMed] [Google Scholar]
- Handel M., Schulz S., Stanarius A., Schreff M., Erdtmann-Vourliotis M., Schmidt H., Wolf G., Hollt V. Selective targeting of somatostatin receptor 3 to neuronal cilia. Neuroscience. 1999;89:909–926. doi: 10.1016/s0306-4522(98)00354-6. [DOI] [PubMed] [Google Scholar]
- Hermosilla R., Schulein R. Sorting functions of the individual cytoplasmic domains of the G protein-coupled vasopressin V(2) receptor in Madin Darby canine kidney epithelial cells. Mol. Pharmacol. 2001;60:1031–1039. doi: 10.1124/mol.60.5.1031. [DOI] [PubMed] [Google Scholar]
- Hildebrandt F., Otto E. Cilia and centrosomes: a unifying pathogenic concept for cystic kidney disease? Nat. Rev. Genet. 2005;6:928–940. doi: 10.1038/nrg1727. [DOI] [PubMed] [Google Scholar]
- Hipkin R. W., Wang Y., Schonbrunn A. Protein kinase C activation stimulates the phosphorylation and internalization of the sst2A somatostatin receptor. J. Biol. Chem. 2000;275:5591–5599. doi: 10.1074/jbc.275.8.5591. [DOI] [PubMed] [Google Scholar]
- Horn F., Bettler E., Oliveira L., Campagne F., Cohen F. E., Vriend G. GPCRDB information system for G protein-coupled receptors. Nucleic Acids Res. 2003;31:294–297. doi: 10.1093/nar/gkg103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton R. M., Cai Z. L., Ho S. N., Pease L. R. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- Hu J., Bae Y. K., Knobel K. M., Barr M. M. Casein kinase II and calcineurin modulate TRPP function and ciliary localization. Mol. Biol. Cell. 2006;17:2200–2211. doi: 10.1091/mbc.E05-10-0935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins P. M., Hurd T. W., Zhang L., McEwen D. P., Brown R. L., Margolis B., Verhey K. J., Martens J. R. Ciliary targeting of olfactory CNG channels requires the CNGB1b subunit and the kinesin-2 motor protein, KIF17. Curr. Biol. 2006;16:1211–1216. doi: 10.1016/j.cub.2006.04.034. [DOI] [PubMed] [Google Scholar]
- Kjelsberg M. A., Cotecchia S., Ostrowski J., Caron M. G., Lefkowitz R. J. Constitutive activation of the alpha 1B-adrenergic receptor by all amino acid substitutions at a single site. Evidence for a region which constrains receptor activation. J. Biol. Chem. 1992;267:1430–1433. [PubMed] [Google Scholar]
- Kohen R., Fashingbauer L. A., Heidmann D. E., Guthrie C. R., Hamblin M. W. Cloning of the mouse 5-HT6 serotonin receptor and mutagenesis studies of the third cytoplasmic loop. Brain Res. Mol. Brain Res. 2001;90:110–117. doi: 10.1016/s0169-328x(01)00090-0. [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Ludwig D. S., Tritos N. A., Mastaitis J. W., Kulkarni R., Kokkotou E., Elmquist J., Lowell B., Flier J. S., Maratos-Flier E. Melanin-concentrating hormone overexpression in transgenic mice leads to obesity and insulin resistance. J. Clin. Invest. 2001;107:379–386. doi: 10.1172/JCI10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W. F., Nonaka S. Cilia: tuning in to the cell's antenna. Curr. Biol. 2006;16:R604–614. doi: 10.1016/j.cub.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Marszalek J. R., Liu X., Roberts E. A., Chui D., Marth J. D., Williams D. S., Goldstein L. S. Genetic evidence for selective transport of opsin and arrestin by kinesin-II in mammalian photoreceptors. Cell. 2000;102:175–187. doi: 10.1016/s0092-8674(00)00023-4. [DOI] [PubMed] [Google Scholar]
- McEwen D. P., Koenekoop R. K., Khanna H., Jenkins P. M., Lopez I., Swaroop A., Martens J. R. Hypomorphic CEP290/NPHP6 mutations result in anosmia caused by the selective loss of G proteins in cilia of olfactory sensory neurons. Proc. Natl. Acad. Sci. USA. 2007;104:15917–15922. doi: 10.1073/pnas.0704140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauli S. M., et al. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat. Genet. 2003;33:129–137. doi: 10.1038/ng1076. [DOI] [PubMed] [Google Scholar]
- Oksenberg D., Havlik S., Peroutka S. J., Ashkenazi A. The third intracellular loop of the 5-hydroxytryptamine2A receptor determines effector coupling specificity. J. Neurochem. 1995;64:1440–1447. doi: 10.1046/j.1471-4159.1995.64041440.x. [DOI] [PubMed] [Google Scholar]
- Pan J., Wang Q., Snell W. J. Cilium-generated signaling and cilia-related disorders. Lab. Invest. 2005;85:452–463. doi: 10.1038/labinvest.3700253. [DOI] [PubMed] [Google Scholar]
- Patel Y. C. Somatostatin and its receptor family. Front. Neuroendocrinol. 1999;20:157–198. doi: 10.1006/frne.1999.0183. [DOI] [PubMed] [Google Scholar]
- Pazour G. J., Witman G. B. The vertebrate primary cilium is a sensory organelle. Curr. Opin. Cell Biol. 2003;15:105–110. doi: 10.1016/s0955-0674(02)00012-1. [DOI] [PubMed] [Google Scholar]
- Pissios P., Bradley R. L., Maratos-Flier E. Expanding the scales: the multiple roles of MCH in regulating energy balance and other biological functions. Endocr. Rev. 2006;27:606–620. doi: 10.1210/er.2006-0021. [DOI] [PubMed] [Google Scholar]
- Praetorius H. A., Spring K. R. Bending the MDCK cell primary cilium increases intracellular calcium. J. Membr. Biol. 2001;184:71–79. doi: 10.1007/s00232-001-0075-4. [DOI] [PubMed] [Google Scholar]
- Qin H., Burnette D. T., Bae Y. K., Forscher P., Barr M. M., Rosenbaum J. L. Intraflagellar transport is required for the vectorial movement of TRPV channels in the ciliary membrane. Curr. Biol. 2005;15:1695–1699. doi: 10.1016/j.cub.2005.08.047. [DOI] [PubMed] [Google Scholar]
- Qu D., Ludwig D. S., Gammeltoft S., Piper M., Pelleymounter M. A., Cullen M. J., Mathes W. F., Przypek R., Kanarek R., Maratos-Flier E. A role for melanin-concentrating hormone in the central regulation of feeding behaviour. Nature. 1996;380:243–247. doi: 10.1038/380243a0. [DOI] [PubMed] [Google Scholar]
- Ren Q., Kurose H., Lefkowitz R. J., Cotecchia S. Constitutively active mutants of the alpha 2-adrenergic receptor. J. Biol. Chem. 1993;268:16483–16487. [PubMed] [Google Scholar]
- Rosenbaum J. L., Witman G. B. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Schulz S., Handel M., Schreff M., Schmidt H., Hollt V. Localization of five somatostatin receptors in the rat central nervous system using subtype-specific antibodies. J. Physiol. Paris. 2000;94:259–264. doi: 10.1016/s0928-4257(00)00212-6. [DOI] [PubMed] [Google Scholar]
- Shimada M., Tritos N. A., Lowell B. B., Flier J. S., Maratos-Flier E. Mice lacking melanin-concentrating hormone are hypophagic and lean. Nature. 1998;396:670–674. doi: 10.1038/25341. [DOI] [PubMed] [Google Scholar]
- Singla V., Reiter J. F. The primary cilium as the cell's antenna: signaling at a sensory organelle. Science. 2006;313:629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- Takeda S., Kadowaki S., Haga T., Takaesu H., Mitaku S. Identification of G protein-coupled receptor genes from the human genome sequence. FEBS Lett. 2002;520:97–101. doi: 10.1016/s0014-5793(02)02775-8. [DOI] [PubMed] [Google Scholar]
- Wheatley D. N., Wang A. M., Strugnell G. E. Expression of primary cilia in mammalian cells. Cell Biol. Int. 1996;20:73–81. doi: 10.1006/cbir.1996.0011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.